Abstract
The purpose of this study was to investigate the molecular and therapeutic effects of small-interfering RNA (siRNA)-mediated c-MYC silencing in cisplatin-resistant ovarian cancer. Statistical analysis of patient’s data extracted from The Cancer Genome Atlas (TCGA) portal showed that the progression free- (PFS) and the overall (OS) survival were decreased in ovarian cancer patients with high c-MYC mRNA levels. Furthermore, analysis of a panel of ovarian cancer cell lines showed that c-MYC protein levels were higher in cisplatin-resistant cells when compared to their cisplatin-sensitive counterparts. In vitro cell viability, growth, cell cycle progression, and apoptosis, as well as in vivo therapeutic effectiveness in murine xenograft models, were also assessed following siRNA-mediated c-MYC silencing in cisplatin-resistant ovarian cancer cells. Significant inhibition of cell growth and viability, cell cycle arrest, and activation of apoptosis, were observed upon siRNA-mediated c-MYC depletion. In addition, single weekly doses of c-MYC-siRNA incorporated into 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG-2000)-based nanoliposomes resulted in significant reduction in tumor growth. These findings identify c-MYC as a potential therapeutic target for ovarian cancers expressing high levels of this oncoprotein.
Keywords: c-MYC, siRNA, nanoliposomes, ovarian cancer, cisplatin
Introduction
The c-myc (v-myc avian myelocytomatosis viral oncogene homolog) proto-oncogene belongs to a family of transcription factors characterized by the basic helix-loop-helix leucine-zipper (bHLHZ) motif which allows binding to specific DNA sequences as multimeric complexes (1,2). c-MYC regulates the expression of genes involved in a myriad of cellular processes including replication, growth, metabolism, differentiation, and apoptosis (1–3). Transcriptional activation by c-MYC involves heterodimer complex formation with its protein partner Max (MYC associated factor X), as well as the recruitment of histone acetyltransferases and other coactivators (1,2,4–7).
Oncogenic c-MYC arises through multiple molecular mechanisms including gene amplification, gene translocation, enhanced transcription for other upstream pathways, dysregulation of mRNA-interacting molecules, and decreased rates of ubiquitin-mediated proteolysis (8–10). Overexpression of c-MYC has been reported in most, if not all, types of human malignancies (8,11,12). In fact, integrated genome analysis of ovarian carcinoma using The Cancer Genome Atlas (TCGA) project revealed that the most common somatic focal amplification encodes eight genes, including the c-MYC gene, which is amplified in 30–60% of human ovarian tumors (13,14). In other tumor types, c-MYC expression levels have been associated with drug resistance (15–26). Current adjuvant chemotherapy for ovarian cancer includes a platinum-based drug such as cisplatin plus a taxane (i.e. paclitaxel) (27). Unfortunately, despite initial response, most patients develop chemoresistant disease, resulting in progressive disease and death (28). Therefore, elucidation of the molecular mechanisms underlying such resistance is imperative to identify novel targets for ovarian cancer therapy. Given the pivotal role of c-MYC in ovarian cancer, its therapeutic targeting in chemoresistance is evident. Here, we examine the biological and therapeutic effects of targeting c-MYC by small-interfering RNAs (siRNAs) in cisplatin-resistant cells and in pre-clinical models of ovarian cancer.
Materials and Methods
Cells and culture conditions
The human ovarian epithelial cancer cells A2780CP20, SKOV3ip1, SKOV3.TR, HEYA8 and HEYA8.MDR were generous gifts from Dr. Anil K. Sood (MD Anderson Cancer Center), and have been described elsewhere (29,30). All cell lines were obtained in 2010 and authenticated in 2013 by Promega and ATCC using Short Tandem Repeat (STR) analysis. A2780 and A2780CIS cells were purchased in 2010 from the European Collection of Cell Cultures (ECACC), which provides authenticated cell lines. All cell lines (A2780, A2780CP20, A2780CIS, SKOV3ip1, SKOV3.TR, HEYA8 and HEYA8.MDR) were thawed in 2013, expanded and cryopreserved in several aliquots. Each aliquot was thawed and cultured for no more than 10–12 passages. Cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 0.1% antibiotic/antimycotic solution in a humidified incubator containing 95% air and 5% CO2 at 37°C. c-MYC-overexpressing clones and cell clones carrying the empty vectors (EV) were cultured in the same media but containing G418 (500 μg/mL). All tumor cell lines were screened for Mycoplasma using the LookOut® Mycoplasma PCR detection kit from Sigma-Aldrich (St. Louis, MO) as described by the manufacturer’s instructions. In vitro assays were performed at 70–85% cell density.
Chemicals, reagents and antibodies
Cisplatin (CIS) and ter-butanol were purchased from Sigma. CIS was reconstituted in 0.9% NaCl. Antibodies against c-MYC, full caspase-3, cleaved caspase-3, full caspase-9, cleaved caspase-9, PARP-1, cyclin D3, cyclin-dependent kinase (CDK) 4, and p27 were purchased from Cell Signaling (Danvers, MA). β-actin monoclonal antibody, and mouse and rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Sigma. DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DSPE-PEG-2000 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL).
Protein extraction and Western blot analysis
Cells were detached using 0.25% Trypsin-EDTA at 37°C and washed with phosphate-buffered saline (PBS). Cell lysates were prepared using ice-cold lysis buffer (1% Triton X, 150 mM NaCl, 25 mM Tris HCl, 0.4 mM NaVO4, 0.4 mM NaF and protease inhibitor cocktail from Sigma), incubated on ice for 30 min, and vortex mixed periodically. Lysates were centrifuged, supernatants were collected, and total protein concentration was determined using Bio-Rad DC Protein Assay reagents (Bio-Rad, Hercules, CA) following the manufacturer’s instructions. Equal amounts of each protein sample (typically 30 or 50 μg per lane) were separated by SDS-PAGE, blotted onto nitrocellulose membranes, blocked with 5% non-fat dry milk, rinsed, and probed with the appropriate dilution of the corresponding primary antibody. Following antibody incubation, the membranes were rinsed, incubated with the corresponding HRP-conjugated secondary antibody, and rinsed again. The bound antibodies were detected using enhanced chemiluminescence substrate followed by autoradiography. Bands were imaged and quantified using a FluorChem system (Alpha Innotech Corporation, San Leandro, CA) and AlphaEaseFC software.
