Abstract
Introduction
The complexity of obesity and onset and susceptibility of cardio-metabolic disorders are still poorly understood and is addressed here through studies of genetic influence on weight gain and increased metabolic risk longitudinally.
Subjects/Methods
Twenty seven previously identified obesity, eating disorder or metabolic risk susceptibility SNPs were tested for association with weight or metabolically related traits longitudinally in 3999 adults participating both in the HUNT2 (1995–97) and HUNT3 (2006–08) surveys. Regression analyses were performed with changes from normal weight to overweight/obesity or from metabolically healthy to adverse developments with regards to blood pressure, glucose, HDL cholesterol, triglycerides or metabolic syndrome as outcomes. Additionally, a sub-sample of 1380 adolescents was included for testing association of nine SNPs with longitudinal weight gain into young adulthood.
Results
The most substantial effect on BMI-based weight gain from normal to overweight/obesity in adults was observed for the DRD2 variant (rs6277)(OR: 0.79, 95% CI: 0.69–0.90, P = 3.9x10-4, adj. P = 0.015). DRD2 was not associated with BMI on a cross-sectional level. In the adolescent sample, FTO (rs1121980) was associated with change to overweight at adulthood in the combined male-female sample (OR: 1.27, 95% CI: 1.09–1.49, P = 3.0x10-3, adj. P = 0.019) and in females (OR: 1.53, 95% CI: 1.23–1.91, P = 1.8x10-4, adj. P = 0.003). When testing for association to longitudinal adverse developments with regard to blood pressure, blood lipids and glucose, only rs964184 (ZNF259/APOA5) was significantly associated to unfavourable triglyceride changes (OR: 1.66, 95% CI: 1.36–2.03, P = 5.7x10-7, adj. P = 0.001). Pleiotropic effects on metabolic traits, however, were observed for several genetic loci cross-sectionally, ZNF259/APOA5, LPL and GRB14 being the most important.
Conclusions
DRD2 exhibits effects on weight gain from normal weight to overweight/obesity in adults, while, FTO is associated to weight gain from adolescence to young adulthood. Unhealthy longitudinal triglyceride development is strongly affected by ZNF259/APOA. Our main finding, linking the DRD2 variant directly to the longitudinal weight gain observed, has not previously been identified. It suggests a genetic pre-disposition involving the dopaminergic signalling pathways known to play a role in food reward and satiety linked mechanisms.
Introduction
Obesity has become a major global health burden [1] with numerous comorbidities such as metabolic syndrome (MetS), type 2 diabetes and cardiovascular disease [2]. Globally, from 1980 to 2008, the prevalence of obese adults has almost doubled with an increase of 4.8% to 9.8% in men and from 7.9% to 13.8% in women, respectively [3]. In Norway, represented by the HUNT study, the prevalence of BMI-based obesity has increased from 7.7% in 1984/86 to 22.1% in 2006/08 in men and from 13.3% to 23.1% in women. Likewise, abdominal obesity has increased markedly within the same time period [4] suggesting a general increased risk of metabolic syndrome, type 2 diabetes and cardiovascular disease in this population.
Obesity and associated MetS are regarded as complex traits influenced by both environmental factors and additive genetic effects. Through twin and family studies obesity heritability estimates for obesity range between 40 and 70% has been estimated [5]. Although GWAS studies based on common genetic variants with small size effects have enabled the identification of some pathologically important genetic markers and molecular pathways [6, 7], large steps forward in elucidating the genetics of obesity are expected with more studies on rare and copy number variants with larger effect sizes [8–10]. Identification of epigenetic and environment and gene interaction (GxE) effects are further thought to contribute to unraveling the “missing heritability”[11].
The metabolic syndrome refers to a combination of traits including increased central obesity, insulin resistance, dyslipidemia and hypertension [12]. The rise in MetS prevalence is postulated to mostly be caused by changes in life style. Even so a moderate to high heritability has been found for all underlying metabolic syndrome traits and data indicate that most of the individual variation observed is due to genetic differences [13]. Genetic pleiotropy [14] has been identified with common genes reported to affect clusters of MetS traits [15, 16].
The wide-ranging genetic variants shown to be implicated in common obesity, suggest that genetic susceptibility manifested in an obesogenic environment do so through complex interactions, also implicated in systems controlling food intake such as food reward and eating behaviour [17, 18]. Several studies have shown that obese individuals behave differently with regard to food stimuli and reward compared to normal weight individuals. Previous investigations have shown that dopamine and leptin signalling pathways may be involved in the stimulation of these [19, 20]. Likewise, several recent reports discuss the common genetic grounds for obesity and drug addiction [17, 21].
Causes that influence weight gain during adulthood are incompletely understood, but thought to be affected by a complex pattern of interactions between genetic susceptibility and lifestyle. Studies have focused on candidate genes’ influence on longitudinal weight change going in both directions (ΔBMI) [22], on genetic implications on weight gain/loss in intervention studies [23] or genetic pre-disposition to weight gain during antipsychotic treatment [24]. There are indications of increased risk of unhealthy weight gain due to reduced dopamine signalling and hence weaker responsiveness to food reward [20], however, very few population based studies address the issue of genetic pre-disposition to unhealthy weight or metabolic change over time.
Investigations of factors influencing longitudinal adverse developments with regard to obesity and cardio-metabolic disorders may provide new insight into processes of timing and susceptibility important for clinical prevention strategies. Through a longitudinal design based on the HUNT Study, our main investigation included 3999 adult individuals in a study of whether genetic variants previously associated with obesity, eating behaviors and metabolic traits influence both weight gain and metabolic adverse developments longitudinally over a time period of 11 years. Our study showed that the DRD2 variant involved in the dopaminergic signalling pathways, affects adverse weight gain in adults. The finding emphasises the importance of molecular mechanisms pre-disposing to food reward and satiety linked processes to be addressed in obesity management initiatives. Pleiotropic effects on metabolic traits were observed for several genetic loci cross-sectionally although very few genetic variants seem to influence both weight gain and adverse metabolic developments.
Materials and Methods
Study population and phenotypic measurements
In the HUNT Study [25] data have been collected in three different time waves, HUNT1 (1984–86), HUNT2 (1995–97) and HUNT3 (2006–08), comprising the entire adult population of the Nord-Trøndelag County in Norway. Our primary study sample included 3999 adults (47.7% men) who participated both in the HUNT2 and in the HUNT3 survey 11 years apart. Comprehensive health data questionnaires, clinical measurements and biological material were collected at both survey attendances. Another longitudinal sub-sample consisting of 1380 individuals was also included. This adolescent sample (Young-HUNT1, 45% males) had an average age of 16.0 years at baseline and 27.2 years at follow-up (HUNT3), described in detail elsewhere [26].
