Abstract
Ependymoma (EPN) in childhood is a brain tumor with substantial mortality. Inflammatory response has been identified as a molecular signature of high-risk Group A EPN. To better understand the biology of this phenotype and aid therapeutic development, transcriptomic data from Group A and B EPN patient tumor samples, and additional malignant and normal brain data, were analyzed to identify the mechanism underlying EPN Group A inflammation. Enrichment of interleukin-6 (IL6) and STAT3 pathway genes were found to distinguish Group A EPN from Group B EPN and other brain tumors, implicating an IL6 activation of STAT3 mechanism. EPN tumor cell growth was shown to be dependent on STAT3 activity, as demonstrated using shRNA knockdown and pharmacologic inhibition of STAT3 that blocked proliferation and induced apoptosis. The inflammatory factors secreted by EPN tumor cells were shown to reprogram myeloid cells and this paracrine effect was characterized by a significant increase in pSTAT3 and IL8 secretion. Myeloid polarization was shown to be dependent on tumor secretion of IL6, and these effects could be reversed using IL6 neutralizing antibody or IL6 receptor-targeted therapeutic antibody tocilizumab. Polarized myeloid cell production of IL8 drove un-polarized myeloid cells to up-regulate CD163 and to produce a number of pro-inflammatory cytokines. Collectively, these findings indicate that constitutive IL6/STAT3 pathway activation is important in driving tumor growth and inflammatory crosstalk with myeloid cells within the Group A EPN microenvironment. Effective design of Group A-targeted therapy for children with EPN may require reversal of this potentially immunosuppressive and pro-tumor pathway.
Keywords: ependymoma, inflammation, interleukin-6, STAT3, myeloid, microenvironment
Introduction
Molecular sub-groups of primary EPN have recently been defined, including two main EPN subgroups arising in the posterior fossa, termed Groups A and B, that confer different biologic phenotypes and clinical courses (1–3). Phenotypically, Group A tumors occur in younger children and are characterized by up-regulation of a variety of pathways, including angiogenesis, mesenchymal cell differentiation, cell proliferation and inflammatory response, that are hallmarks of aggressive tumor biology. Group A designation confers increased recurrence risk and poorer overall outcome. The difference in outcome for Group A and Group B tumors is seen at relapse where most Group B patients can be salvaged by surgery and radiation while most Group A patients will go on to subsequent relapses which almost invariably results in death (3). Group B tumors generally occur in adolescents and young adults, and overexpress genes involved in ciliogenesis, microtubule assembly, and oxidative metabolism.
A recent study by our group identified inflammatory response as the predominant ontology distinguishing Group A from Group B tumors at presentation (normalized enrichment score (NES)=3.98; false discovery rate (FDR) q-value<0.001) (3). Up-regulation of inflammatory genes was also seen in independent Group A cohorts (1,2). In the original study that delineated posterior fossa EPN into Groups A and B, inflammatory response genes were significantly enriched in the Heidelberg-cohort Group A tumors (NES=1.93; FDR q=0.016) (1). A second independent study also identified inflammatory response as the second highest enriched geneset in Group A EPN (FDR q<0.0005) (2). It is widely accepted that many tumors arise from and are promoted by an inflammatory microenvironment (4,5). This hallmark of cancer is thought to be tumorigenic in part due to suppression of host antitumor immunity. Accordingly, support for the existence of an immunosuppressive phenotype in Group A EPN was previously demonstrated by the identification of an exhausted phenotype in tumor-infiltrating T cells (3). Abrogation of detrimental immune phenotypes in cancer has now emerged as a novel goal for experimental therapeutic approaches (6). To identify similar clinical approaches for refractory Group A EPN, this study sought to identify the mechanism driving the inflammatory phenotype of these tumors. We show that a potential molecular mechanism underlying Group A EPN inflammatory phenotype is constitutive activation of the IL6/STAT3 pathway and crosstalk between tumor and immune cells within the tumor microenvironment (TME).
Our study is the first to identify persistent IL6/STAT3 activation as a driver of tumor growth and an associated inflammatory microenvironment in Group A EPN. Targeting the IL6/STAT3 pathway, either directly or by modifying the pro-inflammatory microenvironment, is a potential therapeutic approach to improve patient survival and outcomes in this deadly tumor of childhood.
Methods
Transcriptomic analysis
The primary EPN study cohort consisted of 21 primary Group A and 20 primary Group B posterior fossa EPN and 9 supratentorial EPN. A broader transcriptomic analyses utilized EPN samples obtained at relapse (8 Group A and 6 Group B) and other pediatric and adult brain tumors. This consisted of 17 atypical teratoid/rhabdoid tumors (AT/RT); 14 pediatric and 7 adult high-grade gliomas (HGG); 8 sonic hedgehog, 5 Group 3 and 7 Group 4 medulloblastomas (MED); and 15 pilocytic astrocytoma (PA). Normal brain samples (n=13) were also included in this analysis, obtained from autopsy and epilepsy surgery from both infra- and supratentorial anatomic sites. All patient samples were obtained with consent (COMIRB 95–500). Tumor samples were processed identically at our institution and analyzed using the Human Genome U133plus2 Array (Affymetrix) platform as described previously (3). Microarray .CEL datafiles were background corrected and normalized using the guanine cytosine robust multi array average (gcRMA) algorithm resulting in log2 expression values (7). To reduce error associated with multiple testing, a filtered list was created containing the highest expressed probe across all samples for each gene that possessed multiple probe sets. This list was further filtered to remove probe sets that were expressed below a threshold level that denoted absence of expression in any sample. These microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (8) and are publicly accessible through GEO Series accession number GSE66354 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66354). Primary EPN were assigned to consensus molecular subgroups using nonnegative matrix factorization (NMF) available through the Broad Institute Gene Pattern platform (Supplementary figure 1) (9).
Geneset enrichment analysis (GSEA) was used to examine enrichment of genes in predefined reference sets that are based on biologic knowledge using tools available from the Broad Institute Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb) (10).
Tumor versus myeloid cell transcriptome analysis
To determine the distribution of transcripts of interest between tumor cells and tumor-associated myeloid cells we performed transcriptomic analysis on populations that had been isolated from patient surgical samples by tissue disaggregation and flow cytometric sorting as described previously (3). Briefly, resected tumor was finely minced with a razor and further triturated by vigorous pipetting. A single-cell suspension was obtained by passing the sample through a 70µm cell strainer (Becton Dickinson) of sufficiently large pore size to permit passage of all immune and tumor cells but not clumped tumor cells. Disaggregated cells were viably frozen in standard freezing media containing 10% DMSO and stored in liquid nitrogen for subsequent analysis. Isolation of tumor and myeloid subpopulations was performed by staining disaggregated samples with anti-CD45-FITC and anti-CD11b-APC antibodies (Becton Dickinson clones 2D1 and ICRF44 respectively) followed by flow sorting using a Beckman-Coulter MoFlo XDP-100. RNA was isolated from sorted cells using a microRNeasy kit (Qiagen). Extracted RNA was amplified using the Nugen WT-Ovation One-Direct System (NuGEN Technologies) that reverse transcribes RNA to cDNA, which is amplified during SPIA amplification, a linear isothermal DNA amplification process resulting in end-products of single-strand DNA. Amplified RNA was labeled (NuGEN Encore Biotin Module) and hybridized to Affymetrix Human Genome U133plus2 chips. Microarray .CEL file data were normalized as described above.
Cytokine release assay
Viably frozen disaggregated tumor samples were thawed and flow sorted to separate immune cells from tumor cells as previously described (3). Tumor cells were cultured in 1ml optimem media supplemented with 15% FBS (O15) for 96 hours on ultra-low attachment plates. Media were harvest and immediately frozen at −80°C for later use. A high sensitivity Milliplex Map kit (Millipore) was used to measure the concentration of 13 common cytokines (GM-CSF, IFNγ, IL1β, IL2, IL4, IL5, IL6, IL7, IL8, IL10, IL12 [p70], IL13, and TNFα) per manufacturer’s instructions. IL6 secretion in media from cell lines was also measured using Quantikine ELISA human IL6 (R&D Systems) according to manufacturer’s instructions.