Small-interfering RNAs (siRNAs) and in vitro siRNA transfection
To silence human c-MYC (NM_002467), two siRNAs targeting exon 2 (5′-GCTTGTACCTGCAGGATCT-3′) and exon 3 (5′-CGTCCAAGCAGAGGAGCAA-3′), and a non-silencing negative control siRNA (C-siRNA) were purchased from Sigma. Briefly, A2780CP20, A2780CIS and HEYA8 cells (2×104 cells/mL or 3.5×104 cells/mL) were seeded into 10-cm Petri dishes. Twenty-four hours later, siRNAs were mixed with HiPerFect transfection reagent (Qiagen, Valencia, CA) at 1:2 (A2780CP20 and A2780CIS) or 1:3 (HEYA8) ratio (siRNA: transfection reagent) in serum and antibiotic-free Opti-MEM medium (Life Technologies, Grand Island, NY). The transfection mix was incubated for 20 min at room temperature (RT) and then added to the cells. Cells were incubated at 37°C and collected 24-hr post-transfection to assess the downregulation of c-MYC by Western blot analysis.
Stable transfections
Ectopic c-MYC expression was performed in cisplatin-sensitive A2780 cells. Briefly, A2780 (4.5×104 cells/mL) cells were seeded into 6-well plates. For each well, pcDNA3-cmyc (1.0 μg) (Addgene plasmid 16011, depositor: Wafik El-Deiry) or empty vector (1.0 μg) (pcDNA3.1) (Invitrogen, Life Technologies, Carlsbad, CA) were mixed with MegaTran 1.0 transfection reagent (1:1 v/v) (OriGene, Rockville, MD) and Opti-MEM medium. The mixture was incubated for 10 min at RT and added to the cells. Twenty-four hours later, the medium was replaced with fresh RPMI-1640 (10% FBS, 0.1% antibiotic/antimycotic solution and 500 μg/mL G418 disulfate salt solution). After 2–3 weeks, individual colonies were picked and cultured separately as independent clones.
Cell growth and viability
Cell viability was measured using the Alamar blue dye (Invitrogen) following the manufacturer’s instructions. Briefly, A2780CP20 and A2780 cells (2×104 cells/mL or 3×104 cells/mL) were seeded into 96-well plates. Twenty-four hours later, siRNAs were added to the cells. Seventy-two hours post-transfection, the medium was removed and 95 μL of Alamar blue was added. OD values were obtained spectrophotometrically in a plate reader (BioRad) after a maximum of 4-hr of dye incubation. In all cases, percentages of cell viability were obtained after blank OD subtraction, taking the untreated cells values as a normalization control. For combination treatments (siRNAs + CIS), OD values were obtained at 96-hr post-transfection. For colony formation assays, A2780CP20, A2780CIS and HEYA8 cells (3×104 cells/mL or 4.5×104 cells/mL) were seeded into 6-well plates. Twenty-four hours later, siRNAs were added to the cells. Eight hours post-transfection, 1000 (A2780CP20 and HEYA8) or 2500 (A2780CIS) cells were seeded into 10-cm Petri dishes containing RPMI-1640 (10% FBS and 0.1% antibiotic/antimycotic solution) and incubated at 37°C. Seven days (A2780CP20) or ten days (A2780CIS and HEYA8) later, colony-forming cells were stained with 0.5% crystal violet solution. A2780CP20 and A2780CIS colonies of at least 50 cells were scored under a light microscope (Olympus CKX41) in five random fields with a total magnification of 40X. Visible HEYA8 colonies were counted manually.
Assessment of cell cycle progression and apoptosis by flow cytometry
To assess cell cycle progression, A2780CP20 and HEYA8 cells were transfected with siRNAs for 24-hr. Forty-eight hours post-transfection, cells were collected, washed in ice-cold PBS, fixed with 70% cold ethanol and stored at 4°C. Twenty-four hours later, cells were washed with ice-cold PBS, re-suspended in PI/RNase Staining Buffer (BD Biosciences, San Jose, CA), incubated in the dark for 15 min at RT, and then analyzed by flow cytometry in a Beckman Gallios flow cytometer (Beckman Coulter Inc., Brea, CA). FCS Express software (Beckman Coulter) was used to determine the percentage of cells in each phase of the cell cycle. Apoptosis was measured with the fluorescein isothiocyanate (FITC) Annexin-V apoptosis detection kit (BD Biosciences), which uses Annexin-V and propidium iodide (PI) as the apoptotic and necrotic markers, respectively. Briefly, A2780CP20 and HEYA8 cells were transfected with siRNAs for 24-hr. Seventy-two hours post-transfection, both floating and attached cells were collected, washed in ice-cold PBS, and re-suspended in 1X Binding Buffer. Cells were then incubated in the dark for 15 min at RT with Annexin-V-FITC and/or PI according to the manufacturer’s instructions. Apoptotic cells were analyzed in a Beckman Gallios flow cytometer. Gallios 1.2 software (Beckman Coulter) was used to determine the percentage of apoptotic and necrotic cells.
SiRNA incorporation into DOPC-PEG-nanoliposomes
SiRNAs were mixed with DOPC (1:10 w/w), DSPE-PEG-2000 (5% mol/mol of total lipids) and cholesterol (50% w/w of DOPC) in the presence of excess ter-butanol (30–35). The mixture was frozen in an acetone-dry ice bath and lyophilized. For in vivo administration, the lyophilized powder was hydrated with Ca2+ and Mg2+-free PBS at a concentration of 25 mg/mL to achieve the desired dose of 5 μg of siRNA in 200 μL/injection.
Tumor implantation and drug treatment
Female athymic nude mice (NCr-nu, 6 weeks old) were purchased from Taconic (Hudson, NY). To assess the efficacy of c-MYC silencing in vivo, mice were intraperitoneally (i.p.) injected with A2780CP20 cells (1×106 cells/0.2 mL HBSS). Three weeks after tumor implantation, mice received 2 single doses (i.p.) of C-siRNA-DOPC-PEG-nanoliposomes or c-MYC-siRNA-DOPC-PEG-nanoliposomes (5 μg siRNA/injection). Two days after siRNA injection, animals were sacrificed; the peritoneal tumors were harvested and tissue samples were snap-frozen in liquid nitrogen and stored at −80°C for protein analysis by Western blot with a specific antibody against c-MYC. To evaluate the therapeutic activity of c-MYC siRNAs alone or in combination with cisplatin (CIS) in vivo, A2780CP20 cells (1×106 cells/0.2 mL HBSS) were injected i.p. Seven days later, mice were randomly divided into the following treatment groups (N=10 per group): (a) C-siRNA, (b) CIS alone, (c) c-MYC-siRNA, (d) C-siRNA plus CIS, and (e) c-MYC-siRNA plus CIS. DOPC-PEG-liposomal-siRNAs (5 μg siRNA/injection) and CIS (160 μg/injection) were injected i.p. once a week for 4 weeks. At the end of the treatment, mice were sacrificed and tumors collected. The number of tumor nodules and tumor weight was recorded. The entire peritoneal cavity was examined for tumor metastases.
