Abstract
Genetically engineered mouse models of lung cancer have demonstrated an important role in understanding the function of novel lung cancer oncogenes and tumor suppressor genes identified in genomic studies of human lung cancer. Further, these models are important platforms for pre-clinical therapeutic studies. Here, we generated a mouse model of lung adenocarcinoma driven by mutation of the Discoidin Domain Receptor 2 (DDR2) gene combined with loss of TP53. DDR2L63V;TP53L/L mice developed poorly differentiated lung adenocarcinomas in all transgenic animals analyzed with a latency of 40-50 weeks and a median survival of 67.5 weeks. Mice expressing wild-type DDR2 with combined TP53 loss did not form lung cancers. DDR2L63V; TP53L/L tumors displayed robust expression of DDR2 and immunohistochemical markers of lung adenocarcinoma comparable to previously generated models though also displayed concomitant expression of the squamous cell markers p63 and SOX2. Tumor-derived cell lines were not solely DDR2 dependent and displayed up-regulation of and partial dependence on MYCN. Combined treatment with the multitargeted DDR2 inhibitor dasatinib and BET inhibitor JQ1 inhibited tumor growth in vitro and in vivo. Together, these results suggest that DDR2 mutation can drive lung cancer initiation in vivo and provide a novel mouse model for lung cancer therapeutics studies.
Keywords: Lung cancer, mouse models, experimental therapeutics, oncogenes
INTRODUCTION
Lung cancer is the leading cause of cancer-related death in the United States and worldwide(1). More than 85% of lung cancer patients are diagnosed with non-small-cell lung cancer (NSCLC), including adenocarcinoma (ADC; 50% of lung cancers), squamous cell carcinoma (SCC; 30%) and large-cell carcinoma (10%)(2). Technological advances in recent years including the application of next-generation sequencing (NGS) have allowed researchers to construct large databases describing the molecular features of human lung tumors. These efforts have been accompanied by the generation of a number of new models of lung cancer, notably genetically engineered mouse models (GEMMs), which have facilitated the study of candidate human lung cancer oncogenes and tumor suppressor genes in vivo. These models have been utilized for a number of purposes including studies of tumor formation using one or more genetic alterations found in human cancers, studies of the impact of specific genetic changes on the tumor and its microenvironment and for studies of anti-cancer therapies. EGFR, Kras, BRAF and ERBB2 mutated and translocated EML4-ALK lung adenocarcinoma models have been generated and used for studies providing important insights into the mechanisms of response and resistance to clinically relevant targeted lung cancer therapies(3-7). Lung cancer genome studies continue to nominate an increasing number of candidate lung cancer oncogenes and tumor suppressors in lung adenocarcinoma, and more recently, in lung squamous cell carcinoma. Some candidate genomic alterations from studies of squamous cell lung cancers include amplification of SOX2, PDGFRA and FGFR1 and mutations of KEAP1, STK11, ERBB4 and DDR2 (Discoidin domain receptor 2)(8, 9).
DDR2 is a type I transmembrane receptor tyrosine kinase (RTK). DDR kinases are widely expressed in human tissues, are activated by collagens, and have roles in cell adhesion, migration, proliferation and survival when activated by ligand binding and phosphorylation(10). DDR kinases play a role in cancer progression by regulating the interactions of tumor cells with their surrounding collagen matrix. DDR2 mutations have been reported in multiple tumor types including lung cancer, breast cancer, brain cancer, gynecological cancer and prostate cancer(10). We previously reported the identification of novel somatic mutations in the DDR2 gene at a frequency of 3.8% in a sample set of 290 squamous cell lung cancer samples(11). Overall, 11 mutations were found throughout the entire gene and located in various DDR2 domains, including L63V, I120M, and D125Y within the collagen-binding discoidin 1 domain; L239R and G253C within the discoidin 2 domain; G505S in the cytosolic JM domain; and C580Y, I638F, T765P, G774E, and G774V within the kinase domain. DDR2 L63V and I638F mutations were shown to confer oncogenicity in NIH-3T3 fibroblasts in a colony forming assay in soft agar (11).
DDR2 can interact with multiple proteins resulting in complex signaling processes. Src has been shown to phosphorylate DDR2 resulting in subsequent DDR2 autophosphorylation(12). Thus, DDR2-Src interactions may play a key role in DDR2-intiated signaling. The signaling networks downstream of DDR2 include MAPK(13) and phosphoproteomic studies have nominated SHP-2 as a critical mediator of DDR2 signaling(14). Acquisition of an EMT phenotype in MDCK and human breast epithelial cells has also been shown to induce DDR2 expression(15). Activation of DDR2 regulates the EMT driver SNAIL1 stability by stimulating ERK2 activity, in a Src-dependent manner. DDR2-mediated stabilization of SNAIL1 promotes breast cancer cell invasion and metastasis in vivo(16). However, the transcription factors and mechanisms involved in upregulation of DDR2 during EMT have not been elucidated.
Four kinase inhibitors, dasatinib, imatinib, ponatinib and nilotinib were identified initially as inhibitors of DDR1 and DDR2 by chemical proteomic profiling studies(17, 18), suggesting that tumors with activated DDR2 signaling may be targeted by agents which are already FDA approved for other indications. In pre-clinical studies dasatinib was shown to inhibit proliferation in two lung cancer cell lines with DDR2 mutations both in vitro and in vivo and two case reports of response to dasatinib in lung cancer patients with DDR2 mutations have been published.(11, 19). However, studies have shown that DDR2 inhibition with dasatinib leads to both adaptive(20) and acquired resistance with resistance mechanisms including DDR2 gatekeeper mutation, NF1 loss and activation of parallel RTK pathways including EGFR, IGF1R and MET(13)
While several studies performed in cellular models have suggested that DDR2 mutations may be clinically relevant, data are lacking to prove that DDR2 mutations can drive lung cancer in vivo. Here we report that the conditional over-expression of the DDR2 L63V mutant downstream of a murine CCSP promoter can promote tumorigenesis in genetically engineered mice with a phenotype of lung adenocarcinoma. Additionally MYCN was elevated in the tumors driven by this DDR2 mutation and cell lines derived from the murine tumors were partially dependent on NYMC. We showed that the bromodomain inhibitor JQ1, an inhibitor of MYC-driven malignancies, plus the non-selective DDR2 inhibitor dasatinib as combination therapy could suppress growth of these tumors. Together, these data suggest that DDR2 mutations can contribute to lung cancer formation in vivo.
