Abstract
Transgenic labeling of innate immune cell lineages within the larval zebrafish allows for real-time, in vivo analyses of microbial pathogenesis within a vertebrate host. To date, labeling of zebrafish macrophages has been relatively limited, with the most specific expression coming from the mpeg1 promoter. However, mpeg1 transcription at both endogenous and transgenic loci becomes attenuated in the presence of intracellular pathogens, including Salmonella typhimurium and Mycobacterium marinum. Here, we describe mfap4 as a macrophage-specific promoter capable of producing transgenic lines in which transgene expression within larval macrophages remains stable throughout several days of infection. Additionally, we have developed a novel macrophage-specific Cre transgenic line under the control of mfap4, enabling macrophage-specific expression using existing floxed transgenic lines. These tools enrich the repertoire of transgenic lines and promoters available for studying zebrafish macrophage dynamics during infection and inflammation and add flexibility to the design of future macrophage-specific transgenic lines.
Introduction
The zebrafish has been widely adopted in the study of myriad vertebrate biological processes, including inflammation and infection. Two important features of zebrafish as a model system are the optical transparency of embryos and larvae and the ease with which transgenesis can be accomplished [1–5]. In simple fluorescent reporter zebrafish lines, cell-type specific fluorescent reporter expression combined with the optical transparency of the larvae allows for real-time visualization of specific cell lineages [6–10].
Transgenic lines involving the labeling or manipulation of immune cell lineages can offer an unprecedented view into the dynamics of the vertebrate innate immune system and its interactions with a number of human pathogens [11]. The larval zebrafish has been used to study a growing array of bacterial pathogens, including Mycobacterium [12–19], Pseudomonas [20–22], Salmonella [23, 24], and Listeria spp. [25, 26]. Studies of fungal disease pathogenesis, including infections with Candida albicans [27] and Aspergillus fumigatus [28] have utilized the zebrafish as a model host, as have investigations of Influenza A virus and Chikungunya virus [29, 30]. These infection studies benefit from the ability to visualize host-microbe interactions within a transparent and genetically tractable vertebrate host that can be produced in large numbers.
The larval zebrafish possesses an innate immune system that comprises cell types common to all vertebrate innate immune systems, including those of mammals [31, 32]. This similarity has allowed for the study of host-pathogen interactions with high spatial and temporal resolution, and has been facilitated by the use of transgenic lines with fluorescently labeled innate immune cell types [6, 7, 9, 33]. Macrophages and neutrophils are among the first immune cell lineages to encounter invading pathogens and are particularly well conserved between zebrafish and mammals in both cellular composition and function [34, 35].
However, more than 15 years after the first descriptions of zebrafish macrophages [35, 36], there exist a limited number of transgenic tools for visualizing and manipulating macrophages in live zebrafish. Transgenic lines have been generated using the myeloid differentiation factor pu.1 which labels macrophages, neutrophils and their precursors [31]. Further analysis of the role of pu.1 in zebrafish myelopoiesis identified four putative macrophage-specific genetic markers, all of which exhibited striking transcriptional downregulation upon knockdown of pu.1 [37]. One such marker, mpeg1, was later identified as macrophage specific and transgenes were constructed under the control of upstream regulatory elements of the mpeg1 locus [6]. Two additional transgenic lines have been generated that drive transgene expression in macrophages: one operates under the control of the immunoresponsive gene 1 (irg1) promoter and labels macrophages only upon activation [10]; the other is a bacterial artificial chromosome (BAC) transgenic utilizing the zebrafish macrophage colony stimulating factor (fms) promoter that labels both macrophages and skin xanthophores [38]. However, transgenic lines based on mpeg1 have to date remained the primary tool for real-time, in vivo analyses of global zebrafish macrophage dynamics [34, 39].
Downregulation of endogenous zebrafish mpeg1 expression during infection with M. marinum or S. typhimurium has been previously described [16], and our lab has observed severe attenuation of mpeg1 transgene expression in larval macrophages occurring as early as three days post-infection with M. marinum. We describe the development of a new macrophage-specific mfap4 transgenic line that, in contrast to mpeg1, remains stable during mycobacterial infection. In addition, we describe the generation of a macrophage-specific Cre transgenic line that will allow lineage-tracing and easy combinatorial transgenic approaches to investigating in vivo, real-time macrophage dynamics.
Materials and Methods
Zebrafish handling
Animal research and protocols were approved by the Duke University IACUC A145-14-06. Adult zebrafish were housed under standard conditions using an automated system for water changes and maintenance of pH and conductivity (Aquaneering). Larvae were raised at 28.5°C in incubators. Manipulation and imaging were carried out using tricaine anesthesia as described below. Euthanasia of any severely ill or moribund animals was performed by immersion in water at a temperature below 4°C, as approved by IACUC.
Generation of the mfap4 promoter element
The forward and reverse primers (5’-CATGTTCTCGAGGCGTTTCTTGGTACAGCTGG–3’ and 5’-CATGTTGGATCCCACGATCTAAAGTCATGAAGAAAGA–3’, respectively) were used to amplify a 1.6 kb region immediately upstream of the third codon of the second exon within the mfap4 locus. The BAC clone CH73-131D4 obtained from Children’s Hospital Oakland Research Institute (CHORI) was used as the PCR template. The linear mfap4 promoter amplicon was cloned into the p5E-MCS vector of the Gateway Cloning system via restriction digest and ligation with XhoI/BamHI and T4 DNA Ligase (NEB) as described previously in [14].
Start codon mutagenesis
The p5E-mfap4 Entry Clone was linearized and amplified via inverse PCR using the following primers: 5’-CTTCTCACTCTCTCCTCAACAG–3’ and 5’-GTAAGTTCTGTGGCTGTTTTATTC–3’. The start codon was mutated from ATG to ATT via Gibson Assembly using the linearized backbone and the following single-stranded DNA oligonucleotide: 5’-CTGAGCTGTTGAGGAGAGAGTGAGAAGATTGCAGTAAGTTCTGTGGCTGTTTTATTCC–3’ as described previously in [14].
Construction of the mfap4:tdTomato transgene
The tdTomato fluorescent protein was amplified from pTEC5 (a kind gift of Dr. Lalita Ramakrishnan, University of Cambridge) using the following primers: Forward- 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGACCATGGTGAGCAAGGGCGAGGAG–3’ and Reverse–5’-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACTTGTACAGCTCGTCCAT–3’. Amplified tdTomato sequence was BP cloned into pDONR221 to generate pME-tdTomato.