Characterization of liposomal formulations
Cryo-electron microscopy (Cryo-EM)
The lyophilized liposomal-siRNA powder was resuspended in 500 μL Ca2+ and Mg2+-free PBS by ultrasonic agitation for 15 min. A 3 μL suspension was applied to fenestrated carbon films blotted to form a vicinal layer, then vitrified in liquid ethane at −180°C. Frozen hydrated samples were observed in a FEI Tecnai TF20 electron microscope (FEI, Hillsboro, OR) with a Gatan 626 cryogenic specimen holder (Gatan Inc., Pleasanton, CA), and images recorded on a Falcon2 camera of the same instrument.
Particle size and zeta potential
Light scattering was used to determine the particle size and surface charge (zeta potential) of the liposomal formulations. Liposomes were reconstituted in Ca2+ and Mg2+-free PBS and sonicated for 15–20 min. After sonication, particle size and zeta potential were measured at RT with Zeta Pals (Brookhaven Instruments, Holtsville, NY) and Mobius instruments (Wyatt Technology, Santa Barbara, CA), respectively.
SiRNA encapsulation efficiency and siRNA release
Liposomal-siRNAs were reconstituted in Ca2+ and Mg2+-free PBS, and sonicated for 15–20 min. The mixture was added to an Amicon 50K filter (EMD Millipore, Darmstadt, Germany), and centrifuged at 7500 rpm for 15 min. The eluted fraction was collected to measure the amount of free siRNA using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). 0.1% Triton X-100 was then added to the filter, centrifuged again at 7500 for 15 min, and measured spectrophotometrically. This fraction corresponded to the encapsulated siRNA. All experiments included naked-siRNA and empty liposomes (liposomes without siRNA) as controls. The encapsulation efficiency (%E) was calculated using the following equation:
SiRNA released from liposomes was calculated by dissolving the liposomal-siRNA in 50% FBS for 0-hr, 2-hr, 8-hr, 24-hr, 48-hr and 72-hr at 37°C. Liposomes were centrifuged at 7500 rpm for 15 min. Then 0.1% Triton X-100 was added to an Amicon 50K filter and centrifuged again at 7500 rpm for 15 min. The amount of RNA from the Triton X-100 fraction corresponds to the non-released siRNA portion. Percentage of release at each time point was calculated using the following equation:
Biological half-life (serum stability) and shelf-life
For serum stability, naked-siRNA and liposomal-siRNA were resuspended in 50% FBS and incubated at 37°C. Aliquots were collected at 0-hr, 3-hr, 6-hr, 12-hr and 24-hr, and frozen at each time point. Prior to electrophoresis, 1% Triton X-100 was added to the aliquots and vortexed-mixed for 2 min. 5X loading dye was added and samples were loaded into a 3% agarose gel. Bands were imaged using a FluorChem system (Alpha Innotech) and AlphaEaseFC software. The shelf-life was determined by the conservation of the particle size and surface charge of liposomes at 4°C for 0-weeks, 2-weeks, and 4-weeks. Similarly, the size and charge of the liposomal formulations were measured 0-min, 30-min, 60-min and 120-min at RT after liposomal reconstitution with Ca2+ and Mg2+-free PBS.
In vitro toxicity
In vitro toxicity was measured using the Alamar blue dye (Invitrogen) following the manufacturer’s instructions. Briefly, A2780CP20 cells (2×104 cells/mL) were seeded into 96-well plates. Twenty-four hours later, liposomal formulations (empty liposomes with different DOPC concentrations) were added to the cells. Seventy-two hours post-transfection, the medium was removed and 95 μL of Alamar blue was added. OD values were obtained spectrophotometrically in a plate reader (BioRad) after a maximum of 4-hr of dye incubation. In all cases, percentages of cell viability were obtained after blank OD subtraction, taking the untreated cells values as a normalization control.
In vivo safety study (proinflammatory cytokines and blood chemistry)
Female wild type mice (Balb-c, 4 weeks old) were purchased from Taconic. Serum samples were collected from heart blood and further analyzed for proinflammatory cytokine production by ELISA (Qiagen), and blood chemistry [lactate dehydrogenase (LDH) activity and urea] (Sigma-Aldrich) following the manufacturer’s instructions. Briefly, the immune response was evaluated after 5-hr and 24-hr of i.p. injections (single dose). LDH activity and urea levels were evaluated at 3 weeks (a single injection per day, four consecutive days a week).
Statistical analysis
For in vitro and in vivo experiments, statistical analysis was performed using Student’s t-test for comparing two groups and by ANOVA for multiple group comparisons. P-values of <0.05 were considered statistically significant. GraphPad Prism software (San Diego, CA) was used for graphing and statistical analysis. Ovarian cancer patient data were downloaded and analyzed from the Cancer Genome Atlas Project (TCGA; http://tcga-data.nci.nih.gov/) for “Ovarian Serous Cystadenocarcinoma” (OV). Level 3 Illumina RNASeq “gene.quantification” files were used to extract MYC expression (RPKM: Reads Per Kilobase of exon model per Million mapped reads) values. Statistical analyses were performed in R (version 3.0.1) (http:///www.r-project.org/), and the statistical significance was defined as a P-value of <0.05. The Log-rank test was employed to determine the relationship between c-MYC expression and overall/disease free survival. Kaplan-Meyer method was used to generate survival curves. The entire population was randomly split into training (2/3) and validation (1/3) cohorts. In each cohort, patients were divided into percentiles according to c-MYC mRNA expression. Then using the training set, any cut-off between percentiles of 25th and 75th were considered as statistically significant. The statistical significance was corroborated with the validation set.