MATERIALS AND METHODS
Generation of the DDR2wt and L63V Mutant Mouse Cohort
The full-length DDR2wt cDNA was obtained from Origene and cloned into expression vector pBS31(21). L63V and I638F mutations were generated by site-directed mutagenesis using the Quickchange site directed mutagenesis kit (Strategene) and further verified by DNA sequencing. For the generation of transgenic mice with lung-specific doxycycline-inducible DDR2 expression we used a previously described method(22). The CCSP-rtTA and conditional Trp53-deficient allele (p53L/L) mice were kept at DFCI. DDR2wt and DDR2L63V mice were crossed with CCSP-rtTA and p53L/L mice. Progeny of DDR2wt;p53, DDR2L63V;p53 and DDR2I638F;p53 were genotyped as described in the supplemental methods. The DDR2 mice were fed a doxycycline diet at 6 weeks of age to induce DDR2 expression and intranasally instilled with Ad-Cre (University of Iowa viral vector core) to delete Trp53 as described (23). Mouse health condition was monitored weekly and evaluated by MRI as described previously(6).
All care of experimental animals was in accordance with Harvard Medical School/Dana-Farber Cancer Institute (DFCI) institutional animal care and use committee (IACUC) guidelines. All mice were housed in a pathogen-free environment at a DFCI animal facility and handled in strict accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare.
Establishment of GEMM-derived Primary Cell lines
When DDR2L63V; p53 mice developed lung tumors confirmed by MRI, the mice were sacrificed and lung tumor nodules were harvested, finely minced, and cultured in 100 mm dishes with RPMI 1640/10% FBS/1% pen-strep/2mM L-Glutamine. After 3 passages, frozen stocks of these short-term cultures were prepared, and further characterized by genotyping. DDR2 expression was induced by treating cells with 2 μg/ml DOX every 2 to 3 days and confirmed by western blot analysis. 634, 855 and 858 cells were primary murine cell lines with Kras;p53 mutation established in Wong lab (24). The cells were cultured in RPMI 1640/10% FBS/1% penstrep/2mM L-Glutamine. All cells were cultured at 37°C in a humidified incubator with 5% CO2.
Histology and immunohistochemistry
Mice were sacrificed with CO2; half of the dissected tumors were snap-frozen in liquid nitrogen for preparation of protein lysates and the left lung tissue was fixed in 10% neutral buffered formalin for 24 hours at room temperature, and then transferred to 70% ethanol, embedded in paraffin, and sectioned at 5μm for IHC staining. Hematoxylin and eosin (H&E) stains were performed in the Department of Pathology in Brigham and Women’s Hospital. Immunohistochemistry was performed using previously described methods(23). Anti-DDR2 antibody was from Bethyl Laboratories. All the other antibodies used for dynamic markers are listed in Table 1.
Table 1.
Antibodies used for immunohistochemistry
| Antibodies | Companies | Cat. ID |
|---|---|---|
| TTF1 | DAKO | M3575 |
| p63 | Abcam | ab53039 |
| SOX2 (C70B1) | Cell Signaling | 3728S |
| SPC | Millipore | AB3786 |
| F4/80 | eBioscience | 14-4801-82 |
| MPO | Novus | R-1073 |
| CCSP | Santa Cruz | sc-9772 |
Gene Expression Profiling Analysis
RNA was extracted from snap-frozen Kras;p53 and DDR2L63V;p53 tumor tissue using Trizol (Invitrogen) and further purified by RNeasy MinElute Cleanup Kit (Qiagen). Arrays were preformed at Dana-Farber Cancer Institute facility on Affymetrix mouse Gene1.0ST arrays. Data were preprocessed and normalized using the Genepattern Preprocess module with default parameters followed by Genepattern Comparative Marker Selection(25). GEO accession number for the microarray data reported in this paper is GSE64907.
Cell growth and proliferation assays
DDR2 DOX induced lung cancer cell lines (3941 and 3942) were obtained from DDR2L63V mice and were cultured in RPMI 1640 (Invitrogen) and 10% fetal bovine serum (Gibco). Kras mutated cell lines were previously generated(24) and maintained in the same conditions. DDR2 expression was induced by treating cells with 2 ug/ml DOX every 2 to 3 days.
Proliferation of MYCN and control siRNA tranfected cells was measured in triplicate after 24 and 48 hours using a Vi-CELL Cell Viability Analyzer (Beckman Coulter). Proliferation of cells (634, 855, 858, 3941 DDR2, and 3942 DDR2) was measured with the Cell-Titer-Glo reagent (Promega) per the maufacturer’s instructions. Cells were seeded in triplicate at 1,500 cells per well in 96-well clear-bottomed plates. Single drugs and combinations were added the following day at concentrations of 10nM, 50nM, 100nM, 500nM, 1uM, and 10uM and after 5 days a standard 96-well plate luminometer was used to measure cell proliferation. Comparison of untreated cells with those treated at a given concentration was used to determine percent survival. For proliferation analysis, mean values were calculated from samples in triplicate and SEs were calculated by Microsoft Excel. GraphPad Prism software was used to determine IC50 values.
