Abstract
Although the general pathway of sex pheromone synthesis in moth species has been established, the molecular mechanisms remain poorly understood. The common cutworm Spodoptera litura is an important agricultural pest worldwide and causes huge economic losses annually. The female sex pheromone of S. litura comprises Z9,E11-14:OAc, Z9,E12-14:OAc, Z9-14:OAc, and E11-14:OAc. By sequencing and analyzing the transcriptomic data of the sex pheromone glands, we identified 94 candidate genes related to pheromone biosynthesis (55 genes) or chemoreception (39 genes). Gene expression patterns and phylogenetic analysis revealed that two desaturase genes (SlitDes5 and SlitDes11) and one fatty acyl reductase gene (SlitFAR3) showed pheromone gland (PG) biased or specific expression, and clustered with genes known to be involved in pheromone synthesis in other moth species. Furthermore, 4 chemoreception related genes (SlitOBP6, SlitOBP11, SlitCSP3, and SlitCSP14) also showed higher expression in the PG, and could be additional candidate genes involved in sex pheromone transport. This study provides the first solid background information that should facilitate further elucidation of sex pheromone biosynthesis and transport, and indicates potential targets to disrupt sexual communication in S. litura for a novel pest management strategy.
Introduction
Species-specific sex pheromone-elicited behaviors play a key role in sexual communication and reproduction in most moth species, which therefore serve as a good model to study reproductive isolation in animals, from insects to mammals [1–3]. Moth sex pheromones are biosynthesized and released by specialized sex pheromone glands (PGs) that are located along the inter-segmental membrane between the 8th and 9th abdominal segments of females [4, 5]. In most moths, sex pheromone components are composed of C10 –C18 unsaturated acyclic aliphatic compounds with a functional group such as formyl, hydroxyl, or acyloxyl [6, 7].
Sex pheromone biosynthesis in moths begins with a palmitic or stearic acid moiety that is synthesized de novo in the PG through modifications of the fatty acid biosynthetic pathway [8]. Through a series of enzymatic reactions such as desaturation, chain-shortening reaction, reduction, acetylation, and oxidation, the palmitic or stearic acids are then converted to the final pheromone components in a step-wise manner [5, 9, 10]. Therefore, different enzymes are likely to be involved in the different reactions, and to date, the genes encoding 4 different classes of enzymes that are essential for this pathway have been functionally identified—desaturases (Des), fatty acid reductases (FARs), fatty acid transport proteins (FATPs), and acyl-CoaA-binding proteins (ACBPs). Among these, Des proteins are the most intensively studied class of enzymes involved in moth sex pheromone biosynthesis, which can introduce double bonds into pheromone precursors. Previous studies have demonstrated the broad functional diversity of these enzymes such as Δ5 [11, 12], Δ6 [13], Δ9 [9, 14], Δ11 [15, 16], Δ10–12 [10], and Δ14 desaturase [17]. FARs are responsible for reducing fatty acids to alcohols, and have also been functionally identified in a few moth species, including Bombyx mori [18], Ostrinia nubilalis [19] and 4 Heliothine species [20]. FATPs and ACBPs have been functionally identified to play roles in the production of the B. mori sex pheromone bombykol based on in vivo RNA interference methods [21, 22]. In addition, some other important enzymes that have not been functionally confirmed are postulated to be involved in the sex pheromone biosynthesis pathway. For example, biochemical studies have suggested that acetyltransferase (ACT) and alcohol oxidase (AO) play a role by converting alcohol into acetate ester [23] and by oxidizing alcohol into the corresponding aldehyde component [13, 24], respectively.
The common cutworm Spodoptera litura (Lepidoptera: Noctuidae) is an important agricultural pest worldwide and causes huge economic losses annually. The female sex pheromones of S. litura have been identified as a blend of Z9-E11-14:OAc, Z9-E12-14:OAc, Z9-14:OAc, and E11-14:OAc with a ratio of 100:27:20:27 in China [25]. To date, only 9 chemoreception genes have been identified and functionally characterized, including 5 odorant-binding proteins (OBPs) [26, 27] and 4 pheromone receptors (PRs) [28]. However, the genes involved in the pheromone biosynthesis of S. litura have not been explored. In present study, we constructed a genetic database of the genes expressed in the female PGs of S. litura using the Illumina HiSeq(TM) 2500 sequencing platform. In total, we identified 94 genes that are possibly related to pheromone biosynthesis (55 genes) or chemoreception (39 genes). Furthermore, tissue expression evaluation and phylogenetic analyses were performed to postulate the functions of the identified genes. The results indicated that some of these genes might play crucial roles in the biosynthesis and transport of S. litura sex pheromones and could be as candidates for further functional studies.
Results
Transcriptome Sequencing and Sequence Assembly
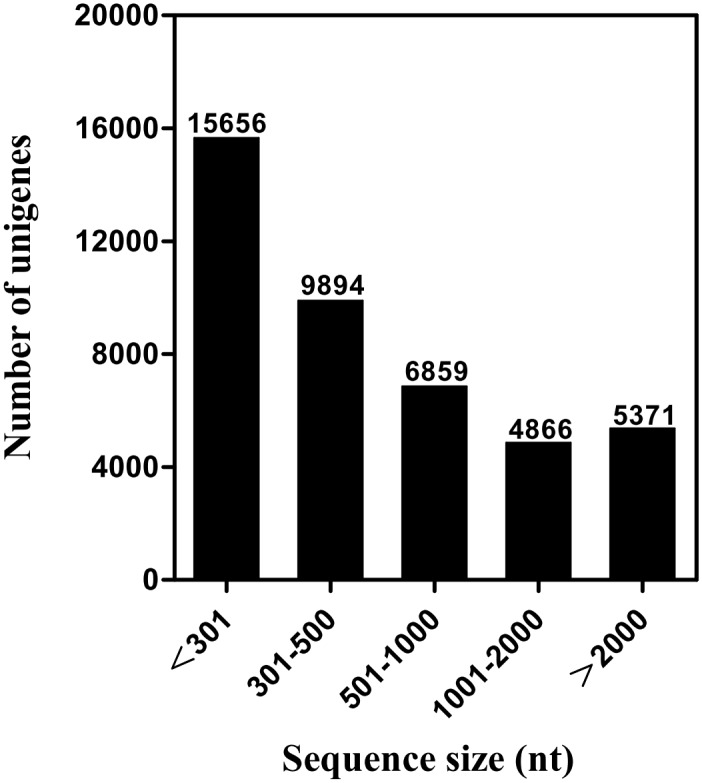
We carried out a next-generation sequencing analysis using a cDNA library constructed from the female PGs of S. litura using the Illumina HiSeq(TM) 2500 platform. The transcriptome sequencing provided approximately 63 million reads (6.3 Gb). After clustering and redundancy filtering, we finally acquired 42,646 unigenes with an N50 length of 2,093 bp (Table 1). We called these 42,646 ones unigenses according to some recently published papers [29, 30], although each of them may not necessarily represents a unique gene. Of the 42,646 unigenes, those with a sequence length of more than 500 bp accounted for 40.08% of the total transcriptome assembly (Fig 1).
Table 1. Summary of S. litura transcriptome assembly.
| Statistics Project | Number |
|---|---|
| Total clean reads | 63,209,172 |
| GC percentage | 44.69% |
| Q20 percentage | 97.38% |
| Total unigene nucleotides | 39,831,030 |
| Total unigene | 42,646 |
| N50 of unigenes (nt) | 2,093 |
| Min length of unigenes (nt) | 201 |
| Median length of unigenes (nt) | 385 |
| Max length of unigenes (nt) | 41,274 |
| Unigenes with homolog in NR | 14,370 |
Fig 1. Distribution of unigene size in the S. litura transcriptome assembly.