Results
Expression of c-MYC in human ovarian cancer patients and ovarian cancer cells
To determine potential clinical relevance in drug-resistant ovarian cancer, c-MYC mRNA levels were correlated with clinical data from ovarian cancer patients. Ovarian cancer patient data were downloaded and analyzed from TCGA. Level 3 Illumina RNASeq “gene.quantification” files were used to extract MYC expression. The entire cohort was separated into two sets, the training set (219 patients) (Figure 1A) and the validation set (110 patients) (Figure 1B). The log-rank test revealed that recurrence of disease (expressed as percentage disease free) occurred significantly (P=0.0277) faster for patients with higher c-MYC expression levels (Figure 1A). These findings were corroborated with further analysis with the validation set (P=0.0289) (Figure 1B). In addition, overall survival (expressed as the percentage survival) was significantly reduced for patients with higher c-MYC expression values (Figure 1C, P=0.0058). Statistical analysis with the validation set cohort (Figure 1D) corroborated these findings (P=0.0138).
Figure 1. Expression of c-MYC in ovarian cancer cells and human tumors.
Level 3 Illumina RNASeq “gene.quantification” files were used to extract MYC expression. Statistical analysis of c-MYC mRNA expression and clinical data from patients with high grade serous ovarian cancer showed that the PFS (A–B) and the OS (C–D) were significantly reduced for patients with higher c-MYC expression levels. (E) Western blot analysis of ovarian cancer cells was performed as described in the “Materials and Methods” section. β-actin was used as a loading control. (F) Densitometric analysis of the intensities of the bands shown in Figure 1E. Fold changes in protein levels were calculated relative to A2780 cisplatin-sensitive cells.
To assess c-MYC protein levels, a panel of multiple ovarian cancer cell lines was evaluated by Western blot analysis. Interestingly, cisplatin-resistant cells (A2780CP20 and A2780CIS) expressed higher levels of c-MYC protein when compared to their sensitive counterparts (A2780) (Figure 1E). Densitometric analysis of the band intensities (Figure 1F) confirmed these findings. The taxane-resistant ovarian cancer cells SKOV3.TR and HEYA8.MDR exhibited similar c-MYC levels when compared to their taxane-sensitive counterparts SKOV3ip1 and HEYA8, respectively. Together, these results suggest that c-MYC is a clinically relevant target for cisplatin-resistant ovarian cancer patients.
Effects of c-MYC silencing on cell growth and viability
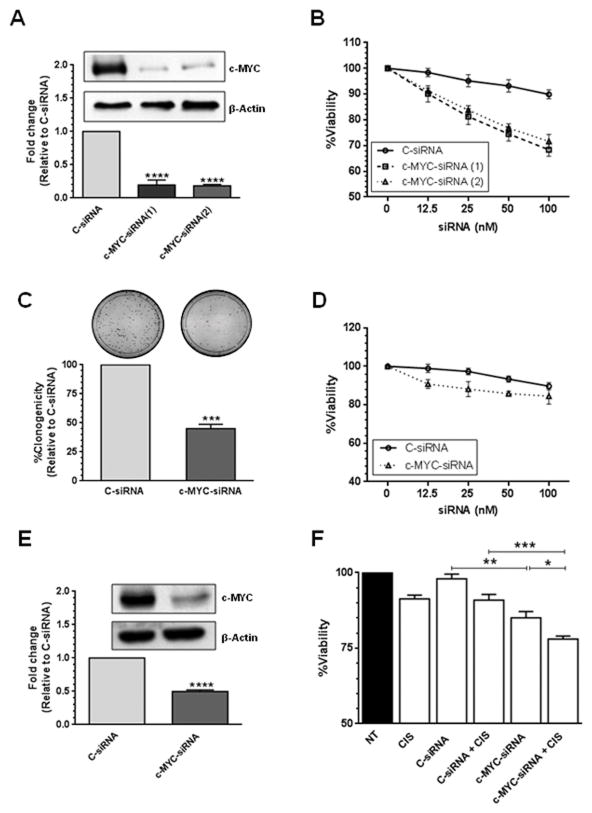
Next, we examined the biological effects of c-MYC silencing in cisplatin-resistant ovarian cancer cells. Western blot analysis confirmed that the two siRNAs used against c-MYC reduced the levels of the c-MYC protein in the A2780CP20 cisplatin-resistant ovarian cancer cell line (Figure 2A). Densitometric analysis of the band intensities showed that both siRNAs decreased c-MYC expression by more than 80% (****P<0.0001) compared with C-siRNA (Figure 2A). Similar results were observed when c-MYC-siRNA was transfected into A2780CIS and HEYA8 ovarian cancer cells (Supplementary Figure 1A and 1C). Dose-dependent inhibition of cell viability was observed after 72-hr of c-MYC-targeted siRNA treatment (Figure 2B). The inhibitory effects on viability were observed even at doses as low as 12.5 nM of c-MYC-targeted siRNAs (Figure 2B). Treatment with c-MYC-siRNA also induced long-term effects in cell growth as evident in colony formation assays (Figure 2C). Transient transfection of c-MYC-siRNA(2) in A2780CP20 cells significantly reduced (55% reduction, ***P<0.001) the number of colonies formed after 7 days in culture, compared with the C-siRNA-transfected cells (Figure 2C). Similarly, transfection of c-MYC-siRNA(2) in A2780CIS and HEYA8 cells significantly reduced (48% and 70% reduction, ****P<0.0001 and ***P<0.001, respectively) the number of colonies formed after 10 days in culture (Supplementary Figure 1B and 1D). On the other hand, silencing c-MYC in A2780 cisplatin-sensitive ovarian cancer cells, which express low c-MYC levels, induced negligible changes in cell viability (Figure 2D). Combination of a low-active c-MYC-siRNA(2) dose (50 nM, Figure 2B and 2E) with a relatively low cisplatin dose (2 μM) induced significant (*P<0.05) additive-like inhibitory effects on the viability of A2780CP20 cells (Figure 2F) compared to c-MYC-siRNA(2) alone. These data suggest that c-MYC levels are associated with the sensitivity of ovarian cancer cells to cisplatin treatment.
Figure 2. SiRNA-based silencing of c-MYC.
