Abstract
Epithelial-restricted with serine box (ESX), a member of the ETS transcription factor family, is elevated and regulates epidermal growth factor receptor (EGFR) in head and neck squamous cell carcinoma (HNSCC). However, the molecular mechanisms that contribute to ESX dysregulation remains to be elucidated. In this study, in silico analysis of the 3′-untranslated region (UTR) of ESX predicted two miR-124 binding sites. Delivery of miR-124 inhibited the 3′UTR ESX-driven reporter activity by 50% (p<0.05) confirming ESX as a direct target of miR-124. Loss of miR-124 was found to a frequent event in HNSCC. miR-124 expression was significantly depleted in the primary tumor compared to matched normal tissue in 100% (12/12) of HNSCC patients; relative mean miR-124 expression of 0.01197 and 0.00118 (p<0.001, n=12) in matched normal adjacent tissue and primary HNSCC tumor, respectively. Over-expression of miR-124 decreased ESX and EGFR levels in miR-124low/ESXhigh/EGFRhigh SCC15 HNSCC cells and reduced cell invasion, migration, proliferation and colony formation. SCC15 cells with miR-124 restoration were less tumorigenic in vivo than miR-control SCC15 cells (70% inhibition, p<0.01). Restoration of miR-124 in SCC15 cells enhanced the anti-proliferative efficacy of the EGFR/Her2 TKIs. Furthermore, recapitulation of EGFR in miR-124 over-expressing SCC15 cells was sufficient to completely block the anti-proliferative effects of lapatinib and afatinib. Taken together, our work provides intriguing evidence that miR-124 is a novel therapeutic approach to reduce ESX/EGFR and may be a tractable strategy to enhance the response rate of HNSCC patients to current anti-EGFR/Her2 therapies.
Keywords: Head and neck cancer, anti-cancer therapeutics, EGFR, ESX, microRNAs
Introduction
Accumulating evidence exists to implicate epithelial-restricted with serine box (ESX), an ETS transcription factor family member, as key player in the tumorigenesis program especially in Her2-positive breast carcinomas. In lung and breast carcinomas, ESX was elevated, in part, through gene amplification (1, 2). Enforced ESX expression induced a transformed phenotype and promoted epithelial to mesenchymal transition in MCF12A mammary epithelial cells (3). Conversely, shRNA-mediated ablation of ESX reduced cell proliferation and anchorage-independent growth in ZR-75-1 and MCF7 breast carcinoma cells (4). ESX expression was reported to be elevated in breast carcinomas compared to normal breast tissue (5). Moreover, ESX was positively associated with Her2 levels in a cohort of breast carcinomas; 93% of Her2-positive tumors had elevated ESX (5). Another group showed that ESX transactivated the Her2 promoter through direct occupancy at a discrete ETS transcriptional response element (6). Over-expression of Her2 enhanced ESX promoter activity in MCF7 cells and chemical inhibition of Her2 decreased ESX promoter activity in SKBR3 Her2-positive breast carcinoma cells (7). Consistent with these findings, Her2 and ESX were shown to synergize to enhance Her2 and ESX transcription (5). These results indicate that a positive ESX/Her2 feedback loop may be critical to initiate and/or promote Her2-positive breast carcinomas.
In head and neck squamous cell carcinoma (HNSCC), Her2 dysregulation is an infrequent event, however, over-expression of epidermal growth factor receptor (EGFR) occurs frequently and associated with increased local recurrence and decreased disease-free survival (8–11). Unlike other solid malignancies, amplification of the EGFR gene is rare and only ranges between 10–15% in HNSCC tumors (12–14). EGFR mRNA expression was shown to be dramatically higher in HNSCC tumors than in normal mucosa (15). Furthermore, EGFR mRNA was over-expressed in 10 HNSCC cell lines without any evidence of EGFR gene amplification or rearrangement (15). These observations suggest that the predominant mechanism for EGFR dysregulation in HNSCC may be at the transcriptional level. Recent work from our laboratory revealed ESX as a transcription factor that modulates EGFR at the transcriptional level (16). Genetic ablation of ESX in ESXhigh/EGFRhigh HNSCC cells resulted in a reduced EGFR promoter activity and mRNA expression (16). Our work showed that ESX is upstream of EGFR to provide a novel molecular mechanism to account for EGFR dysregulation in HNSCC.
There is evidence that ESX is elevated and modulates EGFR in HNSCC (16). However, a mechanism for ESX dysregulation in HNSCC remains to be elucidated. In this study, miR-124 was demonstrated to bind to two distinct 3′-UTR sites in the ESX transcript to negatively regulate ESX. Depletion of miR-124 is a frequent event in HNSCC and restoration of miR-124 reduced ESX and EGFR, an ESX-regulated target, in miR-124low/ESXhigh/EGFRhigh SCC15 HNSCC cells. Enforced over-expression of miR-124 was sufficient to promote a pleiotropic anti-tumor effect in vitro and in vivo and potentiated the efficacy of EGFR/Her2 TKIs in vitro. Recapitulation of EGFR in miR-124 over-expressing HNSCC cells completely blocked the anti-proliferative activities of the EGFR/Her2 TKIs. Our work showed that miR-124 regulates the ESX/EGFR signaling axis in HNSCC. Moreover, we provide evidence that delivery of miR-124 may be a novel therapeutic strategy to reduce ESX/EGFR levels and enhance the efficacy of current EGFR/Her2 TKIs in the HNSCC patient population.