Gene Ontology (GO) Annotation
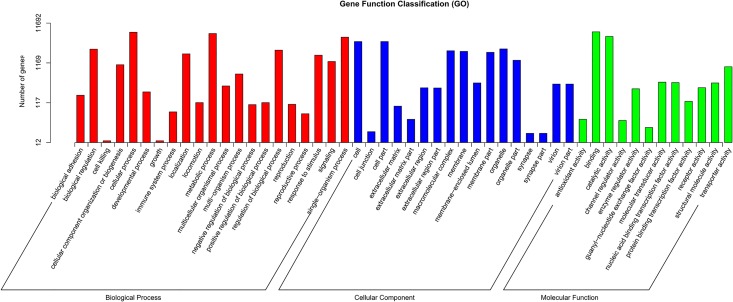
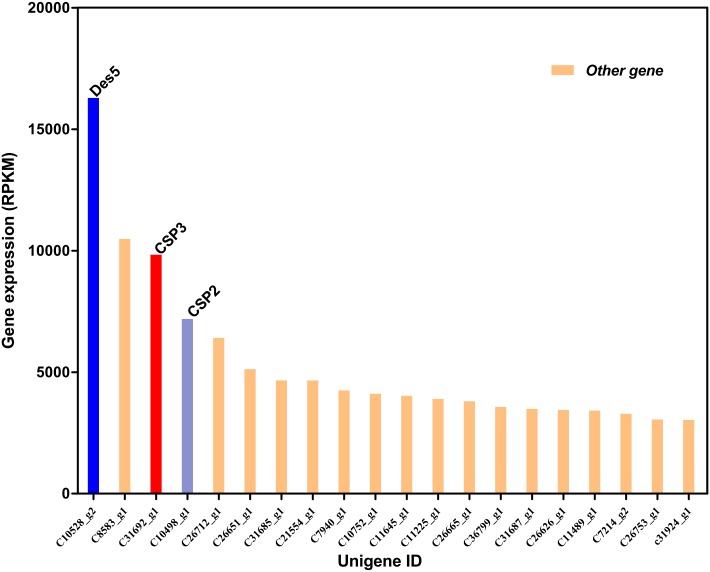
GO annotation was used to classify the transcripts into functional groups according to GO categories. Of the 42,646 unigenes, 11,692 (27.41%) could be annotated based on sequence homology. In the molecular function category, the genes expressed in the PG were mostly enriched to binding, catalytic activity and transporter activity. Cellular and metabolic processes were the most highly represented in the biological process categories, and cell, cell part, and organelle were most abundantly represented in the cellular component category (Fig 2). In addition, the Des gene SlitDes5 was the most abundant of all unigenes, and two CSP genes (SlitCSP3 and SlitCSP2) also showed very high abundance (Fig 3 and S1 Table).
Fig 2. Gene ontology (GO) classification of the S. litura transcripts with Blast2GO program.
Fig 3. Top 20 most abundant transcripts in the S. litura transcriptome dataset.
The genes expression abundance is indicated as the Reads Per Kilobase per Million mapped reads (RPKM) values. The transcript annotation by homologous comparisons with Blastx is indicated in Tables 3 and 4 and S1 Table.
Identification of Putative Genes Related to Sex Pheromone Biosynthesis and Transport
By homology analysis using the reported genes of other moth species in NCBI as queries [31–36], we identified a total of 94 transcripts that belong to gene families that are putatively involved in sex pheromone biosynthesis and transport, including 12 Des, 13 FAR, 23 ACT, 2 ACBP, 4 FATP, 1 ACC, 25 OBP, and 14 CSP genes. In comparison to sequences of other insects, we found that the number of Des genes in S. litura (12) is close to H. assulta (8) [35] and H. virescens (9) [31], but is lower than that of B. mori (24) (GenBank data) and Ephestia cautella (21) [36]. Furthermore, the number of FAR genes in S. litura (13) is the same as that in H. assulta (13) [35] and Agrotis ipsilon (13) [32], but is less than that of H. armigera (18) [35] and E. cautella (28) [36]. The numbers of OBP (25) and CSP genes (14) in S. litura are similar to those in H. armigera (26 and 19) [35] and H. assulta (23 and 16) [35], but are more than those in other moth species [31, 32, 34, 36] (Tables 2–4).
Table 2. The number of sex pheromone biosynthesis and transport related genes in different moths.
| Species | Sex pheromone biosynthesis | Chemoreception | ||||||
|---|---|---|---|---|---|---|---|---|
| Des | FAR | ACT | ACC | ACBP | FATP | OBP | CSP | |
| Spodoptera litura | 12 (9) | 13 (11) | 23 (16) | 1 (0) | 2 (1) | 4 (1) | 25 (2) | 14 (2) |
| Heliothis virescens | 9 | 5 | 3 | 1 | — | — | 9 | 16 |
| Agrotis ipsilon | 5 (2) | 13 (3) | 5 (3) | 2 (2) | — | — | 7 (1) | 8 (1) |
| Chilo suppressalis | 6 (2) | 10 (1) | — | 1 (0) | 3 (0) | 4 (0) | 9 (0) | 10 (2) |
| Helicoverpa armigera | 7 (2) | 18 (12) | — | 2 (1) | — | — | 26 (1) | 19 (5) |
| Helicoverpa assulta | 8 (3) | 13 (11) | — | 2 (1) | — | — | 23 (3) | 16 (2) |
| Ephestia cautella | 21 | 18 | 18 | 6 | — | 5 | 17 | 7 |
Note: Digits in parentheses indicate the number of genes with PG-specific or biased expression. “—” indicates the gene has not been reported. The expression profiles of genes in PGs and other tissues of H. virescens and E. cautella had not been reported. All of the data are based on PGs transcriptome analyses, and the references are [31, 32, 34, 35, 36].
Table 4. The Blastx match of S. litura chemoreception genes.
| Gene Name | Acc. No. | ORF (aa) | Signal Peptide | Complete ORF | Best Blastx Match | ||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Acc. No. | Species | E value | Identity (%) | |||||
| Odorant Binding Protein (OBP) | |||||||||
| OBP1 | KT261647 | 147 | 1–20 | Y | odorant binding protein 6 | AFM77984.1 | Spodoptera exigua | 9.10E-80 | 78 |
| OBP2 | KT261648 | 156 | 1–18 | Y | odorant-binding protein 2 | XP_004923270.1 | Chilo suppressalis | 2.00E-16 | 31 |
| OBP3 | KT261649 | 104 | 1–17 | N | odorant binding protein 11 | AGH70107.1 | Spodoptera exigua | 2.00E-13 | 100 |
| OBP4 | KT261650 | 167 | 1–16 | Y | odorant binding protein | AII00985.1 | Dendrolimus houi | 2.00E-07 | 32 |
| OBP5 | KT261651 | 145 | 1–24 | Y | SexiOBP13 | AGP03459.1 | Spodoptera exigua | 4.00E-79 | 84 |
| OBP6 | KT261652 | 158 | 1–26 | Y | SexiOBP8 | AGP03454.1 | Spodoptera exigua | 2.00E-42 | 58 |
| OBP7 | KT261653 | 213 | 1–18 | Y | odorant-binding protein 19 | AGC92793.1 | Helicoverpa assulta | 3.00E-56 | 54 |
| OBP8 | KT261654 | 184 | 1–20 | Y | odorant binding protein | AII00978.1 | Dendrolimus houi | 7.00E-109 | 92 |
| OBP9 | KT261655 | 239 | 1–19 | Y | odorant binding protein fmxg18C17 | NP_001157372.1 | Bombyx mori | 1.00E-70 | 50 |
| OBP10 | KT261656 | 115 | N | N | odorant-binding protein 1 | AFG72998.1 | Cnaphalocrocis medinalis | 8.00E-71 | 83 |
| OBP11 | KT261657 | 149 | 1–21 | Y | SexiOBP8 | AGP03454.1 | Spodoptera exigua | 4.00E-10 | 35 |
| OBP12 | KT261658 | 74 | N | N | SexiOBP14 | AGP03460.1 | Spodoptera exigua | 3.00E-46 | 90 |
| OBP13 | KT261659 | 129 | 1–17 | Y | odorant binding protein | ADY17884.1 | Spodoptera exigua | 1.00E-56 | 81 |