Western immunoblots
Primary antibodies used were pSTAT3 (Tyr-705) (cat# 9145 clone D3A7), STAT3 (cat# 9139 clone 124H6), suppressor of cytokine signaling 3 (SOCS3) (cat# 2923), myeloid cell leukemia sequence 1 (BCL2-related) (MCL1) (cat# 5453 clone D35A5) (Cell Signaling Technologies, Danvers, MA, USA). Anti-β-actin (cat# 12262 clone 8H10D10) (Cell Signaling) was used as the protein loading control. Membranes were blocked in TBS-Tween 5% milk or bovine serum albumin depending on manufacturer’s recommendation. Bands were visualized with Immobilon Western Chemiluminescent HRP substrate (Millipore) on X-ray film and densitometry measurements were performed. All experiments were performed three times and representative blots are shown.
Cell lines
EPN cell line 811 was established from the 4th recurrence in a patient with metastatic anaplastic EPN that exhibited a Group A phenotype. This unique cell line is positive for chromosome 1q gain (1q+) as was the tumor. 1q+ is known to be a poor risk factor for EPN. This cell line was maintained under normal culture conditions in Optimem media supplemented with 15% FBS (O15) (Invitrogen). Patient samples were obtained with consent (COMIRB#95–500). Glioblastoma (GBM) cell line U87, obtained from ATCC, were cultured in MEM supplemented with 10% FBS, non-essential amino acids, sodium pyruvate, and sodium bicarbonate. Conditioned media (CM) from cell lines, as used for characterization of cytokine release and monocyte polarization, was obtained by harvesting media from a known number of cells after 24 hours incubation. Cell line authentication was performed using short tandem repeat profiling and comparison to known cell line DNA profiles.
shRNA Transfection
A pLKO system (Sigma-Aldrich) was utilized for lentiviral RNAi-mediated STAT3 protein knockdown. TRC numbers for shRNAs used were: STAT3 (20839) STAT3 (20840) STAT3 (20842), non-target (SHC016). Virus was removed from cells after 24 hour incubation. Validation of STAT3 knockdown was performed by Western blot analysis of cells treated identically alongside MTS, tritiated(3H)- thymidine uptake, and apoptosis assays.
MTS assay
Cells were seeded at 4,000 cells per well in 96-well plates (Corning) in antibiotic (AB)-free medium and treated with drug or lentiviral solution the next day. S3I-201 was used to inhibit STAT3 in 811. S3I-201 (Selleck Chemicals) inhibits STAT3:STAT3 complex formation and STAT3 DNA-binding and transcriptional activity and in the original paper inhibited the growth of breast cancer in vivo (11). Cell viability was measured using the MTS assay with CellTiter 96 AQueous One Solution Reagent (Promega) following the manufacturer's protocol 72 hours after drug treatment or 5 days after virus removal. Optical density of each well was measured with a Synergy 2 microplate reader (Biotek) at 490 nm. The proportion of cells per treatment group was normalized to control wells of DMSO control-treated cells or sh non-targeting (NT) control.
3H-thymidine uptake
Cell proliferation as determined by rate of DNA synthesis was measured by 3H-thymidine incorporation. Cells were plated in 96-well plates and treated with a dose range of S3I-201 in triplicate as described above. Twenty hours after treatment, each well was pulsed with 0.5 mCi 3H-thymidine and the plates were incubated at 37°C for 4 hours prior to cell harvest. For shRNA transfections, lentivirus was added to cells as above, and 3H-thymidine was added 5 days after virus removal. Wells were washed with PBS and 6% trichloroacetic acid was added to each well. Wells were washed with 1ml cold 6% trichloroacetic acid after incubation for 1hour at 4°C. The acid precipitate was dissolved overnight in 50µl 0.5N NaOH and transferred to scintillation vials containing 3ml of ScintiSafe-30%. Incorporated radioactivity was measured using a scintillation counter. Mitomycin C blocks proliferation and was used as a negative control for 3H-thymidine incorporation.
IncuCyte apoptosis analysis
Cells were transfected with nuclear locating signal mCherry (NLS-mCh) virus and antibiotic selected for NLS-mCh+ cells. Cells were seeded at 4,000 cells per well in 96-well plates in media. Cells were cultured at 37°C and 5% CO2 and monitored using an Incucyte Zoom (Essen BioScience). CellEvent Caspase-3/7 Green Detection Reagent (Life Technologies) was added the next day and baseline images were taken using 10× objective. For lentiviral knockdown, caspase reagent was added following virus removal. S3I-201 was added after initial baseline images. Images were captured at 4-hour intervals from 4 separate regions per well using a 10× objective over 72 hours. Each experiment was done in triplicate and accumulation of caspase 3/7 over time was normalized to confluence of cells.
Monocyte polarization assays
CD14+ monocytes were isolated from HLA-matched donor peripheral blood by flow sorting using CD14-FITC (Becton Dickinson) and Moflo Astrios EQ flow cytometry sorter. Post-sorted cells were incubated in various conditions for 96 hours on ultra-low attachment plates (Corning). 811-conditioned media (811 CM) was collected off 90% confluent 811 cell line and immediately frozen at −80°C for later use. IL6 neutralizing antibody (R& D Systems) was used at concentration of 50µg/ml and tocilizumab (Genentech) was used at concentration of 1mg/ml. Media were harvested from cells after 96 hours and cytokine concentrations were measured by multiplex bead cytokine analysis as described above. Cells were stained with pSTAT3-PE (BD) according to manufacturer’s instructions and run on Beckman Coulter Gallios 561; analysis was done using FlowJo v10.0.7 (FlowJo).
IL8 functional assay
Media was harvested from 811-polarized monocytes (MonoCM) or non-polarized monocytes (MonoO15) as described above. Fresh CD14+ monocytes were then incubated in MonoCM or MonoO15 for 96 hours. IL8 neutralizing antibody (R&D Systems) was added to MonoCM at 5µg/ml. Cells were collected and myeloid phenotype cell surface markers CD163, HLA-DR and CD64 were analyzed by flow cytometry as previously described (12). Media were harvested and cytokine concentrations were measured as described above. Cytokine concentrations from primed media were subtracted in order to assess additional secretion.
Statistical analyses
Statistical analyses were performed using R bioinformatics, Prism (GraphPad), and Excel (Microsoft) software. For all tests, statistical significance was defined as p-value < 0.05.
Results
STAT3-upregulated genes and IL6 signaling pathway genesets are significantly enriched in Group A EPN
We used non-biased GSEA analysis to identify transcription factor activity that might mechanistically underlie the Group A inflammatory response phenotype. Primary EPN Group A (n=21) and B (n=20) transcriptomic profiles were screened for enrichment of transcriptional programs using MSigDB C3 TFT (transcription factor targets) v.4 motif genesets (n=615) containing genes that share a transcription factor binding site motifs defined in the TRANSFAC database (version 7.4, http://www.gene-regulation.com/). In Group A EPN, transcription factor targets genes of leading cancer inflammatory mediator STAT3 (MSigDB: V$STAT3_01) was the second most enriched of 615 TFT genesets (NES= 2.07; FDR q<0.0001) (Table 1, Supplementary figure 2A). Specific STAT3 TFT motif-containing genes highly upregulated (FC>10) in Group A include chemokine (C-C motif) ligand 2 (CCL2), and v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (MAFF) (Table 2).
Table 1.