Materials and Methods
Cell lines
SCC15 and CAL27 cells were purchased from ATCC (Manass, VA) in 2010. USC-HN2 cells were provided by Dr. Alan L. Epstein (University of Southern California, Los Angeles, CA) in 2012. Human oral keratinocytes (HOK) and neonatal keratinocytes (PK) were purchased from ScienCell Research Laboratories (Carlsbad, CA) in 2012 and Life Technologies (Carlsbad, CA) in 2011, respectively. Primary human tonsillar epithelial cells (HTEC) were provided by Dr. John H. Lee (Sanford Health, Sioux Falls, SD) in 2010. SCC15 cells were grown in a 1:1 mixture of Ham’s F-12 and DMEM supplemented with 10% FBS, 0.4 μg/mL hydrocortisone, 2 mM L-glutamine, 100 mg/mL streptomycin and 100 U/mL penicillin. CAL27 cells were grown in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 mg/mL streptomycin and 100 U/mL penicillin. HTEC and PK cells were grown in keratinocyte serum-free medium (Life Technologies) and HOK cells were grown in complete OKM medium (Life Technologies). Cell lines were authenticated using short tandem repeat profiling every six months by our research group.
miR-124 expression in HNSCC specimens and cell lines
Twelve primary tumors and paired normal adjacent tissues were collected at The Ohio State University James Cancer Hospital from HNSCC patients at the time of surgical resection between 1997 and 2000. All tissues were diagnosed histologically by a board certified pathologist. Written informed consent, as required by the institutional review board, was obtained from all patients. Collected samples were stored immediately in liquid nitrogen at -80°C until analysis. Total RNA was isolated from the frozen HNSCC tumors and matched normal adjacent tissue with TRIzol (Invitrogen, Carlsbad, CA). Similarly, total RNA was extracted from HNSCC cells, HTEC, HOK and PK using TRIzol. Expression of miR-124 was determined using the Applied Biosystems 7900HT Fast Real-Time PCR System with a validated TaqMan assay (Applied Biosystems, Foster City, CA). Gene expression was normalized to RNU48 using the ΔΔCt method.
Luciferase reporter assay
Wildtype ESX-3′UTR was amplified by PCR and ligated into the psiCHECK-2 (Promega). Mutant ESX-3′UTR/psiCHECK-2 was generated using the QuikChange mutagenesis kit (Agilent Technologies, Santa Clara, CA). H293T cells were transfected with 50 ng of psiCHECK-2 (wildtype or mutant ESX-3′UTR) along with pre-miR-124 or pre-miR-control (100 nmol/L; Life Technologies) using Lipofectamine 2000 (Life Technologies). After 48 hours, cells were washed with PBS, resuspended in the lysis buffer (100 mM potassium phosphate pH 7.8, 0.2% Triton X-100, 0.5 mM dithiothreitol), and measured for Firefly/Renilla luciferase activities in a luminometer using the Dual-Light System (Applied Biosystems, Foster City, CA). Renilla luciferase activities were normalized to Firefly luciferase activities to control for transfection efficiency.
Generation of stable cell lines
pCMV-MIR (miR-control) and pCMV-MIR containing the precursor miR-124 sequence (miR-124) were purchased from OriGene Technologies, Inc. (Rockville, MD). Endotoxin-free plasmids were transfected into SCC15 and CAL27 cells using Lipofectamine 2000. Stable polyclonal SCC15/miR-control, CAL27/miR-control, SCC15/miR-124 and CAL27/miR-124 cells were generated by selection in neomycin and assessed for miR-124 expression using real-time PCR. SCC15/miR-124 cells were transfected with pBabe or pBabe/EGFR. Polyclonal SCC15/miR-124/vector and SCC15/miR-124/EGFR cells were generated by selection in puromycin. EGFR protein rescue was confirmed using immunoblot analysis.
Immunoblot analysis
Cells were washed in ice-cold PBS and lysed in buffer containing 1% Triton X-100, 50 mM HEPES, pH 7.4, 10% glycerol, 137 mM NaCl, 10 mM NaF, 100 mM Na3VO4, 10 mM Na4P2O7, 2 mM EDTA, 10 μg/mL leupeptin, and 1 mM PMSF. Whole cell lysates were mixed with Laemmli loading buffer, boiled, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Subsequently, immunoblot analysis was performed using an ESX-specific antibody (GenWay Biotech, San Diego, CA), an EGFR-specific antibody (Cell Signaling Technology, Danvers, MA), a Her2-specific antibody (Santa Cruz Biotechnology, Santa Cruz, CA), a STAT3-specific antibody (Sigma, St. Louis, MO), a phospho-STAT3 (Y705) antibody (Sigma) or a GAPDH-specific antibody (Sigma).
Cell proliferation
Cell proliferation was assessed using the CCK-8 reagent to detect metabolic active cells (Dojindo Inc., Gaithersburgh, MD). The absorbance at 450 nm was quantitated using a microplate reader (Molecular Devices, Sunnyvale, CA). IC50 values were generated using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA).
Cell invasion and migration
Cell invasion was determined as described from the cell invasion assay kit (Chemicon International, Temecula, CA). Cells were harvested and re-suspended in serum-free medium. An aliquot (1 × 105 cells) of the prepared cell suspension was added to the top chamber and 10% FBS was added to the bottom chamber. After 24 hours, non-invading cells were gently removed from the interior of the inserts with a cotton-tipped swab. Invasive cells were stained and visualized. Cell migration was determined using the wound healing assay. Cells were seeded and allowed to grow until confluence. Confluent monolayers were scratched using a sterile pipette tip, washed and incubated in complete medium. To minimalize potential experimental variations due to the difference in the scratch width, each well was examined by microscope and fields with a similar scratch width in the miR-control and miR-124 cells were noted and used for further analysis. Images were captured immediately after scratch and 24 hours post-scratch. Gap distance was measured using ImageJ and % of filled area was calculated for each sample. Mean % of filled area was determined for the miR-control and miR-124 cells and data presented was normalized to the miR-control cells.