| OBP14 | KT261660 | 193 | 1–17 | Y | odorant binding protein 1 | AGR39564.1 | Agrotis ipsilon | 1.00E-72 | 58 |
| OBP15 | KT261661 | 147 | 1–17 | Y | odorant binding protein 6 | AGR39569.1 | Agrotis ipsilon | 8.00E-60 | 76 |
| OBP16 | KT261662 | 137 | 1–19 | Y | antennal binding protein X | CAA05508.1 | Heliothis virescens | 1.00E-62 | 92 |
| OBP17 | KT261663 | 147 | 1–21 | Y | antennal binding protein | ADY17881.1 | Spodoptera exigua | 9.00E-90 | 91 |
| OBP18 | KT261664 | 139 | 1–18 | Y | OBP8 | AEB54589.1 | Helicoverpa armigera | 1.00E-81 | 84 |
| OBP19 | KT261665 | 52 | N | N | odorant binding protein 11 | AGH70107.1 | Spodoptera exigua | 1.00E-32 | 92 |
| OBP20 | KT261666 | 147 | 1–21 | Y | odorant-binding protein | AAR28762.1 | Spodoptera exigua | 1.00E-78 | 93 |
| OBP21 | KT261667 | 240 | 1–20 | Y | odorant binding protein fmxg18C17 | NP_001157372.1 | Bombyx mori | 2.00E-37 | 43 |
| OBP22 | KT261668 | 133 | 1–16 | Y | odorant binding protein 9 | AGH70105.1 | Spodoptera exigua | 1.00E-85 | 95 |
| OBP23 | KT261669 | 142 | 1–21 | Y | odorant binding protein 7 | AGH70103.1 | Spodoptera exigua | 6.00E-93 | 97 |
| OBP24 | KT261670 | 83 | 1–24 | N | SexiOBP12 | AGP03458.1 | Spodoptera exigua | 9.00E-42 | 88 |
| OBP25 | AIS72934.1 | 148 | N | Y | pheromone-binding protein 3 | AIS72934.1 | Spodoptera litura | 3.00E-116 | 100 |
| Chemosensory Protein (CSP) | |||||||||
| CSP1 | KT261672 | 122 | 1–16 | Y | chemosensory protein 10 | AFR92094.1 | Helicoverpa armigera | 1.00E-63 | 91 |
| CSP2 | KT261673 | 128 | 1–18 | Y | chemosensory protein CSP2 | ABM67689.1 | Spodoptera exigua | 3.00E-71 | 96 |
| CSP3 | KT261674 | 123 | 1–18 | Y | chemosensory protein 8 | AGR39578.1 | Agrotis ipsilon | 1.00E-70 | 89 |
| CSP4 | KT261675 | 287 | 1–16 | Y | chemosensory protein | AIW65104.1 | Helicoverpa armigera | 9.00E-125 | 83 |
| CSP5 | KT261676 | 128 | 1–18 | Y | chemosensory protein CSP1 | ABM67688.1 | Spodoptera exigua | 7.00E-82 | 94 |
| CSP6 | KT261677 | 123 | 1–16 | Y | chemosensory protein | AIW65100.1 | Helicoverpa armigera | 2.00E-59 | 81 |
| CSP7 | KT261678 | 113 | 1–16 | Y | sensory appendage protein-likeprotein | AAK14793.1 | Mamestra brassicae | 2.00E-37 | 62 |
| CSP8 | KT261679 | 128 | 1–16 | Y | chemosensory protein | AIU68827.1 | Chilo auricilius | 1.00E-83 | 98 |
| CSP9 | KT261680 | 107 | 1–18 | Y | chemosensory protein 5 | AGR39575.1 | Agrotis ipsilon | 4.00E-53 | 93 |
| CSP10 | KT261681 | 127 | 1–17 | Y | chemosensory protein 6 | AGR39576.1 | Agrotis ipsilon | 3.00E-60 | 87 |
| CSP11 | KT261682 | 78 | 1–19 | N | chemosensory protein 13 | BAG71921.1 | Papilio xuthus | 5.00E-32 | 77 |
| CSP12 | KT261683 | 120 | 1–16 | Y | CSP2 | AEX07265.1 | Helicoverpa armigera | 9.00E-75 | 91 |
| CSP13 | KT261684 | 123 | 1–18 | Y | chemosensory protein 8 | AFR92092.1 | Helicoverpa armigera | 2.00E-37 | 67 |
| CSP14 | AAY26143.1 | 126 | 1–16 | Y | chemosensory protein CSP | AAY26143.1 | Spodoptera litura | 2.00E-75 | 100 |
Of the 94 identified genes (Tables 3 and 4), the sequences of 5 genes were identical to those already deposited in GenBank: 3 SlitDes genes (GenBank Accession No.: AGH12217.1, AGH12217.1 and AGH12218.1), 1 SlitOBP (GenBank Accession No.: AIS72934.1), and 1 SlitCSP (GenBank Accession No.: AAY26143.1), whereas the other 89 transcripts found in the current study were new in S. litura.
Table 3. The Blastx match of S. litura sex pheromone biosynthesis genes.
| Gene Name | Acc. No. | ORF (aa) | Complete ORF | Best Blastx Match | ||||
|---|---|---|---|---|---|---|---|---|
| Name | Acc. No. | Species | E value | Identity (%) | ||||
| Desaturase (Des) | ||||||||
| Des1 | KT261685 | 38 | N | acyl-CoA delta 11 desaturaseacyl | AGR49312.1 | Agrotis ipsilon | 3.00E-11 | 54 |
| Des2 | KT261686 | 376 | Y | desaturase | AAQ74260.1 | Spodoptera littoralis | 0.00E+00 | 99 |
| Des3 | KT261687 | 444 | Y | desaturase | AID66662.1 | Agrotis segetum | 0.00E+00 | 80 |
| Des4 | KT261688 | 335 | Y | desaturase | AID66661.1 | Agrotis segetum | 0.00E+00 | 90 |
| Des5 | AGH12217.1 | 338 | Y | delta 11 desaturase | AGH12217.1 | Spodoptera litura | 0.00E+00 | 100 |
| Des6 | KT261689 | 50 | N | delta 11 desaturase | AGH12217.1 | Spodoptera litura | 2.00E-28 | 68 |
| Des7 | KT261690 | 84 | N | delta–9 desaturase 14–26 | AFO38465.1 | Spodoptera exigua | 1.00E-79 | 100 |
| Des8 | KT261691 | 321 | Y | desaturase | AID66658.1 | Agrotis segetum | 0.00E+00 | 93 |
| Des9 | AGH12218.1 | 353 | Y | delta 9 desaturase | AGH12218.1 | Spodoptera litura | 0.00E+00 | 100 |
| Des10 | KT261692 | 451 | Y | desaturase | AID66663.1 | Agrotis segetum | 0.00E+00 | 90 |
| Des11 | KT261693 | 353 | Y | delta–9 desaturase | AAQ74258.1 | Spodoptera littoralis | 0.00E+00 | 99 |
| Des12 | KT261694 | 296 | N | acyl-CoA Delta(11) | NP_001274329.1 | Bombyx mori | 2.00E-158 | 73 |
| Fatty-Acyl Reductase (FAR) | ||||||||
| FAR1 | KT261695 | 535 | Y | fatty acyl reductase | AID66649.1 | Agrotis segetum | 9.10E-80 | 84 |
| FAR2 | KT261696 | 234 | N | fatty-acyl CoA reductase 6 | XP_004923270.1 | Agrotis ipsilon | 2.00E-133 | 87 |
| FAR3 | KT261697 | 454 | Y | Far1 protein | CDG50833.1 | Spodoptera littoralis | 0.00E+00 | 99 |