RNAi and cell culture
MYCN expression was knocked down using MYCN siRNA (m) obtained from Santa Cruz Biotechnology, INC. Cell lines 3941 DDR2 and 858 were seeded at 2 × 105 cells per well in 6 well tissue culture plates. Control and N-Myc siRNA transfected cells were used for immunoblot analysis and counted at 24 and 48 hours requiring a total of 12 seeded wells. Control siRNA was obtainted from the siRNA Reagent System (Santa Cruz Biotechnology) and cells were transfected according to the manufacturer’s protocol (Santa Cruz Biotechnology). For DDR2 siRNA the same protocol was followed using siRNAs s9761 and s9762 from Santa Cruz Biotechnology. siRNA efficiency was determined by western blot as below or real-time PCR. For real-time PCR total cellular RNA was prepared from the cells by using an RNeasy Mini Kit (Qiagen) and 1.0 μg of the RNA was then reverse transcribed to cDNA using TaqMan Reverse Transcription Reagents (Life Technologies). Mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization of input cDNA. The human NSCLC cell lines NCI-H2286 and HCC-366 were obtained from the ATCC in 2009 and not further authenticated.
Western blotting
Snap-frozen tissues or cells cultured in 6-well plates were homogenized with RIPA buffer containing phosphatase and protease inhibitors (Thermo), lysates were cleared by centrifugation, protein concentration was determined using the Bradford reagent (Bio-Rad), and 100ug of lysate was loaded per sample. Immunoblots were then performed using the Nupage System (Invitrogen) per the manufacturer’s instructions. Primary antibodies used were NMYC (Santa Cruz Biotechnology), DDR2 (Bethyl Laboratories), β-Actin (Sigma), and Vinculin (Sigma). Secondary antibodies used were horseradish peroxidase mouse and rabbit (Pierce) and ECL prime (Thermo Fisher Scientific) was used for protein detection.
Xenografts and in vivo treatment
Nu/Nu mice were purchased from Charles River Laboratories International Inc. 3941-DDR2 was detected as pathogen free in Charles River Laboratories International Inc. and cultured in RPMI1640 with 10% FBS. The cells were washed with serum-free medium and resuspended in serum-free medium mixed with an equal amount of Matrigel (BD Biosciences). Mice were injected with 1 million cells per tumor and 3 locations per mouse. The mice were randomly grouped and treatment was started the second day after inoculation. Each cohort included 5 mice. Dasatinib was dissolved in HKI solution and administered orally at 50 mg/kg daily. HKI solution was prepared as same as the previous publication(22). JQ1 was formulated in 10% DMSO, 90% 10%-Hyroxypropyl Beta Cyclodextrin and given IP route at 50 mg/kg daily. Both oral and IP vehicle were administered to the control group in the same way as the treatment mice. Tumor sizes were monitored twice weekly and volumes were calculated with the formula: (mm3) = length × width × width × 0.5.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism software. Proliferation assays are represented as the mean +/− SD. In vivo experiments are represented as the mean +/− SEM. Significance of in-vivo treatment was assessed by two-way ANOVA and Bonferonni’s multi-comparison test. Survival analysis of DDR2wt;p53 and DDR2L63V;p53 mice was assessed by the log-rank test.
RESULTS
DDR2L63V expression is oncogenic in the lung
Given that somatic DDR2 mutants have been observed in human NSCLC and shown to be potential therapeutic targets in cellular studies, we sought to generate a mouse model of lung cancer driven by mutated DDR2 to examine whether expression of DDR2 mutants could initiate lung tumorigenesis in vivo. To address this question, we selected the L63V and I638F mutations for characterization given that they appeared to be the most oncogenic in NIH-3T3 assays reported previously(11). These DDR2 mutations were introduced in the human DDR2 gene by site directed mutagenesis and cloned into pBS31 flp-in vector followed by a tetO minimal CMV promoter and an ATG and an FRT site(21). DDR2wt, DDR2L63V and DDR2I638F transgenic mice were then generated by injection of the construct into FVB/N fertilized eggs. Progeny were genotyped by PCR. Founders were crossed with CCSP-rtTA as well as conditional Trp53-deficient allele (p53L/L) mice to accelerate tumor formation as no tumors were observed in the DDR2 transgenics alone with two years of observation. The complex model tet-op-DDR2wt;CCSP-rtTA;p53L/L (DDR2wt;p53 hereafter), tet-op-DDR2L63V;CCSP-rtTA;p53L/L (DDR2L63V;p53 hereafter) and tet-op-DDR2I638F;CCSP-rtTA;p53L/L (DDR2I638F;p53 hereafter) were confirmed by genotyping and expanded for subsequent analyses and experiments (Figure 1A). DDR2wt;p53, DDR2L63V;p53 and DDR2I638F;p53 mice were placed on a continuous doxycycline (DOX) diet at 6 weeks of age to induce DDR2 expression, and Adenovirus-Cre intranasally instilled to delete Trp53 on the same day. Lungs of induced mice were visualized by serial MRI every 8 weeks and mice were also sacrificed every 8 weeks for histological examination.
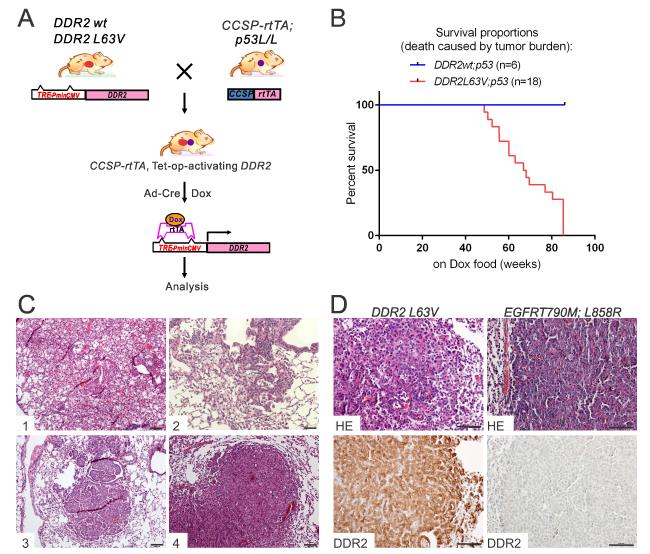
Figure 1. Generation and characterization of the genetically engineered mice with inducible expression of DDR2L63V and TP53 deletion in the lung.