Clonogenic survival
SCC15/miR-control, SCC15/miR-124, SCC15/miR-124/vector or SCC15/miR-124/EGFR cells were treated with lapatinib or afatinib for 72 hours. Cell were harvested, seeded onto 60-cm2 dishes in fresh medium and allowed to grow until visible colonies formed (14 days). Cell colonies were fixed with cold methanol and stained with 0.5% crystal violet.
Cell adhesion
SCC15/miR-control, SCC15/miR-124 cells were seeded onto 24-well plates coated with fibronectin (20 μg/mL) and incubated for 1 hour. Subsequently, the plates were washed 3X with PBS and the remaining attached cells were stained with 0.5% crystal violet.
Tumor growth in nude mice
SCC15/miR-control or SCC15/miR-124 cells (1×106 cells) were implanted into the flanks of nude mice (6–8 weeks; National Cancer Institute, Fredericks, MD). After two weeks, tumors were measured once a week using a digital caliper and tumor volumes were calculated as length × width × (height/2). All animal work performed was in accordance with and approved by the IACUC committee at The Ohio State University.
Immunohistochemical analysis
Resected tumors were fixed in 10% formalin and paraffin-embedded. Slides were incubated in citrate buffer (pH 6.0) for antigen retrieval and immunohistochemical staining was performed using Peroxidase Histostain-Plus Kit (Invitrogen) according to the manufacturer’s protocol. ESX antibody (LifeSpan Biosciences Inc., Seattle, WA) was used at a 1:500 dilution, EGFR antibody (Millipore, Billerica, MA) was used at a 1:10 dilution and Her2 antibody (Santa Cruz Biotechnology) was used at a 1:200 dilution. Slides were counter-stained with hematoxylin and coverslipped using glycerin.
Statistical analysis
Data were analyzed by two-tailed Student’s t-test. p-values of <0.05 were considered significant.
Results
miR-124 is depleted and regulates ESX in HNSCC
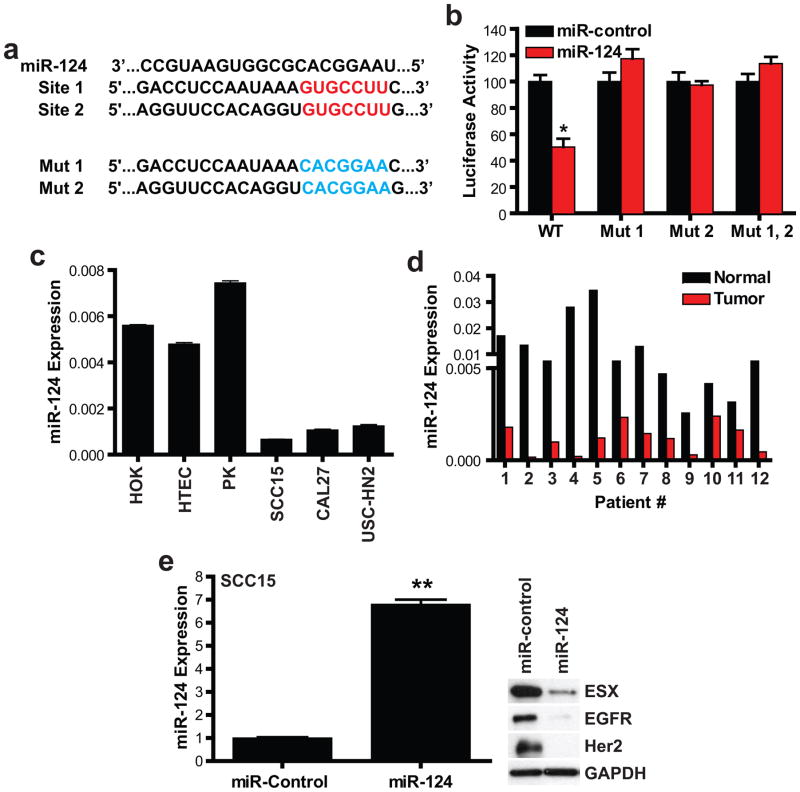
miRs are appreciated to play critical roles in tumorigenesis through regulation of target protein levels (17, 18). We determined if miRs are involved in the regulation of ESX in HNSCC. Interrogation of the 3′-UTR region of ESX using TargetScan predicted two miR-124 binding sites, positions 661–667 and 1646-1552 (Figure 1a). To experimentally validate that miR-124 directly binds to the predicted 3′-UTR sites of ESX, cells were co-transfected with pre-miR-control (100 nmol/L for 48 hours) or pre-miR-124 (100 nmol/L for 48 hours) and a reporter expression vector containing wildtype or mutant 3′-UTR of ESX cloned downstream of a luciferase gene (Figure 1b). pre-miR-124 blocked luciferase activity by 50 ± 2% (p<0.01, n=3) in cells transfected with the full-length wildtype 3′-UTR of ESX. Mutations in any of the two miR-124 sites completely dampened (p<0.01, n=3) the inhibitory effect of pre-miR-124 on luciferase activity. Our work showed that miR-124 regulates ESX through direct binding in the 3′-UTR of the ESX mRNA. Furthermore, occupation of both miR-124 sites appears to be indispensable to modulate ESX levels suggesting an all or none regulatory mechanism.
Figure 1. miR-124 is depleted and regulates ESX in HNSCC.