| FAR4 | KT261698 | 250 | N | fatty acyl reductase | AID66654.1 | Agrotis segetum | 3.00E-164 | 90 |
| FAR5 | KT261699 | 522 | N | fatty acyl-CoA reductase CG5065-like | XP_004925987.1 | Bombyx mori | 0.00E+00 | 49 |
| FAR6 | KT261700 | 520 | Y | fatty-acyl CoA reductase 6 | AGR49316.1 | Agrotis ipsilon | 0.00E+00 | 67 |
| FAR7 | KT261701 | 520 | Y | fatty acyl-CoA reductase CG5065-like | XP_004929961.1 | Bombyx mori | 0.00E+00 | 78 |
| FAR8 | KT261702 | 526 | Y | fatty acyl reductase | AID66652.1 | Agrotis segetum | 0.00E+00 | 89 |
| FAR9 | KT261703 | 231 | N | fatty acyl reductase | AID66654.1 | Agrotis segetum | 5.00E-158 | 88 |
| FAR10 | KT261704 | 624 | Y | fatty acyl reductase | AID66650.1 | Agrotis segetum | 0.00E+00 | 85 |
| FAR11 | KT261705 | 512 | Y | fatty acyl reductase | AID66647.1 | Agrotis segetum | 0.00E+00 | 55 |
| FAR12 | KT261706 | 510 | Y | fatty acyl-CoA reductase | AGR49318.1 | Agrotis ipsilon | 0.00E+00 | 90 |
| FAR13 | KT261707 | 246 | N | fatty acyl reductase | AID66647.1 | Agrotis segetum | 0.00E+00 | 80 |
| Acetyltransferase (ACT) | ||||||||
| ACT1 | KT261708 | 396 | Y | fatty alcohol acetyltransferase | AIN34689.1 | Agrotis segetum | 0.00E+00 | 94 |
| ACT2 | KT261709 | 383 | Y | fatty alcohol acetyltransferase | AIN34698.1 | Agrotis segetum | 0.00E+00 | 96 |
| ACT3 | KT261710 | 719 | Y | fatty alcohol acetyltransferase | AIN34709.1 | Agrotis segetum | 0.00E+00 | 65 |
| ACT4 | KT261711 | 271 | Y | acyltransferase AGPAT2 | AGG54993.1 | Heliothis virescens | 9.00E-178 | 90 |
| ACT5 | KT261712 | 397 | Y | fatty alcohol acetyltransferase | AIN34682.1 | Agrotis segetum | 0.00E+00 | 73 |
| ACT6 | KT261713 | 714 | Y | fatty alcohol acetyltransferase | AIN34685.1 | Agrotis segetum | 0.00E+00 | 99 |
| ACT7 | KT261714 | 276 | N | fatty alcohol acetyltransferase | AIN34682.1 | Agrotis segetum | 1.00E-165 | 90 |
| ACT8 | KT261715 | 180 | Y | acetyltransferase | AGQ45625.1 | Agrotis ipsilo | 1.00E-124 | 97 |
| ACT9 | KT261716 | 471 | Y | fatty alcohol acetyltransferase | AIN34706.1 | Agrotis segetum | 9.00E-177 | 96 |
| ACT10 | KT261717 | 479 | Y | fatty alcohol acetyltransferase | AIN34712.1 | Agrotis segetum | 0.00E+00 | 94 |
| ACT11 | KT261718 | 431 | Y | fatty alcohol acetyltransferase | AIN34699.1 | Agrotis segetum | 0.00E+00 | 95 |
| ACT12 | KT261719 | 477 | Y | fatty alcohol acetyltransferase | AIN34710.1 | Agrotis segetum | 0.00E+00 | 84 |
| ACT13 | KT261720 | 373 | N | fatty alcohol acetyltransferase | AIN34702.1 | Agrotis segetum | 4.00E-25 | 71 |
| ACT14 | KT261721 | 652 | Y | fatty alcohol acetyltransferase | AIN34708.1 | Agrotis segetum | 0.00E+00 | 89 |
| ACT15 | KT261722 | 95 | N | fatty alcohol acetyltransferase | AIN34682.1 | Agrotis segetum | 3.00E-52 | 85 |
| ACT16 | KT261723 | 282 | Y | acyltransferase AGPAT5 | AGG55013.1 | Heliothis subflexa | 4.00E-166 | 89 |
| ACT17 | KT261724 | 359 | Y | fatty alcohol acetyltransferase | AIN34693.1 | Agrotis segetum | 0.00E+00 | 89 |
| ACT18 | KT261725 | 355 | Y | fatty alcohol acetyltransferase | AIN34705.1 | Agrotis segetum | 0.00E+00 | 90 |
| ACT19 | KT261726 | 390 | Y | fatty alcohol acetyltransferase | AIN34704.1 | Agrotis segetum | 0.00E+00 | 88 |
| ACT20 | KT261727 | 330 | Y | fatty alcohol acetyltransferase | AIN34713.1 | Agrotis segetum | 0.00E+00 | 83 |
| ACT21 | KT261728 | 400 | Y | fatty alcohol acetyltransferase | AIN34683.1 | Agrotis segetum | 0.00E+00 | 92 |
| ACT22 | KT261729 | 480 | Y | fatty alcohol acetyltransferase | AIN34694.1 | Agrotis segetum | 0.00E+00 | 83 |
| ACT23 | KT261730 | 504 | Y | fatty alcohol acetyltransferase | EFN73032.1 | Agrotis segetum | 4.00E-128 | 47 |
| Acetyl-CoA Carboxylase (ACC) | ||||||||
| ACC | KT261731 | 2385 | Y | acetyl-CoA carboxylase-like | AID66639.1 | Agrotis segetum | 0.00E+00 | 94 |
| Fatty Acid Transport Protein (FATP) | ||||||||
| FATP1 | KT261734 | 700 | Y | fatty acid transport protein 1 | AII21952.1 | Sesamia inferens | 0.00E+00 | 90 |
| FATP2 | KT261735 | 651 | Y | fatty acid transport protein 2 | AII21953.1 | Sesamia inferens | 0.00E+00 | 89 |
| FATP3 | KT261736 | 661 | Y | fatty acid transport protein 3 | AII21954.1 | Sesamia inferens | 0.00E+00 | 86 |
| FATP4 | KT261737 | 643 | Y | fatty acid transport protein 4 | AII21955.1 | Sesamia inferens | 0.00E+00 | 93 |
| Acyl-CoA Binding Protein (ACBP) | ||||||||
| ACBP1 | KT261732 | 255 | Y | acyl-CoA binding protein 3 | AII21948.1 | Sesamia inferens | 8.00E-141 | 83 |
| ACBP2 | KT261733 | 90 | Y | acyl-CoA binding protein 1 | AII21946.1 | Sesamia inferens | 1.00E-45 | 92 |
Expression Profile of the Putative Genes Related to Sex Pheromone Biosynthesis and Transport
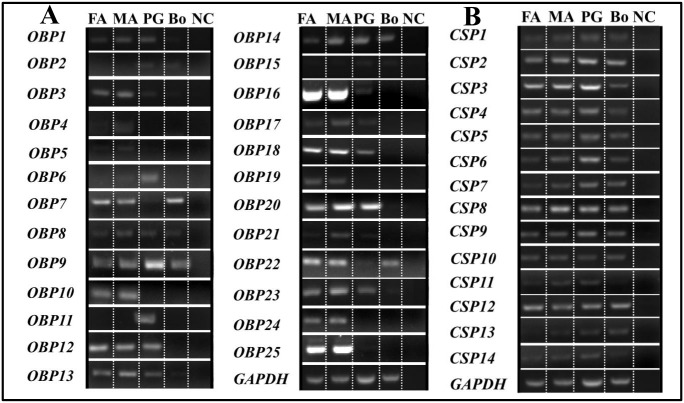
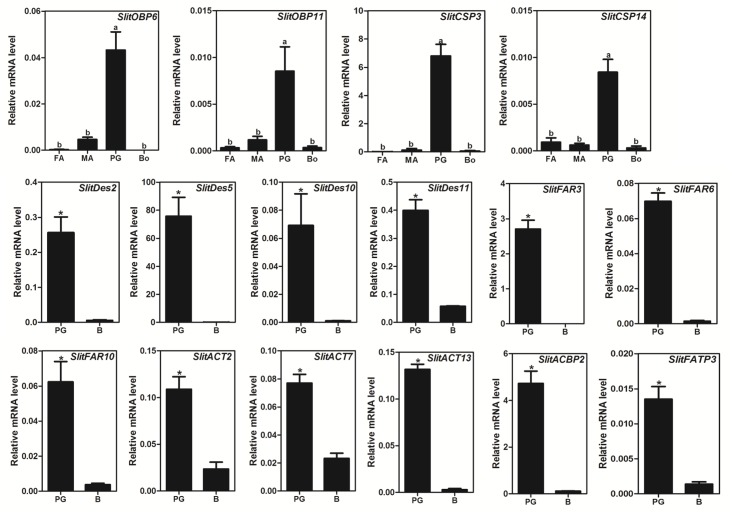
To investigate the general expression profiles of the candidate genes, reverse transcription-polymerase chain reaction (RT-PCR) analyses were conducted for all 94 genes (Figs 4 and 5), and the expression levels of 16 selected genes were further quantified with qPCR (Fig 6) to validate the RT-PCR results. The overall relative expression profiles of these genes in different tissues obtained were similar with the two methods.