All examinations were done by trained nurses or technicians and weight, height and waist circumference (WC) were measured using standardised weight scales and meter bands. Height was measured to the nearest centimetre (cm), weight to the nearest 0.5 kilogram (kg) and body mass index (BMI) was calculated as weight in kg/height in m2. Blood pressure (BP) was measured using a Dinamap 845XT (Critikon) based on oscillometry, automatically three times per minute intervals. The average values of the two last measurements were used in our study. Total cholesterol (TC), high density lipoprotein Cholesterol (HDL-C), blood glucose (GLU) and triglycerides (TG) were measured in non-fasting serum from fresh blood samples at Levanger Hospital, Norway. Details of instruments and procedures are described previously [27, 28]. Overweight and obesity in adults were defined as having a BMI ≥ 25 or 30 kg/m2, respectively, or a waist circumference of ≥ 94 cm 102 cm in men and ≥ 80 cm or ≥ 88 cm in women. Overweight were assumed according to Cole et al. [29] Cases were defined as normal weight at Young-HUNT1 and overweight (BMI ≥25) at HUNT3 while controls were defined as normal weight both at Young-HUNT1 and HUNT3.
Metabolic syndrome (MetS) was defined as having at least three metabolic abnormalities for WC, blood pressure, blood glucose, HDL-cholesterol or triglycerides. Cut-offs were based on the original NCEP—ATP III definition [30]. However, as fasting measures were not available, the cut-offs for glucose and triglycerides were modified to ≥7.0 mmol/l and ≥2.1 mmol/l instead of ≥6.1 mmol/l and ≥1.7 mmol/l, respectively [2, 28]. Elevated blood pressure were defined as systolic blood pressure (SBP) ≥130 mmHg or diastolic blood pressure (DBP) ≥85 mmHg, or antihypertensive drug treatment and elevated blood glucose level as ≥7.0 mmol/l or use of or diabetes medical treatment. Decreased HDL-C levels were defined as <1.0 mmol/l in men or <1.3 mmol/l in women [31].
Pregnant women were excluded both at baseline and follow-up (n = 148 at HUNT2 and n = 6 at HUNT3) in analyses using BMI or WC. Participants treated with anti-hypertensive medication were excluded from cross-sectional and descriptive analyses where blood pressure levels were taken into account. Treatment with blood pressure lowering medication or diabetes medication, were classified as above cut-off respectively where the blood pressure and glucose measurements were used in the longitudinal association analyses.
Ethics
All participants gave a written informed consent. The protocol was in accordance with the Helsinki Declaration approved by the Regional Committee for Ethics in Medical Research and the Norwegian Data inspectorate.
Genotyping
DNA was extracted from peripheral blood leukocytes from EDTA whole blood or blood clots using the Gentra Purgene blood kit (QIAGEN Science, Maryland, USA). The procedure was done manually or automated with an Autopure LS (QIAGEN Science, Maryland, USA) as described by the manufacturer. To estimate associations with anthropometry (obesity/weight/height) and metabolic traits, SNPs were selected based on the most robust findings at the time of study design (year 2012) or on our own investigations [26, 32]. Genotyping was performed at CIGENE using the MassARRAY and iPlex system of the Sequenome genotyping platform (Sequenom, San Diego, CA, USA) in a SNP-multiplex design. The system uses the MALDITOF primer extension assay according to manufacturers’ recommendations. Forty ng DNA was used in the multiplex. Assays were optimised on 384 samples initially which resulted in five SNPs being excluded due to poor genotyping quality (lower call rates than 95%). This left the following 27 SNPs with their nearby gene for analyses: rs1121980 (FTO), rs17782313 (MC4R), rs11084753 (KCTD15), rs10838738 (MTCH2), rs4074134 (BDNF), rs569356 and rs533123 (OPRD1), rs35683 and rs2075356 (GHRL), rs6277 (DRD2), rs10195252 (GRB14), rs10242595 (IL–6), rs1049353 (CNR1), rs3782905 (VDR), rs3828942 (LEP), rs4929984 (H19), rs7180942 (NTRK3), rs8179183 (LEPR), rs890 (NR2B 5073T), rs964184 (ZNF259/APOA5), rs6810075 (ADIPOQ), rs560887 (G6PC2), rs12922394 (CDH13), rs1501299 (Adipoq), rs268 (LPL), rs782590 (SMEK2), rs1042725 (HMGA2) (S1 Table). Individuals with >10% genotype missing were removed. Two negative controls were run per 384-well plate. Samples were run blinded to the laboratory personnel.
Statistical analyses
SNPs were tested for deviation from Hardy Weinberg Equilibrium (HWE) in the total sample (S1 Table). To test cross-sectional SNP effects, linear regression was performed on the cross-sections using BMI, WC, SBP, DBP, GLU, TC, HDL-C and TG as continuous variables. Association with WC was adjusted for height. In addition, age and sex were adjusted for in analyses of the total sample sets and age in the sex stratified analyses. Due to departure from normal distribution by right skewness, the inverse values of glucose and the lg10 values of HDL-C and TG were used. The regression analyses were performed assuming additive models for each SNP. The minor allele was used as reference.
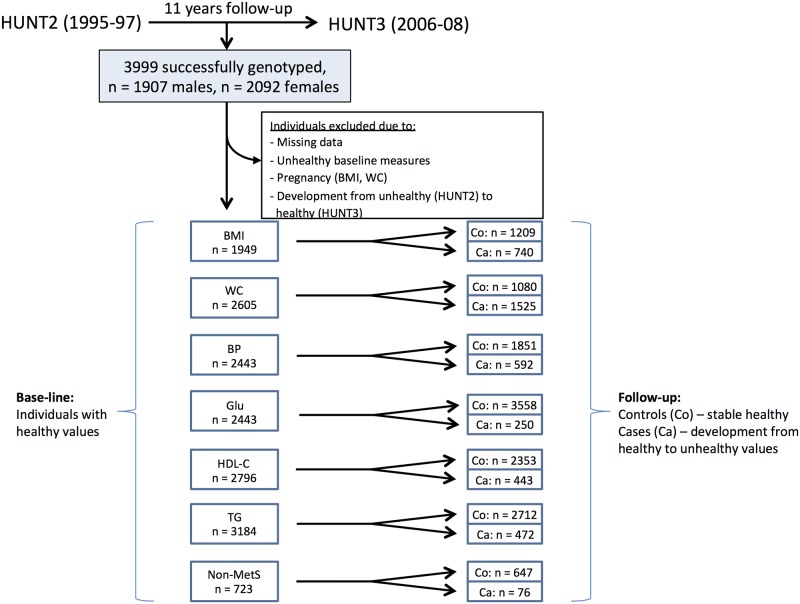
Individual changes over time from HUNT2 to HUNT3 were evaluated using ANOVA repeated measures (SPSS, version 20). Additionally, association between SNPs and changes from healthy to adverse metabolic status (HUNT2 to HUNT3) in individuals with cut-offs defined above for BP/antihypertensive medication, GLU/diabetes medication, HDL-C and TG were tested by logistic regression. Likewise, logistic regression was employed testing associations between genes and longitudinal changes from normal weight to overweight (HUNT2 to HUNT3). Controls were defined as participants categorized as healthy (below cut-off) with regard to the outcome variables both at baseline and follow-up, while cases were defined as those who displayed healthy values at baseline and above cut-off metabolically or categorized as overweight at follow-up. Participants not categorized as cases or controls were excluded from the analyses (study design outlined in Fig 1). PLINK Software was used for genetic analyses [33]. Nominal significance was considered at P<0.05 and for defining sex-specific interactions (SNP*sex). A PLINK-based permutation-based test (max(T)) with 1000 permutations per analysis was used in order to adjust for multiple testing of the SNPs (equals stringency of Bonferroni correction when single SNPs are tested).