(a) Two putative miR-124 binding sites in the 3′-UTR of ESX. Wildtype miR-124 binding sites are highlighted in red and mutant miR-124 binding sites are highlighted in blue. (b) miR-124 directly regulates ESX. HEK293T cells were co-transfected with wildtype or mutant ESX-3′UTR and pre-miR-control or pre-miR-124. ESX-3′UTR Mut 1 contains mutations located at the miR-124 binding site 1. ESX-3′UTR Mut 2 contains mutations located at the miR-124 binding site 2. ESX-3′UTR Mut1, 2 contains mutations located at the miR-124 binding sites 1 and 2. Renilla and Firefly luciferase activities were measured using the Dual-Luciferase Reporter assay system. Renilla luciferase is normalized to Firefly luciferase and data are presented as mean ± SEM. *p<0.01, n=3. (c and d) miR-124 is depleted in HNSCC cell lines and primary HNSCC tumors. Total RNA was extracted from established cell lines and primary HNSCC tumors and paired adjacent normal epithelium from HNSCC patients. miR-124 expression was assessed by qPCR using a validated TaqMan primer set. Data are presented as mean ± SEM. (e) Restoration of miR-124 reduces ESX levels in HNSCC. Stable polyclonal SCC15/miR-control and SCC15/miR-124 cells were generated by antibiotic selection. Total RNA was extracted and measured for miR-124 expression using qPCR. Data is presented and mean ± SEM. **p<0.01, n=3. Whole cell lysates were extracted and immunoblot analyses were performed for ESX, EGFR and Her2.
To explore the relationship between miR-124 and HNSCC, we examined the levels of miR-124 in a panel of HNSCC cell lines and primary HNSCC tumors. As shown in Figure 1c, miR-124 levels were dramatically lower in a panel of HNSCC cell lines, SCC15, CAL27 and USC-HN2 compared to primary epithelial cells, including oral keratinocytes (HOK), tonsillar epithelial cells (HTEC) and skin keratinocytes (PK). All (12/12) HNSCC patients had significantly lower miR-124 expression in the primary tumor compared to patient matched normal adjacent mucosal tissue (Figure 1d). A 90.1% reduction in miR-124 expression in the primary tumor was demonstrated for this patient cohort (p<0.0001, n=12); relative mean miR-124 expression was 0.01197 ± 0.00293 for the adjacent normal epithelium and 0.00118 ± 0.00023 for the primary HNSCC tumors. Our findings demonstrate that miR-124 is depleted and suggest that miR-124 may be a potential tumor-suppressor miR in HNSCC.
Next, we determined if restoration of miR-124 is sufficient to reduce ESX in HNSCC cells. Stable polyclonal miR-124 over-expressing SCC15 cells were generated and assessed for miR-124 expression and ESX levels. SCC15/miR-124 cells showed a 6.8-fold increase in miR-124 expression compared to SCC15/miR-control cells (Figure 1e). Restoration of physiological levels of miR-124 in SCC15 (Figure 1e) and CAL27 (Supplemental Figure 1) HNSCC cells reduced the levels of ESX and two ESX-regulated targets, EGFR and Her2. These results provide initial evidence that depletion of miR-124 is a pathogenic event responsible for ESX dysregulation in HNSCC.
Restoration of miR-124 suppresses the tumorigenicity of HNSCC
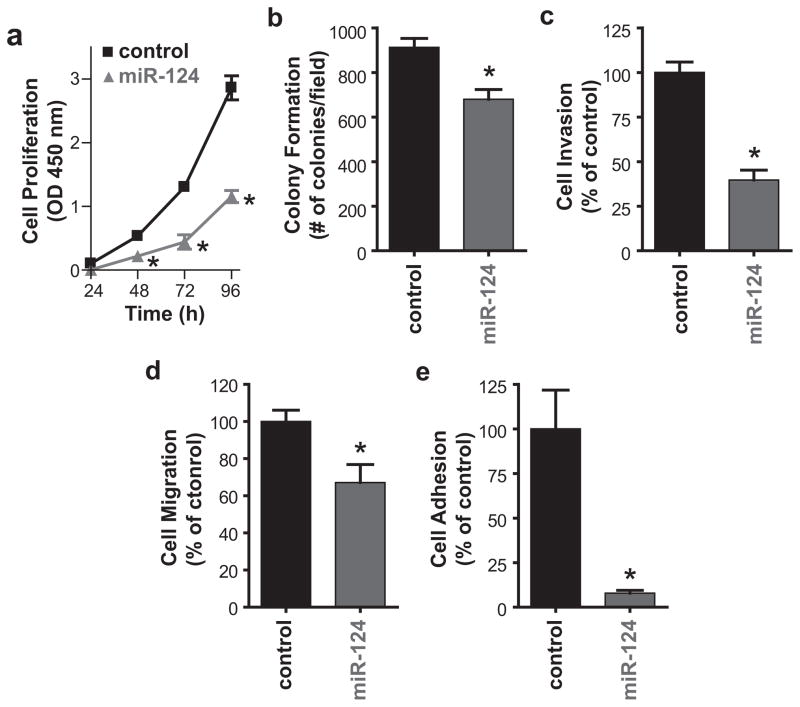
Down-regulation of ESX or EGFR induced a pleotropic anti-tumor effect in HNSCC (16). Our results showed that rescue of miR-124 to physiologic levels in HNSCC cells reduced ESX and EGFR. Therefore, we determined if restoration of miR-124 will promote a global anti-tumor effect. SCC15/miR-124 cells were less proliferative than SCC/miR-control cells (Figure 2a). Cell proliferation was reduced by 59.6% (p<0.01) at 96 hours with miR-124 restoration. In comparison to SCC15/miR-control cells, colony formation, cell invasion and cell migration in SCC15/miR-124 cells was suppressed by 25.7% (p<0.05), 60.4% (p<0.01) and 32.9% (p<0.05), respectively (Figures 2b–d). A recent study showed that miR-124 reduces cell adhesion and motility of oral carcinoma cells through down-regulation of integrin β1 (19). Since fibronectin is a well-recognized ligand for integrin β1, we examined if restoration of miR-124 in SCC15 cells will modulate cell adhesion in fibronectin-coated plates. Cell adhesion was dramatically compromised (92.4% inhibition, p<0.01) in SCC15/miR-124 cells compared to SCC15/miR-control cells (Figure 2e).
Figure 2. Restoration of miR-124 promotes a global anti-tumor effect in HNSCC in vitro.