A. Schema of the DDR2 construct, cross strategy and induction for the conditional transgenic mice.
B. Kaplan-Meyer survival analysis of DDR2;p53 mice following intra-nasal Ad-Cre instillation and DOX administration. All death was caused by tumor burden. Median survival of DDR2L63V;p53= 67.5 weeks.
C. Representative H&E-stained sections derived from tumors arising in the DDR2L63V;p53 mouse model at different time points after DOX and Ad-Cre administration.
D. Immunohistochemical staining of DDR2 on tumor nodules driven by DDR2L63V;p53 and EGFRT 790M-L858R.
Scale bars represent 100 mm for all panels.
Expression of DDR2 was targeted to lung type II alveolar cells by crossing mice carrying club (originally known as Clara) cell secretory protein (CCSP)-regulated reverse tetracycline transactivator (rtTA) transgene, regulated by tetracycline (tet)-responsive elements, which allowed for DDR2 expression in lung pneumocytes after DOX administration. The median survival of DDR2L63V;p53 mice was 67.5 weeks (Figure 1B) and there was no mortality observed in the DDR2wt;p53 or DDR2I638F;p53 models at two years of observation. After 20-30 weeks of DOX treatment, histological examination of DDR2L63V;p53 mice demonstrated the development of focal or diffuse bronchioloalveolar carcinoma (BAC), in some cases preceded by precancerous adenomatous lesions in the airway epithelia (Figure 1C). After 30-40 weeks on DOX, the DDR2L63V;p53 mice developed small intrabronchial carcinomas in the distal (Figure 1C) and proximal (Figure 1C) bronchioloalveolar locations. After 50-60 weeks, all of the tested DDR2L63V;p53 mice developed lung ADC (Figure 1C). In agreement with the pathological observations, MRI images also showed gradual tumor development of DDR2L63V;p53 mice after 40 weeks of DOX induction (Figure S1A). The DDR2wt;p53 and DDR2I638F;p53 models only showed epithelial hyperplasia (Figure S1B); none of tested mice developed malignant tumors within two years. Both immunohistochemical stains (IHC) and western blotting confirmed DDR2 expression in tumor nodules (Figures 1D, S1C and S1D) and lack of p53 (Figure S2A). The majority of DDR2L63V tumors showed marked SPC staining, implying a type II pneumocyte origin, as expected given the use of the CCSP promoter. Interestingly, CCSP staining was nearly negative (Figure S2B).. One possibility is that these tumors are of club cell origin which is then followed by altered differentiation leading to loss of the CCSP expression marker. The same phenomenon was observed previously for the tet-op-EGFR-T790M-L858R;CCSP-rtTA mice(22).
NSCLC driven by DDR2L63V mutant is morphologically adenocarcinoma but with p63 and SOX2 expression
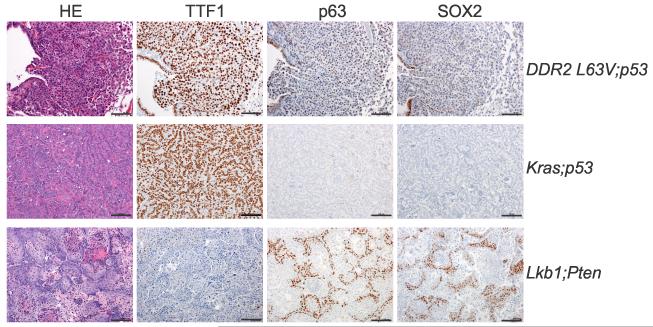
Since somatic DDR2 mutations are more frequently found in human SCC, we explored whether the DDR2L63V model we generated displayed any markers of lung SCC. p63 is a well known marker of squamous differentiation and over-expression of this gene has been consistently identified in lung SCCs by global gene expression profiling or by IHC(26, 27). Several other markers are widely used in the subclassification of lung carcinomas including CK5/6 and TTF1(26-28). Most human SCC cases display a p63+, TTF1− immunophenotype, while most ADC cases show the opposite expression pattern: TTF1+, p63−. SOX2 is a transcription factor reported as an important gene in SCC and is also used as histological marker of human SCC(29, 30). As controls for this analysis we utilized LSL-KrasG12D;p53L/L (Kras;p53 hereafter) genetically engineered mice which develop typical ADC and mice with biallelic inactivation of Lkb1 and Pten which leads to SCC(23) (Figure 2). Interestingly, DDR2L63V tumors displayed typical ADC morphology with strong TTF1 expression but also displayed some p63 and SOX2 staining (Figure 2). This mixed phenotype is not unique to the tumors induced by DDR2L63V and has been previously reported with inducible deletion of the tumor suppressor Lkb1 along with KrasG12D mutant(5).
Figure 2. NSCLC driven by DDR2L63V displays a mixed phenotype.
Immunohistochemical staining of tumor nodules of DDR2L63V;p53 mice, the SCC canonical markers p63 and SOX2, and the ADC canonical marker TTF1 were used to distinguish the tumor types.
Scale bars represent 100 mm for all panels.
MYCN is elevated in DDR2L63V tumors
Given the long latency of the DDR2L63V model we generated two tumor-derived cell lines (3941 and 3942) to study signaling and therapeutics in more detail. Both cell lines demonstrated expression of DDR2 which was augmented by culture in DOX, as would be expected (Figure 3A). Cell line 855 displayed in this Figure is one of several previously described cell lines generated from murine Kras;p53 tumors(24). Interestingly, both the 3941 and 3942 cell lines proliferated well in culture in the presence of or absence of DOX with no discernible difference in phenotype, suggesting that they were not dependent on ongoing DDR2 expression and raising the possibility that additional genomic alterations may be contributing to transformation in the DDR2 mutated mouse model. To support this observation we performed siRNA mediated knock-down of DDR2 in the 3941 and 3942 cell lines. While the degree of knock-down in these lines was only ~50% as measured by real-time PCR (Figure S3A) there was no effect seen on the proliferation of these cell lines or of the Kras;p53 cell line 858 which was selected on the basis of transfectibility (Figure S3B).