Fig 1. Flow diagram of individuals included in the adult longitudinal study.
Overweight/obesity was defined as having a BMI ≥ 25 kg/m2 or as ≥ 94 cm 102 cm (male) and ≥ 80 (female) with regards to waist circumference (WC). Unhealthy blood pressure (BP) was defined as systolic blood pressure ≥130 mmHg or diastolic blood pressure ≥ 85 mmHg, or antihypertensive drug treatment. Unhealthy blood glucose (GLU) level was defined as ≥7.0 mmol/l or use of or diabetes medical treatment and triglyceride (TG) level as ≥2.1 mmol/l (both cut-offs modified due to non-fasting measurements). An unhealthy HDL cholesterol (HDL-C) level was defined below <1.0 mmol/l (male) or <1.3 mmol/l (female). Metabolic syndrome (MetS) phenotype cut-offs were based on the original NCEP—ATP III definition taking into account WC, BP, GLU, HDL-C and TG levels. MetS-cases were those scoring below cut-off for all five measures at baseline, but above cut-off for at least three components at follow-up. Controls scored below cut-offs for all five measures both at base-line and at follow-up.
Results
Study subjects and phenotypic measurements
Descriptive characterisation of the adult longitudinal sample is summarised in Table 1. The sample consisted of 3999 individuals (48% males) with an average age of 35.6 years (HUNT2) at baseline and 46.8 years at follow-up (HUNT3). Comparing baseline and follow-up measures showed significant longitudinal adverse developments for most included weight and metabolic variables, however, in men HDL-C and DBP stayed nearly unchanged, while SBP significantly decreased. In contrast, DBP declined significantly over time in women. Overall overweight (BMI based) changed from 60.3% to 77.2% and 39.6% to 56.7% for men and women, respectively, and WC-based overweight from 28.9% to 62.5% and 35.6% to 77.8% for men and women, respectively. Overall obesity (BMI based) changed from 9.6% to 22.0% in men and from 10.7% to 19.4% in women. WC-based obesity changed from 7.4% to 30.5% in men and 14.7% to 51.3% in women.
Table 1. Descriptive characteristics of the 3999 individuals (male 48%) in the adult longitudinal study (HUNT2, 1995–97) with follow up 11 years later (HUNT3, 2006–08).
| HUNT2 (1995–97) | HUNT3 (2006–08) | P c | ||||
|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | |
| Subject (n) | 1907 | 2092 | 1907 | 2092 | ND | ND |
| Age (years) a | 37.34 (6.0) | 33.98 (5.3) | 48.5 (5.9) | 45.16 (5.3) | ND | ND |
| BMI (kg/m2) a | 25.96 (3.03) | 24.79 (3.93) | 27.56 (3.43) | 26.57 (4.66) | <0.001 | <0.001 |
| Waist circumference a | 89.82 (7.8) | 77.57 (9.53) | 97.04 (9.46) | 88.98 (11.65) | <0.001 | <0.001 |
| Triglycerides (mmol/L) a | 1.87 (1.15) | 1.21 (0.69) | 1.96 (1.27) | 1.28 (0.74) | <0.001 | <0.001 |
| Total cholesterol (mmol/L) a | 5.54 (1.05) | 5.08 (0.99) | 5.64 (1.02) | 5.30 (0.98) | <0.001 | <0.001 |
| HDL cholesterol (mmol/L) a | 1.22 (0.32) | 1.47 (0.35) | 1.22 (0.29) | 1.44 (0.33) | 0.693 | <0.001 |
| Non-fasting glucose (mmol/L) a | 5.18 (0.97) | 4.94 (0.76) | 5.64 (1.41) | 5.26 (1.20) | <0.001 | <0.001 |
| Systolic blood pressure (mm Hg) a | 130.52 (11.68) | 118.47 (11.24) | 129.28 (13.75) | 120.11 (14.37) | <0.001 | <0.001 |
| Diastolic blood pressure (mm Hg) a | 76.14 (8.72) | 70.89 (8.13) | 76.46 (9.61) | 69.81 (9.73) | 0.152 | <0.001 |
| Overweight, BMI ≥ 25 b | 1150 (60.3%) | 780 (39.6%) | 1470 (77.2%) | 1182 (56.7%) | ||
| Obesity, BMI ≥ 30 b | 183 (9.6%) | 211 (10.7%) | 419 (22.0%) | 404 (19.4%) | ||
| Overweight, WC b , d | 552 (28.9%) | 703 (35.6%) | 1191 (62.5%) | 1621 (77.8%) | ||
| Obesity, WC b | 141 (7.4%) | 290 (14.7%) | 582 (30.5%) | 1068 (51.3%) | ||
aVariables expressed as means ± standard deviations.
bVariables expressed as number of individuals and percentages.
c P-value derived from pairwise comparisons (ANOVA). Inverse values for Glucose and Lg10 for HDL cholesterol and Triglyceride measurements in the ANOVA analyses. Waist circumference (WC) overweight, men ≥ 94 cm, women ≥ 80 cm. WC obesity, men ≥ 102 cm, women ≥ 88 cm.
dOverweight at BMI ≥ 25 includes overweight and obese individuals.