The complete transgene was subsequently generated by recombination of p5E-mfap4, pME-tdTomato, and p3E-polyA [4] elements into the destination vector pDestTol2pA2 using the LR Clonase II Plus enzyme mix, resulting in pDestTol2; mfap4:tdTomato.
Construction of the mfap4:tdTomato-CAAX transgene
pME-tdTomato-CAAX was generated as described [14].
The complete transgene was subsequently generated by recombination of p5E-mfap4, pME-tdTomato-CAAX, and p3E-polyA elements into the destination vector pDestTol2UbpA using the LR Clonase II Plus enzyme mix, resulting in pDestTol2; mfap4:tdTomato-CAAX.
Construction of the mfap4:Turquoise2 transgene
Both pME-Turquiose2 and the complete pDestTol2; mfap4:Turquoise2 were generated as described [14].
Construction of the mfap4:iCre:p2A-tdTomato transgene
The mfap4:iCre:p2A-tdTomato transgene was generated through multisite gateway recombination of p5E-mfap4, pME-iCre, and p3E-p2A-tdTomato into pDestTol2UbpA. pME-iCre was generated by amplifying the codon-optimized iCre from pDIRE (Addgene plasmid 26745 from Zeller lab, [40]) with attB1 and attB2 flanked primers: Forward– 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCACCATGGTGCCCAAGAAGAAGAG–3’, Reverse– 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTCGTCCCCATCCTCGAGC–3’ and recombining with pDONR 221 by BP recombination (Life Technologies).
To generate p3E-p2a-tdTomato, tdTomato was amplified from pTEC5 with the forward primer 5’-GGGGACAGCTTTCTTGTACAAAGTGGTTGGATCCTTCAGTCTCGAGATGGTGAGCAAGGGCGAGGAG–3’ and reverse primer 5’-GGGGACAACTTTGTATAATAAAGTTGTTACTTGTACAGCTCGTCCAT–3’ and gateway cloned in a BP cloning reaction into pDONR p2R-P3. The resulting plasmid contained BamHI and XhoI restriction sites upstream of the Tomato start codon. The p3E-Tomato construct was digested with BamHI and XhoI and annealed linkers encoding the porcine teschovirus–1 2A sequence (p2A) [41] were ligated into the plasmid to generate p3E p2A-tdTomato. Annealed linker sequences encoding p2A were as follows: Top strand– 5’- GATCCGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTC–3’ and bottom strand– 5’- TCGAGAGGTCCAGGGTTCTCCTCCACGTCTCCAGCCTGCTTCAGCAGGCTGAAGTTAGTAGCTCCGCTTCCG–3’.
pDestTol2UbpA was generated by amplifying the ubiquitin B poly A signal with the following primers– 5’-TATTATACATAGTTGATATTCTCAGTATCCCCTGCTTA–3’ and 5’-CGACGGCCAGTGAATTATACTTCTCATTTCGCATCTTAT–3’; and amplifying the pDestTol2pA2 backbone with the following primers– 5’-AATTCACTGGCCGTCG–3’ and 5’-ATCAACTATGTATAATAAAGTTGAACGAG–3’, and joining the PCR products with Infusion (Clontech).
The complete transgene was subsequently generated by recombination of p5E-mfap4, pME-iCre, and p3E-p2A-tdTomato elements into the destination vector pDestTol2UbpA using the LR Clonase II Plus enzyme mix, resulting in pDestTol2; mfap4:iCre:p2A-tdTomato.
Construction of mfap4:dLanYFP-CAAX transgene
For Tg(mfap4:dLanYFP-CAAX) xt11 only, the p5E-mfap4 Entry Clone was slightly modified by the inclusion of the p2A sequence at the 3’ end of the promoter element, in-frame with both the upstream mfap4 coding sequence and downstream transgene coding sequence. This sequence was included as an additional mechanism to ensure that any translation of the mfap4 coding sequence would be separated from the transgene polypeptide. No differences in expression were observed between this line and other mfap4 lines. The modified p5E-mfap4 was generated by inverse PCR of p5E-mfap4 using the primers 5’-GGCTGAAGTTAGTAGCTCCGCTTCCCACGATCTAAAGTCATGAAG–3’ and 5’-AGACGTGGAGGAGAACCCTGGACCTGGATCCACTAGTTCTAGAGCGG–3’, followed by Gibson Assembly (NEB) of the linearized p5E-mfap4 amplicon and a single-stranded DNA oligonucleotide 5’- AGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGA–3’.
The sequence of dLanYFP was PCR amplified from pNCS dLanYFP (Allele Biotechnology) with the primers 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGACCATGGTGAGCAAGGGCGAGGAG–3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTAGATCTACTTGTAGAGCTCGTCCATGCCG–3’. These primers resulted in a silent SacI site in the 3’ end of the LanYFP coding sequence and a BglII site downstream of the dLanYFP stop codon. The PCR product was cloned by BP gateway cloning (Life Technologies) into pDONR221 to generate pME-dLanYFP. The resulting pME-dLanYFP construct was digested with SacI and BglII and annealed oligos encoding the human H-Ras prenylation signal [42, 43] were cloned into the plasmid to generate pME-dLanYFP-CAAX. Oligo sequences for the linker are: top strand- 5’-CTACAAGAAGCTGAACCCTCCTGATGAGAGTGGCCCCGGCTGCATGAGCTGCAAGTGTGTGCTCTCCTA–3’, bottom strand- 5’-GATCTAGGAGAGCACACACTTGCAGCTCATGCAGCCGGGGCCACTCTCATCAGGAGGGTTCAGCTTCTTGTAGAGCT–3’.
The complete transgene was subsequently generated by recombination of the modified p5E-mfap4, pME-dLanYFP-CAAX, and p3E-polyA elements into the destination vector pDestTol2pA2 using the LR Clonase II Plus enzyme mix, resulting in pDestTol2; mfap4:dLanYFP-CAAX.
Generation of transgenic zebrafish lines
In all cases, transgenic lines were generated as follows: Single-cell zebrafish embryos were injected with approximately 1nl of a mixture containing 50ng/μl of transgenic construct (i.e., pDestTol2; mfap4:dLanYFP-CAAX), 25ng/μl of Tol2 transposase mRNA, and 0.17% phenol red in 1x Buffer Tango. The Tol2 transposase mRNA was generated via in vitro transcription from T3TS-Tol2 [2] using the mMessage mMachine T3 Kit (Life Technologies). Larvae positive for transgenesis were selected at 48 hours post-fertilization on the basis of detection of transgene fluorescence. Adults from these founder populations were individually outcrossed to wildtype fish, and larvae positive for macrophage fluorescence were retained as F1 populations.