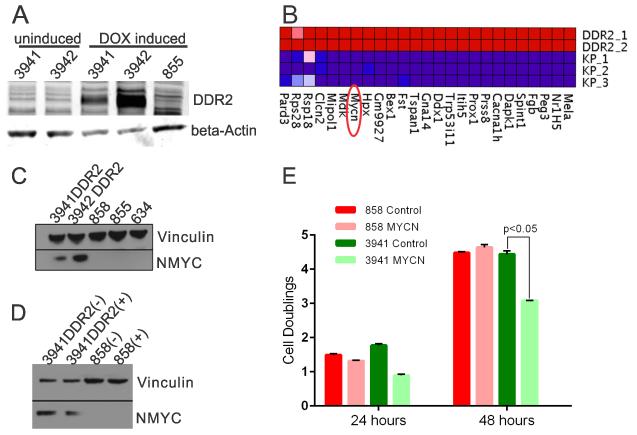
Figure 3. MYCN is elevated in DDR2L63V tumors.
A. Immunoblot analysis of DDR2 levels in uninduced and DOX induced cell lines (3941 and 3942) from DDR2 transgenic tumors. The 855 line was previously derived from a murine lung adenocarcinoma from a Kras;p53 mouse.
B. Microarray expression profiling of mouse DDR2L63V;p53 (3941 and 3942) and Kras;p53 cell lines (634, 855 and 858); Heatmap depicts elevated expression of selected genes in DDR2L63V;p53 lines.
C. Immunoblot analysis of NMYC in DDR2L63V;p53 cell lines vs Kras;p53 cell lines.
D. Immunoblot analysis of NMYC knockdown (“+” indicates siRNA knockdown and “−” a non-targeting siRNA).
E. Proliferation assay of a Kras;p53 cell line (858) compared to the DDR2L63V;p53 cell line (3941) 24 and 48 hours after siRNA mediated NMYC knockdown. Control indicates a non-targeting siRNA control.
To address the possibility that other genetic alterations may be playing an important role in the tumors from the DDR2L63V;p53 mutated mice we performed microarray analyses to compare the gene expression profiles of these two tumor-derived cell lines from the DDR2L63V;p53 mutated mice to lines from three mouse Kras;p53 tumors(24). This was performed after comparative immunoblot analysis of these cell lines did not show any differential phosphorylation of RAS/MAPK or PI3K effectors as compared to the Kras mutated cell lines (data not shown). Of note the cell lines studied were derived from tumors which developed typical ADC on pathology and p53 was conditionally deleted at the same ages. Differential gene expression analysis using the Genepattern module identified 541 genes differentially upregulated in the DDR2L63V tumors as compared to the Kras tumors using a fold-change cut-off of 2.0 and a corrected p value of less than 0.001 (Supplementary Data File). A representative heatmap of the top differentially expressed genes is shown in Figure 3B. Manual inspection of these top differentially expressed genes focused our attention on MYCN, a member of the myc family of proto-oncogenes. The MYC gene is expressed in a wide variety of tissues, while MYCN expression is restricted to early stages of embryonic development, amplification of MYCN is frequently found in a number of advanced-stage tumors, including lung cancer, contributing to a myriad of phenotypes associated with growth, invasion, and drug resistance(31).
In the microarray data, MYCN expression was 37-fold higher in the DDR2L63V tumor-derived cell lines and immunoblotting demonstrated elevated MYCN protein levels in 3941 and 3942 cell lines as compared to Kras lines 634, 855, and 858 (Figure 3C). Analysis of gene expression data from The Cancer Genome Atlas demonstrated a trend towards higher expression of MYCN in DDR2 mutated lung squamous cell carcinomas (mean RSEM 176 versus 127) but this difference was not stastisticaly significant. DDR2 and MYCN expression levels were weakly correlated across the TCGA cohort (Pearson correlation 0.22). We did not note any upregulation of MYCN in two prevously described NSCLC cell lines with DDR2 mutations (NCI-H2286 and HCC-366) as compared to other NSCLC cell lines described in the Cancer Cell Line Encylopedia.
Given increased MYCN expression in the DDR2 mutated lines (Figure 3C), we assessed whether MYCN loss would have an effect on their proliferation. siRNA was used to knock-down MYCN (Figure 3D) and cellular viability was counted after 24 and 48 hours. No substantial difference was observed in the 858 Kras mutated cell line with MYCN depletion. However, we observed a reduction in proliferation of the 3941 DDR2 mutated cell line with MYCN depletion (Figure 3E), indicating partial dependence on MYCN in DDR2L63V mutated cell lines.
NSCLC driven by DDR2L63V is sensitive to dasatinib and JQ1 combination therapy
We next probed the 3941 and 3942 cell lines for sensitivity to two previously reported non-selective DDR2 inhibitors, dasatinib and ponatinib and compared the sensitivity to three previously generated Kras;p53 cell lines (634, 855 and 858). We did not observe any differences in the sensitivity of these cell lines to these two chemical inhibitors (dasatinib: 3941 and 3942 DDR2 IC50 1.96μM and 0.682μM respectively as compared to Kras 634, 855, 858 with IC50 0.484μM, 1.76μM, and 0.557μM respectively. Ponatinib: 3941 and 3942 DDR2 IC50 0.579μM and 0.978μM respectively as compared to Kras 634, 855, 858 IC50 2.26μM, 2.31μM, and 2.57μM respectively.) (Figure S4). These two cell lines were also probed for sensitivity to a previously reported SRC inhibitor sarcatinib given studies linking SRC and DDR2 activity and no differences in sensitivity were observed (saracatinib: 3941 and 3942 DDR2 IC50 2.69μM and 5.49 μM respectively as compared to Kras 634, 855, 858 with IC50 4.69μM, 11.67μM, and 8.86μM respectively) (Figure S4). To control for any intrinsic drug resistence of the 3941 or 3942 cell lines as compared to the Kras mutated lines we compared their sensitivity to etoposide and noted no inherent chemoresistance in the DDR2 mutated lines (3941 and 3942 DDR2 IC50 0.840μM and 0.439μM respectively as compared to Kras 634, 855, 858 with IC50 2.49μM, 1.53μM, and 1.00μM respectively) (Figure S4).