Two different siRNAs targeting exon 2 and exon 3 of the human c-MYC sequence (NM_002467) were used. (A) A2780CP20 cells (2×105) were transfected with 200 nM c-MYC-siRNA. Total protein was isolated from siRNA-transfected cells for Western blot analysis as described in the “Materials and Methods” section. Densitometric analysis of the intensities of the bands was calculated relative to the C-siRNA. Averages ±SEM are shown (****P<0.0001). (B) A2780CP20 cells (2×103) were seeded into 96-well plates and 24-hr later cells were transfected with a serial dilution of C-siRNA or c-MYC-targeted siRNAs. Cell viability was calculated 72-hr post-transfection as described in the “Materials and Methods” section. Percentages were obtained after blank OD subtraction, taking the untreated cells values as a normalization control. Averages ±SEM are shown. (C) A2780CP20 cells (6×104) were seeded into 6-well plates and 24-hr later 100 nM c-MYC-siRNA(2) or 100 nM C-siRNA was added to the cells. Eight hours post-transfection, 1000 cells were seeded into 10-cm Petri dishes. Seven days later, cells were stained and colonies of at least 50 cells were scored under a light microscope. The % of clonogenicity was calculated relative to C-siRNA. Averages ±SEM are shown for three independent experiments (***P<0.001). (D) A2780 cells (3×103) were seeded into 96-well plates and 24-hr later cells were transfected with a serial dilution of C-siRNA or c-MYC-siRNA(2). Cell viability was calculated as described in Figure 2B. Averages ±SEM are shown. (E) A2780CP20 cells (2×105) were transfected with 50 nM c-MYC-siRNA(2). Total protein was isolated from siRNA-transfected cells for Western blot analysis as described in the “Materials and Methods” section. Densitometric analysis of the intensities of the bands was calculated relative to the C-siRNA. Averages ±SEM are shown (****P<0.0001). (F) A2780CP20 cells (2×103) were seeded into 96-well plates and 24-hr later cells were transfected with 50 nM C-siRNA or 50 nM c-MYC-siRNA(2). The next day, the media was replaced by CIS (2 μM final concentration)-containing RPMI-1640 media. Cell viability was calculated 72-hr after CIS treatment (96-hr post-transfection) as previously described. Averages ± SEM are shown (*P<0.05, **P<0.01, ***P<0.001).
Effects of c-MYC silencing on apoptosis and cell cycle progression
We next investigated whether the effects induced by c-MYC-siRNA were due to apoptosis, cell cycle arrest or both. Transfection of A2780CP20 cells with c-MYC-siRNA(2) induced up to 15% (**P<0.01) apoptosis compared with cells treated with C-siRNA (Figure 3A). Western blot analysis showed that siRNA-based c-MYC silencing induced activation of the apoptotic-related molecules caspase-3, caspase-9 and poly(ADP-ribose)polymerase-1 (PARP-1) 72-hr post-transfection (Figure 3B). Furthermore, 25% of the A2780CP20 cells (****P<0.0001) were arrested in the G0/G1 phase, 48-hr after c-MYC-siRNA(2) transfection (Figure 3C). Western blot analysis confirmed that key proteins required for transition from G0/G1 to S phase were also altered following c-MYC silencing in A2780CP20 cells (Figure 3D). Particularly, cyclin D3/cyclin-dependent kinase 4 (CDK4) levels were decreased, while the inhibitory protein of cell cycle progression, p27, increased in cells treated with c-MYC-siRNA(2) compared with untreated (NT) or C-siRNA-treated cells (Figure 3D). Densitometric analysis of the band intensities is shown in Figure 3E. Similar results for apoptosis and cell cycle progression were observed when c-MYC-siRNA(2) was transfected into HEYA8 ovarian cancer cells (Supplementary Figure 1E–F).
Figure 3. Apoptosis and cell cycle progression following c-MYC silencing.
Assessment of cell cycle progression and apoptosis was performed by flow cytometry after siRNA-based c-MYC silencing in cisplatin-resistant cells. (A) A2780CP20 cells (6×104) were seeded into 6-well plates and 24-hr later 200 nM of C-siRNA or 200 nM of c-MYC-siRNA(2) was added to the cells. Seventy-two hours post-transfection, apoptosis was measured with the FITC-apoptosis detection kit as described in the “Materials and Methods” section. Averages ±SEM are shown for two independent experiments (**P<0.01). (B) A2780CP20 cells (2×105) were seeded into 10-cm Petri dishes and transfected as described in Figure 1A. Western blot analysis was performed 72-hr post-transfection for detection of apoptotic related proteins as described in the “Materials and Methods” section. (C) A2780CP20 cells (6×104) were transfected as described in Figure 3A. Forty-eight hours after siRNA transfection, A2780CP20 cells were fixed and cell cycle progression was assessed using PI and the FCS Express software. Averages ±SEM are shown for three independent experiments (**P<0.01, ****P<0.0001). (D) A2780CP20 cells (2×105) were transfected as described in Figure 3B. Western blot analysis was performed 48-hr post-transfection for detection of cell cycle related proteins. (E) Densitometric analysis of the band intensities shown in Figure 3D are expressed relative to C-siRNA-treated cells. Averages ±SEM are shown for three independent experiments (*P<0.05, **P<0.01).
Effect of c-MYC overexpression in the sensitivity of ovarian cancer cells to cisplatin treatment
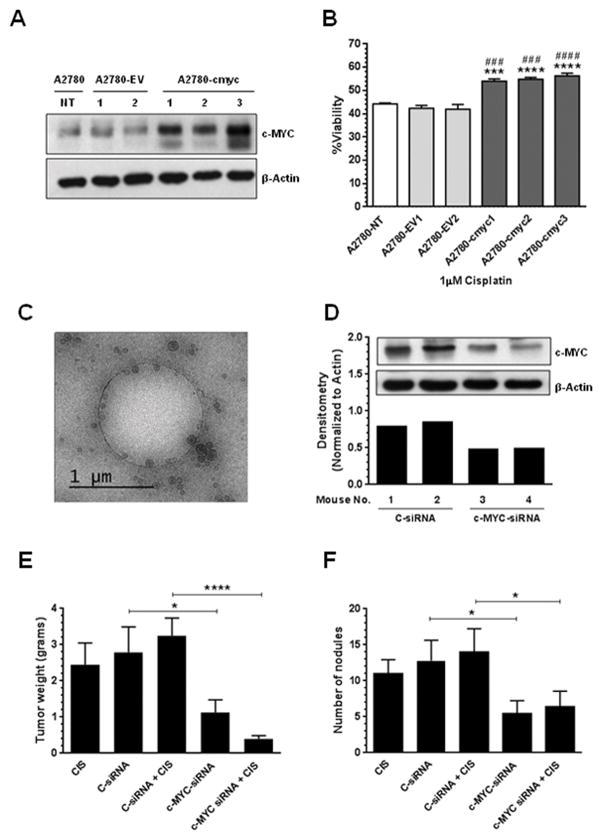
To confirm the role of c-MYC in the cisplatin resistance of ovarian cancer cells, we performed stable c-MYC transfections in A2780 cisplatin-sensitive cells. Figure 4A shows that compared with A2780 untransfected cells (A2780-NT) or with empty vector transfected clones (A2780-EV), A2780-cmyc clones express higher c-MYC protein levels. At concentrations as low as 1 μM of cisplatin, the c-MYC-overexpressing clones were significantly less sensitive to cisplatin treatment compared with the A2780 untransfected cells or with the EV clones (Figure 4B). These data suggest that c-MYC contributes to the cisplatin-resistant phenotype observed in ovarian cancer cells.