M. marinum larval infections, imaging, and quantitation
The transgenic zebrafish lines Tg(mpeg1:tdTomato-CAAX) xt3 or Tg(mfap4:dLanYFP-CAAX) xt11 were crossed to generate double transgenic larvae. Larvae positive for both red and yellow fluorescence were anesthetized in 160 μg/ml Tricaine (MS–222, Sigma Aldrich) and injected via the caudal vein at 48 hours post-fertilization with 100–200 M. marinum expressing the Cerulean fluorescent protein. Larvae were imaged at 1 day post-infection and 5 days post-infection in all three fluorescent channels as well as transmitted light. All imaging for this experiment was carried out using a Zeiss Observer Z1 inverted microscope. Quantitation of fluorescence was carried out using Zen Image Processing software (Zeiss).
Confocal images were acquired on a Leica SP8 Confocal Microscope using a 25x water-immersion lens.
M. marinum adult infections and frozen tissue sections
Adult infections, tissue preparation, and frozen sectioning were performed as described in [14].
Results
Construction of macrophage-specific transgenic lines based on mfap4.
To overcome the limitation in long-term imaging of macrophage/M. marinum interactions imposed by the loss of mpeg1 transgene expression, we developed novel transgenic lines based on another of the original four spi1-controlled genes, microfibrillar-associated protein 4 (mfap4) [37]. The turquoise version of this line has been reported previously by our laboratory [14], but here we describe its characterization and the development of additional tools that expand the repertoire of macrophage-specific tools. To generate an mfap4 promoter element likely to recapitulate the endogenous, macrophage-specific expression pattern, we amplified ~1.6 kb of genomic sequence at the 5’ end of the mfap4 locus. Specifically, the promoter element includes 1.5 kb of sequence upstream of the mfap4 transcription start site, continues through the entire first intron and terminates at the second codon of the second exon (Fig 1A). We elected to retain sequences to direct splicing of the transgene, as mRNA export has been shown to be enhanced following a splicing event [44–46]. In order to ensure translation initiation at the start site of the inserted transgene, we mutated the mfap4 start codon in the promoter element from ATG to ATT. 5’ RACE indicated a single initiation codon of the endogenous mfap4 transcript pool, and so alteration of this ATG likely abrogates any translation of any mfap4-specific sequence retained at the 3’ end of the promoter element of mfap4 transgenes (data not shown).
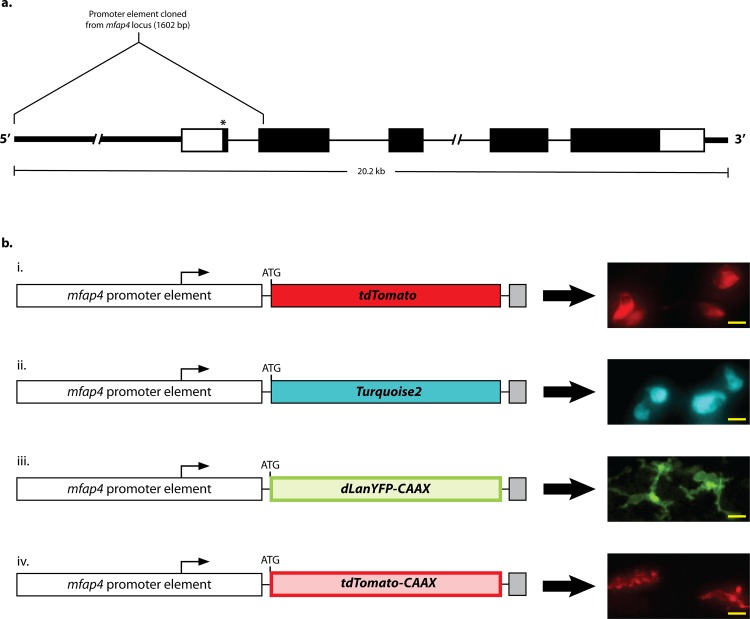
Fig 1. mfap4 transgene design.
a. Schematic of the mfap4 promoter sequence with respect to the full endogenous mfap4 locus. The full promoter element spans 1,602 bp. Thick lines represent genomic sequence flanking the 5’ and 3’ UTRs. Empty and solid boxes represent UTRs and coding regions, respectively. Thin lines represent introns. The asterisk denotes the location of the start codon mutation (ATG>ATT). b. Schematics of mfap4 transgenes and examples of their resulting expression. The bent arrows mark transcription start sites; “ATG” marks the beginning of the fluorescent protein ORF. White boxes depict the mfap4 promoter as the 5’ Element of an expression construct generated via the Gateway Recombination System; colored boxes depict Middle Elements containing fluorescent protein ORFs; grey boxes depict 3’ Elements containing a poly-adenylation signal. The lines connecting each element represent small linker sequences present as a result of the recombination process. i,ii. Expression of cytosolic tdTomato and Turquoise2 fluorescent proteins, respectively. iii,iv. Expression of membrane localized dLanYFP and tdTomato, respectively. The addition of the CAAX prenylation motif to the C-terminus of either fluorescent protein allows macrophage membrane projections to be easily visualized. Scale bars = 10 μm. Expression data representative of more than 50 animals for each construct.
We cloned the amplified promoter as a 5’ Element of the Multisite Gateway Recombination System to facilitate simple, rapid generation of an array of mfap4-controlled transgenic constructs through modular assembly of the promoter element with different transgenes [47, 48]. In this manner we generated several independent transgenic zebrafish lines each expressing either the tdTomato (red) [49], dLanYFP (yellow/green) [50], or Turquoise2 (cyan) [51] fluorescent proteins: Tg(mfap4:tdTomato) xt12 and Tg(mfap4:tdTomato-CAAX) xt6, Tg(mfap4:dLanYFP-CAAX) xt11, and Tg(mfap4:Turquoise2) xt27, respectively. Each of these lines exhibited robust expression in cells that appeared to be macrophages (Fig 1B); this is especially apparent for the transgenic lines expressing fluorescent proteins with a C-terminal CAAX motif, resulting in targeting of the transgene to the plasma membrane [42, 43]. In these two lines, long membrane projections characteristic of motile macrophages are always observed (Fig 1Biii and 1Biv). We also observed a modest increase in fluorescence signal in the yolk of two tdTomato-expressing transgenic lines as compared to the yolk of wildtype larvae, but this limited yolk fluorescence was absent in other mfap4 lines indicating that it is not a general property of the promoter (S1 Fig).