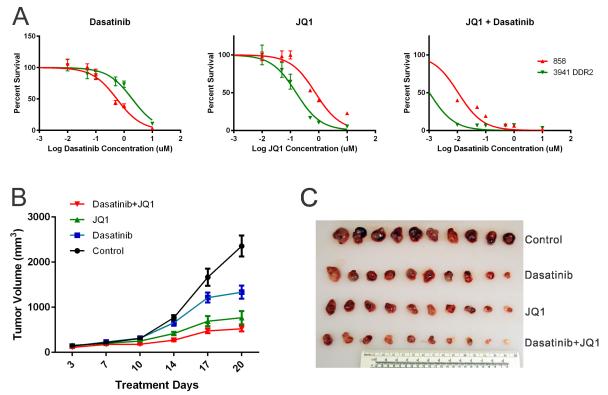
Given evidence that MYCN knock-down in DDR2 mutated cell lines decreased cellular proliferation, we reasoned that combined inhibiton of DDR2 and NYMC might further suppress proliferation of these models. To this end, we assessed the impact of the compound JQ1, a prototype bromodomain and extra-terminal (BET) bromodomain inhibitor which has been previously shown to suppress MYC activity(32, 33), in the 3941 and 858 cell lines. We observed enhanced sensitivity of 3941 to JQ1 alone (3941 DDR2 IC50 of 160nM as compared to Kras 858 IC50 of 761nM) and to the combination of JQ1 and Dasatinib as compared to 858 (3941 DDR2 IC50 of 1.0nM as compared to Kras 858 IC50 of 10.8nM). (Figure 4A) To assess this combination treatment in a more physiological setting, we performed a xenograft study where Nu/Nu mice were injected with 3941 DDR2 cells and treated with vehicle, dasatinib, JQ1, or dasatinib plus JQ1 combination. Tumor formation was monitored twice weekly for 3 continuous weeks. Consistent with our previous observation(11), dasatinib showed anti-tumor efficacy on DDR2L63V mouse model as compared to vehicle control. JQ1 also showed anti-tumor efficacy; however, the combination treatment of JQ1 and dasatinib displayed the most potent effect with growth inhibition of all tested tumors (Figure 4B, C, S5, p<0.001; individual tumor responses are shown in S5). While they were not as sensitive as the murine lines to JQ1 alone JQ1 potentiated the effects of dasatinib in the DDR2 mutated NSCLC cell lines NCI-H2286 and HCC366 with IC50 values of 10 and 14 nM respectively for dasatinib with 1 μM JQ1, a one-log reduction from their typical dasatinib IC50 (Figure S6).
Figure 4. NSCLC driven by DDR2L63V is sensitive to dasatinib and JQ1 combination therapy.
A. Proliferation of Kras;p53 cell lines 858, and DDR2L63V;p53 cell line 3941 grown for 5 days in the presence of varying concentrations of dasatinib, JQ1 and a combination of 1μM JQ1 and varying dasatinib concentrations.
B, C. Tumor volume measurement in a xenograft study of the DDR2L63V;p53 (3941) lung cancer cell line. Nu/Nu mice were treated with dasatinib, JQ1, and dasatinib plus JQ1 combination for 3 weeks following tumor formation. B. Average tumor volumes; C. Harvested tumor nodules sizes;
DISCUSSION
Here, we have presented a novel genetically engineered mouse model of lung cancer driven by L63V mutation in DDR2. These studies were motivated by prior work suggesting that DDR2 mutations may be oncogenic and confer sensitivity to FDA approved tyrosine kinase inhibitors including dasatinib, imatinib, nilotinib and ponatinib. Since all of the prior work was completed in lung cancer cell lines or other cellular models, we sought to explore whether mutated DDR2 could drive lung cancer formation in an organism and observed a phenotype of lung adenocarcinoma with a relatively long latency. The adenocarcinoma phenotype was not surprising given our use of the CCSP promoter to drive expression of mutated DDR2 in cell types known to be precursors for lung adenocarcinoma as well as prior reports of DDR2 mutations in lung adenocarcinoma(34). The recent publication of several models of mouse lung SCCs also suggests that an inflammatory insult and/or a genetic lesion driving squamous metasplasia is required for SCC formation(23, 35), neither of which was present in this model.
One of the challenges in advancing DDR2 mutations as a potential therapeutic target in lung cancer has been the lack of recurrent point mutations in the gene as well as the infrequency of DDR2 mutations overall. Studies in cell lines have suggested that only a subset of patient-derived mutations confer gain-of-function phenotypes and the ability to discern “driver” from “passenger” events in DDR2 has not been well-established in an experimental system with conflicting reports in some cases such as DDR2 I638F which was not oncogenic in our mouse model nor in prior proteomic studies but was transforming in some cellular systems(11, 14). Our results, therefore, call into question whether I638F is really a gain-of-function mutation or is operating via a different mechanism as compared to L63V. Interestingly, two cases of response to dasatinib in patients with DDR2 S768R mutations have been reported, suggesting that at least some patient-derived mutations are likely to be therapeutic targets, though dasatinib therapy has been associated with toxicity in lung cancer patients(36). In this work we established a single point mutation (L63V) of DDR2 as oncogenic in vivo and the generation of additional cellular and animal models will be necessary to discern which DDR2 mutations are truly oncogenic given that it is likely that many DDR2 mutations will be passenger events given the high background mutation rate of lung cancers. This work does, however, serve as a proof of concept of the oncogenicity of mutated and not wild-type DDR2 in a mouse model.