Enriched genesets in Group A EPN using GSEA analysis.a
| NAME | NES | NOM p-val |
FDR q-val |
FWER p-val |
|---|---|---|---|---|
| MSigDB C3: motif genesets: transcription factor targets | ||||
| GGGNNTTTCC_V$NFKB_Q6_01 | 2.16 | <0.0001 | <0.0001 | <0.0001 |
| V$STAT3_01 | 2.06 | <0.0001 | <0.0001 | <0.0001 |
| V$NFKAPPAB_01 | 1.99 | <0.0001 | 0.000468 | 0.001 |
| TGASTMAGC_V$NFE2_01 | 1.96 | <0.0001 | 0.000351 | 0.001 |
| V$SRF_01 | 1.93 | <0.0001 | 0.000281 | 0.001 |
| V$NFKB_Q6_01 | 1.93 | <0.0001 | 0.000234 | 0.001 |
| V$NFKB_C | 1.92 | <0.0001 | 0.000201 | 0.001 |
| V$PEA3_Q6 | 1.90 | <0.0001 | 0.000176 | 0.001 |
| V$CREL_01 | 1.90 | <0.0001 | 0.000156 | 0.001 |
| V$AP1_Q4 | 1.89 | <0.0001 | 0.000140 | 0.001 |
| Experimentally determined STAT3 geneset | ||||
| STAT3-upregulated (Yu et al [REF]) | 2.33 | <0.0001 | <0.0001 | <0.0001 |
| MSigDB C2: curated genesets: BioCarta | ||||
| BIOCARTA_NKT_PATHWAY | 1.97 | <0.0001 | 0.00684 | 0.00400 |
| BIOCARTA_INFLAM_PATHWAY | 1.95 | <0.0001 | 0.00342 | 0.00400 |
| BIOCARTA_PML_PATHWAY | 1.92 | <0.0001 | 0.00290 | 0.00500 |
| BIOCARTA_LAIR_PATHWAY | 1.86 | <0.0001 | 0.0124 | 0.0270 |
| BIOCARTA_KERATINOCYTE_PATHWAY | 1.83 | 0.00338 | 0.0191 | 0.0540 |
| BIOCARTA_COMP_PATHWAY | 1.82 | <0.0001 | 0.0168 | 0.0570 |
| BIOCARTA_IL2RB_PATHWAY | 1.82 | 0.00166 | 0.0146 | 0.0570 |
| BIOCARTA_CDMAC_PATHWAY | 1.82 | <0.0001 | 0.0149 | 0.0660 |
| BIOCARTA_IL6_PATHWAY | 1.81 | <0.0001 | 0.0155 | 0.0770 |
| BIOCARTA_NFKB_PATHWAY | 1.80 | 0.00337 | 0.0158 | 0.0870 |
The top 10 enriched genesets as ranked by NES are shown for transcription factor target motif and BioCarta genesets.
NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate; FWER, family-wise error rate.
Table 2.
Significantly upregulated genes in Group A EPN from enriched STAT3 and IL-6 pathway genesets.
| Gene symbol |
Gene title | FC | p-value | q-value |
|---|---|---|---|---|
| MSigDB C3: motif genesets: transcription factor targets: V$STAT3_01 genes | ||||
| CCL2 | chemokine (C-C motif) ligand 2 | 21.62 | 1.39E-08 | 3.67E-06 |
| IRF1 | interferon regulatory factor 1 | 8.66 | 8.48E-08 | 1.11E-05 |
| ICAM1 | intercellular adhesion molecule 1 | 8.32 | 2.11E-07 | 2.00E-05 |
| MAFF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | 15.87 | 7.78E-07 | 5.19E-05 |
| SERPING1 | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 | 3.11 | 6.45E-06 | 0.000264 |
| CISH | cytokine inducible SH2-containing protein | 2.24 | 2.45E-05 | 0.000737 |
| APBA1 | amyloid beta (A4) precursor protein-binding, family A, member 1 | 1.93 | 0.00496 | 0.0393 |
| Experimentally determined STAT3 upregulated genes (Yu et al [REF]) | ||||
| IL8 | interleukin 8 | 24.13 | 5.22E-09 | 2.17E-06 |
| CCL2 | chemokine (C-C motif) ligand 2 | 21.62 | 1.39E-08 | 3.67E-06 |
| SOCS3 | suppressor of cytokine signaling 3 | 14.61 | 1.88E-08 | 4.35E-06 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 5.49 | 3.65E-08 | 6.76E-06 |
| ICAM1 | intercellular adhesion molecule 1 | 8.32 | 2.11E-07 | 2E-05 |
| CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39); YKL-40 | 40.27 | 2.87E-07 | 2.51E-05 |
| VEGFA | vascular endothelial growth factor A | 4.71 | 9.77E-07 | 6.07E-05 |
| PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase); COX-2 | 10.57 | 2.82E-06 | 0.000142 |
| STAT3 | signal transducer and activator of transcription 3 (acute-phase response factor) | 1.63 | 3.72E-06 | 0.000176 |
| MCL1 | myeloid cell leukemia sequence 1 (BCL2-related) | 1.65 | 7.74E-06 | 0.00031 |
| VIM | vimentin | 1.41 | 8.05E-06 | 0.000318 |
| IL6 | interleukin 6 (interferon, beta 2) | 13.41 | 1.02E-05 | 0.000381 |
| BIRC3 | baculoviral IAP repeat-containing 3 | 5.94 | 6.29E-05 | 0.001533 |
| MMP9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 7.96 | 6.32E-05 | 0.00154 |
| JAK3 | Janus kinase 3 | 3.56 | 6.54E-05 | 0.00157 |
| CXCL10 | chemokine (C-X-C motif) ligand 10 | 7.40 | 0.000143 | 0.00281 |
| LIF | leukemia inhibitory factor (cholinergic differentiation factor) | 3.88 | 0.000153 | 0.00298 |
| MMP2 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 2.59 | 0.00111 | 0.0134 |
| CCL5 | chemokine (C-C motif) ligand 5 | 2.28 | 0.00405 | 0.0340 |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 2.09 | 0.00414 | 0.0345 |
| IL1B | interleukin 1, beta | 3.10 | 0.0064 | 0.0474 |
| MSigDB C2: curated genesets: BioCarta: IL6_PATHWAY genes | ||||
| MAP2K1 | mitogen-activated protein kinase kinase 1 | 1.80 | 3.65E-06 | 0.000174 |
| STAT3 | signal transducer and activator of transcription 3 (acute-phase response factor) | 1.63 | 3.72E-06 | 0.000176 |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | 2.51 | 5.35E-06 | 0.000226 |
| IL6 | interleukin 6 (interferon, beta 2) | 13.41 | 1.02E-05 | 0.000381 |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | 2.01 | 4.38E-05 | 0.00114 |
| JAK3 | Janus kinase 3 | 3.56 | 6.54E-05 | 0.00157 |
| ELK1 | ELK1, member of ETS oncogene family | 1.42 | 0.000123 | 0.00251 |
| SRF | serum response factor (c-fos serum response element-binding transcription factor) | 1.77 | 0.000234 | 0.00409 |
| MAPK3 | mitogen-activated protein kinase 3 | 1.53 | 0.000782 | 0.0104 |
| JUN | jun proto-oncogene | 1.70 | 0.000952 | 0.0120 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 3.99 | 0.00140 | 0.0159 |
FC, fold change
We further characterized the involvement of STAT3 in Group A EPN by using GSEA to measure enrichment of experimentally determined STAT3-upregulated geneset (n=36), created from a review of the role of STAT3 in cancer inflammation and immunity (13) (Supplementary table 1). This geneset was enriched in Group A to a greater extent (NES=2.33; FDR q<0.001) than the STAT3 TFT geneset (Table 1, Supplementary figure 2B). Experimentally determined STAT3 up-regulated genes in Group A (Table 2) were largely discrete from those identified as STAT3 TFTs with the exception of CCL2 and ICAM1. Notable STAT3 up-regulated genes (FC>10) in Group A include chitinase 3-like 1 (CHI3L1, AKA YKL-40), interleukin-8 (IL8), CCL2, suppressor of cytokine signaling 3 (SOCS3), IL6 and prostaglandin-endoperoxide synthase 2 (PTGS2, AKA COX-2). Collectively these data suggest activation of the STAT3 signaling pathway in Group A EPN.