Genetic associations with longitudinal change from normal weight to overweight
To investigate whether the genetic variants included in the study were associated with longitudinal changes from normal weight to overall overweight/obesity (BMI ≥25), a logistic regression model was employed. The sample included 740 cases (353 males, 387 females) with normal weight at HUNT2 and overweight/obesity at HUNT3 and 1209 controls (404 males, 805 females) with normal weight at both time points (Fig 1). The DRD2 (Dopamine receptor D2)-variant (rs6277) was highly significant where the C-allele displayed a protective effect towards overweight development (OR: 0.79, 95% CI: 0.69–0.90, P = 3.9x10-4, adj. P = 0.015 (Table 2). Other gene variants were only significant at nominal significance levels.
Table 2. Association between SNPs and the longitudinal changes from normal to overweight/obesity (BMI ≥25) in the adult (HUNT2 to HUNT3) and the adolescent to adult subsamples (Young-HUNT1 to HUNT3).
| HUNT2→HUNT3 | ||||||||
| cases: 740, controls: 120 | ||||||||
| Sample | SNP | Gene | Ref. allele/ other allele | OR | L95 | U95 | P | Pa |
| Combined | rs560887 | G6PC2 | A/G | 0.90 | 0.78 | 1.05 | 0.18 b | 1.00 |
| Male | 1.12 | 0.89 | 1.41 | 0.35 | 1.00 | |||
| Female | 0.78 | 0.65 | 0.96 | 0.01 | 0.36 | |||
| Combined | rs2075356 | GHRL | C/T | 0.91 | 0.73 | 1.13 | 0.37 b | 1.00 |
| Male | 0.61 | 0.43 | 0.88 | 8.0x10 -3 | 0.16 | |||
| Female | 1.13 | 0.86 | 1.49 | 0.37 | 1.00 | |||
| Combined | rs268 | LPL | G/A | 1.58 | 1.06 | 2.34 | 0.02 | 0.47 |
| Male | 1.77 | 0.88 | 3.54 | 0.11 | 0.96 | |||
| Female | 1.49 | 0.92 | 2.42 | 0.11 | 0.95 | |||
| Combined | rs4929984 | H19 | A/C | 0.86 | 0.75 | 0.98 | 0.02 | 0.41 |
| Male | 0.88 | 0.71 | 1.07 | 0.20 | 1.00 | |||
| Female | 0.85 | 0.72 | 1.00 | 0.05 | 0.79 | |||
| Combined | rs4074134 | BDNF | A/G | 0.84 | 0.71 | 0.99 | 0.04 | 0.64 |
| Male | 0.83 | 0.64 | 1.07 | 0.15 | 0.99 | |||
| Female | 0.86 | 0.69 | 1.06 | 0.16 | 0.99 | |||
| Combined | rs10838738 | MTCH2 | G/A | 1.17 | 1.02 | 1.34 | 0.03 | 0.56 |
| Male | 1.16 | 0.92 | 1.44 | 0.21 | 1.00 | |||
| Female | 1.20 | 1.00 | 1.43 | 0.05 | 0.79 | |||
| Combined | rs6277 | DRD2 | C/T | 0.79 | 0.69 | 0.90 | 3.9x10 -4 | 0.02 |
| Male | 0.82 | 0.67 | 1.00 | 0.05 | 0.72 | |||
| Female | 0.77 | 0.65 | 0.92 | 3.3x10-3 | 0.07 | |||
| Young-HUNT1→HUNT3 | ||||||||
| cases: 553 controls: 827 | ||||||||
| Combined | rs1121980 | FTO | T/C | 1.27 | 1.09 | 1.49 | 3.0x10 -3 b | 0.04 |
| Male | 1.06 | 0.84 | 1.32 | 0.63 | 1.00 | |||
| Female | 1.53 | 1.23 | 1.91 | 1.8x10 -4 | <0.01 | |||
| Combined | rs17782313 | MC4R | C/T | 1.21 | 1.02 | 1.44 | 0.03 | 0.22 |
| Male | 1.16 | 0.91 | 1.49 | 0.22 | 0.89 | |||
| Female | 1.26 | 0.98 | 1.63 | 0.07 | 0.51 | |||
In the adult sample, cases (n = 740) were defined with normal weight at HUNT2 and overweight at HUNT3 while controls (n = 1209) were defined as normal weight both at HUNT2 and HUNT3. Overweight at adolescents were assumed according to Cole et al (2001). Cases were defined as normal weight at Young-HUNT1 and overweight (BMI ≥25) at HUNT3 while controls were defined as normal weight both at Young-HUNT1 and HUNT3. All measures were age adjusted and combined samples were additionally sex-adjusted. Empirical P-values were corrected for multiple testing by 1000 permutations.
Pa—P-values after multiple testing. Only results with a nominal significant P-value (P<0.05, underlined) at any of the measures included are shown. Significant results after multiple testing are shown in bold.
bSex interaction P<0.05.
To explore potential associations between the genetic variants and the change from normal weight to overweight/obesity (men ≥ 94 cm, women ≥ 80 cm) and normal weight to obesity (men ≥ 102 cm, women ≥ 88 cm) based on WC as outcome, logistic regression models were used. There were 1525 individuals (678 males, 847 females) in our sample that developed overweight /obesity between HUNT2 and HUNT3 and 632 obese cases (202 males, 421 females) while 1080 controls (673 males, 407 females) had WC below cut-off for overweight at both time points (Fig 1, S2 Table). The resulting associations were not generally comparable to the BMI-based results neither with regards to SNPs being identified nor the effect sizes displayed. The DRD2 was on the border of significance with the same direction of effect in females as identified for BMI-based change to overweight. Cross-sectionally, the FTO (fat mass and obesity associated) variant (rs1121980) was the only marker being significantly associated in sex and age-adjusted linear regression models with BMI after multiple testing (S3 Table).
To address whether the DRD2 showed the same longitudinal change association pattern as identified in the adults, we performed a nearly corresponding logistic regression analysis on the adolescent sub-sample. This sub-sample consisted of 553 cases (300 males and 253 females) that went from normal weight (weight categories according to Cole et al. [29]) to overweight/obese at adulthood and 827 controls (331 males and 496 females) with normal weight at both time points. Of the nine genetic variants that were included in this analysis (rs1121980 (FTO), rs17782313 (MC4R, melanocortin 4 receptor), rs11084753 (KCTD15, potassium channel tetramerization domain containing 15), rs10838738 (MTCH2, mitochondrial carrier 2), rs4074134 (BDNF- brain-derived neurotrophic factor), rs569356 (OPRD1- opioid receptor, delta 1), rs35683 (GHRL, ghrelin), rs6277 (DRD2) and rs10195252 (GRB14—growth factor receptor-bound protein 14)), only the FTO variant (rs1121980) was significant after multiple testing in the combined male-female sample (OR: 1.27, 95% CI: 1.09–1.49, P = 3.0x10-3, adj. P = 0.019). Additionally, sex-interaction was identified with a female-only effect observed in the stratified analyses (OR: 1.53, 95% CI: 1.23–1.91, P = 1.8x10-4, adj. P = 0.003) (Table 2).