(a) Cell proliferation. Cell proliferation was measured using the CCK-8 reagent to detect metabolic active cells. Data are from three independent experiments and presented as mean ± SEM. *p<0.01, n=3. (b) Colony formation. Colonies were stained with crystal violet and counted. Data are from three independent experiments and presented as mean ± SEM. *p<0.05, n=3. (c) Cell invasion. Cell invasion was assessed using the Modified Boyden chamber invasion assay with Matrigel basement membrane. Invasive cells were counted, normalized to SCC15/miR-control and presented as mean ± SEM from three independent experiments. *p<0.01, n=3. (d) Cell migration. Cell migration was determined using the wound healing assay. % of filled area is calculated, normalized to SCC15/miR-control and presented as mean ± SEM from three independent experiments. *p<0.05, n=3. (e) Cell adhesion. Cells were plated on fibronectin-coated plates and subsequently washed with PBS. The remaining attached cells were stained with 0.5% crystal violet and counted. Data are from three independent experiments and presented as mean ± SEM. *p<0.01, n=3.
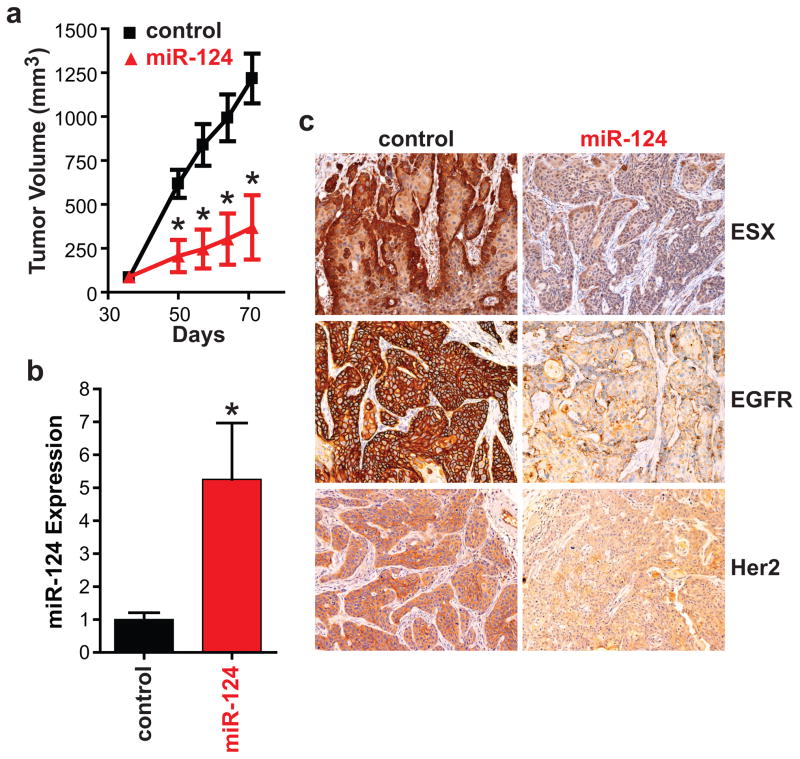
To determine the effect of miR-124 restoration on in vivo tumorigenicity, SCC15/miR-control and SCC15/miR-124 cells were implanted into the flanks of athymic nude mice (Figure 3). SCC15/miR-124 cells were less tumorigenic in vivo than SCC15/miR-control cells. Mean tumor volume was 3.3-fold (p<0.01, n=7) higher in mice bearing SCC15/miR-control tumors than in mice bearing SCC15/miR-124 tumors at 71 days post-tumor cell implantation (Figure 3a). At the end of the protocol, tumor were resected and analyzed for miR-124 expression and ESX, EGFR and Her2 levels. In Figure 3b, miR-124 was higher (5.3-fold increase, p<0.01) in SCC15/miR-124 tumors compared to SCC15/miR-control tumors demonstrating that miR-124 restoration was maintained long-term in vivo. Immunohistochemical analyses of the tumors showed that ESX, EGFR and Her2 levels are lower in SCC15/miR-124 tumors (Figure 3c). Our results indicate that restoration of miR-124 promotes a global anti-tumor effect in vitro and in vivo.
Figure 3. Restoration of miR-124 suppresses the tumorigenicity of HNSCC in vivo.
(a) Tumor growth. SCC15/miR-control or SCC15/miR-124 cells were injected subcutaneously to the flanks of the nude mice. Tumors were measured using a caliper and tumor volumes were calculated. Data is presented as mean ± SEM. *p<0.01, n=7. (b) Intratumoral miR-124 expression. SCC15/miR-control and SCC15/miR-124 tumors were resected and total RNA was isolated. miR-124 expression was measured using qPCR. Data is presented as mean ± SEM. *p<0.01. (c) Intratumoral ESX, EGFR and Her2 levels. ESX, EGFR and Her2 levels was assessed by standard immunohistochemistry. A representative image is shown for the SCC15/miR-control and SCC15/miR-124 tumors.