It is known that IL6 drives oncogenic activation of STAT3, increasing cell proliferation, survival and invasion while suppressing antitumor immunity (13). Persistent IL6/STAT3 pathway activation is involved in various inflammation-associated cancers, most notably colorectal (14), gastric (15) and liver (16) cancers. Given the high level of IL6 gene expression we hypothesized that IL6 may be driving activation of STAT3 in Group A EPN. Enrichment of MSigDB C2 BioCarta pathway v.4 curated genesets (n=217) (http://www.biocarta.com/genes/index.asp) were assessed using GSEA and showed that, in agreement with our hypothesis, the BioCarta IL6 pathway geneset was among the 10 highest enriched pathways in Group A (NES=1.81; FDR q=0.015) (Tables 1 and 2, Supplementary figure 2C). Of note, the BioCarta cytokines and inflammatory response geneset (BIOCARTA_INFLAM_PATHWAY) was the second most enriched BioCarta pathway geneset (NES=1.95; FDR q=0.0034) (Supplementary figure 2D).
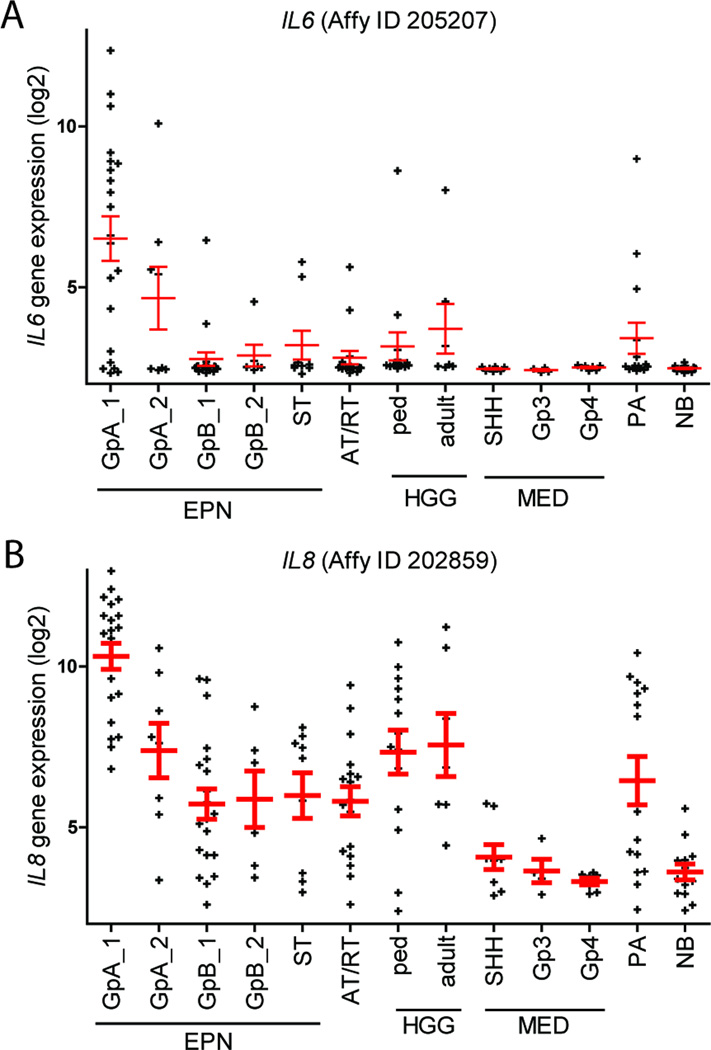
A broader examination of Group A EPN associated IL6/STAT3 pathway genes identified by GSEA gene expression was next performed in EPN from recurrences and supratentorial sites, as well as other common pediatric and adult brain tumors and normal brain (NB). Recurrent Group A EPN showed reduced expression of IL6, consistent with the decreased inflammatory phenotype observed at recurrence that was previously identified (3). IL6 expression was significantly higher in primary Group A EPN than in all other brain tumor types (Figure 1A). IL6 expression was higher in Group A EPN than in both adult GBM (FC= 7.0; p=0.039) and MED (FC=16.5;p=2.5×10−6) which is notable as constitutive IL6/STAT3 activation has been identified in these tumor types (17,18).
Figure 1. Key inflammatory cytokine genes IL6 and IL8 are upregulated in Group A EPN versus a broad range of brain tumors and normal brain tissues.
(A) IL6 and (B) IL8 in Group A EPN compared to recurrent EPN specimens, other pediatric and adult brain tumors and normal brain tissues. Abbreviations: GrpA, Group A EPN (GrpA_2 and GrpB_2 are first recurrences); ST, supratentorial; AT/RT, atypical teratoid/rhabdoid tumor; HGG, high grade glioma; ped, pediatric; MED, medulloblastoma; SHH, sonic hedgehog; Gp3, Group 3; PA, pilocytic astrocytoma; NB, normal brain
Similar to IL6, IL8 was significantly overexpressed compared to recurrent Group A EPN (FC=7.6, p=0.0016), to all other brain tumor types (FC=22-fold higher than all other tumor samples combined; p=4.3×10−11) and to NB (FC=104; p=1.4×10−13) (Figure 1B). Thus, overexpression of key inflammatory mediators IL6 and IL8 appear to be hallmarks of the Group A EPN transcriptome in the context of not only EPNs as a whole, but of all brain tumors. The apparent restriction of IL6 and IL8 gene expression to Group A EPN in this broad transcriptomic analysis provide further evidence that an IL6/STAT3 signaling pathway underlies the inflammatory signaling pathway present in Group A EPN.
Key Group A EPN IL6/STAT3 inflammation signature genes are differentially distributed between tumor and tumor-infiltrating myeloid cells
Prior studies of EPN immunobiology have identified that tumor-infiltrating immune cells can significantly contribute to the overall gene expression profiles of surgical tumor samples (19). A subset of genes associated with longer progression-free survival in EPN were found to be restricted to tumor-infiltrating myeloid cells that contribute up to 25% of the cellular content of surgical tumor specimens (12). The contribution of tumor infiltrating myeloid cells to the inflammatory TME, through the IL6/STAT3 activation, is well established (13,20). We, therefore, sought to determine the contribution of tumor-infiltrating myeloid cells to the Group A EPN IL6/STAT3 gene expression signature. Surgical samples from two posterior fossa EPNs were disaggregated and flow-sorted to isolate myeloid cells (CD45+, CD11b+) and tumor cells (CD45−). Transcriptomic analysis was performed, and the relative expression of Group A IL6/STAT3 inflammation signature genes between these TME cellular compartments was measured (Supplementary figure 3). Of the 34 genes identified by GSEA as enriched in the STAT3 TFT, STAT3-upregulated and IL6 pathway genesets (Table 2), IL8, ICAM1, PTGS2 and IL1B were on average more than 10-fold higher in the myeloid than in the tumor cell compartments. Conversely, CHI3L1 gene expression was 67-fold higher in the tumor compartment. CHI3L1 has a proposed role in driving the Group A EPN-specific mesenchymal phenotype (2). Thus tumor-infiltrating myeloid cells contribute to the IL6/STAT3 inflammatory gene expression signature in the Group A EPN.