Figure 4. In vitro effects of c-MYC overexpression, and in vivo therapeutic efficacy of liposomal c-MYC-siRNA.
A2780 cells (9×104) were stably transfected with an empty vector (EV) or with a c-MYC-containing vector as described in the “Materials and Methods” section. (A) Western blot analysis was performed as previously described. Compared with untrasfected cells or with empty vector clones, the c-MYC overexpressing clones showed higher c-MYC protein levels. (B) Stable transfected A2780 cells were exposed to CIS (1 μM final concentration)-containing RPMI-1640 media. Cell viability was calculated as described in the “Materials and Methods” section. Averages ± SEM are shown relative to A2780-EV1 (***P<0.001, ****P<0.0001) or to A2780-EV2 (###P<0.001, ####P<0.0001). Lyophilized liposomal-siRNAs were reconstituted in Ca2+ and Mg2+-free PBS. (C) Cryo-EM shows that the majority of the particles are small unilamellar vesicles in the 100–150 nm range. The hole in the image is a fenestration in the carbon support, which measures 1.2 microns in diameter. Nude mice were injected i.p. with A2780CP20 cells and randomly allocated in the groups described in the “Materials and Methods” section. (D) Western blot analysis shows that c-MYC-siRNA-DOPC-PEG treatment reduced c-MYC protein levels in vivo. Therapy began one (1) week after tumor cell inoculation. (E) Mean tumor weight and (F) number of nodules was recorded after 4 weeks. Averages ±SEM are shown (*P<0.05, ****P<0.0001).
Characterization of liposomal-siRNA formulations
Particle size and zeta potential of liposomal-siRNA formulations were evaluated by dynamic light scattering, which showed that the liposomes used in this study were slightly negative, and around 100–150 nm in diameter (Supplementary Tables 1, 2 and 3). The percentage of cholesterol induced changes in the size but not in the surface charge (zeta potential) of the liposomal formulations (Supplementary Table 1). The efficiency of siRNA encapsulation was slightly higher for liposomes with 50% cholesterol (w/w DOPC) as compared with liposomes with 25% cholesterol (w/w DOPC) (Supplementary Table 1). The kinetics of siRNA release from liposomes with 25% cholesterol was slower in the first hours compared with liposomes with 50% cholesterol (Supplementary Figure 2A). However, the kinetics of siRNA release was constant over time for liposomes with 50% cholesterol compared with liposomes with 25% cholesterol. For these reasons, liposomes with 50% cholesterol were used for further studies. A cryo-EM micrograph confirmed the particle size (100–150 nm) and showed that the majority of the liposomes are small unilamellar vesicles (Figure 4C). The size and charge of the reconstituted liposomal-siRNA formulations were stable over 2-hr at RT (Supplementary Table 2) and 4-weeks at 4°C (Supplementary Table 3). The ability of the liposomes to protect the stability of the siRNA from serum nucleases was evaluated in vitro. Results showed that siRNA degradation occurred faster for naked-siRNA compared to liposomal-siRNA (Supplementary Figure 2B). Furthermore, the liposomal-siRNA formulation was not toxic in vitro even at DOPC concentrations as high as 50 μM (Supplementary Figure 2C). In vivo studies showed that a single injection of empty liposomes or liposomal-siRNA formulations did not induce early (5-hr) or late (24-hr) immune response (Supplementary Figure 3A–B). Repeated doses of liposomal-siRNA formulations (a single injection per day for 4 days, during 3 weeks) were not toxic for mice as indicated by the LDH activity or urea levels (Supplementary Figure 3C–D), which were similar to the control group (saline solution, only). No weight loss was noted during the treatment period (Supplementary Figure 3E).
Therapeutic effect of DOPC-PEG-liposomal siRNAs
DOPC-PEG-cholesterol-based nanoliposomes were used for in vivo siRNA delivery. First, we assessed whether the c-MYC silencing was effective in vivo. Nude mice bearing A2780CP20 tumors were injected i.p. with 2 single doses of 5 μg of DOPC-PEG-liposomal-C-siRNA or 5 μg of DOPC-PEG-liposomal-c-MYC-siRNA. Two days post-injection, mice were sacrificed and tumors were dissected. Western blot analysis confirmed that DOPC-PEG-liposomal-c-MYC-siRNA reduced the levels of c-MYC protein at two days post-injection (Figure 4D) compared with control groups. Next, the anti-tumor effects of c-MYC-siRNA as compared to C-siRNA were tested in A2780CP20 tumor-bearing mice. Tumor weight and number of nodules were assessed in five treatment groups (10 mice each): (a) C-siRNA, (b) CIS alone, (c) c-MYC-siRNA, (d) C-siRNA plus CIS, and (e) c-MYC-siRNA plus CIS. All DOPC-PEG-liposomal siRNAs (5 μg siRNA/injection) and CIS (160 μg/injection) treatments were injected i.p. once per week for 4 weeks. No weight loss was noted during the treatment period (Supplementary Figure 3F). Cisplatin treatment by itself did not induce a significant effect on tumor growth (Figure 4E). On the other hand, decreased tumor weight was observed in the c-MYC-siRNA group (*P<0.05) compared with the C-siRNA group (Figure 4E). In addition, c-MYC-siRNA induced a decrease in the number of tumor nodules (*P<0.05) compared with the C-siRNA group (Figure 4F).