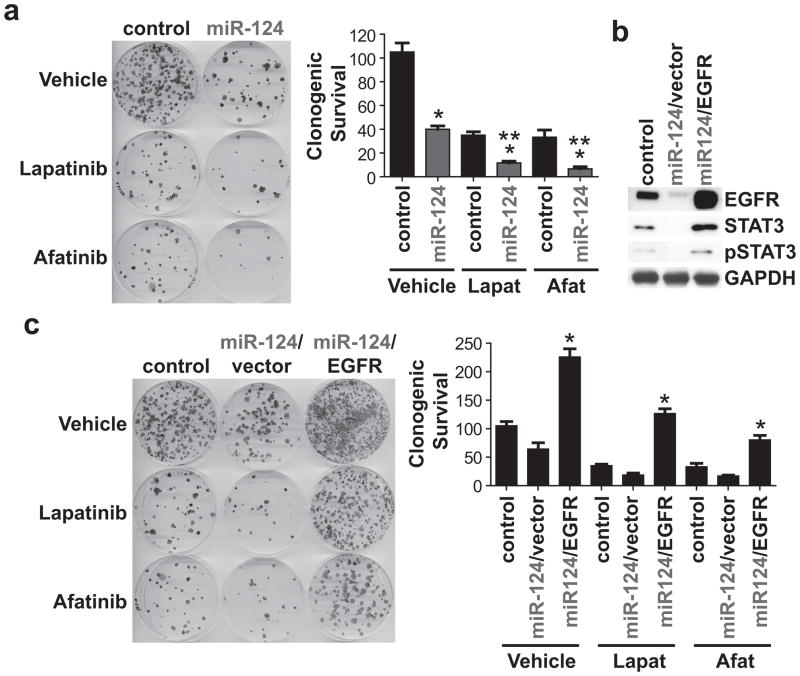
miR-124 modulates the ESX/EGFR axis to potentiate the anti-tumor efficacy of EGFR/Her2 TKIs
Recent work from our laboratory showed that a small molecule ESX inhibitor reduces EGFR/Her2 levels and enhances the anti-tumor effect of afatinib, an EGFR/Her2 TKI, in vitro and in vivo (16). Since ectopic miR-124 decreased ESX, EGFR and Her2 levels, we determined if miR-124 is sufficient to potentiate the anti-tumor effect of lapatinib and afatinib, two FDA-approved EGFR/Her2 TKIs (www.fda.gov). Lapatinib and afatinib inhibited SCC15 cell proliferation in a dose-dependent manner with IC50 values of 4.8 μmol/L and 2.4 μmol/L, respectively. Single-agent lapatinib or afatinib (IC50 dose) was active and inhibited clonogenic survival of SCC15/miR-control cells by 66.7% (p<0.01) and 68.3% (p<0.01), respectively (Figure 4a). SCC15/miR-124 cells were more responsive than SCC15/miR-control cells to both EGFR/Her2 TKIs. Lapatinib suppressed clonogenic survival of SCC15/miR-124 cells by 88.9% (p<0.01) and afatinib reduced clonogenic survival of SCC15/miR-124 cells by 93.6% (p<0.01). Moreover, in another miR-124low/ESXhigh HNSCC cell line, restoration of miR-124 in CAL27 cells potentiated the activity of afatinib to reduced clonogenic survival (Supplemental Figure 1).
Figure 4. miR-124 modulates the ESX/EGFR axis to potentiate the anti-tumor efficacy of EGFR/Her2 TKIs.
(a) Restoration of miR-124 potentiates the activity of afatinib and lapatinib. SCC15/miR-control and SCC15/miR-124 cells were treated with lapatinib or afatinib at the IC50 dose. Colonies were stained with crystal violet. Data is normalized to vehicle-treated SCC15/miR-control cells and presented as mean ± SEM. *p<0.01, SCC15/miR-control vs. SCC15/miR-124 for vehicle, lapatinib, or afatinib, n=3. **p<0.01 SCC15/miR-124/vehicle, SCC15/miR-control/lapatinib or SCC15/miR-control/afatinib vs. SCC15/miR-124/lapatinib or SCC15/miR-124/afatinib, n=3. (b) Ectopic over-expression of EGFR in SCC15/miR-124 cells. Stable polyclonal SCC15/miR-124/vector and SCC15/miR-124/EGFR cells were generated by antibiotic selection. Whole cell lysates were extracted and immunoblot analyses were performed for EGFR, STAT3 and pSTAT3. (c) Recapitulation of EGFR in SCC15/miR-124 cells promotes resistance to EGFR/Her2 TKIs. SCC15/miR-control, SCC15/miR-124/vector and SCC15/miR-124/EGFR cells were treated with lapatinib or afatinib at the IC50 dose. Colonies were stained with crystal violet. Data is normalized to vehicle-treated SCC15/miR-control cells and presented as mean ± SEM. *p<0.01, SCC15/miR-124/vector vs. SCC15/miR-124/EGFR for vehicle, lapatinib, or afatinib, n=3.
Since EGFR/Her2 TKIs target EGFR and Her2 with high specificity, we hypothesized that down-regulation of EGFR/Her2 may be critical to potentiate the efficacy of lapatinib and afatinib in SCC15/miR-124 cells. We focused on EGFR since EGFR is almost universally dysregulated in HNSCC (20). SCC15/miR-124 cells were transfected to over-express EGFR to determine if EGFR rescue would be sufficient to alter the phenotype of SCC15/miR-124 cells to be refractory to EGFR/Her2 TKIs. SCC15/miR-124/EGFR cells were confirmed to have higher EGFR levels than SCC15/miR-124/vector cells (Figure 4b). Recapitulation of EGFR completely rendered SCC15/miR-124 cells non-responsive to lapatinib and afatinib (Figure 4c). These findings show that simultaneous inhibition of EGFR protein levels, in this case via the miR-124/ESX axis, and EGFR kinase activity, with EGFR/Her2 TKIs, is a highly active therapeutic approach to optimally ablate HNSCC cells.