Genetic associations with longitudinal change towards adverse measures of blood pressure, blood glucose, HDL cholesterol and triglycerides
In order to examine potential genetic effects with regards to change from healthy to adverse metabolic status over time, logistic regression models were used in the same manner as for the studies of genetic effects on disadvantaged weight developments. The four different metabolic trait sub-samples included were each composed of stable healthy controls for the measure in question and cases which were healthy at HUNT2, but had developed to an adverse state at HUNT3 defined by cut-offs defined in Materials and Methods. For the blood pressure analysis 592 cases (275 males, 317 females) and 1851 (536 males, 1315 females) controls were included. The glucose based analysis involved 250 cases (164 males, 86 females) and 3558 controls (1637 males, 1921 females), HDL cholesterol 443 cases (170 males, 273 females) and 2353 controls (1107 males, 1246 females) and the triglyceride set-up 472 cases (299 males, 173 females) and 2712 controls (998 males, 1714 females)(Fig 1). Only one variant, rs964184 (ZNF259/APOA5, zinc finger protein 259/apolipoprotein A-V) was significantly associated with unfavourable triglyceride changes even after multiple testing (OR: 1.66, 95% CI: 1.36–2.03, P = 5.7x10-7, adj. P = 0.001)(S4 Table).
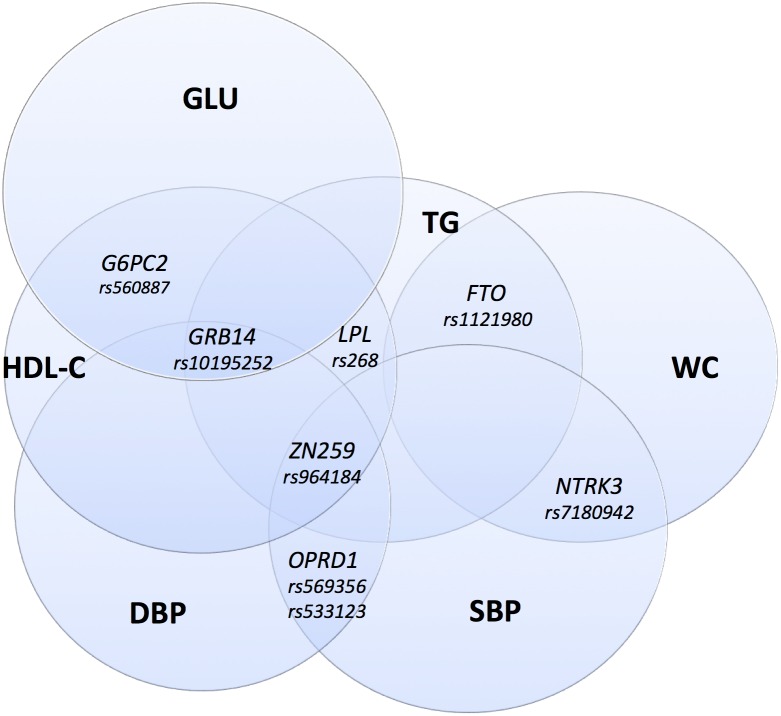
Cross-sectional linear regression analyses were performed testing associations between SNPs and metabolic traits at the HUNT2 and HUNT3 time points separately. The markers ZNF259/APOA5, LPL (lipoprotein lipase) and GRB14 displayed typical pleiotropic effects (S3 Table) further illustrated in Fig 2. The effects of rs964184 (ZNF259/APOA5) on TC, HDL-C and TG were significant sex-specifically with larger effects in males than in females (S5 Table).
Fig 2. Cross-sectional associations between SNPs and metabolic syndrome (MetS) components.
MetS-components: waist circumference (WC), HDL cholesterol (HDL-C), triglycerides (TG), glucose (GLU), systolic and diastolic blood pressure (SBP, DBP) at baseline (HUNT2) and follow-up (HUNT3). The following markers included were all significantly (P<0.05) associated to at least one trait cross-sectionally: ZNF259/APOA5 (rs964184), G6PC2 (glucose-6-phosphatase catalytic 2, rs560887), LPL (rs268), GRB14 (rs10195252), FTO (rs1121980), OPRD1 (rs569356 and rs533123) and NTRK3 (neurotrophic tyrosine kinase receptor type 3, rs7180942).
Genetic associations with longitudinal change from healthy to MetS positive
To explore the possibility of genetic effects influencing the longitudinal change from a healthy metabolic status to scoring positive for MetS, logistic regression was performed on 76 cases (32 males and 44 females) against 647 controls. Despite few cases, four variants displayed nominal significance. The rs560887 (G6PC2) was nominally significant in the combined sample and the rs533123 (OPRD1), rs1049353 (CNR1, cannabinoid receptor 1) and rs10242595 (IL6, interleukin 6) showed associations in males (significant sex interaction identified for IL6 and nearly for CNR1 (P = 0.06)). Tests for associations cross-sectionally, displayed significant results after adjustment for multiple testing with rs964184 (ZNF259/APOA5) at HUNT3 (OR: 1.39, 95% CI: 1.14–1.71, P = 1.6x10-3, adj. P = 0.031)(S6 Table).
Discussion
In this study, genetic variants near or within 24 genes previously associated with obesity, eating behaviors and metabolic traits were used to investigate potential influence both on weight gain and metabolic adverse development over time. The study included an adult sample of 3999 and an adolescent sample of 1380 individuals whom all had participated in two HUNT surveys performed 11 years apart. The availability of prospective data and DNA enabled us to study genetic effects at two different time points as well as investigating genetic effects with potential influence on the change to overweight or an impaired metabolic status.
The DRD2 variant was shown to influence BMI-based weight gain from normal to overweight/obesity in adults, but was not associated to BMI on a cross-sectional level. This finding was not replicated in the adolescent sub-sample. However, FTO showed significant association both in the combined male-female sample and in females separately. Only the ZNF259/APOA5 variant was significantly associated to impaired metabolic changes and only with regards to triglyceride levels. Cross-sectionally, pleiotropic effects on metabolic traits were observed for several genetic variants, ZNF259/APOA5, LPL and GRB14 being the most important.