Discussion
In this study, we showed that high levels of c-MYC are associated with faster recurrence and poor overall survival of patients with high grade serous ovarian cancer, and with cisplatin resistance in ovarian cancer cells. Another key finding was that siRNA-based silencing of c-MYC inhibits cell growth in vitro and reduces tumor growth in xenograft models of cisplatin-resistant ovarian cancer. c-MYC, an oncoprotein highly abundant in several types of cancer, is considered an undruggable molecule by virtue of its flat protein surface (36,37). Thus, the evidence we present here suggests that siRNA-based c-MYC targeting is a therapeutic modality for ovarian cancer patients expressing high c-MYC levels, including those that are resistant to cisplatin treatment.
The c-MYC transcription factor, which regulates 15% of all human genes, plays an important role in a myriad of biological processes including cell growth and proliferation, cell cycle progression, apoptosis, angiogenesis, senescence and genomic instability (1–3,38,39). In addition, c-MYC regulates the expression of not only a particular group of genes but acts in concert with RNA polymerase and transcription factors as a universal amplifier of gene expression in embryonic stem cells (40) and tumor cells (41). In fact, c-MYC amplification has been reported in multiple malignancies including ovarian cancer (14). In other tumor types, c-MYC expression levels have been associated with drug resistance (15–26). For instance, Sakamuro and co-workers have shown that c-MYC oncoprotein increases cisplatin resistance by decreasing production of the c-MYC inhibitor bridging integrator 1 (BIN1) (16). Our findings related to the role of ectopic expression of c-MYC in decreasing the sensitivity of ovarian cancer cells to cisplatin treatment are in agreement with these reports.
The therapeutic effects of siRNA-based c-MYC silencing in cisplatin-resistant ovarian cancer cells have not been fully addressed. Current adjuvant chemotherapy for ovarian cancer includes cisplatin and paclitaxel; unfortunately, the majority of the patients develop chemoresistance which leads to therapeutic failure (27,28). Thus, our study provides further evidence that c-MYC is a plausible target for ovarian cancer patients with high c-MYC expression levels. Moreover, the findings that the c-MYC-targeted siRNA did not affect the viability of cells with low c-MYC protein levels, suggests that c-MYC could be considered as a potential biomarker and an indicative of chemotherapy response. Nevertheless, the results that siRNA-mediated c-MYC silencing induced more pronounced effects in c-MYC downregulation than in cell growth and viability confirm previous findings (42) that molecular pathways other than c-MYC upregulation contribute to the cisplatin resistance of ovarian cancer cells. These results suggest that therapies targeting different cell survival pathways should be more beneficial than targeting a single pathway in advanced and drug resistant ovarian cancer.
We have shown that c-MYC siRNA-based silencing induces short- and long-term effects in cell growth and viability. These effects were associated with both apoptosis induction, and cell cycle arrest. Future studies should determine the c-MYC-regulated anti-apoptotic genes associated with the cisplatin resistance in ovarian cancer cells. In agreement with previous studies (14,43), one of the major cell cycle inhibitory proteins, p27, was increased following c-MYC depletion. Similarly, decreased levels of CDK4 and cyclin D3 following c-MYC silencing probably occurred by the ability of c-MYC to transcriptionally regulate the expression of these proteins (44).
In conclusion, we present evidence here that DOPC-PEG-liposomal c-MYC-targeted siRNA alone or in combination with chemotherapy is efficacious in preclinical models. Further pharmacodynamic, pharmacokinetic, and tissue distribution studies will be required prior to clinical translation.
Supplementary Material
Acknowledgments
Grant Support:
This project was supported partially by the NIH/NCI 1K22CA166226-01A1 and institutional seed funds from the University of Puerto Rico Comprehensive Cancer Center (P.E. Vivas-Mejía); and the National Institutes of Health, Minority Biomedical Research Support (MBRS) RISE Grant Number R25GM061838 (J.M. Reyes-González). This project was also supported partially by CA016672, P50 CA083639, U54 CA151668, UH2 TR000943, the Blanton-Davis Ovarian Cancer Research Program, the RGK Foundation, the Gilder Foundation, and the Betty Anne Asche Murray Distinguished Professorship (A.K. Sood).
Abbreviations List
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DSPE-PEG-2000
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]
- CIS
Cisplatin
Footnotes
Conflict of Interest: No conflict of interest is declared by the authors.
References
- 1.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 3.Bui TV, Mendell JT. Myc: Maestro of MicroRNAs. Genes Cancer SAGE Publications. 2010;1:568–75. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoepfler PS. Myc goes global: new tricks for an old oncogene. Cancer Res. 2007;67:5061–3. doi: 10.1158/0008-5472.CAN-07-0426. [DOI] [PubMed] [Google Scholar]
- 5.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–62. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–74. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 7.Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan H-M, et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–80. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–30. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24:1236–41. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–30. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 12.Baker VV, Borst MP, Dixon D, Hatch KD, Shingleton HM, Miller D. c-myc amplification in ovarian cancer. Gynecol Oncol. 1990;38:340–2. doi: 10.1016/0090-8258(90)90069-w. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prathapam T, Aleshin A, Guan Y, Gray JW, Martin GS. p27Kip1 mediates addiction of ovarian cancer cells to MYCC (c-MYC) and their dependence on MYC paralogs. J Biol Chem. 2010;285:32529–38. doi: 10.1074/jbc.M110.151902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torigoe T, Izumi H, Ishiguchi H, Yoshida Y, Tanabe M, Yoshida T, et al. Cisplatin resistance and transcription factors. Curr Med Chem Anticancer Agents. 2005;5:15–27. doi: 10.2174/1568011053352587. [DOI] [PubMed] [Google Scholar]