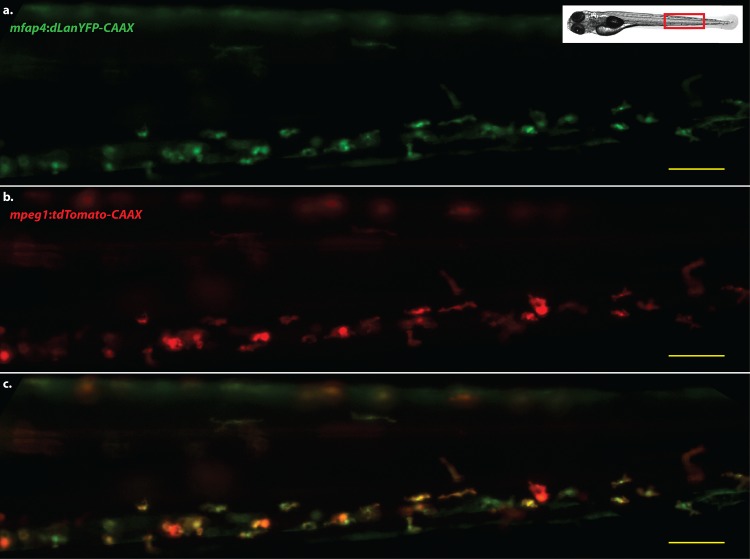
Subsequent populations of these lines derived from stable, germ-line transmitting genomic integrations exhibited transgene expression detectable in a specific population of cells by 36 hours post-fertilization (hpf), consistent with the timing of macrophage development and accumulation of sufficient fluorescent protein to yield a signal [35]. Fluorescence is greatly increased by 48 hpf, and can be detected easily via epifluorescence microscopy at relatively low magnification (e.g., 5x objective lens/0.25 NA). Expression remains robust in the adult and continues to be restricted to cells that appear to be macrophages to at least three months of age, including cells infected with M. marinum (S2 Fig). Because transgenic lines based on mpeg1 have been previously characterized as macrophage-specific [6], we sought to determine the specificity of the mfap4 promoter by crossing Tg(mfap4:dLanYFP-CAAX) xt11 with Tg(mpeg1:tdTomato-CAAX) xt3. We observed essentially complete colocalization of expression of both fluorophores (Fig 2), indicating that mfap4 transgenics exhibit an expression pattern that is highly restricted to zebrafish macrophages.
Fig 2. mfap4 transgene expression colocalizes with the expression of mpeg1 transgenes.
Representative image of the caudal hematopoietic tissue of a double transgenic larva, four days post-fertilization. a. Cells expressing dLanYFP via the Tg(mfap4:dLanYFP-CAAX) xt11 transgene. b. Cells expressing the tdTomato fluorescent protein via the Tg(mpeg1:tdTomato) xt3 transgene. c. Merged image of a,b. Scale bars = 50 μm. Images representative of at least 40 animals.
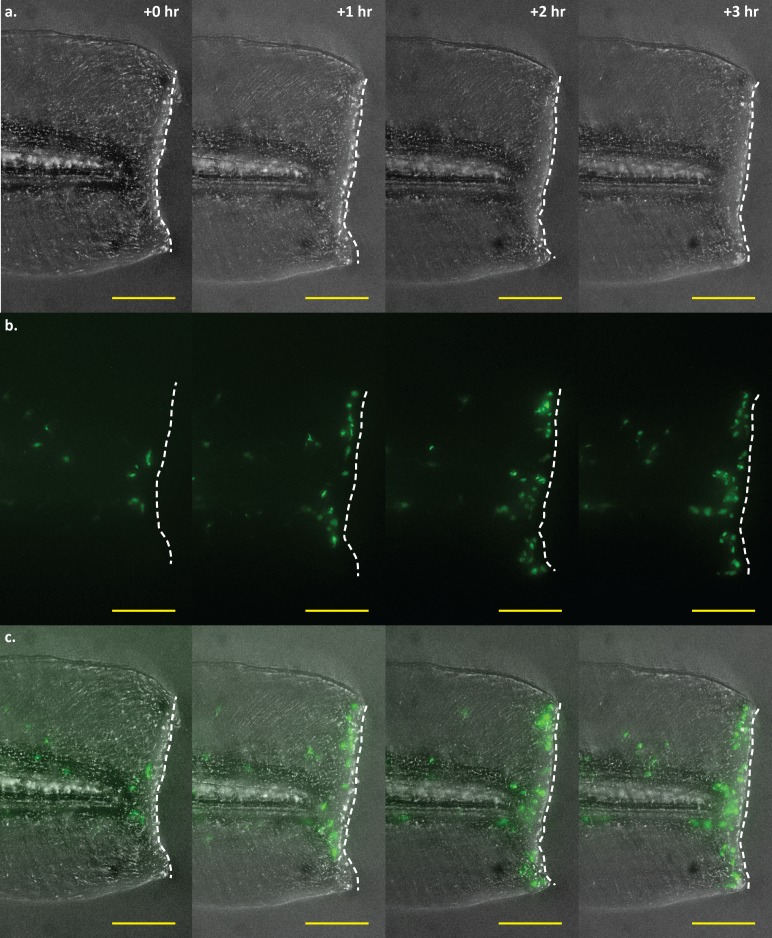
We then sought to demonstrate the utility of mfap4 transgenics for in vivo time-lapse imaging of zebrafish larvae, as well as confirm standard macrophage behaviors of the labeled cell population. We wounded Tg(mfap4:dLanYFP-CAAX) xt11 larvae by excising a small portion of the developing tail fin, and imaged the region over several hours to track the movements of the labeled cell population (Fig 3). Fluorescent cells can be seen tracking toward and accumulating at the edge of the wound, a behavior consistent in both timing and activity for macrophages upon introduction of an inflammatory stimulus [52]. Thus, mfap4 driven fluorescent proteins and the added CAAX motif do not appear to alter macrophage behavior and, therefore, permit the visualization of membrane projections in migrating macrophages.
Fig 3. Time-lapse imaging of a tail wound of an mfap4:dLanYFP-CAAX transgenic larva.
Time-lapse imaging of macrophage recruitment to the site of a wound at the posterior end of the larval tail. Macrophage movements may be visualized in real-time via fluorescence imaging of mfap4 transgenic larvae. Shown here are four representative frames of such an experiment, showing macrophage recruitment to the wound edge at 0, 1, 2, and 3 hours post-wounding. a. Brightfield detailing the extent and location of the wound. The wound edge is highlighted as a dashed white line. b. Macrophages visualized via dLanYFP fluorescence. The number of cells present proximal to and along the site of the wound can be seen increasing over time as macrophages track toward the damaged tissue. c. Merge of brightfield and fluorescence channels. Scale bars = 100 μm. Images representative of 12 animals from two independent biological replicates.
