Abstract
Melanoma is a very aggressive skin cancer with increasing incidence worldwide. MicroRNAs are small, noncoding RNAs that regulate gene expression of targeted gene(s). The hallmark of cancer model outlined by Hanahan and Weinberg offers a meaningful framework to consider the roles of microRNAs in melanoma development and progression. In this systematic review of the literature, we associate what is known about deregulation of microRNAs and their targeted genes in melanoma development with the hallmarks and characteristics of cancer. The diagnostic and therapeutic potential of microRNAs for future melanoma management will also be discussed.
Keywords: MITF, BRAF, miR-137, miR-18b, miR-214
melanoma is a malignancy of melanocytes, a cell type that produces melanin pigment in the skin to protect skin from ultra violet radiation (81). The incidence of melanoma is increasing at a rapid rate of ∼3% annually, faster than any other cancer in the United States (50). The lifetime risk of developing invasive melanoma as an adult in the United States is 1 in 58 overall (75). While the survival rate of early-stage melanoma is reported to be as high as 90%, late-stage metastatic melanoma has a survival rate of only 10% (83) with a recurrence risk up to 60% (24). Thus, early diagnosis is very important for improving survival outcomes, and effective therapies for malignant melanoma are urgently needed.
Recent publications support an important role for microRNAs (miRNAs) in melanoma disease development (see reviews Refs. 24, 26, 42, 87). MiRNAs are short (∼22 nucleotide), noncoding RNAs that can regulate gene expression posttranscriptionally by pairing to the 3′-untranslated region (UTR), the 5′-UTR, or the coding region of an mRNA (29, 45, 54). They act to reduce targeted protein expression in two general ways, namely by targeting mRNA degradation and/or by inhibiting mRNA translation (4). To better understand the molecular mechanisms underlying malignant melanoma growth and metastasis, many investigators to date have focused on elucidating the deregulated miRNAs specific to melanoma. Current research also aims to reveal potential prognostic and therapeutic roles for these miRNAs in order to improve survival outcomes in people with melanoma.
The hallmarks of cancer provide a useful framework for understanding the complex biology of cancer (31, 32). In contrast to normal cells, cancer cells express novel capabilities that support malignant growth. A number of reports have described the effects of miRNA deregulation in the development of cancer cells. Herein, we associate some of the miRNAs known to be altered in melanoma with the corresponding characteristics of cancer. In particular, cancer cells and tissues demonstrate the characteristics of sustaining proliferative growth signaling, evading growth suppressor signaling, resisting cell death (antiapoptosis), enabling replicative immortality (antisenescence), inducing angiogenesis, activating invasion and metastasis, genome instability and mutation, and tumor-promoting inflammation (31, 32). These eight basic features are crucial for a given tissue type to generate and sustain a viable population of cancer cells (32).
Extensive reports summarizing the role(s) of various miRNAs in melanomagenesis have been published recently (24, 26, 42, 87), but a functional review of miRNA with respect to the hallmarks in melanoma development has not yet been illustrated. In this review, we present selected miRNAs that are dysregulated in melanoma with regard to their associated cancer hallmark (Table 1).
Table 1.
MiRNA deregulation with respect to affected hallmarks of cancer capabilities
| Hallmarks of Cancer | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sustaining Proliferative Signaling | Evading Growth Suppressors | Resisting Cell Death | Enabling Replicative Mortality | Inducing Angiogenesis | Activating Invasion and Metastasis | Genome Instability and Mutation | Tumor Promoting Inflammation | |
| ↑ | miR-7 | miR-195 | miR-290–295 | miR-1908 | miR-214 | miR-29 | ||
| miR-17 | miR-221 | miR-199a-5p | miR-21 | |||||
| miR-18 | miR-222 | miR-199a-3p | ||||||
| miR-19 | ||||||||
| miR-92a | ||||||||
| miR-106b ∼ 25 | ||||||||
| miR-222 | ||||||||
| ↓ | miR-let7 | miR-18b | miR-18b | miR-34a | miR-let-7a | miR-let-7a† | ||
| miR-10 | miR-26a | miR-203 | miR-9 | miR-let-7b | ||||
| miR-22 | miR-137 | miR-205 | miR-137 | miR-18b | ||||
| miR-26a | miR-193b | miR-125b | miR-148 | miR-146a* | ||||
| miR-30 | miR-205 | miR-182 | miR-193b | |||||
| miR-34 | miR-196a | miR-203 | ||||||
| miR-125a | miR-211 | miR-205 | ||||||
| miR-137 | miR-340 | |||||||
| miR-193b | miR-573 | |||||||
| miR-205 | ||||||||
| miR-211 | ||||||||
The lists of the microRNAs (miRNA, miR) here are those discussed in this paper and are not meant to be a comprehensive list. Up and down arrows indicate up- and downregulation of miRNA in melanoma.
The miRNA is mutated in melanoma, resulting in decreased expression of this miRNA. †A mutation has been reported in the KRAS gene located in the binding site of this miRNA, which allowed this KRAS variant to escape the regulation of this miRNA.
While most of this review is focused on direct relationships between a given miRNA and its target gene, it is important to recognize that the role of miRNAs in melanoma development is much more complex than can be described in such a linear manner with one miRNA regulating one gene. On one hand, a single miRNA can influence multiple cancer hallmarks either by regulating a single gene which is involved in multiple hallmarks [i.e., miR-18b through p53 (17)] or by regulating multiple gene targets at once(6, 19, 53) (Fig. 1A). On the other hand, multiple miRNAs can converge and target one specific gene to fine tune the protein expression of an important gene in melanocyte and melanoma (6, 28, 30, 78) (Fig. 1B). Moreover, multiple miRNAs can target a specific signaling pathway to cooperatively maximize silencing of that signaling pathway (69) (Fig. 1C). Finally, multiple miRNAs can be regulated by a single oncogene, and these miRNAs can function combinatorially in multiple hallmarks of cancer, and the functional interactions among these miRNAs can be complex with additive interference as well as latency effects (see below in sustaining proliferative signaling on the BRAF-regulated miRNAs) (15). Thus, acquired cancer hallmark features can occur through the combined deregulation of multiple miRNAs. In summary, multiple ways of miRNA deregulation lead to multiple levels of gene control with various contributions to cancer hallmarks in melanoma. Therefore, understanding miRNA deregulation will provide insight for future development of diagnostic tools and therapeutic strategies.
Fig. 1.
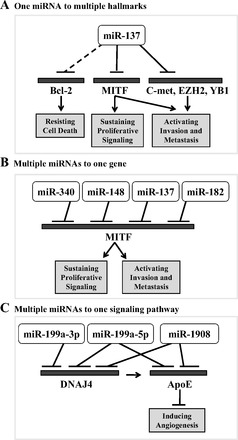
Complexity of microRNA (miRNA, also miR) regulation in melanoma. Examples of various ways that miRNAs regulate expression of gene(s) involved in cancer hallmark characteristics. A: miR-137 is an example of how a single miRNA can regulate many genes, enabling melanoma cells to acquire multiple hallmark functions (6, 53). B: micropthalmia-associated transcription factor (MITF) is an example of one gene regulated by multiple miRNAs to fine tune the control of gene expression (6, 28, 30, 78). C: ApoE signaling pathway is an example of multiple miRNAs regulating multiple genes within one signaling pathway in order to maximize inhibition of this pathway (69).