Fig 4. Expression patterns of sex pheromone biosynthesis related genes, using RT-PCR.
(A) Expression of Des genes. (B) Expression of FAR genes. (C) Expression of ACT genes. (D) Expression of ACBP, FATP and ACC genes. GAPDH gene was used as a positive control and NC (no cDNA template) as a negative control. PG, female pheromone glands; B, whole insect body without PGs.
Fig 5. Expression patterns of sex pheromone chemoreception related genes, using RT-PCR.
(A) Expression of OBP genes. (B) Expression of CSP genes. GAPDH gene was used as a positive control and NC (no cDNA template) as a negative control. PG, female pheromone glands; B, whole insect body without PGs.
Fig 6. Relative expression levels of 16 pheromone biosynthesis and chemoreception releated genes, using qPCR.
FA, female antennae; MA, male antennae; PG, female pheromone glands; Bo, whole insect body without PGs and antennae. The relative expression level is indicated as mean ± SE (N = 3). Different capital letters mean significant difference between tissues (P < 0.05, ANOVA, LSD); the “*” indicates significant difference between male and female (P < 0.05, Student t-test).
Most of the pheromone biosynthesis-related genes (>70%) displayed PG-biased or specific expression, and 4 chemoreception related genes (SlitOBP6, SlitOBP11, SlitCSP3, and SlitCSP14) were also more highly expressed in the PG than in other tissues of S. litura (Table 2).
Desaturase (Des) and Fatty Acyl Reductase (FAR)
The results of RT-PCR and qPCR showed that 9 of the 12 Des genes showed PG-biased or specific expression, which is greater than the proportion reported for other moth species (Table 2); the other Des genes, SlitDes7, SlitDes9, and SlitDes12, were detected in both the PG and other parts of the body (Table 2, Figs 4A and 6). Similar to H. assulta [35], 11 of the 13 SlitFAR genes (except for SlitFAR4 and SlitFAR5) displayed PG-biased or specific expression (Table 2, Figs 4B and 6).
Acetyltransferases (ACT)
Over half of the 23 newly identified SlitACT genes were predominantly or specifically expressed in the PG. SlitACT16 expression was detected to be highly biased in the body, whereas 6 ACT genes (SlitACT1, 8, 9, 10, 21, and 22) displayed similar expression levels in the PG and body (Table 2, Figs 4C and 6).
Acyl-CoA Binding Protein (ACBP), Fatty Acid Transport Protein (FATP) and Acetyl-CoA Carboxylase (ACC)
Of the 2 SlitACBP and 3 SlitFATP genes, only SlitACBP2 and SlitFATP3 were highly expressed in the PG, whereas the SlitACC gene encoding an ACC with 94% identity to the ACC of Agrotis segetum (GenBank Accession No.: AID66639.1) did not show PG-biased or specific expression (Table 2, Figs 4D and 6).
Odorant Binding Protein (OBP) and Chemosensory Protein (CSP)
We identified a total of 25 OBP genes from the PG of S. litura in this study, including 1 PBP (OBP25) and 24 other OBPs. Of the 25 SlitOBPs, 4 genes (SlitOBP8, SlitOBP9, SlitOBP13, and SlitOBP14) displayed a very wide range of tissue distribution in all 4 tissues examined, whereas 8 other genes (SlitOBP3, SlitOBP4, SlitOBP5, SlitOBP10, SlitOBP16, SlitOBP19, SlitOBP24, and SlitOBP25) were expressed predominately or specifically in the adult antennae. Furthermore, 2 genes (SlitOBP6 and SlitOBP11) were expressed at much higher levels in the PG than in other tissues.
Unlike the SlitOBPs, most of the SlitCSPs were expressed at similar levels among the 4 tissues examined; only SlitCSP3 and SlitCSP14 showed significantly higher expression in the PG than in other tissues (Table 2, Figs 5 and 6).
Phylogenetic Analyses
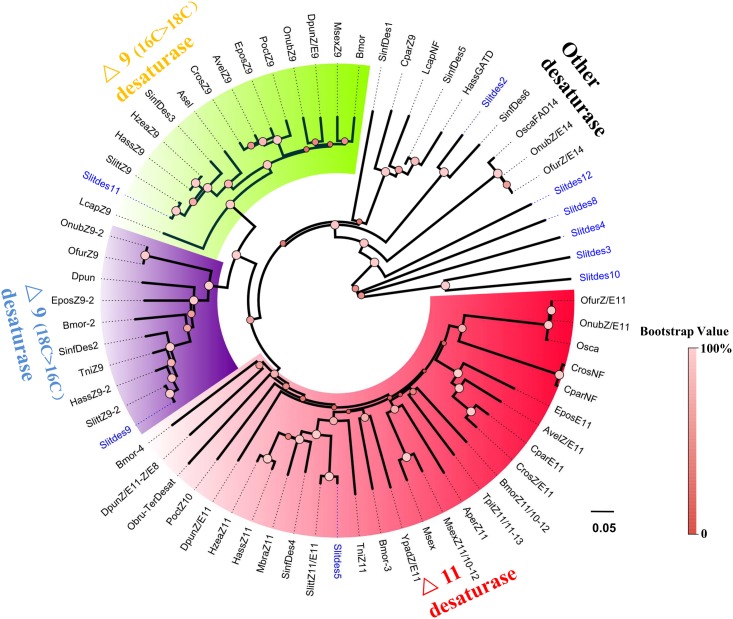
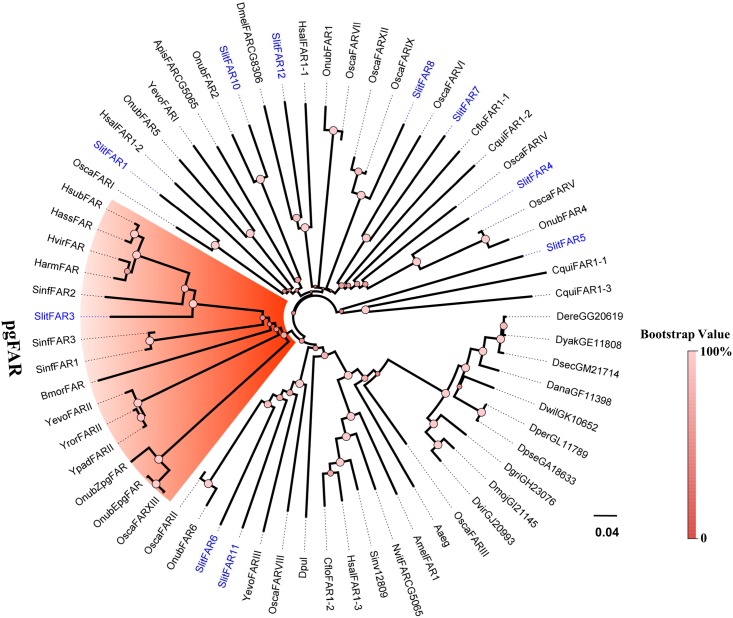
To assign putative functions to these different genes, phylogenetic analyses were conducted for each group of the enzymes. A phylogenetic tree of Des sequences (Fig 7) showed that 3 SlitfDes genes clearly clustered in 3 different groups of insect desaturases: Δ11-desaturase (SlitDes5), Δ9-desaturase (18C>16C) (SinfDes9), and Δ9-desaturase (16C>18C) (SlitDes11). In the FAR phylogenetic tree, only SlitFAR3 clustered within the lepidopteran pgFAR group that contains previously identified FARs known to be involved in moth sex pheromone biosynthesis [20] (Fig 8).