An association between the dopamine receptor variant, DRD2, and overall weight change from normal to an overweight status in adults has not been reported previously. Dopaminergic signalling pathways are known to be involved in the regulation of food intake and energy expenditure through pathways involved in food reward and satiety [34] and the dopamine receptor genes (DRD2 and DRD4) that impact dopamine signaling capacity have recently through functional resonance imaging (fMRI) been shown to moderate the predictive risk of unhealthy weight gain [20]. In our study, other variants previously associated with eating behavior such as GHRL and BDNF also displayed nominal significant associations to weight gain with identified sex-interaction and effect only observed in males for the GHRL. The observed genetic effect exerted by variants involved in neuronal pathways support the increasing amount of evidence pointing towards comparable behaviors involved in food and drug addiction as well as in reward linked mechanisms [17, 35]. As suggested by Volkow et al. [17] “genes that modulate executive control, including self-control, may help counteract the risk for overeating in food-rich environments”, which may well be the case also for the genes identified in this study. Comparable association results concerning adverse abdominal change, was in our study not significant after multiple testing. However, the same directed effect as was observed for adverse changes in BMI-based weight measures was shown for DRD2 and another reward-related locus, OPRD1.
The association with reward-linked variants identified in the adult longitudinal sample was not replicated in the adolescent longitudinal sub-sample. The FTO obesity-risk allele was, however, shown to be associated with overweight development from adolescence to young adulthood. The different genetic effects identified in the adult and adolescent samples could be due to cohort related dissimilarities in general or that FTO may actually have stronger effects at a younger age. Previously, FTO has been shown to have its peak strength at the age of 20 before the effect weakens during adulthood [36]. The FTO gene is known for affecting appetite and satiety [37] and very recently this gene was reported to affect neural activity in homeostatic and brain reward regions [38] both supporting its important role in eating behavior. Interestingly, in our study the effect of FTO on longitudinal overweight development displayed a strong sex-interaction with the risk only being significant in females. This female-specific FTO effect has also been reported previously related to obesity, insulin sensitivity and glucose levels in children [39].
Recent research has shed light on how genetic variants influence combinations of components of MetS in a pleotropic fashion [16, 40]. In our study we replicated this finding for several of the genetic variants included. The G-allele of rs964184 located near the APOA5-A4-C3-A1 gene complex, affected HDL-C and TG levels at both time points in a direction consistent with an adverse metabolic development. However, the same allele displayed protective effects on blood pressure levels with highest effects on SBP. Sex-interactions were observed for this variant when associated to TG, TC and HDL-C where the effect was male-specific in the two latter. Male specific effects of the ZNF259/APOA5 variant on systolic blood pressure has also previously been documented [41]. Association between this variant and metabolic syndrome was identified in our study which has also been documented in a recent meta-analysis [42]. Similarly, associations between ZNF259/APOA5 and increased levels of HDL-C and TG both in children and adults have been reported [42–45], as confirmed here at both time points in our adult sample. An interesting finding in our study was the strong sex interaction (P = 0.004) observed with the development from normal to abdominal obesity where the G-allele in the rs964184 (ZNF259/APOA5) revealed a risk effect in males while the opposite was the case in females. As mentioned, sex-interaction with regards to this locus has been observed before and is suggested to explain sex differences in lipid levels and their heritability [46]. The overlapping association between lipid levels and waist circumference noticed for this locus, could imply common pathophysiology between obesity and lipids traits as hypothesized previously, [46].
Lipoprotein lipase (LPL) is a key enzyme in lipoprotein metabolism and a major candidate gene for coronary heart disease. The previous findings of genetic association between LPL variants (here rs268) and blood lipids such as TG, HDL-C and TC [47–50], was confirmed in our investigation. Additionally, the interrelation that seems to exist between ZNF259/APOA5 and LPL variants reported previously [47, 49], was also verified. In agreement with what was shown in the meta-analysis by Kristiansson and colleagues [42], the genetic association with MetS components seems to be lipid-driven and glucose seems to be less correlated with the other blood based MetS components.
Our main investigation comprised a population of 3999 adult individuals that could be investigated at two time points and be followed over a time period of 11 years. This quite large population sample strengthens the findings achieved on the cross-sections. However, most of the longitudinal models were underpowered due to the decision of addressing only the negatively developed outcomes. This may have precluded the identification of more significant findings. Another possible limitation of the study is the non-fasting lipid measures which may have had an effect on the case categorisation. However, recent investigations suggest that non-fasting lipid profiles change minimally in response to food intake [31]. In addition, both triglycerides and glucose cut-offs were increased according to previous investigations comparing non-fasting and fasting levels values which should preclude potentially misclassified individuals. A strength in this study was that anthropometric and clinical measurements were done by trained personnel avoiding the pitfall of under- or mis-reporting weight related measures [51] and by that underestimating the true weight gain.
The very interesting and not previously identified direct association between DRD2 and weight gain needs to be replicated in other longitudinal studies. As large longitudinal cohorts are difficult to find we have not been able to replicate the finding at present. However, ongoing longitudinal studies such as the Norwegian Tromsø Study [52] and the Dutch study Lifelines [53] may be candidates for replications at a later stage.