SUSTAINING PROLIFERATIVE SIGNALING
A primary hallmark of cancer is sustained proliferative growth signaling. Normal cells must receive exogenous growth factors to enter the cell cycle. However, cancer cells can demonstrate growth factor-independent cell cycle replication when a tumor cell acquires mutations in downstream signaling pathways that allow constitutive activation of signaling, when a tumor cell develops the ability to make its own growth factors or when a tumor cell develops alterations in transmembrane channels or intracellular signaling pathways that enhance its sensitivity to growth factors (31). Below we review several studies to provide examples of how miRNA deregulation can contribute to melanoma cells acquiring sustained proliferating signaling.
The activation of the RAS-RAF-MEK-ERK pathway is very important to many functions that promote melanoma development, including sustained proliferative signaling (82). Multiple alterations in melanoma can lead to activation of this signaling pathway. Activating mutations of BRAF or NRAS are found in 50–70% or 15% of melanomas, respectively (15, 18, 59), and activating mutations in receptor tyrosine kinases upstream of this pathway have also been reported (20). The RAS-RAF-MEK-ERK pathway is clearly a central mechanism disrupted in melanoma; therefore, it comes as no surprise that miRNAs are also implicated in the regulation of this pathway through altering gene expression and loss and gain of miRNA expression (62, 82).
The miR-let-7 family, originally called let-7 (79), encompasses a group of miRNAs that may regulate similar targets, and several of the miR-let-7 family members are known to have decreased expression in melanoma. Muller and Bosserhoff (62) showed that miR-let-7a is downregulated in malignant cell lines compared with melanocytes, and they also demonstrated that miR-let-7a reduces NRAS expression in melanoma cells using transfection experiments. Concordantly, transfection of synthetic inhibitors of miR-let-7a resulted in increased expression of both NRAS and β-integrin in melanoma cells that act together to increase cell proliferation and invasive cellular activity, two activities central to the hallmarks of cancer model (62).
In addition, other members of the miRNA family of let-7 have been shown to be frequently lost in melanoma; these include miR-let-7b, d, e, and g (77). One of the central roles of miR-let-7b is to regulate cyclins D1–3, important positive regulators of proliferative signaling (77, 90). In addition to miR-let-7b, miR-193b has also been reported to directly target cyclin D1 in melanoma, and it has been validated as a direct target of both miRNAs with immunoblot and luciferase reporter assays (12, 77, 90). Moreover, miR-let-7b family and miR-193b are downregulated in malignant melanoma (12, 77, 90). Reduced expressions of these miRNAs allow for cyclin D1 overexpression, which blocks the tumor suppressor function of Rb and promotes proliferative capacity in melanoma.
Even though there is no published evidence to date for miRNAs that can target BRAF, MEK, or ERKs in melanoma, a recent report by Couts and coworkers (15) elegantly demonstrated that activated BRAF can control a network of 20+ miRNAs in melanoma cell lines and affect proliferation capacity in these cells. While activated BRAF increases the expression of many miRNAs including miR-7, miR-17, miR-18, miR-19, miR-92a, miR-106b∼25, and miR-222, it also decreases expressions of other miRNAs including miR-let7i, miR-10, miR-22, miR-26a, miR-30, miR-34, miR-125a, and miR-211 (15). Interestingly, coexpressing experiments with various combinations of these miRNAs indicate complex functional interactions among these miRNAs, including additive, interference and latency effects. However, the sum effect of these BRAF-regulated miRNAs is to induce the proliferation and metastasis of melanoma (15). This comprehensive study sheds light on the complicated interplay of a network of oncogenes, miRNAs, their mRNA targets, and functional interactions among the miRNAs.
Furthermore, deregulating E2F family members such as E2F1 and E2F5 has been shown to promote cell cycle progression in melanoma (16, 65). Dar and colleagues (16) demonstrated that overexpressing miR-205 decreases E2F1 and E2F5 in melanoma cell lines, and endogenous miR-205 is downregulated in melanoma tissue compared with benign nevi. Moreover, two different groups demonstrated that injecting miR-205 (expression vectors or mimics) in melanoma cell lines or in melanoma xenografts in mice reduces proliferation and tumor growth (16, 65). Taken together, these data explain instances where downregulation of miR-205 contributes to melanoma proliferation, in part through deregulation of E2F1 and E2F5 proteins (16, 65).
Another example of miRNAs regulating proliferation is miR-137, which is downregulated in stage IV patient sample-derived melanoma cell lines compared with normal human melanocytes (53). Investigators have demonstrated that miR-137 targets c-Met and YB1 and confirmed that miR-137 targets micropthalmia-associated transcription factor (MITF) and EZH2 (6, 53). Functional studies showed that miR-137 inhibits proliferation in melanoma cell lines and that this antiproliferation effect is mediated by MITF, EZH2, c-Met, and YB1; however, the role of each target other than MITF was shown to vary by cell line (Fig. 1A) (53).
These examples show that deregulation of miRNAs can directly alter the expression of the central gene involved in the RAS/RAF/MEK signaling cascade (miR-let-7a targeting NRAS) or can behave as downstream mediators of this signaling (BRAF-regulated miRNAs) to promote proliferation of melanoma cells. Additionally, miRNAs can alter the important transcription factors involved in cell cycle regulation (miR-205 targeting E2F1 and E2F5) to provide sustained proliferative signaling. Furthermore, miRNAs like miR-137 regulate genes associated with multiple hallmarks, including proliferative signaling (53).
EVADING GROWTH SUPPRESSORS
The rate of normal cell proliferation is maintained within a homeostatic range by balanced growth stimulatory and growth inhibitory signals. Antigrowth (tumor suppressor) genes typically work either temporarily by blocking cell cycle entrance or permanently by forcing cells into a senescent state (discussed below). For cancer cells to thrive, they must acquire the capability to evade these antigrowth signals and to sustain the progrowth signals (31). Some important tumor suppressors that lose function in melanoma are p53, Wee1, and p27 (7, 17, 22, 36), and miRNA deregulation is one of the mechanisms by which deregulation occurs.
One of the most studied tumor suppressors to date is p53, the “guardian of the genome” (44). p53 is commonly mutated in tumors; however, it is rarely mutated in melanoma. Rather, the expression of p53 in melanoma has been shown to be downregulated, in part by miRNAs (17). Dar and coworkers (17) showed that miR-18b can upregulate p53 expression through decreasing p53's inhibitor MDM2, and expression of miRNA-18b is downregulated in melanoma compared with that in benign nevi (17). Deregulation of miRNA-18b leads to a decrease in p53 expression and activity in melanoma. Additionally, adding miR-18b back into human melanoma cells results in significant decrease of proliferation, and injecting miR-18b into a melanoma xenograft mouse model also decreases tumor volume significantly. This extensive study revealed that deregulation of miR-18b regulates growth suppression capacity of cells through indirect regulation of p53 expression (17).
Wee1 kinase is a mitotic G2 gatekeeper that has been implicated in tumorigenesis. Its roles in various cancers seem to be cell-type dependent, and Wee1 kinase likely carries a tumor suppressor function in melanoma (7). Bhattacharya et al. (7) demonstrated that miR-195 plays a role in regulating cellular gatekeeping action of Wee1. Analyses of the expression in primary melanomas and melanoma metastases showed an inverse correlation between Wee1 and miR-195 expression levels, with downregulated Wee1 and upregulated miR-195 in distant metastases. In addition, miR-195 was found to be upregulated in highly aggressive cell lines compared with less aggressive cell lines, and miR-195 was found to reduce stress-related cell cycle arrest (7). Moreover, overexpression of miR-195 reduced Wee1 expression, and reporter gene analysis confirmed miR-195 targeting of the Wee1 3′-UTR in vitro (7). Thus, upregulation of miR-195 reduced Wee1 expression, resulting in evasion of growth inhibition (7).