Fig 7. Phylogenetic tree of insect desaturase (Des).
The S. litura translated genes are shown in blue. Accession numbers are given in S2 Table. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values at the nodes are results of bootstrap with 1000 replicates.
Fig 8. Phylogenetic tree of insect fatty acid redutase (FAR).
The S. litura translated genes are shown in blue. Accession numbers are given in S2 Table. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values at the nodes are results of bootstrap with 1000 replicates.
The OBP phylogenetic tree showed that SlitOBP25 (SlitPBP3) was clustered into the PBP/GOBP clade. The other SlitOBPs and all SlitCSPs were clustered with at least one lepidopteran orthologous gene (S1 and S2 Figs).
Discussion
In the present study, we sequenced and analyzed the transcriptome of the PG of S. litura. Among the 42,646 unigenes identified, only 27.41% could be annotated to one or more GO terms, which is similar to other lepidopteran species [37–39], indicating that a large number of S. litura genes are either non-coding or are homologous with genes that do not have any GO term. Importantly, we identified 89 novel genes that are involved in sex pheromone biosynthesis and transport in S. litura. Our results not only provide an important foundation for further elucidation of the molecular mechanisms of sex pheromone metabolism but also provide general insight into insect physiology and development of a novel pest control strategy [40].
S. litura has 4 sex pheromone components: Z9,E11-14:OAc, Z9,E12-14:OAc, Z9-14:OAc, and E11-14:OAc (at a ratio of 100:27:20:27) in China [25]. According to some previous studies related to the biosynthesis pathway of Δ11- or Δ9-containing sex pheromones in several moth species, the defined pathway involves a step of Δ11 or Δ9 desaturation that is catalyzed by Δ11 or Δ9 desaturase, respectively [16, 41–44]. On the other hand, Liu et al. [45] reported that a Δ9 desaturase participates in the production of the Δ11-containing pheromone component by introducing a Δ9-double bond at 14:CoA, followed by carbon chain elongation to Δ11–16:CoA. In sum, these findings indicate that the Δ11 and Δ9 desaturases are likely responsible for introduction of the Δ11 and Δ9-double bonds in all 4 pheromones of S. litura. In this study, we obtained 9 desaturase genes showing PG-biased or specific expression. Further phylogenetic analysis showed that SlitDes5 was clearly assigned to the Δ11 desaturase group, and was most closely related to Z/E11 of S. littoralis (GenBank Accession No. Q6US81). SlitDes9 and SlitDes11 were allocated to the Δ9 (18C>16C) and Δ9 (16C>18C) desaturase groups, respectively, but only SlitDes11 displayed a PG-biased expression pattern; the other genes were allocated to other desaturase groups. Therefore, SlitDes5 and SlitDes11 are very likely involved in the desaturation step from saturated acids (14C) to unsaturated acids, with a double bond introduced at the 11th and 9th positions of the carbon chain, respectively. To date, there is no report on Δ12 desaturase genes in moths; therefore, the type of enzyme that S. litura uses to introduce a double bond at the 12th position needs to be further studied.
In the process of sex pheromone biosynthesis, once the specific unsaturated fatty acid precursors are produced, they will be converted into corresponding alcohols by FAR, which has been demonstrated in different moth species [18–20, 46]. In this study, we identified 11 FAR genes with PG-biased or specific expression, but only SlitFAR3 was clustered in the moth pgFAR group, suggesting that this gene plays a crucial role in the biosynthesis of precursor alcohols. ACTs are essential in the biosynthesis of acyloxyl components, although no related gene had been functionally characterized to date. In the present study, 15 ACT genes were found to be highly or specifically expressed in the PG by RT-PCR and qPCR, indicating that these genes may participate in the progress of ACT to produce corresponding acetate ester. FATPs have been functionally confirmed to bind to and transport fatty acids across the insect hemolymph into PG cells for pheromone biosynthesis in B. mori [22] and Eilema japonica [47], and ACBPs have been functionally confirmed to serve as carriers or cellular deposits for the acyl-CoAs used in pheromone biosynthesis [22]. Therefore, the PG-biased expression of SlitFATP3 and SlitACBP2 may indicate that they play a similar role in the pheromone biosynthesis of S. litura.
Some previous studies identified the presence of chemosensilla on the ovipositor [48, 49], which may function in the chemoreception of plant odors, ovipositor-deterring pheromones, and sex pheromones, suggesting that female moths may receive and transport pheromone compounds or their precursors via their ovipositor [35, 50–52]. This finding further suggests that there may be a feedback loop in the moth’s PG (including the ovipositor) to achieve accurate control of the biosynthesis pathway and release of sex pheromones. To date, many studies have demonstrated the role of OBPs and CSPs in the binding and transportation of hydrophobic molecules, including plant volatiles, sex pheromones and their precursors [50, 53–57]; therefore, these two proteins appear to be essential for the sex pheromone biosynthesis pathway. Similarly to other moths [32, 34, 35], we identified a total of 39 genes in the S. litura PG, including 25 OBP and 14 CSP genes, but only 4 genes (SlitOBP6, SlitOBP11, SlitCSP3, and SlitCSP14) showed PG-biased expression, indicating that these genes may play important roles in the binding and transport of sex pheromone compounds and plant volatiles.
In conclusion, through sequencing and transcriptome analyses, we obtained an extensive set of putative genes that may be related to the biosynthesis and transport of the sex pheromone of S. litura. As the first step towards understanding the functions of these genes, we conducted a comprehensive and comparative examination of gene expression patterns and conducted a phylogenetic analysis with sequences of other species. We identified a number of genes with PG-biased or specific expression, indicating their involvement in the biosynthesis and transport of the sex pheromone. Further studies are needed to explore the functions of these genes with integrated functional studies.
Materials and Methods
Insects Rearing
S. litura were reared on an artificial diet comprising wheat germ flour and soybean flour, and were sexed as pupae and kept separately in cages for eclosion. The rearing conditions were 27°C, with a 14-h light:10-h dark photoperiod, and 65 ± 5% relative humidity. Adults were provided with a cotton swab dipped in 10% honey solution that was renewed daily.
Tissue Collection
For transcriptome sequencing, 20–25 PGs (with the ovipositor) were collected from 3-day-old virgin female adults at 6–7 h into the scotophase, since 3-day-old moths show particularly high mating activity [25]. For the tissue expression study, 25–30 female antennae (FA), 25–30 male antennae(MA), 20–25 PGs (with the ovipositor), 10–15 whole insect body without PGs(B), and 10–15 whole insect body without PGs and antennae (Bo) were also collected under the same conditions. All samples were immediately frozen and stored at −70°C until use.
cDNA Library Construction
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). cDNA library construction and Illumina sequencing of the samples were performed at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The mRNA was purified from 3 μg of total RNA using oligo (dT) magnetic beads and fragmented into short sequences in the presence of divalent cations at 94°C for 5 min. Then, the first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by synthesis of the second-strand cDNA using RNaseH and DNA polymerase I. After end repair and ligation of adaptors, the products were amplified by PCR and purified using the QIAquick PCR Purification Kit to create a cDNA library, and the library quality was assessed on the Agilent Bioanalyzer 2100 system.
Clustering and Sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq(TM) 2500 platform, and paired-end reads were generated.
De novo Assembly of Short Reads and Gene Annotation
Clean short reads were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from the raw reads. Transcriptome de novo assembly was carried out with these short reads in the assembling program Trinity (r20140413p1) [58, 59] with min_kmer_cov set to 2 by default, and all other parameters at the default settings. The resulting sequences of Trinity were deemed to be unigenes. Unigenes larger than 150 bp were first aligned to protein databases by BlASTX, including Nr, Swiss-Prot, KEGG, and COG (e-value < 10−5), to retrieve proteins with the highest sequence similarity to the obtained unigenes along with their protein functional annotations. Then, we used the Blast2GO program [60] to get the GO annotation of the unigenes, and the GO functional classification was obtained using WEGO software [61].