The mfap4 promoter produces stable transgene expression throughout the course of larval infection with M. marinum
The zebrafish larva has become increasingly utilized for performing real-time, in vivo analyses of mycobacterial disease pathogenesis within a vertebrate host [14–16]. Such studies are greatly enhanced by the ability to track macrophages during infection, as macrophages are the primary cell type involved in initial interactions with pathogenic mycobacteria [13, 53]. However, it has been previously observed that mpeg1 expression in zebrafish macrophages becomes attenuated in the presence of M. marinum [16], and we also observed a marked reduction in the fluorescence signal within infected macrophages of mpeg1 transgenic lines, often to the point that many cells within the macrophage population were no longer detectable by fluorescence (Fig 4).
Fig 4. mpeg1-mediated fluorophore expression is attenuated in infected cells.
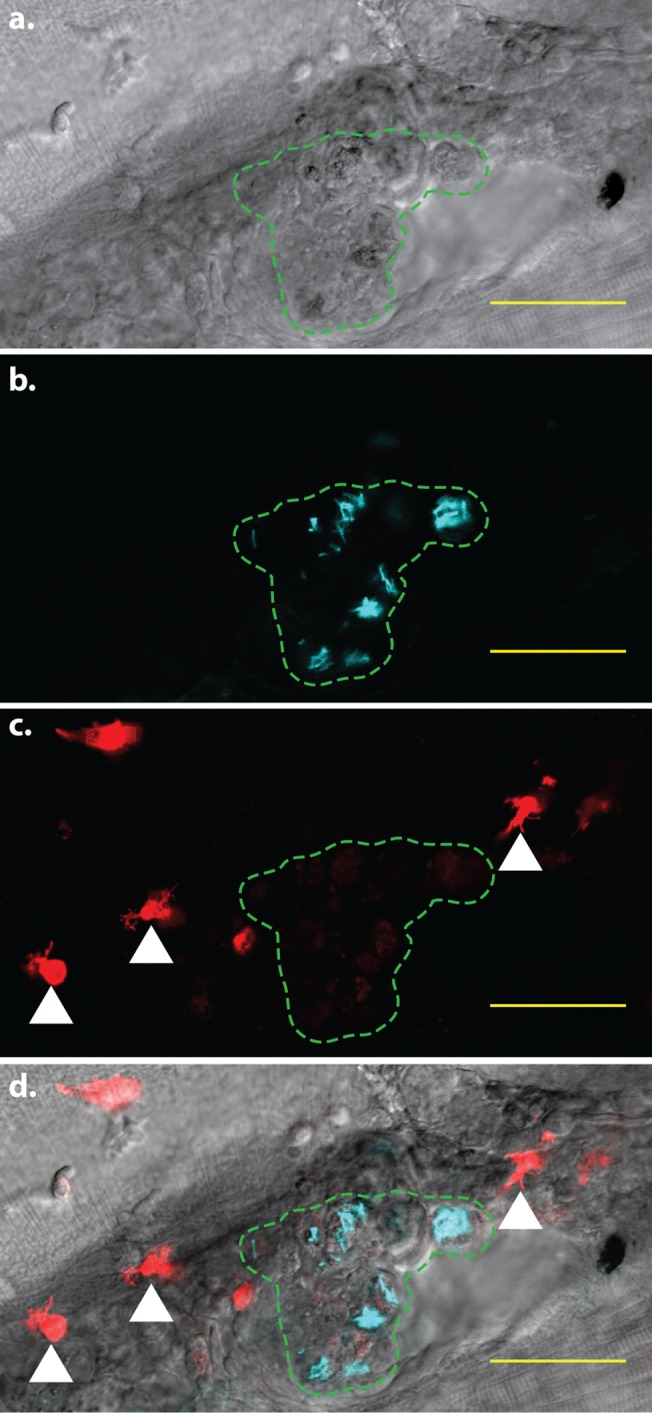
Confocal image of a larval granuloma, 5 days post-infection. The region containing infected cells is indicated by the green dashed line. Within each cell, M. marinum expressing the Cerulean fluorescent protein (cyan signal) are clearly visible. In addition, faint tdTomato fluorescence is visible within the infected cells. White arrowheads indicate examples of bright, uninfected cells exhibiting normal levels of tdTomato fluorescence. a. Brightfield detailing the granuloma in which mpeg1 fluorescent protein expression is attenuated. b. Fluorescent M. marinum in the same area c. Macrophages expressing the tdTomato fluorescent protein. d. Merged image of a-c. Scale bars = 50 μm.
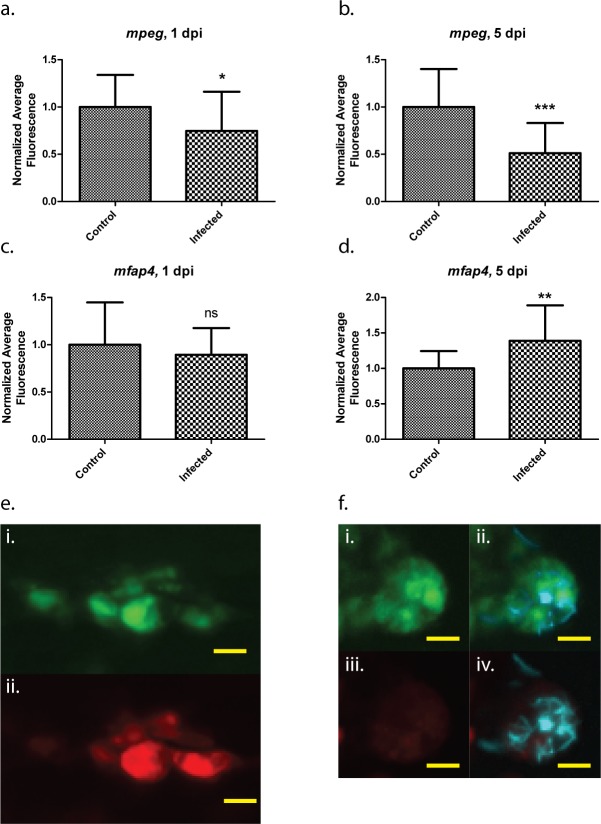
We confirmed via DIC imaging that most of these macrophages were still alive and intact (data not shown), suggesting that expression of the mpeg1 transgene itself is downregulated by infection. Similar experiments using mfap4 transgenics, however, suggested that the expression of mfap4 transgenes was independent of infection status. In order to determine the relative stability of mfap4 transgene expression in infected zebrafish larvae, the transgenic lines Tg(mpeg1:tdTomato-CAAX)xt3 and Tg(mfap4:dLanYFP-CAAX)xt11 were crossed to generate larvae simultaneously expressing both fluorescent proteins in all macrophages. The double transgenic larvae were separated into two groups: the first was infected intravenously with M. marinum and the other injected with vehicle. Fluorescence intensity of either fluorophore was measured for a randomly-selected subset of infected macrophages from the infected pool, and on macrophages from the same region within the uninfected pool. For both day 1 and day 5 post-infection, the fluorescence intensity of the mpeg1-mediated fluorophore was significantly decreased relative to controls, whereas expression of the mfap4-mediated fluorophore remained stable and even exhibited a modest increase in expression level at 5 dpi compared to controls (Fig 5). These data include both solitary and granuloma-associated infected macrophages. Notably, infected macrophages in several larvae exhibited barely detectable mpeg1 transgene fluorescence, while those same macrophages exhibited robust mfap4 transgene fluorescence in the same cell. The same disparity was not observed among macrophages in uninfected larvae, in which the fluorescence of both fluorophores remained relatively stable. We also observed that expression of either transgene appeared unperturbed in uninfected macrophages within infected larvae, although this was not assessed quantitatively. These data indicate that the altered transgene expression may be largely cell-autonomous, requiring the presence of intracellular M. marinum.