In summary, we report for the first time in a population based study, a significant association between the DRD2 gene and an overall weight gain from normal weight to overweight/obese status in adults. The same locus did not affect weight at a cross-sectional level indicating it to influence the behaviour that affects weight gain longitudinally. Interestingly, other variants also known to influence reward/addiction associated processes or eating related behaviour such as OPRD1, BDNF, GHRL and CNR1, also revealed evidence of being associated with overweight/obesity or development to an adverse metabolic status. Several metabolically related factors are influenced by common genes in a seemingly blood lipid-driven process where glucose appeared to be less correlated with the other blood based MetS components. The ZNF259/APOA5 is an important marker in this respect, because it in addition to associate strongly to various blood lipid levels cross-sectionally at both time points investigated, also pre-dispose to a highly significant longitudinal pre-disposition to increased triglyceride levels. The identification of addiction/reward process related genes with regards to weight gain and increased metabolic risk development, state the importance of better understanding the complex neurobiology of body weight regulation in future prevention strategies.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The Nord-Trøndelag Health Study (The HUNT study) is collaboration between HUNT Research Center (Faculty of Medicine, Norwegian University of Science and Technology NTNU), Nord-Trøndelag County Council, Central Norway Health Authority and Norwegian Institute of Public Health. Our study was supported by The Norwegian Research Council and the Liaison Committee between the Central Norway Regional Health Authority and NTNU. K.K. researched data and wrote the manuscript. J.H and K.H. reviewed/edited the manuscript. T.L.H. contributed to discussion and reviewed/edited manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Norwegian Research Council (http://www.forskningsradet.no/), The Liaison Committee between the Central Norway Regional Health Authority and NTNU (https://www.ntnu.no/dmf/rad/samorg). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–43. Epub 2000/04/15. 10.1038/35007508 . [DOI] [PubMed] [Google Scholar]
- 2. Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS, et al. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA. 2003;289(1):76–9. Epub 2002/12/31. . [DOI] [PubMed] [Google Scholar]
- 3. Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377(9765):557–67. Epub 2011/02/08. 10.1016/s0140-6736(10)62037-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Midthjell K, Lee CM, Langhammer A, Krokstad S, Holmen TL, Hveem K, et al. Trends in overweight and obesity over 22 years in a large adult population: the HUNT Study, Norway. Clin Obes. 2013;3(1–2):12–20. Epub 2013/08/13. 10.1111/cob.12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne). 2012;3:29 Epub 2012/05/31. 10.3389/fendo.2012.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loos RJ. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26(2):211–26. Epub 2012/04/14. 10.1016/j.beem.2011.11.003 . [DOI] [PubMed] [Google Scholar]
- 7. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. Epub 2010/10/12. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falchi M, El-Sayed Moustafa JS, Takousis P, Pesce F, Bonnefond A, Andersson-Assarsson JC, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet. 2014. Epub 2014/04/02. 10.1038/ng.2939 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flannick J, Thorleifsson G, Beer NL, Jacobs SB, Grarup N, Burtt NP, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46(4):357–63. Epub 2014/03/04. 10.1038/ng.2915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46(3):294–8. Epub 2014/01/28. 10.1038/ng.2882 . [DOI] [PubMed] [Google Scholar]
- 11. Schwenk RW, Vogel H, Schurmann A. Genetic and epigenetic control of metabolic health. Mol Metab. 2013;2(4):337–47. Epub 2013/12/12. 10.1016/j.molmet.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alberti KG, Zimmet P, Shaw J. Metabolic syndrome–-a new world-wide definition. A Consensus Statement from the International Diabetes Federation. Diabet Med. 2006;23(5):469–80. Epub 2006/05/10. 10.1111/j.1464-5491.2006.01858.x . [DOI] [PubMed] [Google Scholar]
- 13. van Dongen J, Willemsen G, Chen WM, de Geus EJ, Boomsma DI. Heritability of metabolic syndrome traits in a large population-based sample. J Lipid Res. 2013;54(10):2914–23. Epub 2013/08/07. 10.1194/jlr.P041673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011;12(3):204–13. Epub 2011/02/19. 10.1038/nrg2949 . [DOI] [PubMed] [Google Scholar]
- 15. Kullo IJ, de Andrade M, Boerwinkle E, McConnell JP, Kardia SL, Turner ST. Pleiotropic genetic effects contribute to the correlation between HDL cholesterol, triglycerides, and LDL particle size in hypertensive sibships. Am J Hypertens. 2005;18(1):99–103. Epub 2005/02/05. 10.1016/j.amjhyper.2004.09.002 . [DOI] [PubMed] [Google Scholar]
- 16. Andreassen OA, McEvoy LK, Thompson WK, Wang Y, Reppe S, Schork AJ, et al. Identifying common genetic variants in blood pressure due to polygenic pleiotropy with associated phenotypes. Hypertension. 2014;63(4):819–26. Epub 2014/01/08. 10.1161/hypertensionaha.113.02077 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Volkow ND, Wang GJ, Tomasi D, Baler RD. The addictive dimensionality of obesity. Biol Psychiatry. 2013;73(9):811–8. Epub 2013/02/05. 10.1016/j.biopsych.2012.12.020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alsio J, Olszewski PK, Levine AS, Schioth HB. Feed-forward mechanisms: addiction-like behavioral and molecular adaptations in overeating. Front Neuroendocrinol. 2012;33(2):127–39. Epub 2012/02/07. 10.1016/j.yfrne.2012.01.002 . [DOI] [PubMed] [Google Scholar]
- 19. Carpenter CL, Wong AM, Li Z, Noble EP, Heber D. Association of dopamine D2 receptor and leptin receptor genes with clinically severe obesity. Obesity (Silver Spring). 2013;21(9):E467–73. Epub 2013/05/15. 10.1002/oby.20202 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stice E, Yokum S, Bohon C, Marti N, Smolen A. Reward circuitry responsivity to food predicts future increases in body mass: moderating effects of DRD2 and DRD4. Neuroimage. 2010;50(4):1618–25. Epub 2010/02/02. 10.1016/j.neuroimage.2010.01.081 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heber D, Carpenter CL. Addictive Genes and the Relationship to Obesity and Inflammation. Mol Neurobiol. 2011;44(2):160–5. 10.1007/s12035-011-8180-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CG, et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41,504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;60(5):1637–44. Epub 2011/03/15. 10.2337/db10-1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delahanty LM, Pan Q, Jablonski KA, Watson KE, McCaffery JM, Shuldiner A, et al. Genetic predictors of weight loss and weight regain after intensive lifestyle modification, metformin treatment, or standard care in the Diabetes Prevention Program. Diabetes Care. 2012;35(2):363–6. Epub 2011/12/20. 10.2337/dc11-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hong CJ, Liou YJ, Bai YM, Chen TT, Wang YC, Tsai SJ. Dopamine receptor D2 gene is associated with weight gain in schizophrenic patients under long-term atypical antipsychotic treatment. Pharmacogenet Genomics. 2010;20(6):359–66. Epub 2010/04/09. . [DOI] [PubMed] [Google Scholar]