Expression Abundance Analysis of the Unigenes
The expression abundance of these unigenes were calculated based on the reads per kilobase per million mapped reads (RPKM) method [62], using the formula: RPKM (A) = (10,00,000 × C × 1,000)/(N × L), where RPKM (A) is the abundance of gene A, C is the number of reads that uniquely aligned to gene A, N is the total number of reads that uniquely aligned to all genes, and L is the number of bases in gene A. The RPKM method is able to eliminate the influence of different gene lengths and sequencing discrepancy in the calculation of expression abundance.
RNA Isolation and cDNA Synthesis
Total RNA was extracted with the SV 96 Total RNA Isolation System (Promega, Madison, WI, USA) following the manufacturer’s instructions, in which DNaseI digestion was included to avoid genomic DNA contamination. RNA quality was checked with a spectrophotometer (NanoDropTM 1000, Thermo Fisher Scientific, USA). The single-stranded cDNA templates were synthesized from 1 μg of total RNA from various tissue samples using the PrimeScript RT Master Mix (TaKaRa, Dalian, China).
Sequence Analyses
The open reading frames (ORFs) of the putative chemosensory genes were predicted by using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Similarity searches were performed using the NCBI-BLAST network server (http://blast.ncbi.nlm.nih.gov/). Putative N-terminal signal peptides of SlitOBPs and SlitCSPs were predicted by Signal IP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) [63].
Phylogenetic Analyses
The phylogenetic trees were constructed for phylogenetic analyses of SlitOBPs, SlitCSPs, SlitDes and SlitFAR, based on these genes (the signal peptides of sequences had been removed of the putative chemosensory genes) and the sequences of other insects. The OBP dataset contained 25 sequences from S. litura, 19 from Manduca sexta [37, 64], 15 from S. littoralis [64], 23 from Sesamia inferens [65], and 43 from B. mori [66]. The CSP dataset contained 14 sequences from S. litura, 14 from M. sexta [37], 9 from S. littoralis [64], 13 from S. inferens [65], and 14 from B. mori [67]. The Des dataset contained 12 sequences from S. litura and 59 from other insects [33]. The FAR dataset contained 13 sequences from S. litura and 56 from other insects [33]. The names and accession numbers of the genes used for phylogenetic tree construction are listed in S2 Table. Amino acid sequences were aligned with ClustalX 1.83 [68], and unrooted trees were constructed by MEGA5.0 [69] using the neighbor-joining method, with Poisson correction of distances. Node support was assessed using a bootstrap procedure base on 1000 replicates.
Reverse Transcription-PCR Analyses
Gene-specific primers across the ORFs of predicted chemosensory genes were designed using Primer Premier 5.0 (PREMIER Biosoft International, CA, USA). The sequences of these primers are listed in S3 Table. PCR experiments including negative controls (no cDNA template) were carried out under the following conditions: 94°C for 4 min, 30–35 cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 40 sec, and final extension for 10 min at 72°C. The reactions were performed in a total volume of 25 μL, containing 12.5 μL of 2×EasyTaq PCR SuperMix (TransGene, Beijing, China), 0.4 μM for each primer, 1 μL of sample cDNA (15 ng/μL), 9.5 μL of sterilized H2O. PCR products were analyzed by electrophoresis on 1.5% w/v agarose gel in TAE buffer (40 mM Tris-acetate, 2 mM Na2EDTA·H2O). The gene encoding S. litura glyceraldehyde-3-phosphate dehydrogenase (SlitGAPDH) (GenBank Accession No.: HQ012003.2) was used as a reference gene for checking the integrity of the cDNA template. Each PCR reaction was done at least twice.
Quantitative Real Time-PCR Validation
The expression profile of 16 putative genes with PG-enriched or specific expression was evaluated to validate the accuracy of the RT-PCR results using quantitative real time-PCR (qPCR) experiments. The qPCR was performed on an ABI 7300 (Applied Biosystems, Foster City, CA, USA) using a mixture of 10 μL 2× SYBR Green PCR Master Mix, 0.4 μL each primer (10 μM), 2.5 ng of sample cDNA, and 6.8 μL sterilized ultrapure H2O. The reaction programs were at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec, and 60°C for 31 sec. The results were analyzed using the ABI 7300 analysis software SDS 1.4. The qPCR primers (S3 Table) were designed using Beacon Designer 7.9 (PREMIER Biosoft International, CA, USA). The mRNA levels were measured by qPCR using SYBR Premix ExTaq (TaKaRa, Dalian, Liaoning, China). Subsequently, fluorescence was measured throughout a 55–95°C melting curve in order to detect a single gene-specific peak and to check the absence of primer dimer peaks; single and discrete peaks were detected for all primers tested. Negative controls were non-template reactions (replacing cDNA with H2O).
The expression levels of 16 genes were calculated relative to the reference gene SlitGAPDH (GenBank Accession No.: HQ012003.2) and SlitEF (elongation factor–1 alpha) (GenBank Accession No.: DQ192234.1) using the Q-Gene method in the Microsoft Excel-based software Visual Basic [70, 71] For each sample, three biological replications were performed with each biological replication measured in three technique replications.
Statistical Analyses
Data (mean ± SE) form various samples were subjected to one-way nested analysis of variance (ANOVA) followed by the least significant difference test (LSD) for mean comparison, and two-sample analysis was performed by the Student t-test using SPSS Statistics 17.0 software (SPSS Inc., Chicago, IL, USA).
Supporting Information
The S. litura translated genes are shown in blue. Accession numbers are given in S2 Table. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values at the nodes are results of bootstrap with 1000 replicates.
(TIF)
The S. litura translated genes are shown in blue. Accession numbers are given in S2 Table. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values at the nodes are results of bootstrap with 1000 replicates.
(TIF)
(XLS)
(XLS)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files. All genes are available from the NCBI-GenBank database (accession number: KT261647- KT261737).
Funding Statement
This work was supported by grants from Natura Science Fund of Education Department of Anhui province, China (No.: KJ2013A234), the National Natural Science Foundation of China (No.: 31501647) and Doctoral Scientific Research Foundation of Huaibei Normal University, China (No.: 1500969).