While we observed lung cancer formation at a high penetrance in the DDR2 L63V model the latency was long and the fact that we were able to grow tumor derived cell lines in the absence of DOX induction suggests that while DDR2 mutants can play a role in lung cancer initiation there are likely to be other important cooperating oncogenic alterations which facilitate ongoing proliferation of DDR2 mutated tumors. This idea is in agreement with human cancer-derived cell line studies which suggest that a number of oncogenic pathways are activated in DDR2 mutated cell lines and can substitute for DDR2 in the context of DDR2 inhibition by small molecule inhibitors. While these studies have largely focused on compensatory RTK pathways our observation that MYCN can play a role in DDR2 mutated tumors also argues that while DDR2 mutations can facilitate tumor formation specific and selective inhibition of DDR2 may not be sufficient to eradicate DDR2-driven cancers. It is likely that additional genomic alterations beyond overexpression of MYCN are relevant in DDR2 mutated lung cancers as the TCGA datasets show overlap with KRAS mutation and NKX2-1 amplification in the limited number of samples with DDR2 mutation. Limited sequence analysis of the DDR2 mutated nodules in this report also indicated that other mutations are present along with DDR2 and include mutations in Kras (codon 61), Fgfr2, Irs2, Csf1r and Daxx, suggesting that DDR2 mutations are not sufficient alone to drive tumorigenesis and require cooperating events.
In conclusion, our work extends prior cellular studies which have demonstrated that DDR2 mutations are found in non-small cell lung cancers and that a subset of these mutations are oncogenic both in culture systems and in the context of a transgenic mouse model. Our model can facilitate additional studies of the role of mutated DDR2 in lung cancer as well as serve as a platform for therapeutic studies aimed at targeting DDR2.
Supplementary Material
Acknowledgments
FINANCIAL SUPPORT
This work was supported in part by United against Lung Cancer and Susan Spooner Research Fund to K.-K. Wong; NIH grants NCI P01 CA154303 to K.-K. Wong and M. Meyerson; NCI K08 CA163677 to P.S. Hammerman.
Footnotes
CONFLICT OF INTEREST
P.S.H. reports consulting fees from Molecular MD.
K.K. Wong: Consultant/Advisory Board of AstraZeneca and Janssen Pharmaceuticals, Stock ownership in Gatekeeper Pharmaceuticals
M.M: Founder and stock ownership of Foundation Medicine, research support from Bayer and ownership interest in LabCorp.
All other authors report no relevant conflicts of interest
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. doi: 10.3322/caac.21166. PubMed PMID: 23335087. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–46. doi: 10.1038/nrc3775. doi: 10.1038/nrc3775. PubMed PMID: 25056707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–8. doi: 10.1101/gad.943001. doi: 10.1101/gad.943001. PubMed PMID: 11751630; PubMed Central PMCID: PMC312845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji H, Wang Z, Perera SA, Li D, Liang MC, Zaghlul S, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67(10):4933–9. doi: 10.1158/0008-5472.CAN-06-4592. doi: 10.1158/0008-5472.CAN-06-4592. PubMed PMID: 17510423. [DOI] [PubMed] [Google Scholar]
- 5.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–10. doi: 10.1038/nature06030. doi: 10.1038/nature06030. PubMed PMID: 17676035. [DOI] [PubMed] [Google Scholar]
- 6.Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9(6):485–95. doi: 10.1016/j.ccr.2006.04.022. doi: 10.1016/j.ccr.2006.04.022. PubMed PMID: 16730237. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010;70(23):9827–36. doi: 10.1158/0008-5472.CAN-10-1671. doi: 10.1158/0008-5472.CAN-10-1671. PubMed PMID: 20952506; PubMed Central PMCID: PMC3043107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. doi: 10.1038/nature11404. PubMed PMID: 22960745; PubMed Central PMCID: PMC3466113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y, Hammerman PS, Kim J, Yoon JA, Lee Y, Sun JM, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol. 2014;32(2):121–8. doi: 10.1200/JCO.2013.50.8556. doi: 10.1200/JCO.2013.50.8556. PubMed PMID: 24323028; PubMed Central PMCID: PMC4062710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valiathan RR, Marco M, Leitinger B, Kleer CG, Fridman R. Discoidin domain receptor tyrosine kinases: new players in cancer progression. Cancer Metastasis Rev. 2012;31(1-2):295–321. doi: 10.1007/s10555-012-9346-z. doi: 10.1007/s10555-012-9346-z. PubMed PMID: 22366781; PubMed Central PMCID: PMC3351584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1(1):78–89. doi: 10.1158/2159-8274.CD-11-0005. doi: 10.1158/2159-8274.CD-11-0005. PubMed PMID: 22328973; PubMed Central PMCID: PMC3274752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang K, Kim JH, Kim HJ, Park IS, Kim IY, Yang BS. Tyrosine 740 phosphorylation of discoidin domain receptor 2 by Src stimulates intramolecular autophosphorylation and Shc signaling complex formation. J Biol Chem. 2005;280(47):39058–66. doi: 10.1074/jbc.M506921200. doi: 10.1074/jbc.M506921200. PubMed PMID: 16186108. [DOI] [PubMed] [Google Scholar]