Tumor cell secretion of IL6 is associated with STAT3 phosphorylation in Group A EPN
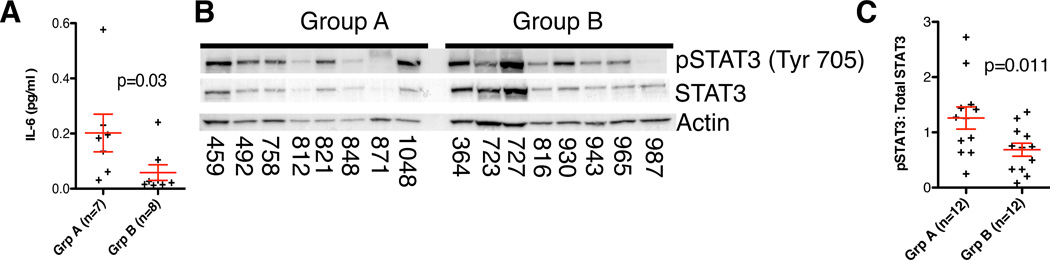
We isolated viable tumor cells, excluding other cells in the TME, from EPN clinical samples (7 Group A; 8 Group B) by tissue disaggregation and flow-sorting to remove all immune cell types as described above. Tumor cells were cultured in 1ml serum-supplemented media (O15) for 96 hours. Multiplexed cytokine profiling revealed that IL6 was the only cytokine that was significantly higher in Group A versus Group B EPN (FC=3.5; p=0.031) (Figure 2A). Of the remaining cytokines, only IL10 was >2-fold higher in Group A than in Group B with a trend toward significance (p=0.12) (Supplementary figure 4).
Figure 2. Group A EPN patients have higher levels of IL6 production and a higher ratio of pSTAT3 than Group B patients.
(A) Concentration of IL6 secretion from flow-sorted disaggregated tumor samples is higher in group A (FC=3.5; p=0.031, 1-tailed t-test). Cells were cultured for 96hours in O15 and normalized to 106 cells (B) Representative Western blot showing expression of pSTAT3 and Total STAT3 in patient samples. (C) Western blot validation of pSTAT3 activation in surgical samples from 12 Group A and 12 Group B EPN patients. The ratio of pSTAT3 to Total STAT3 was determined by densitometric measurement of western blot bands. This demonstrated a higher ratio of STAT3 phosphorylation in samples from group A EPN patients (FC=1.84; p=0.012).
Presence of significantly higher STAT3 transcripts and STAT3-regulated transcripts is evidence of STAT3 activity; hence we evaluated STAT3 phosphorylation as a more direct indicator of STAT3 activity in EPN patient samples. Phosphorylation of STAT3-Tyrosine 705 by upstream signaling results in dimerization and translocation to the nucleus resulting in transcription of STAT3-target genes. The ratio of tyrosine 705-phosphorylated (pSTAT3) to total STAT3 protein, as measured by Western blot analysis, was used as a measure of STAT3 activity in patient tumor samples (12 Group A; 12 Group B) (Figure 2B shows a representative blot). Densitometric quantification of pSTAT3 and total STAT3 revealed a significantly higher ratio of phosphorylation in Group A (FC=1.84; p=0.011) (Figure 2C).
Constitutively activated STAT3 in EPN has pro-proliferative and anti-apoptotic roles
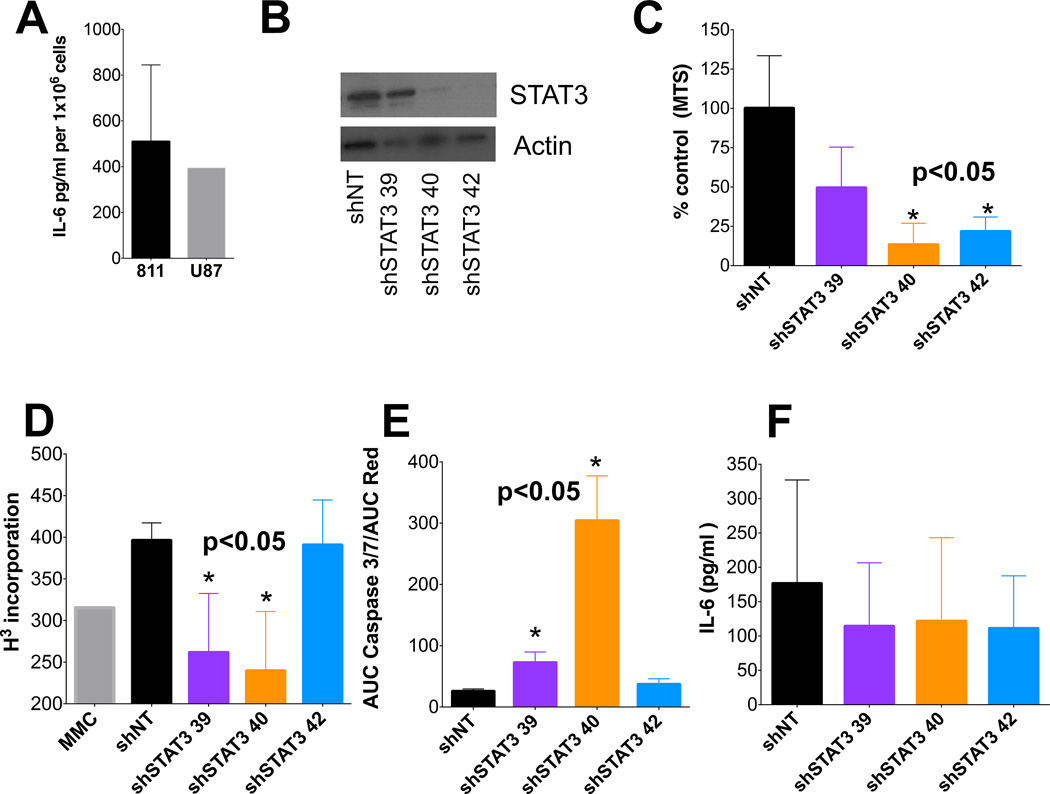
Having identified a potentially critical role for constitutively activated STAT3 in EPN, we examined proliferation and survival of EPN cells after loss of STAT3. Functional studies were performed using an EPN cell line that was recently established from the 4th recurrence in a child with metastatic posterior fossa anaplastic EPN that exhibited a Group A phenotype. This cell line (811) was validated by karyotype analysis and was shown to harbor chromosome 1q-gain (1q+), a known poor-risk factor for EPN (21). Characterization of these EPN cells demonstrated that 811 secreted comparable amounts of IL6 as secreted by GBM cell line U87, in which aberrant IL6/STAT3 pathway activation had previously been identified and associated with invasiveness (22) (Figure 3A).
Figure 3. EPN cell line 811 is sensitive to STAT3 knockdown leading to apoptosis.
(A) Concentration of IL6 secretion from newly created pediatric EPN cell lines and control glioblastoma (GBM) cell line. (B) Western blot validation of STAT3 knockdown in 811 cell line (representative blot). All experiments using shRNA knockdown were validated by western blots. (C–D) Inhibition of cell proliferation by STAT3 knockdown was assessed by MTS and 3H-thumidine incorporation in 811 cells. Graphs are representative experiments. MMC = mitomycin C; (E) Real-time quantification of cleaved Caspase 3/7, representative of apoptosis, over 160 hours of STAT3 knockdown. Graph shows accumulation of cleaved caspase 3/7 normalized to cell numbers. (F) IL6 secretion by 811 cells after STAT3 knockdown. * denotes p-value <0.05 calculated using 2-tailed paired t-test.
Loss of STAT3 through shRNA knockdown (Figure 3B) resulted in significant reduction in proliferation of 811 cells than non-targeting (NT) control shRNA (shNT) as measured by both cell-viability measurement (MTS) and DNA synthesis (3H-thymidine) (Figure 3C,D). Pharmacologic STAT3 inhibition (S3I-201) showed that 811 cells were more sensitive (viability IC50=108µM; DNA synthesis IC50=86µM) than U87 cells (viability IC50=140µM; DNA synthesis IC50=225µM) (Supplementary figure 5A,B). The clinical relevance of these S3I-201 IC50 values are unknown, as S3I-201 has not yet been tested in a clinical trial, rather this inhibitor provides proof-of-principle for therapeutic targeting of STAT3. STAT3 inhibition did not affect cell cycle in 811 cells (Supplementary figure 5C). However, shRNA knockdown of STAT3 resulted in activation of apoptosis, with significant increase of cleaved caspase 3/7 compared to shNT (Figure 3E) and significant apoptosis was demonstrated using S3I-201 at >IC50 concentration (Supplementary figure 5D). Collectively, these data support the hypothesis that, in EPN with constitutively activated STAT3, this transcription factor has both pro-survival and anti-apoptotic roles.