MiR-221 and miR-222 have been shown to target p27 (22, 23, 36). Often downregulated in melanoma, p27 is another important cell cycle regulator that induces cell cycle arrest when it binds and blocks the function of cyclin D1 (49). Felicetti et al. (22) demonstrated that miR-221 and -222 directly target p27 using miR-221 and/or -222 transfection experiments in melanoma cell lines. Additionally, they also demonstrated that miR-221 and -222 antagomirs inhibit tumor progression in melanoma cells, and miR-221- and -222-expressing cells are correlated with increased tumor volume in mice (22). Moreover, miR-221 is upregulated in melanoma compared with melanocytes (36). Interestingly, miR-221 also targets c-KIT, as evident by 3′-UTR reporter assays (36). Progressive loss of c-KIT occurs as melanoma gains aggressiveness (10), and wild-type c-KIT plays an important role in melanocyte differentiation through regulating MITF and tyrosinase; both are melanocyte differentiation genes. Although the roles of MITF in melanoma development is stage dependent, these findings indicate a substantial role for miR-221 and -222 in allowing cells to evade the growth suppressor actions of p27 and c-KIT and to participate in this characteristic hallmark of the cancer phenotype (22, 23, 36).
In sum, these examples demonstrate that deregulation of miRNAs can contribute to evading growth suppression through regulation of inhibitors or activators of this hallmark.
RESISTING CELL DEATH
Programmed cell death, such as apoptosis and autophagy, can act as counterbalances to cancer cell proliferation (31). Acquired resistance to cell death signaling is very important for melanoma progression and melanoma resistance to treatment, and miRNA deregulation has been linked to this hallmark in melanoma.
In addition to its function in growth suppression, p53 also mediates proapoptotic events in the context of DNA stress and oncogene activation (reviewed in Ref. 46). As mentioned before, downregulation of miR-18b indirectly leads to downregulation of p53 in melanoma, which also regulates apoptotic signals in melanoma and other cancers (17). Data from this study suggest that the antiapoptotic role of miR-18b in melanoma is not limited to p53, as miR-18b also affects expression of antiapoptotic Bcl-2 and Bcl-XL gene expression, as well as p53 and p53-induced proapoptotic protein, PUMA. This evidence points to a significant role for deregulation of miR-18b in the resisting cell death capability, in addition to the loss of tumor suppressor capability described above (17).
Bcl-2 family members play essential roles in regulating apoptotic signals, and the commonly known antiapoptotic family members include Bcl-2, Bcl-XL, and Mcl-1. Chen et al. (13) identified miR-193b as a miRNA targeting Mcl-1 in melanoma, and deregulation of miR-193b contributes to melanoma's resistance to cell death through altered regulation of Mcl-1. In support of that, malignant melanoma samples display a lower level of miR-193b and a higher level of Mcl-1 compared with benign nevi, and miR-193b expression is inversely correlated with Mcl-1 expression in these samples (13). Moreover, targeting Bcl-2 antiapoptotic members using small molecules such as ABT-737 has also been investigated as a potential strategy for melanoma treatment (33, 60, 73, 74). However, Mcl-1 is an antiapoptotic member of the Bcl-2 family that is not targeted by ABT-737 and has been implicated in the development of resistance to ABT-737-induced apoptosis (33). Chen et al. (13) found that overexpressing miR-193b restores ABT-737 sensitivity and induces apoptosis through downregulating Mcl-1 expression. Thus, downregulation of miR-193b allows Mcl-1 expression at levels that support acquired evasion to cell death and contributes to melanoma cell resistance to drug treatment such as ABT-737 (13).
Similar to p53, E2F1 also can contribute to multiple hallmarks of cancers. In addition to its role in regulating cell cycle progression mentioned above, E2F1 is a strong apoptotic regulator in the context of DNA damage (1). Since miR-205 can regulate E2F1, one would expect that miR-205 also can regulate apoptotic signals (1). In fact, a recent study elucidated a rather complex, E2F1-p73/DNp73-miR-205 mediated, “autoregulatory circuit” that induces cell death resistance and a chemo-resistant phenotype in malignant melanoma (1). Furthermore, they also showed that downregulation of miR-205 confers cell death resistance through the deregulation of multiple gene targeting, including E2F1, Bcl-2, ABCA2, and ABCA5 (1, 16). As mentioned above, miR-205 treatment reduces melanoma growth in vitro and in vivo (16, 65). In sum, miRNA-205 downregulation has been associated with preventing cell death that contributes to melanoma development and chemo-resistance (1, 65).
We recently identified that miR-26a was strongly downregulated in melanoma compared with normal melanocytes (80). We also found that putting miR-26a back in to melanoma cells can induce apoptotic cell death in five out of seven melanoma cell lines. Further mechanistic studies with 3′-UTR luciferase reporter assays and immunoblot analyses showed that miR-26a can regulate the silencer of death domain protein (SODD) and regulate apoptosis (80). Thus, downregulation of miR-26a may contribute to melanoma resistance to cell death in part through deregulation of the SODD protein.
Although deregulation of cell death pathways beyond apoptosis, such as autophagy and necrosis, also can contribute to melanoma development, the roles of miRNAs in these pathways are not very well defined yet. One study suggested a cluster of miRNAs, miR-290–295, that targets autophagy-related genes in a melanoma cell line and alters melanoma cells' resistance to glucose starvation-induced autophagy (14). Further investigation is needed to define whether deregulation of miRNAs also helps melanoma cells acquire hallmark capacities through altering these cell death pathways.
In brief, the examples discussed above illustrate instances where the deregulation of single miRNAs or clusters of miRNAs in melanoma cells can lead to acquired cell death resistance resulting in an increase of melanoma progression and a decrease in therapeutic response of melanoma. This can be achieved through upregulation of prosurvival members of the Bcl-2 family (Bcl-2, BcL-X, Mcl-1), as well as through downregulation of various inhibitors of apoptosis (1, 13, 16, 17, 80).
ENABLING REPLICATIVE IMMORTALITY THROUGH EVASION OF SENESCENCE
Normal cells carry out a limited number of cell cycles before they enter into a functional but nonproliferative state called senescence, whereas cancer cells gain the ability to avoid senescence in order to continue replicating (31, 32). A few miRNAs have been shown to regulate senescence in melanoma cells, including miR-34a, miR-203, miR-205, and miR-125b (16, 27, 66, 67, 72). In cells with antisenescent capabilities, miRNA deregulation has been shown to affect telomere length and irreversible tumor suppressor inhibition and/or oncogene activation. A few examples discussed herein are miRNA regulation of telomere homeostasis, E2F/Rb or c-Jun pathways (8, 16, 27, 66, 67, 72).
One of the main regulatory mechanisms of limited replicative capacity is telomere shortening, and it is viewed as “a clocking device that determines the limited replicative potential of normal cells” (31). Cancer cells can achieve immortalization through increasing telomere length, directly or indirectly (31, 32). Recently, Boon et al. (8) found that miR-34a can induce telomere shortening by directly targeting PNUTS (also known as PPP1R10). Interestingly, inactivation of miR-34a by aberrant CpG methylation has been reported in 43.2% of melanoma cell lines and 62.5% of primary melanoma (52), and expressing miR-34a alone is sufficient to induce senescence in primary melanocytes (72). Moreover, miR-34a can also be induced by p53, and p53 is commonly downregulated in melanoma (34). Together, these results suggest that downregulation of miR-34a in melanoma may be achieved through multiple ways, and this likely helps melanoma cells evade senescence and gain immortality.