- 16.Pyndiah S, Tanida S, Ahmed KM, Cassimere EK, Choe C, Sakamuro D. c-MYC suppresses BIN1 to release poly(ADP-ribose) polymerase 1: a mechanism by which cancer cells acquire cisplatin resistance. Sci Signal. 2011;4:ra19. doi: 10.1126/scisignal.2001556. [DOI] [PubMed] [Google Scholar]
- 17.Lin C-P, Liu J-D, Chow J-M, Liu C-R, Liu HE. Small-molecule c-Myc inhibitor, 10058-F4, inhibits proliferation, downregulates human telomerase reverse transcriptase and enhances chemosensitivity in human hepatocellular carcinoma cells. Anticancer Drugs. 2007;18:161–70. doi: 10.1097/CAD.0b013e3280109424. [DOI] [PubMed] [Google Scholar]
- 18.Leonetti C, Biroccio A, Benassi B, Stringaro A, Stoppacciaro A, Semple SC, et al. Encapsulation of c-myc antisense oligodeoxynucleotides in lipid particles improves antitumoral efficacy in vivo in a human melanoma line. Cancer Gene Ther. 2001;8:459–68. doi: 10.1038/sj.cgt.7700326. [DOI] [PubMed] [Google Scholar]
- 19.Biroccio A, Benassi B, Amodei S, Gabellini C, Del Bufalo D, Zupi G. c-Myc down-regulation increases susceptibility to cisplatin through reactive oxygen species-mediated apoptosis in M14 human melanoma cells. Mol Pharmacol. 2001;60:174–82. doi: 10.1124/mol.60.1.174. [DOI] [PubMed] [Google Scholar]
- 20.Sklar MD, Prochownik EV. Modulation of cis-platinum resistance in Friend erythroleukemia cells by c-myc. Cancer Res. 1991;51:2118–23. [PubMed] [Google Scholar]
- 21.Leonetti C, Biroccio A, Candiloro A, Citro G, Fornari C, Mottolese M, et al. Increase of cisplatin sensitivity by c-myc antisense oligodeoxynucleotides in a human metastatic melanoma inherently resistant to cisplatin. Clin Cancer Res. 1999;5:2588–95. [PubMed] [Google Scholar]
- 22.Citro G, D’Agnano I, Leonetti C, Perini R, Bucci B, Zon G, et al. c-myc antisense oligodeoxynucleotides enhance the efficacy of cisplatin in melanoma chemotherapy in vitro and in nude mice. Cancer Res. 1998;58:283–9. [PubMed] [Google Scholar]
- 23.Xie X-K, Yang D-S, Ye Z-M, Tao H-M. Recombinant antisense C-myc adenovirus increase in vitro sensitivity of osteosarcoma MG-63 cells to cisplatin. Cancer Invest. 2006;24:1–8. doi: 10.1080/07357900500449520. [DOI] [PubMed] [Google Scholar]
- 24.Knapp DC, Mata JE, Reddy MT, Devi GR, Iversen PL. Resistance to chemotherapeutic drugs overcome by c-Myc inhibition in a Lewis lung carcinoma murine model. Anticancer Drugs. 2003;14:39–47. doi: 10.1097/00001813-200301000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Walker TL, White JD, Esdale WJ, Burton MA, DeCruz EE. Tumour cells surviving in vivo cisplatin chemotherapy display elevated c-myc expression. Br J Cancer. 1996;73:610–4. doi: 10.1038/bjc.1996.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizutani Y, Fukumoto M, Bonavida B, Yoshida O. Enhancement of sensitivity of urinary bladder tumor cells to cisplatin by c-myc antisense oligonucleotide. Cancer. 1994;74:2546–54. doi: 10.1002/1097-0142(19941101)74:9<2546::aid-cncr2820740924>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 27.Gamarra-Luques CD, Goyeneche AA, Hapon MB, Telleria CM. Mifepristone prevents repopulation of ovarian cancer cells escaping cisplatin-paclitaxel therapy. BMC Cancer. 2012;12:200. doi: 10.1186/1471-2407-12-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzog TJ. Recurrent ovarian cancer: how important is it to treat to disease progression? Clin Cancer Res. 2004;10:7439–49. doi: 10.1158/1078-0432.CCR-04-0683. [DOI] [PubMed] [Google Scholar]
- 29.Vivas-Mejia PE, Rodriguez-Aguayo C, Han H-D, Shahzad MMK, Valiyeva F, Shibayama M, et al. Silencing survivin splice variant 2B leads to antitumor activity in taxane--resistant ovarian cancer. Clin Cancer Res. 2011;17:3716–26. doi: 10.1158/1078-0432.CCR-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vivas-Mejia P, Benito JM, Fernandez A, Han H-D, Mangala L, Rodriguez-Aguayo C, et al. c-Jun-NH2-kinase-1 inhibition leads to antitumor activity in ovarian cancer. Clin Cancer Res. 2010;16:184–94. doi: 10.1158/1078-0432.CCR-09-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E, et al. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 2010;70:3687–96. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw TJ, Senterman MK, Dawson K, Crane CA, Vanderhyden BC. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol Ther. 2004;10:1032–42. doi: 10.1016/j.ymthe.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Landen CN, Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol Cancer Ther. 2010;9:3186–99. doi: 10.1158/1535-7163.MCT-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landen CN, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 35.Landen CN, Merritt WM, Mangala LS, Sanguino AM, Bucana C, Lu C, et al. Intraperitoneal delivery of liposomal siRNA for therapy of advanced ovarian cancer. Cancer Biol Ther. 2006;5:1708–13. doi: 10.4161/cbt.5.12.3468. [DOI] [PubMed] [Google Scholar]
- 36.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–70. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 37.Horiuchi D, Anderton B, Goga A. Taking on Challenging Targets: Making MYC Druggable. Am Soc Clin Oncol Educ Book. 2014;34:e497–502. doi: 10.14694/EdBook_AM.2014.34.e497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vervoorts J, Lüscher-Firzlaff J, Lüscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281:34725–9. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 39.Frenzel A, Lovén J, Henriksson MA. Targeting MYC-Regulated miRNAs to Combat Cancer. Genes Cancer. 2010;1:660–7. doi: 10.1177/1947601910377488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Echevarría-Vargas IM, Valiyeva F, Vivas-Mejía PE. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS One. 2014;9:e97094. doi: 10.1371/journal.pone.0097094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Agnano I, Valentini A, Fornari C, Bucci B, Starace G, Felsani A, et al. Myc down-regulation induces apoptosis in M14 melanoma cells by increasing p27(kip1) levels. Oncogene. 2001;20:2814–25. doi: 10.1038/sj.onc.1204392. [DOI] [PubMed] [Google Scholar]
- 44.Qi Y, Tu Y, Yang D, Chen Q, Xiao J, Chen Y, et al. Cyclin A but not cyclin D1 is essential for c-myc-modulated cell-cycle progression. J Cell Physiol Wiley Subscription Services, Inc., A Wiley Company. 2007;210:63–71. doi: 10.1002/jcp.20816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.