We observed an S3I-201 dose-dependent reduction of pSTAT3 pathway protein levels by Western blot (Supplementary figure 5E). While total STAT3 remained relatively constant up to 500µM of S3I-201, phosphorylation was completely inhibited above 125µM. STAT3 transcriptional targets SOCS3 and MCL-1 were similarly reduced in a dose-dependent manner. STAT3 inhibition did not, however, reduce secretion of IL6 suggesting that IL6 regulation is upstream of STAT3 in the signaling pathway (Figure 3F, Supplementary figure 5F).
EPN-secreted IL6 induces CD14+ monocytes to secrete key pro-inflammatory cytokine IL8
Transcriptomic analyses of Group A EPN identified that tumor-infiltrating myeloid cells contributed strongly to the inflammatory phenotype and specifically IL8 expression, a hallmark of the IL6/STAT3 inflammation signature geneset, was highly enriched in tumor-infiltrating myeloid cells (Supplementary figure 3). By multiplex cytokine analysis, IL6 was the predominant cytokine secreted by 811 cells, being 55-fold higher than IL8, the next highest-secreted cytokine (Supplementary figure 6A). No other cytokines were detected apart from trace amounts of GM-CSF, IFNγ, IL13, and IL7 (Supplementary figure 6A).
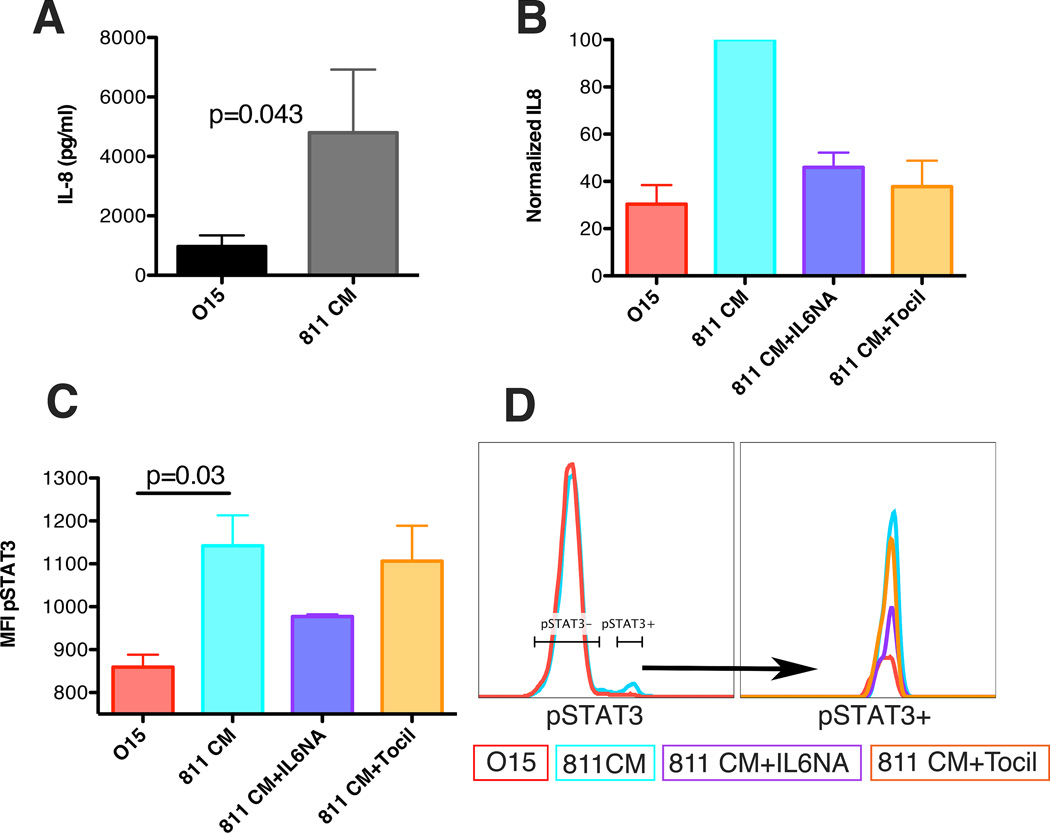
We hypothesized that IL6 secreted at high levels by EPN in the TME polarize tumor-infiltrating myeloid cells to an inflammatory phenotype in a paracrine fashion, resulting in activation of myeloid cell STAT3 and up-regulation of transcriptional targets including IL8. To evaluate this, CD14+ monocytes were isolated from tumor HLA-matched peripheral blood mononuclear cells by flow sorting and were immediately cultured in 250µl of either (A) 811CM, or (B) O15 control media, for 96 hours. Supernatant from the CD14+ monocyte cultures was then subject to multiplex cytokine analysis. Background cytokine levels in CM were subtracted from CD14+ monocyte/CM culture cytokine levels to identify changes in monocyte-derived cytokines. There was a 9.2-fold increase (p=0.043) in IL8 secretion from CD14+ cells treated with 811CM compared to that with media control alone (Figure 4A). The remaining 12 cytokines tested did not show any significant differences (data not shown).
Figure 4. EPN cell line 811 can induce immune suppressive phenotype in HLA-matched donor CD14+ peripheral blood mononuclear cells.
(A) IL8 secretion from HLA-matched donor CD14+ monocytes incubated for 96hours in either control media (O15) or media collected from 811 cell line (811 CM). IL8 concentration in pg/ml per 30,000 cells in 250ml. Experiments were completed in triplicate. P-Value calculated using 1 tailed t-test (B) IL8 secretion from CD14+ monocytes incubated for 96hours in O15, 811 CM, 811 CM plus 50µg/ml IL6 neutralizing Ab (811 CM+IL6NA), 811 CM plus 100µg/ml tocilizumab (811 CM+Tocil). Experiments were run in triplicate and normalized to 811 CM-cultured monocytes for each experiment. (C) Median Fluorescent Intensity (MFI) of pSTAT3 from CD14+ monocytes incubated. Representative flow plot of (left) pSTAT3 for 811CM and control-treated CD14+ monocytes, and (right) representative flow plot of pSTAT3-positive peak in all treatment conditions. P-value calculated using a 1 tailed t-test.
To determine whether IL8 production in CD14+ monocytes was induced by EPN-derived IL6, the experiment was repeated with the addition of IL6 neutralizing antibody (IL6NA) or IL6 receptor neutralizing antibody tocilizumab. In both cases, production of IL8 by monocytes was significantly lowered by greater than 75% (Figure 4B). Furthermore, flow cytometric measurement of pSTAT3 demonstrated significant induction of STAT3 phosphorylation (Tyr-705) in CD14+ monocytes and this was reversed with addition of IL6NA (Figure 4C,D). EGF, a potential alternative activator of STAT3, was shown not to drive STAT3 phosphorylation or IL8 release, as EGFR-blocking antibody cetuximab did not reverse this phenotype in 811 CM-treated myeloid cells (Supplementary figures 6B,C). Together, these data support our hypothesis that Group A EPN secretes IL6, which stimulates production of pro-inflammatory IL8 by myeloid cells via activation of STAT3 in the tumor microenvironment in a paracrine fashion.