Deregulation of E2F family is essential for cancer development, and its members also play a role in senescence and thus in affecting cancer cell's capacity for immortality. E2F1, E2F5, and E2F3a allow a cell to avoid senescence by activating cell replication, whereas E2F3b induces senescence by inhibiting replication (16, 66). Two miRNAs have been reported to regulate E2F family members and senescence in melanoma, miR-205 and miR-203 (16, 65, 66).
As mentioned before, miR-205 has been shown to target E2F1 and E2F5 and to be downregulated in melanoma tissue compared with benign nevi (16), and ectopic expression of miR-205 induces a senescent phenotype in melanoma cells in vitro, through downregulating E2F1 and E2F5 (16, 65). MiR-203 has also been reported to be downregulated in human melanoma cells (16, 66), and ectopic miR-203 overexpression in melanoma cells also induces a senescent phenotype in vitro (66). 3′-UTR reporter assays confirmed that miR-203 targets E2F3a and E2F3b, and the oncogenic action of E2F3a outweighs the suppressive action of E2F3b in melanoma, as downregulation of miR-203 results in evading senescence (66).
MiR-125b has recently been linked to capability of evading senescence in melanoma development (40). Kappelmann and colleagues (40) demonstrated that miR-125b is downregulated in melanoma tissue samples and cell lines compared with normal melanocytes. Transfection experiments and luciferase assays support c-Jun, a key controller of tumor progression, as a gene target of miR-125b (40). In addition, overexpression of miR-125b induces a senescent phenotype and downregulation of miR-125b reverses that phenotype in melanoma cells in vitro (27). Together, these data suggest that loss of miR-125b may contribute to melanoma cells' ability to evade senescence.
INDUCING ANGIOGENESIS
As the cancer grows larger, the surrounding vasculature needs to grow to maintain nutrient and oxygen availability and to prevent waste and carbon dioxide accumulation. This occurs through a process called angiogenesis. In normal cells, there are counterbalanced angiogenic and antiangiogenic signaling mechanisms. Just as angiogenesis can be turned on in adults under circumstances like wound healing, angiogenesis can be abnormally activated in cancer to support the growing cell population (32).
Using an elegant, systematic, in vivo selection-based approach, a recent study identified a set of miRNAs (miR-1908, miR-199a-5p, and miR-199a-3p) that drive melanoma metastasis in part through promoting angiogenesis (69). Additional transcriptomic profiling and 3′-UTR luciferase assays identified that these miRNAs cooperatively target ApoE and DNAJA4 (a protein that also activates ApoE) and leads to maximal silencing of ApoE signaling (Fig. 1C) (69). Further functional studies indicated that overexpressing any of these three miRNAs is sufficient to increase lung metastatic colonization in a mouse model and silencing of each miRNA causes significant decrease of lung metastasis. Finally, these miRNAs are upregulated in the primary melanomas that metastasized compared with those that had not (69).
This study thus identified not only multiple miRNAs, but also a novel ApoE signaling pathway involved in angiogenesis for melanoma (69). This is an excellent example of how multiple miRNAs can cooperatively target a single important signaling pathway to promote melanoma development.
ACTIVATING INVASION AND METASTASIS
For cancer cells to extend beyond the borders of a distinct primary tumor, they must acquire the ability to invade adjacent tissue and to metastasize to different nonadjacent tissue via the bloodstream or lymphatics (32). Metastasis is an extremely complex process that involves not only the cancer cells acquiring multiple new abilities but also the tumor microenvironment's altering its function to permit and even promote cancer cells to invade and survive at the distant site (32). The ability of miRNAs to regulate numerous target genes makes them especially interesting in the multicomponent process of metastasis, but this field is still in its early stage with limited understanding of miRNA's roles in the multiple components of melanoma metastasis. Thus, this section will focus on the miRNAs contributing to melanoma cell's alteration in adhesion, migration, and invasion, and below are examples of miRNAs and targets for each of these critical components of metastasis. Interestingly, some miRNAs, such as miR-214 (70), play critical roles in melanoma metastasis through regulating more than one component of this process.
Adhesion
MiRNAs have been implicated in altering melanoma cell adhesion through regulating adhesion molecules directly or indirectly by regulating transcription factors controlling their transcription. Integrins are important in cell adhesion, and the increase in some of them correlates with increased metastatic potential. It had long been known that integrin-β3 is frequently overexpressed in melanoma. However, comprehensive studies were not able to uncover the mechanism of integrin-β3 overexpression until Muller and Bosserhoff (62) showed that integrin-β3 is a target of miR-let-7a, which is highly expressed in melanocytes but frequently lost in melanoma. In addition, this group has also shown that another miRNA, miR-196a, is involved in adhesion through its regulation of HOX-C8, a transcription factor regulating several proteins involved in adhesion (cadherin-11, calponin-1, and osteopontin) (61). MiR-196 is downregulated in melanoma cell lines and tissue samples compared with normal melanocytes. Moreover, functional studies demonstrated that loss of miR-196a allows increased invasion in vitro, probably through increased expression of HOX-C8 and thus HOX-C8-regulated adhesion proteins (61).
Deregulation of other miRNAs can also alter melanoma metastasis through regulating adhesion molecules, such as miR-573 and miR-214. A recent study links altered miR-573 expression to altered expression of additional cellular adhesion molecules including MCAM. MiRNA-573 overexpression reduces invasive potential, and subsequent MCAM overexpression is able to replicate the malignant phenotype (88).
A better characterized miRNA involved in melanoma adhesion and metastasis is miR-214, which was identified as a significantly upregulated miRNA in more aggressive melanoma in a melanoma progression model (70). This upregulation of miR-214 in highly aggressive melanoma has been validated in patient samples, and one of its direct targets is adhesion molecule integrin-β3 (70). However, miR-214 can regulate other components of metastasis in addition to cell adhesion, and these functions will be discussed below in more detail.
Migration
Penna et al. (70) also demonstrated that overexpressing miR-214 increases melanoma migration in vitro as well as extravasation from blood vessels and lung metastasis formation in vivo. Recently, the same group further elegantly illustrated the mechanism of how miR-214 coordinates melanoma metastasis through directly repressing TFAP2, resulting in upregulation of ALCAM (70, 71). TFAP2, an AP-2 family member, is a transcription factor that controls ALCAM expression transcriptionally. Interestingly, TFAP2 also upregulates miR-148 expression transcriptionally, and miR-148b can directly inhibit ALCAM in these cells. Knockdown of ALCAM or overexpressing of TFAP2 or miR-148b in miR-214-overexpressing melanoma cells abolishes the effects of miR-214. Furthermore, the expression patterns of miR-214, ALCAM, and miR-148b in human melanoma specimens are consistent with all the studies of function. These studies nicely demonstrated the critical role of miR-214 in melanoma metastasis.
Altered miR-9 expression can also affect migration in melanoma. Expression of miR-9 is downregulated in metastatic melanoma compared with in primary melanoma, and miR-9 is thought to target NF-KB1 directly. NF-KB1 is thereby upregulated in melanoma, which activates Snail1 expression. Snail1 is ultimately responsible for downregulating E-cadherin, which supports tumor metastasis. Functional studies performed by Liu and colleagues (51) revealed that miR-9 knockdown induces coordinated Snail1 upregulation and E-cadherin downregulation in vitro. Liu et al. (51) also tested the function of miR-9 in a mouse xenograft model and found that miR-9 overexpression significantly inhibits melanoma tumor growth and metastasis.
Invasion
As discussed above, miR-214 also plays a critical role in the melanoma metastasis including extravasation.