- 13.Beauchamp EM, Woods BA, Dulak AM, Tan L, Xu C, Gray NS, et al. Acquired resistance to dasatinib in lung cancer cell lines conferred by DDR2 gatekeeper mutation and NF1 loss. Mol Cancer Ther. 2014;13(2):475–82. doi: 10.1158/1535-7163.MCT-13-0817. doi: 10.1158/1535-7163.MCT-13-0817. PubMed PMID: 24296828; PubMed Central PMCID: PMC3946067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwai LK, Payne LS, Luczynski MT, Chang F, Xu H, Clinton RW, et al. Phosphoproteomics of collagen receptor networks reveals SHP-2 phosphorylation downstream of wild-type DDR2 and its lung cancer mutants. Biochem J. 2013;454(3):501–13. doi: 10.1042/BJ20121750. doi: 10.1042/BJ20121750. PubMed PMID: 23822953; PubMed Central PMCID: PMC3893797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107(35):15449–54. doi: 10.1073/pnas.1004900107. doi: 10.1073/pnas.1004900107. PubMed PMID: 20713713; PubMed Central PMCID: PMC2932589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW, et al. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol. 2013;15(6):677–87. doi: 10.1038/ncb2743. doi: 10.1038/ncb2743. PubMed PMID: 23644467; PubMed Central PMCID: PMC3794710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110(12):4055–63. doi: 10.1182/blood-2007-07-102061. doi: 10.1182/blood-2007-07-102061. PubMed PMID: 17720881. [DOI] [PubMed] [Google Scholar]
- 18.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035–44. doi: 10.1038/nbt1328. doi: 10.1038/nbt1328. PubMed PMID: 17721511. [DOI] [PubMed] [Google Scholar]
- 19.Pitini V, Arrigo C, Di Mirto C, Mondello P, Altavilla G. Response to dasatinib in a patient with SQCC of the lung harboring a discoid-receptor-2 and synchronous chronic myelogenous leukemia. Lung Cancer. 2013;82(1):171–2. doi: 10.1016/j.lungcan.2013.07.004. doi: 10.1016/j.lungcan.2013.07.004. PubMed PMID: 23932362. [DOI] [PubMed] [Google Scholar]
- 20.Bai Y, Kim JY, Watters JM, Fang B, Kinose F, Song L, et al. Adaptive Responses to Dasatinib-Treated Lung Squamous Cell Cancer Cells Harboring DDR2 Mutations. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-0505. doi: 10.1158/0008-5472.CAN-14-0505. PubMed PMID: 25348954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44(1):23–8. doi: 10.1002/gene.20180. doi: 10.1002/gene.20180. PubMed PMID: 16400644. [DOI] [PubMed] [Google Scholar]
- 22.Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12(1):81–93. doi: 10.1016/j.ccr.2007.06.005. doi: 10.1016/j.ccr.2007.06.005. PubMed PMID: 17613438. [DOI] [PubMed] [Google Scholar]
- 23.Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25(5):590–604. doi: 10.1016/j.ccr.2014.03.033. doi: 10.1016/j.ccr.2014.03.033. PubMed PMID: 24794706; PubMed Central PMCID: PMC4112370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;3(8):870–9. doi: 10.1158/2159-8290.CD-13-0015. doi: 10.1158/2159-8290.CD-13-0015. PubMed PMID: 23715154; PubMed Central PMCID: PMC3753578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22(15):1924–5. doi: 10.1093/bioinformatics/btl196. doi: 10.1093/bioinformatics/btl196. PubMed PMID: 16709585. [DOI] [PubMed] [Google Scholar]
- 26.Kim MJ, Shin HC, Shin KC, Ro JY. Best immunohistochemical panel in distinguishing adenocarcinoma from squamous cell carcinoma of lung: tissue microarray assay in resected lung cancer specimens. Annals of diagnostic pathology. 2013;17(1):85–90. doi: 10.1016/j.anndiagpath.2012.07.006. doi: 10.1016/j.anndiagpath.2012.07.006. PubMed PMID: 23040737. [DOI] [PubMed] [Google Scholar]
- 27.Fatima N, Cohen C, Lawson D, Siddiqui MT. Combined double CK5/P63 stain: useful adjunct test for diagnosing pulmonary squamous cell carcinoma. Diagn Cytopathol. 2012;40(11):943–8. doi: 10.1002/dc.21678. doi: 10.1002/dc.21678. PubMed PMID: 21472873. [DOI] [PubMed] [Google Scholar]
- 28.Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(10):1348–59. doi: 10.1038/modpathol.2011.92. doi: 10.1038/modpathol.2011.92. PubMed PMID: 21623384. [DOI] [PubMed] [Google Scholar]
- 29.Brcic L, Sherer CK, Shuai Y, Hornick JL, Chirieac LR, Dacic S. Morphologic and clinicopathologic features of lung squamous cell carcinomas expressing Sox2. American journal of clinical pathology. 2012;138(5):712–8. doi: 10.1309/AJCP05TTWQTWNLTN. doi: 10.1309/AJCP05TTWQTWNLTN. PubMed PMID: 23086772. [DOI] [PubMed] [Google Scholar]
- 30.Tsuta K, Tanabe Y, Yoshida A, Takahashi F, Maeshima AM, Asamura H, et al. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2011;6(7):1190–9. doi: 10.1097/JTO.0b013e318219ac78. doi: 10.1097/JTO.0b013e318219ac78. PubMed PMID: 21623236. [DOI] [PubMed] [Google Scholar]
- 31.Bell E, Chen L, Liu T, Marshall GM, Lunec J, Tweddle DA. MYCN oncoprotein targets and their therapeutic potential. Cancer Lett. 2010;293(2):144–57. doi: 10.1016/j.canlet.2010.01.015. doi: 10.1016/j.canlet.2010.01.015. PubMed PMID: 20153925. [DOI] [PubMed] [Google Scholar]
- 32.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–73. doi: 10.1038/nature09504. doi: 10.1038/nature09504. PubMed PMID: 20871596; PubMed Central PMCID: PMC3010259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19(22):6183–92. doi: 10.1158/1078-0432.CCR-12-3904. doi: 10.1158/1078-0432.CCR-12-3904. PubMed PMID: 24045185; PubMed Central PMCID: PMC3838895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ford CE, Lau SK, Zhu CQ, Andersson T, Tsao MS, Vogel WF. Expression and mutation analysis of the discoidin domain receptors 1 and 2 in non-small cell lung carcinoma. Br J Cancer. 2007;96(5):808–14. doi: 10.1038/sj.bjc.6603614. doi: 10.1038/sj.bjc.6603614. PubMed PMID: 17299390; PubMed Central PMCID: PMC2360060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629. doi: 10.1038/ncomms5629. doi: 10.1038/ncomms5629. PubMed PMID: 25115923. [DOI] [PubMed] [Google Scholar]
- 36.Brunner AM, Costa DB, Heist RS, Garcia E, Lindeman NI, Sholl LM, et al. Treatment-related toxicities in a phase II trial of dasatinib in patients with squamous cell carcinoma of the lung. J Thorac Oncol. 2013;8(11):1434–7. doi: 10.1097/JTO.0b013e3182a47162. doi: 10.1097/JTO.0b013e3182a47162. PubMed PMID: 24128713; PubMed Central PMCID: PMC3801424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.