IL8-mediated signaling between monocytes perpetuates inflammatory signaling in the tumor microenvironment
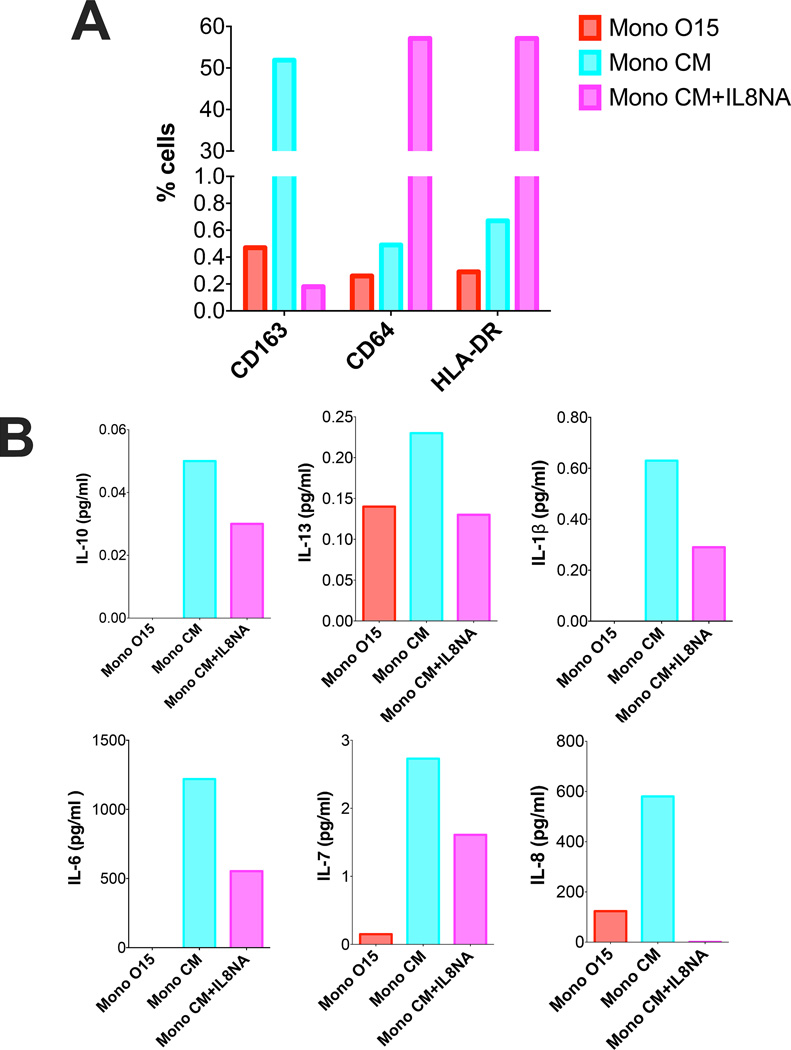
IL8 is a potent chemokine that has a variety of roles in inflammation. While its most common function is to induce neutrophil infiltration, IL8 can also promote tumor-associated macrophages to secrete more cytokines and growth factors that further induce proliferation and tumor invasion (23). IL8 was the only cytokine we found to be significantly upregulated by monocytes in response to tumor cell paracrine signaling (Figure 4A). We hypothesized that IL8 production by tumor-polarized monocyte could further polarize monocytes in a paracrine fashion. To test this hypothesis, freshly sorted CD14+ monocytes were cultured in media harvested from CD14+ monocytes that had been primed in either tumor-conditioned media (MonoCM) or control media (MonoO15) for 96 hours. Strikingly, a significant upregulation of CD163 was observed in cells cultured in MonoCM compared to those cultured in control media (Figure 5A). Additionally MonoCM-induced secretion of a number of common pro-inflammatory cytokines IL10, IL13, IL1β, IL6, IL7 and IL8 (Figure 5B). Addition of IL8 neutralizing antibody to MonoCM prevented CD163 upregulation and was accompanied with upregulation of both HLA-DR and Fc-receptor CD64 expression (Figure 5A). All pro-inflammatory cytokine production was reduced with the addition of IL8 neutralizing antibody to MonoCM (Figure 5B).
Figure 5. IL8 secreted by EPN-polarized macrophages induces further M2-like polarization of monocytes.
CD14+ monocytes were incubated in O15 or 811 CM as previously described in figure 4. Media were then used to incubate another set of CD14+ monocytes (MonoO15, MonoCM). IL8 neutralizing antibody (MonoCM+IL8NA) was used at 5µg/ml. (A) Flow cytometry evaluation of cell surface markers on CD14+ monocytes after incubation in media collected from EPN tumor-primed monoctyes. Graphs are percentage myeloid cells. (B) Concentration of cytokines secreted by monocytes from panel A. Cytokine already present in the media from initial polarization was subtracted to reflect additional cytokine production in pg/ml.
Collectively these studies provide evidence that EPN tumor cells induce paracrine polarization of monocytes through secretion of IL6. This inflammatory phenotype is then amplified between monocytes through up-regulated IL8 production.
Discussion
Inflammatory response has been identified as a predominant transcriptomic phenotype in Group A EPN (1–3). The present study identifies tumor IL6/STAT3 pathway activation and crosstalk with myeloid cells as a potential mechanism underlying this phenotype. IL6/STAT3-mediated crosstalk between tumor and immune cells, resulting in inflammation, has been well documented in a number of tumors (20). Persistent IL6/STAT3 pathway activation is involved in various inflammation-associated cancers, most notably colorectal (14), gastric (15) and liver (16) cancers. It is known that oncogenic activation of STAT3, driven by IL6, increases cell proliferation, survival and invasion as well as suppressing antitumor immunity (13). Tumor-derived IL6 affects the differentiation of myeloid lineages, including macrophages and dendritic cells, through STAT3 activation in these immune cell types (24). STAT3 activation in tumor-associated myeloid cells results in expression of pro-cancer inflammatory mediators and upregulation of angiogenic factors and growth factors, leading to increased tumor growth in a paracrine fashion (20). In this study, EPN tumor cells were shown to release IL6, which activated myeloid cell STAT3 in a paracrine fashion. STAT3 activation resulted in increased IL8 production which further polarizes myeloid cells to an inflammatory phenotype.
In the central nervous system, oncogenic IL6/STAT3 signaling has been documented predominantly in GBM, the most common malignant brain tumor in adults, as important in proliferation, survival and invasiveness of tumor cells and as a key regulator of the immunosuppressive microenvironment (25–29). GBM have been demonstrated to harbor pro-tumor microglia/macrophages that are polarized by glioma stem cell-secreted factors (30). This crosstalk has been attributed to STAT3 activation (31). Therapeutic targeting of STAT3 has also shown promise in the most common malignant pediatric brain tumor, medulloblastoma (MED; refs.17,32). To our knowledge the present study represents the only demonstration of such cross-talk in EPN. Given that IL6 and IL8 gene expression is elevated in Group A EPN when compared to other brain tumors, including GBM and MED, it emphasizes the particular importance of IL6/STAT3 signaling and immune crosstalk in the biology of this tumor.
The EPN Group A IL6/STAT3 activation geneset includes critical effectors of the key phenotypic characteristics of Group A EPN (1–3), namely inflammation (IL8, CCL2, PTGS2 (COX2)), mesenchymal transition (CHI3L1), invasiveness (TIMP1, MMP2, MMP9), angiogenesis (VEGFA), and anti-apoptosis (MCL1, BIRC3). Thus, constitutive IL6/STAT3 signaling may underlie a number of the characteristic oncogenic phenotypes of Group A EPN, representing a “molecular hub” as has previously been ascribed to STAT3 in gliomas (33). A recent epigenomic study of EPN revealed that Group A, unlike Group B, exhibits a CpG island methylator phenotype (CIMP), suggesting epigenetic modifiers as rational therapeutic candidates in this molecular subgroup (34). Both IL6 and STAT3 have been shown to drive hypermethylation, potentially linking the IL6/STAT3 pathway with CIMP in this subgroup (35–37). Given these potentially pivotal roles of IL6/STAT3 in Group A EPN tumor biology, therapeutic exploration of STAT3 and/or IL6 inhibition is warranted.