Some oncogenes that support invasion include enhancer of zeste homolog 2 (EZH2), MITF, c-MET, and Y box-binding protein 1 (YB1). Luo and colleagues (53) showed that miR-137 levels are downregulated in melanoma associated with short survival compared with long survival, and miR-137 levels are downregulated in melanoma compared with normal human melanocytes. These researchers also showed that miR-137 regulates the oncogenes mentioned above, i.e., EZH2, MITF, c-MET, and YB1 (Fig. 1A). It was found that knocking out any and/or all of the named oncogenes with siRNAs is sufficient to reproduce the cancer phenotype associated with miR-137 downregulation. miR-137 mimic inhibits invasive behavior in melanoma cell lines in vitro (53). This study indicates that miR-137 regulates invasive capabilities and that the loss of miR-137 in late-stage melanoma may lead to a poor outcome and reduced survival.
As mentioned above, miR-137 regulates MITF, an oncogene of particular interest, because it is a master regulator of differentiation, proliferation, and survival in melanocyte development. MITF demonstrates the concept of phenotypic plasticity and is a well-studied key regulator of invasive melanoma (5). Loss of more than one miRNA has been found to upregulate MITF expression in melanoma (Fig. 1B). Bemis and colleagues (6) first showed that miR-137, located in a genomic hot spot for melanoma susceptibility, regulates MITF. Similarly, depressed levels of miR-148, miR-182, and miR-340 have been correlated with high levels of MITF and a proliferative phenotype (28, 30, 78). The activation of MITF through miRNA downregulation adds to the complexity of MITF expression and function. A common phenomenon occurs in cancer cells and other rapidly replicating cells, which is the use of alternative polyA termination sites within the UTR (30). It has been well studied by several groups that miRNA-binding sites may be eliminated by the use of an alternative polyA termination site in the UTR (55, 56). Goswami et al. (28) took this finding a step further and also showed that RNA binding proteins may interfere with miRNA regulation by binding to a region of RNA harboring a miRNA binding site, thereby blocking access of the miRNA to the mRNA target.
The miRNA regulation of MITF is even more complicated because MITF is not only regulated by miRNAs, but it is also a transcriptional regulator that affects the expression of another miRNA, miR-211, located in the intron of an MITF controlled gene, TRPM1 (47, 57, 58).However, under certain conditions, miR-211 can also affect MITF expression through inhibiting Brn2 expression, adding further complexity to regulation of MITF (9).
GENOME INSTABILITY AND MUTATION
Genome instability is one of the major enabling hallmarks in many cancers, and it refers to an increased frequency of alterations in the genome of cancer cells. The loss or gain of genetic material and somatic mutations facilitates cells' acquiring other hallmark capabilities and allows for accelerated cancer development and progression (64). This hallmark is not well characterized in melanoma, and examples of direct regulation of genome instability by miRNAs in melanoma are limited. However, it is likely that miRNAs regulate cellular processes that are required for the DNA damage response and DNA repair pathways (89).
The ability of the genome to control its integrity relies on intact cell maintenance mechanisms (32). DNA damage checkpoint genes (p53 and ATM) and genes regulating the cell cycle progression (CDKN2A, cyclin D, and Rb/E2F1) have been implicated in cancer cell genomic instability (64). Being called the guardian of the genome, p53 plays a central role in maintaining cell genome stability. As mentioned before, p53 is rarely mutated in melanoma but commonly downregulated in melanoma. This is in part indirectly caused by the downregulation of miR-18b in melanoma (17). With regard to cell cycle regulatory genes, deregulation of miR-let-7b and miR-193b has been implicated in an increase of cyclin D1 (12, 77, 90), and miR-205 and miR-203 have been shown to regulate E2F family members (16, 65, 66). Thus, deregulation of these miRNAs is also likely to alter the genomic stability of melanoma cells.
Genomic instability also contributes to a cancer cell's ability to evade miRNA regulation. First, miRNAs known to function in a variety of cancers including melanoma are located in regions of genomic instability as are their regulatory proteins such as Dicer1 (5, 92). Second, somatic mutations including those in targets of miRNAs, miRNA regulators, and the miRNAs themselves (although fairly rare) have been reported in melanoma and other cancers (2, 6, 41). Single nucleotide mutations in miRNAs, called MIRSNPs, can alter the miRNA expression levels or the miRNA-mRNA binding affinity, thereby reducing targeted protein inhibition (91). These changes in miRNA-regulated gene expression have been shown to yield acquired cancer capabilities. Yamashita et al. (91) investigated a miR-146a MIRSNP and found that the rs2910164 G-to-C polymorphism was correlated with a more aggressive proliferative, migratory, and invasive melanoma phenotype. Mutations may also occur within the 3′-UTR miRNA-mRNA binding regions, thereby altering expression of that gene through altering a miRNA regulating effect (93). The KRAS variant, shown to correlate with increased risk for melanoma (11), is thought to contain a 3′-UTR single nucleotide polymorphism (SNP) within the miR-let-7a binding site and to confer resistance to the inhibitory effects of miR-let-7a regulation (41). While it is not specific to melanoma, Kundu and coworkers (41) recently identified many molecular consequences of the KRAS variant on cancer cell lines, and those findings may inform future melanoma-specific studies. Interestingly, SNPs and point mutations in miRNAs may offer prognostic medical value as they have been linked to disease susceptibility (48) and therapeutic response (2) and may be predictive of disease (24) in multiple cancers.
In summary, miRNAs might contribute to genomic instability, and genomic instability may also help cancer cells evade control by miRNAs.
TUMOR-PROMOTING INFLAMMATION
Inflammatory cells are often attracted to sites where neoplasms are growing. There have been many clues that, while the immune system attempts to control the growth of cancer cells, the cells and microenvironment acquire the ability to evade immune surveillance mechanisms. Moreover, the inflammatory cells have a paradoxical role as they release progrowth, -survival, -angiogenesis, -invasion, and -metastasis factors that help the tumor cells acquire the capabilities necessary for advanced malignant progression (32). Research supports the notion that miRNAs are involved in these inflammatory processes that promote melanoma development.
On one hand, inflammatory cytokines can cause miRNA deregulation. For example, interferon-γ is a proinflammatory cytokine that is known to upregulate the miR-29 family in melanoma (76), which in turn may activate further immune response. On the other hand, deregulated miRNAs may activate the immune response, and miR-29a and miR-21 have both been shown to directly bind Toll-like receptors, thereby activating prometastatic inflammation (21). While much remains unclear about the connection between inflammation and cancer, these studies illustrate instances where miRNAs act as mediators. Furthermore, the finding of this direct binding of miRNAs to Toll-like receptors is the very beginning of a new and developing field to determine the role of extracellular RNA, which includes many miRNAs (3). Many miRNAs have been associated with a subset of extracellular vesicles also known as exosomes. The role of these extracellular miRNAs is yet to be completely defined, but this is a rapidly growing and interesting area of miRNA biology in melanoma (68). The idea that miRNAs may act in a manner similar to cytokines is novel, the literature is compelling, and this may provide a new area of therapeutic intervention in the future.
APPLICATIONS OF miRNAS IN DEVELOPING MELANOMA DIAGNOSTIC BIOMARKERS AND THERAPIES
With the significant difference between survival rates in early malignant melanoma (90%) vs. stage III and stage IV metastatic melanoma (10%), there is an urgent need for earlier detection and better therapy for melanoma (83). Understanding how miRNAs are deregulated throughout the development of melanoma can lead to better biomarkers or better treatment strategies for melanoma (63).