Fig 5. Differential regulation of mfap4 and mpeg1 transgenes during mycobacterial infection.
Average fluorescence values of either mpeg1:tdTomato-CAAX or mfap4:dLanYFP-CAAX fluorescent proteins relative to uninfected, age-matched controls. Five randomly chosen infected macrophages per larva (“Infected”), or five randomly chosen macrophages within uninfected larvae (“Control”) were assessed. 20 larvae were analyzed for each of the infected and control groups at 1 dpi as well as for the Infected group at 5 dpi; 19 larvae were analyzed for the Control group at 5 dpi. Both tdTomato fluorescence (a,b) and dLanYFP fluorescence (c,d) were measured for the same pool of cells from each larva. e. Representative image of mfap4 transgene expression (i) and mpeg1 transgene expression (ii) within the same group of cells in an uninfected Control larva, 5 dpi. f. Representative image of mfap4 transgene expression (i) and mpeg1 transgene expression (iii) within the same group of cells in an Infected larva, 5 dpi; also shown are merged images of fluorescent M. marinum (cyan signal) with the YFP (ii) and tdTomato (iv) channels. Note the almost total loss of mpeg1-mediated fluorescence within the infected group of cells, while mfap4 transgene fluorescence remains robust. Scale bars = 10 μm. * p < .05, ** p < .005, *** p = .0002. Student’s t-test with Welch’s correction for unequal variances. Error bars indicate +/- SD.
The mfap4:iCre:p2a-tdTomato transgenic line allows for macrophage-specific expression of novel and pre-existing Cre-compatible transgenes
We have observed that individual transgenes present in zebrafish often exhibit diminishing levels of expression as the transgene design increases in length and complexity (e.g., constructs expressing fluorophore-tagged host proteins, or constructs containing a viral 2A peptide to direct separation of two discrete protein products from a single open reading frame [41]). The lower expression level of longer transcripts is often seen despite the use of stable, lineage-specific promoter elements. To overcome this apparent limitation, as well as to enhance the utility and scope of the mfap4 promoter, we generated the transgenic line Tg(mfap4:iCre:p2A-tdTomato) xt8, resulting in macrophage-specific expression of a codon-optimized Cre recombinase (iCre) [54]; tdTomato serves to identify larvae positive for transgenesis. To assess the efficiency and cell specificity of this system, we crossed the zebrafish line Tg(β-actin2:loxP-DsRed-STOP-loxP-EGFP) s928 [55] with Tg(mfap4:iCre-p2A-tdTomato) xt8. Progeny that contained both transgenes were identified on the basis of macrophage-specific EGFP expression, indicating successful removal of the DsRed-STOP cassette in macrophages. These larvae thus exhibited strong EGFP expression confined to macrophages but under the control of the zebrafish β-actin2 promoter (Fig 6). We noted that progeny of this cross have decreased numbers of fluorescent cells, particularly within the CHT region, as compared to Tg(mfap4:dLanYFP-CAAX) xt11 larvae (Fig 6).
Fig 6. mfap4:iCre:p2A-tdTomato mediates macrophage-specific gene expression of loxP-containing transgenes.
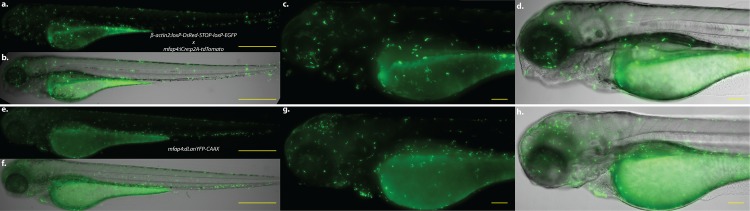
a-d. Zebrafish larva at 3 dpf expressing both Tg(β-actin2:loxP-DsRed-STOP-loxP-EGFP) s928 and Tg(mfap4:iCre:p2A-tdTomato) xt8 ; a,b. 50x magnification of GFP fluorescence and merge with brightfield, respectively. c,d. 200x magnification of the head, yolk sac, and anterior portion of the gut. e-h. Zebrafish larva at 3 dpf expressing Tg(mfap4:dLanYFP-CAAX) xt11 for comparison. e,f. 50x magnification of dLanYFP fluorescence and merge with brightfield, respectively. g,h. 200x magnification of the head, yolk sac, and anterior portion of the gut. Note that in both larvae, fluorescent protein expression is restricted to macrophages all along the body, with a particular enrichment in the head. Both larvae also exhibit expression within the caudal hematopoietic tissue (CHT). The larvae in a-d show a reduced number of fluorescent cells in the nascent macrophage population that predominates in the CHT region, and modestly reduced fluorescent cell population number in regions where mature macrophages have migrated throughout the body. Scale bars for 50x images = 500 μm; scale bars for 200x images = 100 μm.