Discussion
MicroRNAs (miRs) constitute a family of small non-coding RNAs generally 18–22 nucleotides long. miRs bind to the 3′-UTR of target mRNA transcripts to negatively regulate the protein levels of target genes, either through inhibition of target gene translation or enhance degradation of target gene mRNA (21–23). Extensive research over the past decade has revealed that miRs can regulate diverse cellular processes and impart oncogenic or tumor-suppressive actions (23–27). Current literature indicates that miR-124 modulates a cadre of pro-oncogenic targets in several solid malignancies. miR-124 is down-regulated in a panel of gilomas and restoration of miR-124 directly reduces Snail homolog 2 to suppress tumorigenicity in part through depletion of the cancer stem cell population (28). Another study reported that miR-124 controls angiogenesis, chemosensitivity and proliferation by targeting R-Ras and N-Ras in glioma cells (29). In breast carcinoma, miR-124 expression is suppressed and inversely associated with histologic grade (30). Slug, a regulator of E-cadherin and epithelial-to-mesenchymal transition (EMT), was demonstrated to be a direct target for miR-124 in breast carcinoma cells (30). Consistent with the finding, an independent group showed that miR-124 inhibits TGF-α-induced EMT by modulating Slug in DU145 prostate carcinoma cells (31). Moreover, miR-124 was shown to negatively regulate signal transducers and activators of transcription 3 (STAT3) to reduce the tumorigenicity of colorectal and hepatocellular carcinoma cells (32, 33). Taken together, there is accumulating evidence that miR-124 functions as a candidate tumor-suppressor miR and loss of miR-124 may be a critical pathogenetic event common to induce tumor initiation and/or progression in various solid malignancies.
Previous work from our laboratory demonstrated that ESX is elevated in HNSCC; however, a molecular mechanism for this observation remains to be defined (16). In this study, depletion of miR-124 was revealed to be a mechanism responsible for high ESX levels in HNSCC. Loss of miR-124 was shown to be a frequent event and in fact, all of the HNSCC patients in our small cohort (12/12) showed dramatically reduced miR-124 in the primary tumor compared to adjacent normal epithelium. Our work showed that miR-124 binds to the 3′-UTR of the ESX transcript to negatively regulate ESX levels. Recapitulation of miR-124 to physiological relevant levels was sufficient to reduce ESX in miR-124low/ESXhigh HNSCC cells and induce a global anti-tumor effect, including cell proliferation, invasion and migration.
The use of tyrosine kinase inhibitors, including afatinib and lapatinib, to target EGFR have been studied and continue to be under clinical investigation for HNSCC. In a phase II study, afatinib demonstrated comparable activity to cetuximab in platinum-refractory recurrent/metastatic HNSCC patients; response rates of 8.1% and 9.7% for afatinib and cetuximab, respectively (34). Moreover, a recent global phase III study reported that afatinib was more active than methotrexate, a chemotherapeutic that is used globally in the recurrent/metastatic HNSCC setting, as second-line treatment in recurrent/metastatic HNSCC (35). Progression-free survival was 2.6 months for the afatinib arm and 1.7 months for the methotrexate arm (p=0.030; hazard ratio 0.80) (35). Separate phase III trials in HNSCC are ongoing to assess the efficacy of single-agent afatinib or lapatinb in the adjuvant setting to reduce the risk of disease recurrence and progression (36). Even though there is evidence that afatinib has activity in the recurrent/metastatic HNSCC setting, the clinical benefit with this molecularly-targeted agent is quite modest suggesting that the kinase-independent actions of EGFR may be able to compensate for kinase activity blockade and thus, a select population of HNSCC cells may utilize a non-canonical EGFR pathway to maintain survival. Therefore, therapeutic strategies to block kinase-independent EGFR actions need to be developed and use in combination with EGFR/Her2 TKIs to maximally dampen the EGFR signaling cascade.
A major advance in our study is that miR-124 restoration reduces EGFR/Her2 protein levels and potentiates the activity of EGFR/Her2 TKIs, afatinib and lapatinib, in HNSCC. miR-124 and the ESX transcription factor are known to directly regulate the expression and/or levels of multiple genes. Thus, it was impressive that rescue of a single ESX target gene, EGFR, completely rendered SCC15/miR-124 cells resistant to EGFR/Her2 TKIs. Our results provide initial evidence that EGFR dosage may drive HNSCC responsiveness to current EGFR/Her2 TKIs. Interestingly, in breast carcinoma cells, miR-124 was reported to regulate several components in the EGFR-driven cell cycle progression signaling network directly (V-Akt murine thymoma viral oncogene homolog 2, p38 mitogen-activated protein kinase and STAT3) and indirectly (retinoblastoma, cyclin-dependent kinase 2, cyclin-dependent kinase 4 and Src homolog 2 domain containing transforming protein 1) (37). We showed that SCC15/miR-124 cells has reduced total and phosphorylated STAT3 levels compared to SCC15/miR-control cells. However, the other direct and indirect miR-124 targets have yet to be confirmed in HNSCC cells. The possibility that miR-124 can regulate the EGFR signaling network at multiple layers suggests that delivery of miR-124 may be an efficient therapeutic strategy to globally dampen the hyperactive EGFR signal in HNSCC. In any event, our work showed that targeting EGFR levels and/or signaling network by restoring miR-124 augments the efficacy of EGFR/Her2 TKIs. Further development of the combination regimen of miR-124 and EGFR/Her2 TKI is warranted and should be prioritized for the miR-124low/ESXhigh/EGFRhigh HNSCC population.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grants R01CA135096 (Q. Pan) and R01CA140667 (A.K. Mapp and Q. Pan); The Michelle Theado Memorial Grant from the Joan Bisesi Fund for Head and Neck Oncology Research (Q. Pan); Slomin Family Foundation (Q. Pan and T.N. Teknos); and Arthur G. James Cancer Hospital and Richard J. Solove Research Institute, The Ohio State University Comprehensive Cancer Center (Q. Pan and T.N. Teknos).
Footnotes
Authors declare no competing financial interests in relation to the work described.