Diagnostic Biomarkers
Here are a few examples of potential miRNA serum diagnostic/prognostic biomarkers for developing noninvasive and earlier detection of melanoma (24, 39). Kanemaru and colleagues (39) investigated miR-221, known to be overexpressed in malignant melanoma cells, as a potential biomarker. They found that serum miR-221 levels indeed increase significantly from healthy human controls to primary melanoma subjects and then again from primary melanoma to stage I–IV malignant melanoma subjects. Moreover, a longitudinal study revealed that miR-221 levels decrease after surgical melanoma removal and increase again with disease recurrence. Thus, miR-221 may be very useful for monitoring patients with known melanoma diagnoses (39).
Friedman and coworkers (24) performed a miRNA regression analysis comparing serum samples from patients with melanoma against serum samples from patients with other cancers, systemic inflammatory disease, and healthy volunteers. They found that miR-182 and -221 are specific for melanoma compared with all three controls and that miR-182, -221, and -15b are predictive for metastasis (24). The same group also revealed five signature miRNAs (miR-150, miR-15b, miR-199a-5p, miR-33a, miR-424) that predict high vs. low recurrent-free survival groups (24). These examples indicate instances where serum miRNA biomarkers offer diagnostic and prognostic information, and the hope is that these miRNAs may translate to improved patient care and outcomes.
Therapies
MiRNA-based therapies have been regarded as an exciting new area of drug development for several reasons. Deregulation of miRNAs has been implicated in many diseases, and miRNA mimics and inhibitors are small, which make them good candidates for drug development (63). The first published clinical trial with miRNA-targeted therapy was to treat hepatitis C virus (HCV) (38). In HCV, miR-122 binds to the virus and protects it from degradation. A drug called miravirsen is an antisense sequence that was designed to bind to and sequester miR-122 and thus acts as an inhibitor of miR-122. The drug was injected subcutaneously five times per week for 29 days, and the results revealed a dose-dependent reduction in HCV RNA (38).
MiRNA-based therapies have also been considered as an exciting new class of anticancer drugs since some miRNAs can affect multiple oncogenic pathways simultaneously. The first clinical trial for treating cancers using miRNA is just recently under way as a Phase I trial by Mirna Therapeutics, and miR-34 mimic as a replacement of miRNA will be delivered intravenously to the patients (http://clinicaltrials.gov/ct2/show/NCT01829971). Many studies aim to find miRNA targets for development of melanoma treatment, using antisense or a replacement approach, and the target can be used either as a single agent or as an adjuvant to other drugs. Below are a few examples of potential miRNA targets for developing therapy.
Given that many miRNAs are upregulated in melanoma, a similar inhibitor therapy strategy to the antisense to miR-122 of HCV treatment might offer therapeutic benefit for patients with melanoma. Furthermore, Huynh and coworkers (35) investigated administration of antisense-miR-182 molecules against melanoma liver metastasis in a mouse model and found that the antisense-miR-182 decreases metastatic function and overall tumor burden with no evidence of significant toxicity.
A few reports that replace miRNA in mouse models support the potential of miRNA replacement therapy for combating melanoma. After showing that miR-18b is downregulated in melanoma and indirectly inhibits p53, Dar and colleagues (17) injected high-miR-18b-expressing melanoma cells subcutaneously into mice tumors, resulting in a significant decrease in the tumor cell volume and growth compared with control. Similarly, after showing that miR-573 is downregulated in melanoma and targets MCAM, Wang et al. (88) replaced miR-573 in a murine xenograft model, thereby reducing invasive potential and tumor growth compared with the control group. These experiments show that by altering the levels of a miRNA, expression of target genes can also be altered, and therapeutic benefit may follow.
Replacing miRNAs targeting Mcl-1 have been shown to improve melanoma cell response to chemotherapy agents ABT-263 and ABT-737 (13, 43). By administering miRNAs that target Mcl-1 in conjunction with chemotherapy drugs, an improved therapeutic response can be achieved (13, 43). Chen and coworkers (13) found that a single miRNA-193b targets Mcl-1 directly and that miR-193b upregulation sensitizes melanoma cells to ABT-737. In addition, Lam and colleagues (43) used a systematic screen and identified a panel of 19 miRNAs that, when administered with ABT-263, sensitized melanoma cells to ABT-263-induced apoptosis in vitro. Of these, 12 were associated with reduced Mcl-1 expression, and 10 miRNAs were shown to directly target Mcl-1. These studies are examples where miRNA as a drug adjuvant can improve current melanoma drug therapies.
Challenges of MiRNA-Based Therapy
Reports have been published recently to summarize the therapeutic potential as well as the challenges of miRNA-based therapy (25, 37, 63, 84–86), and they support the idea that a better understanding of the following items is necessary to make miRNA-based therapy a reality: toxicity in normal tissues, improved delivery systems that support reagent stability, as well as clearance mechanisms for reagents to be delivered. Since the effects of miRNAs on gene expression can be complex as discussed earlier, miRNA-based therapy might affect gene expression of unanticipated targets and cause off-target toxicity. Thus, more has to be learned about these toxic off-target effects of miRNA therapies. As the effects of miRNA-based therapy are further illustrated using appropriate preclinical in vivo models, better approaches can be developed to minimize this toxicity. In addition, various delivery platforms are being studied, including liposomal, viral, polymer, and nanoparticle-based systems. The ultimate goal is to find delivery systems that stabilize the reagents to be delivered effectively to diseased/cancerous tissues and limit toxicity to normal tissues.
As the pharmacokinetics are better understood and the therapeutic delivery systems are improved, miRNA-based therapy has already entered clinical trials with promising preliminary results (38). The potential for miRNA-based therapies to improve clinical outcomes for patients has energized drug companies and researchers alike. With the on-going studies to address the challenges discussed above, one would hope that patients will experience the benefits of miRNA-based therapy in the near future.
DISCUSSION/FUTURE DIRECTIONS
MiRNAs offer an exciting, fast growing, and challenging field because they give a new direction and robust potential for improving outcomes in cancer and other diseases. While the hallmarks of cancer model offers a framework for organizing the complex miRNA-mediated biological events that have been shown to contribute to melanoma development, many of the cancer hallmark-related miRNAs and characteristics are still not well defined, and better understanding is needed. Other hallmarks and characteristics are emerging to better describe acquired cell function in cancer, and these include deregulating cellular energetics and avoiding immune destruction (32). In addition, complex interactions between cancer cells and tumor microenvironment also play important roles in cancer development (32). Moreover, there is still much to be learned about the role of miRNAs within the complex networks discussed herein that drive cancer development and progression. In further study of these areas, one should always consider the potential role of miRNAs.
To date, miRNAs have been implicated as potential biomarkers, as targeted therapeutics, and as adjuvants to current therapies for melanoma. While much has been learned about miRNAs in melanoma development, more studies are needed to progress toward clinical application of these miRNA biomarkers and therapeutics.
GRANTS
This work was supported in part by a Veterans Administration merit grant from the Department of Veterans Affairs (Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development) to D. A. Norris and by a Southwestern Skin Cancer SPORE Pilot project to Y. G. Shellman.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: P.E.B., L.B., and Y.G.S. prepared figures; P.E.B., L.B., and Y.G.S. drafted manuscript; P.E.B., L.B., D.A.N., and Y.G.S. edited and revised manuscript; Y.G.S. conception and design of research; Y.G.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We apologize to all those colleagues whose important work is not cited because of space consideration and our focus for this review.