Discussion
These data establish the mfap4 promoter element as a stable, robust genetic tool for driving macrophage-specific transgene expression in zebrafish. As a component of the Multisite Gateway Recombination system for generating transgenic constructs, this new promoter facilitates the generation of multiple transgenic constructs and subsequent zebrafish lines for which expression is restricted to macrophages. Transgenes driven by the mfap4 promoter exhibit robust, stable expression independent of larval infection status with M. marinum, making it particularly useful for the study of interactions between pathogenic mycobacteria and a vertebrate host. Beyond simple visualization of macrophages within zebrafish larvae, transgenes other than fluorescent proteins may be expressed in a macrophage-specific manner, facilitating the study of host factors that are involved in restriction or permissiveness to intracellular mycobacteria.
Using additional genetic tools and common breeding techniques (i.e., mfap4-driven Cre recombinase expression), more complex transgenic lines may be generated to further enhance transgene expression in a macrophage-restricted manner. The stronger fluorescent protein expression of these recombined transgenes may be particularly useful for long-term time-lapse imaging requiring high temporal resolution, as much lower exposure times/intensities may be used for each image resulting in significantly less photobleaching and potential phototoxicity. Furthermore, we anticipate that this system will be advantageous for driving macrophage-restricted expression of transgenes designed to rescue or overexpress biologically active proteins.
We note that the mfap4:iCre transgenic line led to strong expression in macrophages, but overall produced a reduced number of labeled macrophages in the CHT region. Because the CHT is the major site of myelopoiesis at this age, this disparity may reflect a delay in the accumulation of Cre recombinase in nascent macrophages. Alternatively, the efficiency of Cre-mediated recombination for the partner transgene tested may be less than 100 percent.
Overall, we anticipate that this promoter element as well as the repertoire of macrophage-specific transgenic lines we have established will be useful in the study of the vertebrate innate immune system. In particular, stable expression from mfap4 transgenes during infection will enable long-term imaging and manipulation of macrophages as they interact with intracellular pathogens. In addition, these transgenes should be compatible with FACS [37, 56] of macrophage populations, both uninfected and infected, from whole animals, facilitating future studies into the modulation of the macrophage transcriptome either by pathogen or host during infection with a variety of pathogens.
Supporting Information
All larvae shown are 3 dpf. Each panel pairs a representative transgenic animal with a non-transgenic animal to provide a direct comparison of background fluorescence in the relevant excitation/emission channel. a. Tg(mfap4:tdTomato) xt12. b. Tg(β-actin2:loxP-DsRed-STOP-loxP-EGFP) s928 X Tg(mfap4:iCre:p2A-tdTomato) xt8. c. Tg(mfap4:dLanYFP-CAAX)xt11. d. Tg(mfap4:tdTomato-CAAX) xt6. Scale bars = 500 μm.
(TIF)
a. Frozen 20 μm section of 3 month old adult Tg(mfap4:tdTomato-CAAX) xt6 zebrafish liver tissue, 2 weeks post-infection (wpi) with approximately 400 c.f.u. of M. marinum expressing the Cerulean (cyan) fluorescent protein. i. Macrophages expressing tdTomato. The stellate morphology expected of Kupffer cells, specialized resident macrophages of the liver, can be seen. ii. M. marinum. iii. DAPI. iv. Merge of i-iii. b. Frozen 20 μm section of kidney tissue from the same fish. i. Macrophages expressing tdTomato. ii. M. marinum. iii. DAPI. iv. Merge of i-iii. For both a and b, tdTomato (macrophage) and Cerulean (M. marinum) fluorescence signals are derived directly from fixed fluorescent protein; other than DAPI, no additional staining was performed. Scale bars = 50 μm.
(TIF)
Acknowledgments
We would like to thank D. Sisk, J. Saelens, and M. Matty for technical assistance and critical review of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by an American Cancer Society Postdoctoral Fellowship PF-13-223-01-MPC (MRC), a Mallinckrodt Scholar Award, a Searle Scholar Award, and a New Innovator Award from the National Institutes of Health, 1DP2OD008614 (DMT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236(11):3077–87. 10.1002/dvdy.21354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLOS genetics. 2006;2(11):e169 10.1371/journal.pgen.0020169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mechanisms of development. 2002;118(1–2):91–8. [DOI] [PubMed] [Google Scholar]
- 4. Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236(11):3088–99. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 5. Kikuta H, Kawakami K. Transient and stable transgenesis using tol2 transposon vectors. Methods in molecular biology. 2009;546:69–84. 10.1007/978-1-60327-977-2_5 [DOI] [PubMed] [Google Scholar]
- 6. Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117(4):e49–56. 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC developmental biology. 2007;7:42 10.1186/1471-213X-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132(23):5199–209. 10.1242/dev.02087 [DOI] [PubMed] [Google Scholar]
- 9. Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. Journal of leukocyte biology. 2006;80(6):1281–8. 10.1189/jlb.0506346 [DOI] [PubMed] [Google Scholar]
- 10. Sanderson LE, Chien AT, Astin JW, Crosier KE, Crosier PS, Hall CJ. An inducible transgene reports activation of macrophages in live zebrafish larvae. Developmental and comparative immunology. 2015. 10.1016/j.dci.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 11. Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Current opinion in immunology. 2010;22(1):10–9. 10.1016/j.coi.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cellular microbiology. 2008;10(5):1027–39. 10.1111/j.1462-5822.2008.01133.x [DOI] [PubMed] [Google Scholar]
- 13. Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell host & microbe. 2007;2(1):29–39. 10.1016/j.chom.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oehlers SH, Cronan MR, Scott NR, Thomas MI, Okuda KS, Walton EM, et al. Interception of host angiogenic signalling limits mycobacterial growth. Nature. 2015;517(7536):612–5. 10.1038/nature13967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tobin DM, Vary JC Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140(5):717–30. 10.1016/j.cell.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benard EL, Racz PI, Rougeot J, Nezhinsky AE, Verbeek FJ, Spaink HP, et al. Macrophage-expressed perforins mpeg1 and mpeg1.2 have an anti-bacterial function in zebrafish. Journal of innate immunity. 2015;7(2):136–52. 10.1159/000366103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Torraca V, Masud S, Spaink HP, Meijer AH. Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. Disease models & mechanisms. 2014;7(7):785–97. 10.1242/dmm.015594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17(6):693–702. [DOI] [PubMed] [Google Scholar]