We have previously shown that immunity affects outcome in pediatric EPN (19). Subsequently, we characterized immunologic differences in posterior fossa EPN, identifying an immunosuppressed phenotype in Group A when compared to Group B (3). Specifically, T cells derived from Group A tumors did not respond to stimulation with appropriate cytokine release and appeared to have an exhausted phenotype compared to those derived from Group B tumors, a finding also seen in GBM (38). Thus, in Group A EPN there is an association of IL6/STAT3-mediated inflammation with an immunosuppressed phenotype. Numerous mechanisms have been demonstrated to link inflammation to immunosuppression, such as generation of myeloid-derived suppressive cells (39) and immunosuppressive T-cell subsets (40). STAT3 has been shown to inhibit expression of antitumor TH1-type immune-stimulating molecules, and promote expression of immunosuppressive factors, and is therefore likely drives immunosuppression in Group A EPN (41). With the recent success of cancer therapies that inhibit immunosuppressive factors, most notably CTLA-4 and PD-1 (42,43), identification of a tumor-associated myeloid cell-mediated immunosuppressive mechanism in Group A EPN has particular clinical relevance. Reversal of myeloid cell-mediated immunosuppressive phenotypes for therapeutic gain has been achieved by T-cell activation strategies such as IL12 treatment that results in reprogramming of tumor-associated myeloid cells (44). Alternatively, directly targeting tumor-associated myeloid cells has shown promise, as achieved by CSF-1R inhibition in GBM (45). The ability of IL6 receptor antagonist tocilizumab, which is FDA-approved for treatment of rheumatoid arthritis (46), to abrogate IL6/STAT3-mediated tumor/myeloid cell crosstalk in the present study provides yet another therapeutic approach. Group A EPN has a poor outcome and little evidence that it is responsive to standard chemotherapy. Our work is highly suggestive that in the realm of pediatric brain tumors this specific subgroup of EPN would be a strong candidate for the development of immunotherapy.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 CA140614) and the Tanner Seebaum Foundation. The Flow Cytometry Core Facility receives direct funding support from the National Cancer Institute through the University of Colorado Cancer Center Support Grant (P30CA046934). We would like to thank Dr. Hideho Okada (UCSF) for assistance with this study.
Footnotes
Conflict of Interest: None
References
- 1.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wani K, Armstrong TS, Vera-Bolanos E, Raghunathan A, Ellison D, Gilbertson R, et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012;123:727–738. doi: 10.1007/s00401-012-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman LM, Donson AM, Nakachi I, Griesinger AM, Birks DK, Amani V, et al. Molecular sub-group-specific immunophenotypic changes are associated with outcome in recurrent posterior fossa ependymoma. Acta Neuropathol. 2014;127:731–745. doi: 10.1007/s00401-013-1212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 8.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griesinger AM, Birks DK, Donson AM, Amani V, Hoffman LM, Waziri A, et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J Immunol. 2013;191:4880–4888. doi: 10.4049/jimmunol.1301966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belluco C, Nitti D, Frantz M, Toppan P, Basso D, Plebani M, et al. Interleukin-6 blood level is associated with circulating carcinoembryonic antigen and prognosis in patients with colorectal cancer. Ann Surg Oncol. 2000;7:133–138. doi: 10.1007/s10434-000-0133-7. [DOI] [PubMed] [Google Scholar]
- 15.Ashizawa T, Okada R, Suzuki Y, Takagi M, Yamazaki T, Sumi T, et al. Clinical significance of interleukin-6 (IL-6) in the spread of gastric cancer: role of IL-6 as a prognostic factor. Gastric Cancer. 2005;8:124–131. doi: 10.1007/s10120-005-0315-x. [DOI] [PubMed] [Google Scholar]
- 16.Giannitrapani L, Cervello M, Soresi M, Notarbartolo M, La Rosa M, Virruso L, et al. Circulating IL-6 and sIL-6R in patients with hepatocellular carcinoma. Ann New York Acad Sci. 2002;963:46–52. doi: 10.1111/j.1749-6632.2002.tb04093.x. [DOI] [PubMed] [Google Scholar]
- 17.Xiao H, Bid HK, Jou D, Wu X, Yu W, Li C, et al. A Novel Small Molecular STAT3 Inhibitor, LY5 Inhibits Cell Viability, Cell Migration, And Angiogenesis in Medulloblastoma Cells. J Biol Chem. 2015;290:3418–3429. doi: 10.1074/jbc.M114.616748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFarland BC, Hong SW, Rajbhandari R, Twitty GB, Jr, Gray GK, Yu H, et al. NF-kappaB-induced IL-6 ensures STAT3 activation and tumor aggressiveness in glioblastoma. PLoS One. 2013;8:e78728. doi: 10.1371/journal.pone.0078728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donson AM, Birks DK, Barton VN, Wei Q, Kleinschmidt-Demasters BK, Handler MH, et al. Immune gene and cell enrichment is associated with a good prognosis in ependymoma. J Immunol. 2009;183:7428–7440. doi: 10.4049/jimmunol.0902811. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 21.Mendrzyk F, Korshunov A, Benner A, Toedt G, Pfister S, Radlwimmer B, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Li G, Deng L, Liu Q, Dai J, Shen J, et al. IL-6 augments the invasiveness of U87MG human glioblastoma multiforme cells via up-regulation of MMP-2 and fascin-1. Oncol Rep. 2010;23:1553–1559. doi: 10.3892/or_00000795. [DOI] [PubMed] [Google Scholar]
- 23.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 24.Park SJ, Nakagawa T, Kitamura H, Atsumi T, Kamon H, Sawa S, et al. IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J Immunol. 2004;173:3844–3854. doi: 10.4049/jimmunol.173.6.3844. [DOI] [PubMed] [Google Scholar]
- 25.Haftchenary S, Luchman HA, Jouk AO, Veloso AJ, Page BD, Cheng XR, et al. Potent Targeting of the STAT3 Protein in Brain Cancer Stem Cells: A Promising Route for Treating Glioblastoma. ACS Med Chem Lett. 2013;4:1102–1107. doi: 10.1021/ml4003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assi H, Espinosa J, Suprise S, Sofroniew M, Doherty R, Zamler D, et al. Assessing the role of STAT3 in DC differentiation and autologous DC immunotherapy in mouse models of GBM. PLoS One. 2014;9:e96318. doi: 10.1371/journal.pone.0096318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain SF, Kong LY, Jordan J, Conrad C, Madden T, Fokt I, et al. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67:9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 28.Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong LY, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita M, Zhu X, Sasaki K, Ueda R, Low KL, Pollack IF, et al. Inhibition of STAT3 promotes the efficacy of adoptive transfer therapy using type-1 CTLs by modulation of the immunological microenvironment in a murine intracranial glioma. J Immunol. 2008;180:2089–2098. doi: 10.4049/jimmunol.180.4.2089. [DOI] [PubMed] [Google Scholar]
- 30.Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Graeber MB. The molecular profile of microglia under the influence of glioma. Neuro Oncol. 2012;14:958–978. doi: 10.1093/neuonc/nos116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang MY, Lee HT, Chen CM, Shen CC, Ma HI. Celecoxib suppresses the phosphorylation of STAT3 protein and can enhance the radiosensitivity of medulloblastoma-derived cancer stem-like cells. Int J Mol Sci. 2014;15:11013–11029. doi: 10.3390/ijms150611013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foran E, Garrity-Park MM, Mureau C, Newell J, Smyrk TC, Limburg PJ, et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res. 2010;8(4):471–481. doi: 10.1158/1541-7786.MCR-09-0496. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A. 2005;102:6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee H, Zhang P, Herrmann A, Yang C, Xin H, Wang Z, et al. Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci U S A. 2012;109:7765–7769. doi: 10.1073/pnas.1205132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Gallina G, Dolcetti L, Serafini P, De Santo C, Marigo I, Colombo MP, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116:2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 42.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerkar SP, Goldszmid RS, Muranski P, Chinnasamy D, Yu Z, Reger RN, et al. IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. J Clin Invest. 2011;121:4746–4757. doi: 10.1172/JCI58814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genovese MC, McKay JD, Nasonov EL, Mysler EF, da Silva NA, Alecock E, et al. Interleukin-6 receptor inhibition with tocilizumab reduces disease activity in rheumatoid arthritis with inadequate response to disease-modifying antirheumatic drugs: the tocilizumab in combination with traditional disease-modifying antirheumatic drug therapy study. Arthritis Rheum. 2008;58:2968–2980. doi: 10.1002/art.23940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.