- 25. Krokstad S, Langhammer A, Hveem K, Holmen T, Midthjell K, Stene T, et al. Cohort Profile: The HUNT Study, Norway. Int J Epidemiol. 2012. Epub 2012/08/11. 10.1093/ije/dys095 . [DOI] [PubMed] [Google Scholar]
- 26. Kvaloy K, Kulle B, Romundstad P, Holmen TL. Sex-specific effects of weight-affecting gene variants in a life course perspective–-The HUNT Study, Norway. Int J Obes (Lond). 2013;37(9):1221–9. Epub 2013/01/16. 10.1038/ijo.2012.220 . [DOI] [PubMed] [Google Scholar]
- 27. Begum G, Davies A, Stevens A, Oliver M, Jaquiery A, Challis J, et al. Maternal undernutrition programs tissue-specific epigenetic changes in the glucocorticoid receptor in adult offspring. Endocrinology. 2013;154(12):4560–9. Epub 2013/09/26. 10.1210/en.2013-1693 . [DOI] [PubMed] [Google Scholar]
- 28. van Vliet-Ostaptchouk JV, Nuotio ML, Slagter SN, Doiron D, Fischer K, Foco L, et al. The prevalence of metabolic syndrome and metabolically healthy obesity in Europe: a collaborative analysis of ten large cohort studies. BMC Endocr Disord. 2014;14(1):9 Epub 2014/02/04. 10.1186/1472-6823-14-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ. 2000;320(7244):1240–3. Epub 2000/05/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA. 2001;285(19):2486–97. Epub 2001/05/23. . [DOI] [PubMed] [Google Scholar]
- 31. Sidhu D, Naugler C. Fasting time and lipid levels in a community-based population: a cross-sectional study. Arch Intern Med. 2012;172(22):1707–10. Epub 2012/11/14. 10.1001/archinternmed.2012.3708 . [DOI] [PubMed] [Google Scholar]
- 32. Cuypers KF, Loos RJ, Kvaloy K, Kulle B, Romundstad P, Holmen TL. Obesity-susceptibility loci and their influence on adiposity-related traits in transition from adolescence to adulthood–-the HUNT study. PLoS One. 2012;7(10):e46912 Epub 2012/10/25. 10.1371/journal.pone.0046912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Z, Hao CJ, Li CG, Zang DJ, Zhao J, Li XN, et al. Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons. PLoS Genet. 2014;10(2):e1004124 Epub 2014/02/20. 10.1371/journal.pgen.1004124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stice E, Figlewicz DP, Gosnell BA, Levine AS, Pratt WE. The contribution of brain reward circuits to the obesity epidemic. Neurosci Biobehav Rev. 2013;37(9 Pt A):2047–58. Epub 2012/12/15. 10.1016/j.neubiorev.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hardy R. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19:545–52. 10.1093/hmg/ddp504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang T, Qi Q, Li Y, Hu FB, Bray GA, Sacks FM, et al. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr. 2014;99(5):1126–30. Epub 2014/03/14. 10.3945/ajcn.113.082164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karra E. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest. 2013;123:3539–51. 10.1172/JCI44403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jacobsson JA, Danielsson P, Svensson V, Klovins J, Gyllensten U, Marcus C, et al. Major gender difference in association of FTO gene variant among severely obese children with obesity and obesity related phenotypes. Biochem Biophys Res Commun. 2008;368(3):476–82. Epub 2008/02/06. 10.1016/j.bbrc.2008.01.087 . [DOI] [PubMed] [Google Scholar]
- 40. van Vliet-Ostaptchouk JV, den Hoed M, Luan J, Zhao JH, Ong KK, van der Most PJ, et al. Pleiotropic effects of obesity-susceptibility loci on metabolic traits: a meta-analysis of up to 37,874 individuals. Diabetologia. 2013;56(10):2134–46. Epub 2013/07/06. 10.1007/s00125-013-2985-y . [DOI] [PubMed] [Google Scholar]
- 41. Yamada Y, Ando F, Shimokata H. Association of the genetic variants of APOA5 and PRKCH with hypertension in community-dwelling Japanese individuals. Mol Med Rep. 2008;1(3):407–14. Epub 2008/05/01. . [PubMed] [Google Scholar]
- 42. Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS, et al. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5(2):242–9. Epub 2012/03/09. 10.1161/circgenetics.111.961482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7(12):e51954 Epub 2012/12/20. 10.1371/journal.pone.0051954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen Y, Xi B, Zhao X, Cheng H, Hou D, Wu L, et al. Common genetic variants associated with lipid profiles in a Chinese pediatric population. Hum Genet. 2013;132(11):1275–85. Epub 2013/07/09. 10.1007/s00439-013-1332-1 . [DOI] [PubMed] [Google Scholar]
- 45. Zhang X, Qi Q, Bray GA, Hu FB, Sacks FM, Qi L. APOA5 genotype modulates 2-y changes in lipid profile in response to weight-loss diet intervention: the Pounds Lost Trial. Am J Clin Nutr. 2012;96(4):917–22. Epub 2012/08/24. 10.3945/ajcn.112.040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walia GK, Gupta V, Aggarwal A, Asghar M, Dudbridge F, Timpson N, et al. Association of common genetic variants with lipid traits in the Indian population. PLoS One. 2014;9(7):e101688 Epub 2014/07/06. 10.1371/journal.pone.0101688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ariza MJ, Sanchez-Chaparro MA, Baron FJ, Hornos AM, Calvo-Bonacho E, Rioja J, et al. Additive effects of LPL, APOA5 and APOE variant combinations on triglyceride levels and hypertriglyceridemia: results of the ICARIA genetic sub-study. BMC Med Genet. 2010;11:66 Epub 2010/05/01. 10.1186/1471-2350-11-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Franceschini N, Carty C, Buzkova P, Reiner AP, Garrett T, Lin Y, et al. Association of genetic variants and incident coronary heart disease in multiethnic cohorts: the PAGE study. Circ Cardiovasc Genet. 2011;4(6):661–72. Epub 2011/11/02. 10.1161/circgenetics.111.960096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu Y, Dolle ME, Imholz S, van 't Slot R, Verschuren WM, Wijmenga C, et al. Multiple genetic variants along candidate pathways influence plasma high-density lipoprotein cholesterol concentrations. J Lipid Res. 2008;49(12):2582–9. Epub 2008/07/29. 10.1194/jlr.M800232-JLR200 . [DOI] [PubMed] [Google Scholar]
- 50. Sagoo GS, Tatt I, Salanti G, Butterworth AS, Sarwar N, van Maarle M, et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am J Epidemiol. 2008;168(11):1233–46. Epub 2008/10/17. 10.1093/aje/kwn235 . [DOI] [PubMed] [Google Scholar]
- 51. Park JY, Mitrou PN, Keogh RH, Luben RN, Wareham NJ, Khaw KT. Effects of body size and sociodemographic characteristics on differences between self-reported and measured anthropometric data in middle-aged men and women: the EPIC-Norfolk study. Eur J Clin Nutr. 2011;65(3):357–67. Epub 2010/12/24. 10.1038/ejcn.2010.259 . [DOI] [PubMed] [Google Scholar]
- 52. Jacobsen BK, Eggen AE, Mathiesen EB, Wilsgaard T, Njolstad I. Cohort profile: the Tromso Study. Int J Epidemiol. 2012;41(4):961–7. Epub 2011/03/23. 10.1093/ije/dyr049 ; PubMed Central PMCID: PMCPmc3429870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scholtens S, Smidt N, Swertz MA, Bakker SJ, Dotinga A, Vonk JM, et al. Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int J Epidemiol. 2014. Epub 2014/12/17. 10.1093/ije/dyu229 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.