REFERENCES
- 1.Alla V, Kowtharapu BS, Engelmann D, Emmrich S, Schmitz U, Steder M, Putzer BM. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell Cycle 11: 3067–3078, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcaroli JJ, Quackenbush KS, Powell RW, Pitts TM, Spreafico A, Varella-Garcia M, Bemis L, Tan AC, Reinemann JM, Touban BM, Dasari A, Eckhardt SG, Messersmith WA. Common PIK3CA mutants and a novel 3′ UTR mutation are associated with increased sensitivity to saracatinib. Clin Cancer Res 18: 2704–2714, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 108: 5003–5008, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, function. Cell 116: 281–297, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Bell RE, Levy C. The three M's: melanoma, microphthalmia-associated transcription factor and microRNA. Pigm Cell Melanoma Res 24: 1088–1106, 2011. [DOI] [PubMed] [Google Scholar]
- 6.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res 68: 1362–1368, 2008. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya A, Schmitz U, Wolkenhauer O, Schönherr M, Raatz Y, Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene 32: 3175–3183, 2013. [DOI] [PubMed] [Google Scholar]
- 8.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature 495: 107–110, 2013. [DOI] [PubMed] [Google Scholar]
- 9.Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA, Hayward NK. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigm Cell Melanoma Res 24: 525–537, 2011. [DOI] [PubMed] [Google Scholar]
- 10.Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ, Chmielowski B, Lutzky J, Pavlick AC, Fusco A, Cane L, Takebe N, Vemula S, Bouvier N, Bastian BC, Schwartz GK. KIT as a therapeutic target in metastatic melanoma. JAMA 305: 2327–2334, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan E, Patel R, Nallur S, Ratner E, Bacchiocchi A, Hoyt K, Szpakowski S, Godshalk S, Ariyan S, Sznol M, Halaban R, Krauthammer M, Tuck D, Slack FJ, Weidhaas JB. MicroRNA signatures differentiate melanoma subtypes. Cell Cycle 10: 1845–1852, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol 176: 2520–2529, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Zhang X, Lentz C, Abi-Daoud M, Pare GC, Yang X, Feilotter HE, Tron VA. miR193b regulates Mcl-1 in melanoma. Am J Pathol 179: 2162–2168, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Liersch R, Detmar M. The miR-290–295 cluster suppresses autophagic cell death of melanoma cells. Sci Rep 2: 808, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couts KL, Anderson EM, Gross MM, Sullivan K, Ahn NG. Oncogenic B-Raf signaling in melanoma cells controls a network of microRNAs with combinatorial functions. Oncogene 32: 1959–1970, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem 286: 16606–16614, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, Nosrati M, Sagebiel R, Miller JR, 3rd, Kashani-Sabet M. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst 105: 433–442, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature 417: 949–954, 2002. [DOI] [PubMed] [Google Scholar]
- 19.Deng Y, Deng H, Bi F, Liu J, Bemis LT, Norris D, Wang XJ, Zhang Q. MicroRNA-137 targets carboxyl-terminal binding protein 1 in melanoma cell lines. Int J Biol Sci 7: 133–137, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Easty DJ, Gray SG, O'Byrne KJ, O'Donnell D, Bennett DC. Receptor tyrosine kinases and their activation in melanoma. Pigm Cell Melanoma Res 24: 446–461, 2011. [DOI] [PubMed] [Google Scholar]
- 21.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 109: E2110–E2116, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Care A. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res 68: 2745–2754, 2008. [DOI] [PubMed] [Google Scholar]
- 23.Felicetti F, Errico MC, Segnalini P, Mattia G, Care A. MicroRNA-221 and -222 pathway controls melanoma progression. Exp Rev Anticancer Ther 8: 1759–1765, 2008. [DOI] [PubMed] [Google Scholar]
- 24.Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, Berman RS, Shapiro RL, Pavlick AC, Hernando E, Baker A, Shao Y, Osman I. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med 10: 155, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9: 775–789, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glud M, Gniadecki R. MicroRNAs in the pathogenesis of malignant melanoma. J Eur Acad Dermatol Venereol 27: 142–150, 2013. [DOI] [PubMed] [Google Scholar]
- 27.Glud M, Manfe V, Biskup E, Holst L, Dirksen AM, Hastrup N, Nielsen FC, Drzewiecki KT, Gniadecki R. MicroRNA miR-125b induces senescence in human melanoma cells. Melanoma Res 21: 253–256, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Goswami S, Tarapore RS, Teslaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J Biol Chem 285: 20532–20540, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Gu W, Wang X, Zhai C, Zhou T, Xie X. Biological basis of miRNA action when their targets are located in human protein coding region. PLoS One 8: e63403, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haflidadottir BS, Bergsteinsdottir K, Praetorius C, Steingrimsson E. miR-148 regulates Mitf in melanoma cells. PLoS One 5: e11574, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 100: 57–70, 2000. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011. [DOI] [PubMed] [Google Scholar]
- 33.Hartman ML, Czyz M. Anti-apoptotic proteins on guard of melanoma cell survival. Cancer Lett 331: 24–34, 2013. [DOI] [PubMed] [Google Scholar]
- 34.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huynh C, Segura MF, Gaziel-Sovran A, Menendez S, Darvishian F, Chiriboga L, Levin B, Meruelo D, Osman I, Zavadil J, Marcusson EG, Hernando E. Efficient in vivo microRNA targeting of liver metastasis. Oncogene 30: 1481–1488, 2011. [DOI] [PubMed] [Google Scholar]
- 36.Igoucheva O, Alexeev V. MicroRNA-dependent regulation of cKit in cutaneous melanoma. Biochem Biophys Res Commun 379: 790–794, 2009. [DOI] [PubMed] [Google Scholar]
- 37.Ishida M, Selaru FM. miRNA-based therapeutic strategies. Curr Anesthesiol Rep 1: 63–70, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med 368: 1685–1694, 2013. [DOI] [PubMed] [Google Scholar]
- 39.Kanemaru H, Fukushima S, Yamashita J, Honda N, Oyama R, Kakimoto A, Masuguchi S, Ishihara T, Inoue Y, Jinnin M, Ihn H. The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker. J Dermatol Sci 61: 187–193, 2011. [DOI] [PubMed] [Google Scholar]
- 40.Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 32: 2984–2991, 2013. [DOI] [PubMed] [Google Scholar]
- 41.Kundu ST, Nallur S, Paranjape T, Boeke M, Weidhaas JB, Slack FJ. KRAS alleles: the LCS6 3′UTR variant and KRAS coding sequence mutations in the NCI-60 panel. Cell Cycle 11: 361–366, 2012. [DOI] [PubMed] [Google Scholar]
- 42.Kunz M. MicroRNAs in melanoma biology. Adv Exp Med Biol 774: 103–120, 2013. [DOI] [PubMed] [Google Scholar]
- 43.Lam LT, Lu X, Zhang H, Lesniewski R, Rosenberg S, Semizarov D. A microRNA screen to identify modulators of sensitivity to BCL2 inhibitor ABT-263 (navitoclax). Mol Cancer Ther 9: 2943–2950, 2010. [DOI] [PubMed] [Google Scholar]
- 44.Lane DP. Cancer. p53, guardian of the genome. Nature 358: 15–16, 1992. [DOI] [PubMed] [Google Scholar]
- 45.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19: 1175–1183, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9: 749–758, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, Chen PH, Li S, Fletcher AL, Yokoyama S, Scott KL, Garraway LA, Song JS, Granter SR, Turley SJ, Fisher DE, Novina CD. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell 40: 841–849, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Duan R, Kooy F, Sherman SL, Zhou W, Jin P. Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet 46: 358–360, 2009. [DOI] [PubMed] [Google Scholar]