References
- 1.Liu E, Thor A, He M, Barcos M, Ljung BM, Benz C. The HER2 (c-erbB-2) oncogene is frequently amplified in in situ carcinomas of the breast. Oncogene. 1992;7(5):1027–32. [PubMed] [Google Scholar]
- 2.Tymms MJ, Ng AY, Thomas RS, Schutte BC, Zhou J, Eyre HJ, et al. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene. 1997;15(20):2449–62. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- 3.Schedin PJ, Eckel-Mahan KL, McDaniel SM, Prescott JD, Brodsky KS, Tentler JJ, et al. ESX induces transformation and functional epithelial to mesenchymal transition in MCF-12A mammary epithelial cells. Oncogene. 2004;23(9):1766–79. doi: 10.1038/sj.onc.1207391. [DOI] [PubMed] [Google Scholar]
- 4.Walker DM, Poczobutt JM, Gonzales MS, Horita H, Gutierrez-Hartmann A. ESE-1 is Required to Maintain the Transformed Phenotype of MCF-7 and ZR-75–1 Human Breast Cancer Cells. Open Cancer Journal. 2010;3:77–88. [Google Scholar]
- 5.Benz CC, O’Hagan RC, Richter B, Scott GK, Chang CH, Xiong X, et al. HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene. 1997;15(13):1513–25. doi: 10.1038/sj.onc.1201331. [DOI] [PubMed] [Google Scholar]
- 6.Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, et al. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene. 1997;14(13):1617–22. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- 7.Neve RM, Ylstra B, Chang CH, Albertson DG, Benz CC. ErbB2 activation of ESX gene expression. Oncogene. 2002;21(24):3934–8. doi: 10.1038/sj.onc.1205503. [DOI] [PubMed] [Google Scholar]
- 8.Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, et al. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90(11):824–32. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. European journal of cancer. 2001;37 (Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 10.Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62(24):7350–6. [PubMed] [Google Scholar]
- 11.Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11(16):5856–62. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 12.Licitra L, Mesia R, Rivera F, Remenar E, Hitt R, Erfan J, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Oncol. 2011;22(5):1078–87. doi: 10.1093/annonc/mdq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24(25):4170–6. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 14.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25(16):2164–70. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 15.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53(15):3579–84. [PubMed] [Google Scholar]
- 16.Zhang M, Taylor CE, Piao L, Datta J, Bruno PA, Bhave S, et al. Genetic and chemical targeting of epithelial-restricted with serine box reduces EGF receptor and potentiates the efficacy of afatinib. Mol Cancer Ther. 2013;12(8):1515–25. doi: 10.1158/1535-7163.MCT-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 18.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt S, Jones AV, Hinsley EE, Whawell SA, Lambert DW. MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS letters. 2011;585(1):187–92. doi: 10.1016/j.febslet.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol. 2006;24(17):2666–72. doi: 10.1200/JCO.2005.04.8306. [DOI] [PubMed] [Google Scholar]
- 21.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–7. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 22.Fabian MR, Sundermeier TR, Sonenberg N. Understanding how miRNAs post-transcriptionally regulate gene expression. Prog Mol Subcell Biol. 50:1–20. doi: 10.1007/978-3-642-03103-8_1. [DOI] [PubMed] [Google Scholar]
- 23.Nikitina EG, Urazova LN, Stegny VN. MicroRNAs and human cancer. Exp Oncol. 34(1):2–8. [PubMed] [Google Scholar]
- 24.Corsini LR, Bronte G, Terrasi M, Amodeo V, Fanale D, Fiorentino E, et al. The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin Ther Targets. 16(Suppl 2):S103–9. doi: 10.1517/14728222.2011.650632. [DOI] [PubMed] [Google Scholar]
- 25.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 13(6):e249–58. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 26.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 482(7385):347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Dou W, He L, Liang S, Tie J, Liu C, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 28.Xia J, Wu Z, Yu C, He W, Zheng H, He Y, et al. miR-124 inhibits cell proliferation in gastric cancer through down-regulation of SPHK1. J Pathol. 227(4):470–80. doi: 10.1002/path.4030. [DOI] [PubMed] [Google Scholar]
- 29.Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro-oncology. 2014;16(10):1341–53. doi: 10.1093/neuonc/nou084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, et al. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34(3):713–22. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin W, Pan Y, Zheng X, Li D, Bu J, Xu C, et al. MicroRNA-124 regulates TGF-alpha-induced epithelial-mesenchymal transition in human prostate cancer cells. Int J Oncol. 2014;45(3):1225–31. doi: 10.3892/ijo.2014.2506. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Lu Y, Yue X, Li H, Luo X, Wang Y, et al. MiR-124 suppresses growth of human colorectal cancer by inhibiting STAT3. PloS one. 2013;8(8):e70300. doi: 10.1371/journal.pone.0070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Yue X, Cui Y, Zhang J, Wang K. MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochemical and biophysical research communications. 2013;441(4):873–9. doi: 10.1016/j.bbrc.2013.10.157. [DOI] [PubMed] [Google Scholar]
- 34.Seiwert TY, Fayette J, Cupissol D, Del Campo JM, Clement PM, Hitt R, et al. A randomized, phase II study of afatinib versus cetuximab in metastatic or recurrent squamous cell carcinoma of the head and neck. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2014;25(9):1813–20. doi: 10.1093/annonc/mdu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machiels JP, Haddad RI, Fayette J, Licitra LF, Tahara M, Vermorken JB, et al. Afatinib versus methotrexate as second-line treatment in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck progressing on or after platinum-based therapy (LUX-Head & Neck 1): an open-label, randomised phase 3 trial. The Lancet Oncology. 2015;16(5):583–94. doi: 10.1016/S1470-2045(15)70124-5. [DOI] [PubMed] [Google Scholar]
- 36.Burtness B, Bourhis JP, Vermorken JB, Harrington KJ, Cohen EE. Afatinib versus placebo as adjuvant therapy after chemoradiation in a double-blind, phase III study (LUX-Head & Neck 2) in patients with primary unresected, clinically intermediate-to-high-risk head and neck cancer: study protocol for a randomized controlled trial. Trials. 2014;15:469. doi: 10.1186/1745-6215-15-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlmann S, Mannsperger H, Zhang JD, Horvat EA, Schmidt C, Kublbeck M, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.