- 19. Clay H, Volkman HE, Ramakrishnan L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity. 2008;29(2):283–94. 10.1016/j.immuni.2008.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, et al. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cellular microbiology. 2009;11(5):755–68. 10.1111/j.1462-5822.2009.01288.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clatworthy AE, Lee JS, Leibman M, Kostun Z, Davidson AJ, Hung DT. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infection and immunity. 2009;77(4):1293–303. 10.1128/IAI.01181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(18):7622–7. 10.1073/pnas.0702386104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Sar AM, Musters RJ, van Eeden FJ, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cellular microbiology. 2003;5(9):601–11. [DOI] [PubMed] [Google Scholar]
- 24. van der Sar AM, Stockhammer OW, van der Laan C, Spaink HP, Bitter W, Meijer AH. MyD88 innate immune function in a zebrafish embryo infection model. Infection and immunity. 2006;74(4):2436–41. 10.1128/IAI.74.4.2436-2441.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levraud JP, Disson O, Kissa K, Bonne I, Cossart P, Herbomel P, et al. Real-time observation of listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infection and immunity. 2009;77(9):3651–60. 10.1128/IAI.00408-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menudier A, Rougier FP, Bosgiraud C. Comparative virulence between different strains of Listeria in zebrafish (Brachydanio rerio) and mice. Pathologie-biologie. 1996;44(9):783–9. [PubMed] [Google Scholar]
- 27. Brothers KM, Newman ZR, Wheeler RT. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryotic cell. 2011;10(7):932–44. 10.1128/EC.05005-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berthier E, Lim FY, Deng Q, Guo CJ, Kontoyiannis DP, Wang CC, et al. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLOS Pathogens. 2013;9(4):e1003289 10.1371/journal.ppat.1003289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gabor KA, Goody MF, Mowel WK, Breitbach ME, Gratacap RL, Witten PE, et al. Influenza A virus infection in zebrafish recapitulates mammalian infection and sensitivity to anti-influenza drug treatment. Disease models & mechanisms. 2014;7(11):1227–37. 10.1242/dmm.014746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palha N, Guivel-Benhassine F, Briolat V, Lutfalla G, Sourisseau M, Ellett F, et al. Real-time whole-body visualization of Chikungunya Virus infection and host interferon response in zebrafish. PLoS pathogens. 2013;9(9):e1003619 10.1371/journal.ppat.1003619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berman JN, Kanki JP, Look AT. Zebrafish as a model for myelopoiesis during embryogenesis. Experimental hematology. 2005;33(9):997–1006. 10.1016/j.exphem.2005.06.010 [DOI] [PubMed] [Google Scholar]
- 32. Renshaw SA, Trede NS. A model 450 million years in the making: zebrafish and vertebrate immunity. Disease models & mechanisms. 2012;5(1):38–47. 10.1242/dmm.007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–8. 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen Chi M, Laplace-Builhe B, Travnickova J, Luz-Crawford P, Tejedor G, Phan QT, et al. Identification of polarized macrophage subsets in zebrafish. eLife. 2015;4 10.7554/eLife.07288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126(17):3735–45. [DOI] [PubMed] [Google Scholar]
- 36. Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Developmental biology. 2001;238(2):274–88. 10.1006/dbio.2001.0393 [DOI] [PubMed] [Google Scholar]
- 37. Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116(3):e1–11. 10.1182/blood-2010-01-262873 [DOI] [PubMed] [Google Scholar]
- 38. Gray C, Loynes CA, Whyte MK, Crossman DC, Renshaw SA, Chico TJ. Simultaneous intravital imaging of macrophage and neutrophil behaviour during inflammation using a novel transgenic zebrafish. Thrombosis and haemostasis. 2011;105(5):811–9. 10.1160/TH10-08-0525 [DOI] [PubMed] [Google Scholar]
- 39. Shiau CE, Kaufman Z, Meireles AM, Talbot WS. Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. PLOS ONE. 2015;10(1):e0117513 10.1371/journal.pone.0117513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nature methods. 2010;7(11):893–5. 10.1038/nmeth.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus–1 in human cell lines, zebrafish and mice. PloS one. 2011;6(4):e18556 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. The EMBO journal. 1991;10(13):4033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63(1):133–9. [DOI] [PubMed] [Google Scholar]
- 44. Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes & development. 2005;19(13):1512–7. 10.1101/gad.1302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen H. UAP56- a key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB reports. 2009;42(4):185–8. [DOI] [PubMed] [Google Scholar]
- 46. Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(9):3386–91. 10.1073/pnas.0800250105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome research. 2004;14(10B):2111–20. 10.1101/gr.2512204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome research. 2000;10(11):1788–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature biotechnology. 2004;22(12):1567–72. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- 50. Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature methods. 2013;10(5):407–9. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nature communications. 2012;3:751 10.1038/ncomms1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mathias JR, Dodd ME, Walters KB, Yoo SK, Ranheim EA, Huttenlocher A. Characterization of zebrafish larval inflammatory macrophages. Developmental and comparative immunology. 2009;33(11):1212–7. 10.1016/j.dci.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annual review of pathology. 2012;7:353–84. 10.1146/annurev-pathol-011811-132458 [DOI] [PubMed] [Google Scholar]
- 54. Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32(1):19–26. [DOI] [PubMed] [Google Scholar]
- 55. Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–5. 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hsu K, Traver D, Kutok JL, Hagen A, Liu TX, Paw BH, et al. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104(5):1291–7. 10.1182/blood-2003-09-3105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All larvae shown are 3 dpf. Each panel pairs a representative transgenic animal with a non-transgenic animal to provide a direct comparison of background fluorescence in the relevant excitation/emission channel. a. Tg(mfap4:tdTomato) xt12. b. Tg(β-actin2:loxP-DsRed-STOP-loxP-EGFP) s928 X Tg(mfap4:iCre:p2A-tdTomato) xt8. c. Tg(mfap4:dLanYFP-CAAX)xt11. d. Tg(mfap4:tdTomato-CAAX) xt6. Scale bars = 500 μm.
(TIF)
a. Frozen 20 μm section of 3 month old adult Tg(mfap4:tdTomato-CAAX) xt6 zebrafish liver tissue, 2 weeks post-infection (wpi) with approximately 400 c.f.u. of M. marinum expressing the Cerulean (cyan) fluorescent protein. i. Macrophages expressing tdTomato. The stellate morphology expected of Kupffer cells, specialized resident macrophages of the liver, can be seen. ii. M. marinum. iii. DAPI. iv. Merge of i-iii. b. Frozen 20 μm section of kidney tissue from the same fish. i. Macrophages expressing tdTomato. ii. M. marinum. iii. DAPI. iv. Merge of i-iii. For both a and b, tdTomato (macrophage) and Cerulean (M. marinum) fluorescence signals are derived directly from fixed fluorescent protein; other than DAPI, no additional staining was performed. Scale bars = 50 μm.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.