- 49.Li W, Sanki A, Karim RZ, Thompson JF, Soon Lee C, Zhuang L, McCarthy SW, Scolyer RA. The role of cell cycle regulatory proteins in the pathogenesis of melanoma. Pathology 38: 287–301, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. J Invest Dermatol 129: 1666–1674, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Kumar SM, Lu H, Liu A, Yang R, Pushparajan A, Guo W, Xu X. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-kB1-Snail1 pathway in melanoma. J Pathol 226: 61–72, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7: 2591–2600, 2008. [DOI] [PubMed] [Google Scholar]
- 53.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, Diederichs S, Eichmuller SB. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol 133: 768–775, 2013. [DOI] [PubMed] [Google Scholar]
- 54.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA 104: 9667–9672, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majoros WH, Ohler U. Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics 8: 152, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, Ray A, Perera RJ. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One 5: e13779, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res 64: 509–516, 2004. [DOI] [PubMed] [Google Scholar]
- 59.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med 355: 51–65, 2006. [DOI] [PubMed] [Google Scholar]
- 60.Miller LA, Goldstein NB, Johannes WU, Walton CH, Fujita M, Norris DA, Shellman YG. BH3 mimetic ABT-737 and a proteasome inhibitor synergistically kill melanomas through Noxa-dependent apoptosis. J Invest Dermatol 129: 964–971, 2009. [DOI] [PubMed] [Google Scholar]
- 61.Mueller DW, Bosserhoff AK. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int J Cancer 129: 1064–1074, 2011. [DOI] [PubMed] [Google Scholar]
- 62.Muller DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27: 6698–6706, 2008. [DOI] [PubMed] [Google Scholar]
- 63.Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93: 98–104, 2013. [DOI] [PubMed] [Google Scholar]
- 64.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228, 2010. [DOI] [PubMed] [Google Scholar]
- 65.Noguchi S, Iwasaki J, Kumazaki M, Mori T, Maruo K, Sakai H, Yamada N, Shimada K, Naoe T, Kitade Y, Akao Y. Chemically modified synthetic microRNA-205 inhibits the growth of melanoma cells in vitro and in vivo. Mol Ther 21: 1204–1211, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noguchi S, Mori T, Otsuka Y, Yamada N, Yasui Y, Iwasaki J, Kumazaki M, Maruo K, Akao Y. Anti-oncogenic microRNA-203 induces senescence by targeting E2F3 protein in human melanoma cells. J Biol Chem 287: 11769–11777, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nyholm AM, Holst LMB, Lerche C, Manfe V, Biskup E, Glud M, Hastrup N, Thomsen BM, Pless V, Rosbjerg A, Wulf HC, Gniadecki R. MiR-125b induces cellular senescence in melanocytes and malignant melanoma. J Invest Dermatol 131: S107, 2011. [Google Scholar]
- 68.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18: 883–891, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 151: 1068–1082, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pitta C, Pinatel E, Stadler MB, Provero P, Bernengo MG, Osman I, Taverna D. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J 30: 1990–2007, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Penna E, Orso F, Cimino D, Vercellino I, Grassi E, Quaglino E, Turco E, Taverna D. miR-214 coordinates melanoma progression by upregulating ALCAM through TFAP2 and miR-148b downmodulation. Cancer Res 73: 4098–4111, 2013. [DOI] [PubMed] [Google Scholar]
- 72.Ren G, Feng J, Datar I, Yeung AH, Saladi SV, Feng Y, de la Serna I, Yeung KC. A micro-RNA connection in BRaf(V600E)-mediated premature senescence of human melanocytes. Int J Cell Biol 2012: 913242, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reuland SN, Goldstein NB, Partyka KA, Cooper DA, Fujita M, Norris DA, Shellman YG. The combination of BH3-mimetic ABT-737 with the alkylating agent temozolomide induces strong synergistic killing of melanoma cells independent of p53. PLoS One 6: e24294, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reuland SN, Goldstein NB, Partyka KA, Smith S, Luo Y, Fujita M, Gonzalez R, Lewis K, Norris DA, Shellman YG. ABT-737 synergizes with Bortezomib to kill melanoma cells. Biol Open 1: 92–100, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rigel DS, Russak J, Friedman R. The evolution of melanoma diagnosis: 25 years beyond the ABCDs. CA Cancer J Clin 60: 301–316, 2010. [DOI] [PubMed] [Google Scholar]
- 76.Schmitt MJ, Philippidou D, Reinsbach SE, Margue C, Wienecke-Baldacchino A, Nashan D, Behrmann I, Kreis S. Interferon-gamma-induced activation of signal transducer and activator of transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun Signal 10: 41, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18: 549–557, 2008. [DOI] [PubMed] [Google Scholar]
- 78.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA 106: 1814–1819, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol 244: 170–179, 2002. [DOI] [PubMed] [Google Scholar]
- 80.Shellman Y, Reuland S, Smith S, Bemis L, Goldstein N, Almeida A, Partyka K, Norris D. Therapeutic potential of miR-26a and its target SODD in treating melanoma. Pigm Cell Melanoma Res 25: 888, 2012. [Google Scholar]
- 81.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev 84: 1155–1228, 2004. [DOI] [PubMed] [Google Scholar]
- 82.Solus JF, Kraft S. Ras, Raf, and MAP kinase in melanoma. Adv Anat Pathol 20: 217–226, 2013. [DOI] [PubMed] [Google Scholar]
- 83.Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice DA, Higgs BW, Richman L, Jallal B, Ranade K, Yao Y. A novel oncogenic role for the miRNA-506–514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 31: 1558–1570, 2012. [DOI] [PubMed] [Google Scholar]
- 84.Tagliaferri P, Rossi M, Di Martino MT, Amodio N, Leone E, Gulla A, Neri A, Tassone P. Promises and challenges of microRNA-based treatment of multiple myeloma. Curr Cancer Drug Targets 12: 838–846, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. The therapeutic potential of MicroRNAs in cancer. Cancer J 18: 275–284, 2012. [DOI] [PubMed] [Google Scholar]
- 86.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res 110: 496–507, 2012. [DOI] [PubMed] [Google Scholar]
- 87.Voller D, Ott C, Bosserhoff A. MicroRNAs in malignant melanoma. Clin Biochem 46: 909–917, 2013. [DOI] [PubMed] [Google Scholar]
- 88.Wang HF, Chen H, Ma MW, Wang JA, Tang TT, Ni LS, Yu JL, Li YZ, Bai BX. miR-573 regulates melanoma progression by targeting the melanoma cell adhesion molecule. Oncol Rep 30: 520–526, 2013. [DOI] [PubMed] [Google Scholar]
- 89.Wang Y, Taniguchi T. MicroRNAs and DNA damage response: implications for cancer therapy. Cell Cycle 12: 32–42, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu D, Tan J, Zhou M, Jiang B, Xie H, Nie X, Xia K, Zhou J. Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett 4: 941–946, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamashita J, Iwakiri T, Fukushima S, Jinnin M, Miyashita A, Hamasaki T, Makino T, Aoi J, Masuguchi S, Inoue Y, Ihn H. The rs2910164 G>C polymorphism in microRNA-146a is associated with the incidence of malignant melanoma. Melanoma Res 23: 13–20, 2013. [DOI] [PubMed] [Google Scholar]
- 92.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 103: 9136–9141, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ziebarth JD, Bhattacharya A, Cui Y. Integrative analysis of somatic mutations altering microRNA targeting in cancer genomes. PLoS One 7: e47137, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