References
- 1. Wyatt TD Pheromones and Animal Behaviour: Communication by Smell and Taste Cambridge University Press; 2003. [Google Scholar]
- 2. Wyatt TD. Fifty years of pheromones. Nature. 2009;457:262–3. Epub 2009/01/17. 10.1038/457262a . [DOI] [PubMed] [Google Scholar]
- 3. Smadja C, Butlin RK. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity. 2009;102(1):77–97. 10.1038/hdy.2008.55 . [DOI] [PubMed] [Google Scholar]
- 4. Raina AK, Wergin WP, Murphy CA, Erbe EF. Structural organization of the sex pheromone gland in Helicoverpa zea in relation to pheromone production and release. Arthropod Struct Dev. 2000;29(4):343–53. Epub 2007/12/20. S1467-8039(01)00014-7 [pii]. . [PubMed] [Google Scholar]
- 5. Tillman JA, Seybold SJ, Jurenka RA, Blomquist GJ. Insect pheromones–-an overview of biosynthesis and endocrine regulation. Insect Biochem Mol Biol. 1999;29(6):481–514. Epub 1999/07/16. S0965174899000168 [pii]. . [DOI] [PubMed] [Google Scholar]
- 6. Jurenka R. Insect pheromone biosynthesis. Topics in current chemistry. 2004;239:97–132. 10.1007/b95450 . [DOI] [PubMed] [Google Scholar]
- 7. Morse D, Meighen EA. Pheromone biosynthesis: enzymatic studies in Lepidoptera Pheromone biochemistry Orlando Fl, 1987:Acandemic Press, IncPrestwich GD, Blomquist GJ. pp. 212–5. [Google Scholar]
- 8. Ando T, Inomata S-I, Yamamoto M. Lepidopteran sex pheromones In The Chemistry of Pheromones and Other Semiochemicals I (Schulz S, ed). 2004:Springer, Berlin/Heidelberg: pp. 51–96. [DOI] [PubMed] [Google Scholar]
- 9. Park HY, Kim MS, Paek A, Jeong SE, Knipple DC. An abundant acyl-CoA (Delta9) desaturase transcript in pheromone glands of the cabbage moth, Mamestra brassicae, encodes a catalytically inactive protein. Insect Biochem Mol Biol. 2008;38(5):581–95. Epub 2008/04/15. S0965-1748(08)00026-X [pii] 10.1016/j.ibmb.2008.02.001 . [DOI] [PubMed] [Google Scholar]
- 10. Moto K, Suzuki MG, Hull JJ, Kurata R, Takahashi S, Yamamoto M, et al. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc Natl Acad Sci U S A. 2004;101(23):8631–6. Epub 2004/06/03. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foster SP, Roelofs WL. Sex pheromone biosynthesis in the tortricid moth, Ctenopseustis herana (Felder & Rogenhofer). Arch Insect Biochem Physiol. 1996;33:135–47. [Google Scholar]
- 12. Hagström AK, Albre J, Tooman LK, Thirmawithana AH, Corcoran J, Löfstedt C, et al. A novel fatty acyl desaturase from the pheromone glands of Ctenopseustis obliquana and C. herana with specific Z5-Desaturase activity on myristic acid. J Chem Ecol. 2014;40(1):63–70. Epub 2014/01/11. 10.1007/s10886-013-0373-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang HL, Lienard MA, Zhao CH, Wang CZ, Lofstedt C. Neofunctionalization in an ancestral insect desaturase lineage led to rare Delta6 pheromone signals in the Chinese tussah silkworm. Insect Biochem Mol Biol. 2010;40(10):742–51. Epub 2010/08/10. S0965-1748(10)00164-5 [pii] 10.1016/j.ibmb.2010.07.009 . [DOI] [PubMed] [Google Scholar]
- 14. Liu W, Ma PW, Marsella-Herrick P, Rosenfield CL, Knipple DC, Roelofs W. Cloning and functional expression of a cDNA encoding a metabolic acyl-CoA delta 9-desaturase of the cabbage looper moth, Trichoplusia ni . Insect Biochem Mol Biol. 1999;29(5):435–43. Epub 1999/06/25. S096517489900020X [pii]. . [DOI] [PubMed] [Google Scholar]
- 15. Jeong SE, Rosenfield CL, Marsella-Herrick P, Man You K, Knipple DC. Multiple acyl-CoA desaturase-encoding transcripts in pheromone glands of Helicoverpa assulta, the oriental tobacco budworm. Insect Biochem Mol Biol. 2003;33(6):609–22. Epub 2003/05/29. S0965174803000432 [pii]. . [DOI] [PubMed] [Google Scholar]
- 16. Fujii T, Ito K, Tatematsu M, Shimada T, Katsuma S, Ishikawa Y. Sex pheromone desaturase functioning in a primitive Ostrinia moth is cryptically conserved in congeners' genomes. Proc Natl Acad Sci U S A. 2011;108(17):7102–6. Epub 2011/03/30. 1019519108 [pii] 10.1073/pnas.1019519108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roelofs WL, Rooney AP. Molecular genetics and evolution of pheromone biosynthesis in Lepidoptera. Proc Natl Acad Sci U S A. 2003;100(16):9179–84. Epub 2003/07/24. 10.1073/pnas.1233767100 1233767100 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moto K, Yoshiga T, Yamamoto M, Takahashi S, Okano K, Ando T, et al. Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori . Proc Natl Acad Sci U S A. 2003;100(16):9156–61. Epub 2003/07/23. 10.1073/pnas.1531993100 1531993100 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lassance JM, Groot AT, Lienard MA, Antony B, Borgwardt C, Andersson F, et al. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010;466(7305):486–9. Epub 2010/07/02. nature09058 [pii] 10.1038/nature09058 . [DOI] [PubMed] [Google Scholar]
- 20. Hagstrröm AK, Liénard MA, Groot AT, Hedenström E, Löfstedt C. Semi-selective fatty acyl reductases from four Heliothine moths influence the specific pheromone composition. PLoS One. 2012;7(5):e37230 Epub 2012/05/23. 10.1371/journal.pone.0037230PONE-D-12-05220 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohnishi A, Hull JJ, Matsumoto S. Targeted disruption of genes in the Bombyx mori sex pheromone biosynthetic pathway. Proc Natl Acad Sci U S A. 2006;103(12):4398–403. Epub 2006/03/16. 0511270103 [pii] 10.1073/pnas.0511270103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohnishi A, Hashimoto K, Imai K, Matsumoto S. Functional characterization of the Bombyx mori fatty acid transport protein (BmFATP) within the silkmoth pheromone gland. J Biol Chem. 2009;284(8):5128–36. Epub 2008/12/30. M806072200 [pii] 10.1074/jbc.M806072200 . [DOI] [PubMed] [Google Scholar]
- 23. Jurenka RA, Roelofs WL. Characterization of the acetyltransferase used in pheromone biosynthesis in moths: Specificity for the Z isomer in tortricidae. Insect Biochem. 1989;19(7):639–44. [Google Scholar]
- 24. Teal PEA, Tumlinson JH. The role of alcohols in pheromone biosynthesis by two noctuid moths that use acetate pheromone components. Arch Insect Biochem Physiol. 1987;4:261–9. [Google Scholar]
- 25. Sun F, Hu YY, Du JW. The sex pheromone communication system of Spodoptera litura (Fabricius). Acta Entomologica Sinica. 2002;45(3):404–7. [Google Scholar]
- 26. Liu NY, He P, Dong SL. Binding properties of pheromone-binding protein 1 from the common cutworm Spodoptera litura . Comp Biochem Physiol B Biochem Mol Biol. 2012;161(4):295–302. Epub 2011/12/07. 10.1016/j.cbpb.2011.11.007 S1096-4959(11)00233-8 [pii]. . [DOI] [PubMed] [Google Scholar]
- 27. Liu NY, Liu CC, Dong SL. Functional differentiation of pheromone-binding proteins in the common cutworm Spodoptera litura . Comp Biochem Physiol A Mol Integr Physiol. 2013;165(2):254–62. Epub 2013/03/20. S1095-6433(13)00074-3 [pii] 10.1016/j.cbpa.2013.03.016 . [DOI] [PubMed] [Google Scholar]
- 28. Zhang J, Yan S, Liu Y, Jacquin-Joly E, Dong S, Wang G. Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem Senses. 2015;40(1):7–16. Epub 2014/10/27. 10.1093/chemse/bju052 bju052 [pii]. . [DOI] [PubMed] [Google Scholar]
- 29. Liu Y, Gu S, Zhang Y, Guo Y, Wang G. Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS One. 2012;7(10):e48260 Epub 2012/10/31. 10.1371/journal.pone.0048260 PONE-D-12-23655 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li SW, Yang H, Liu YF, Liao QR, Du J, Jin DC. Transcriptome and Gene Expression Analysis of the Rice Leaf Folder, Cnaphalocrosis medinalis . PLoS One. 2012;7(11):e47401 Epub 2012/11/28. 10.1371/journal.pone.0047401 PONE-D-11-23279 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens . BMC Genomics. 2010;11:29 Epub 2010/01/16. 1471-2164-11-29 [pii] 10.1186/1471-2164-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu SH, Wu KM, Guo YY, Pickett JA, Field LM, Zhou JJ, et al. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics. 2013;14(1):636 Epub 2013/09/24. 1471-2164-14-636 [pii] 10.1186/1471-2164-14-636 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang YN, Xia YH, Zhu JY, Li SY, Dong SL. Putative pathway of sex pheromone biosynthesis and degradation by expression patterns of genes identified from female pheromone gland and adult antenna of Sesamia inferens (Walker). J Chem Ecol. 2014;40(5):439–51. Epub 2014/05/13. 10.1007/s10886-014-0433-1 . [DOI] [PubMed] [Google Scholar]
- 34. Xia YH, Zhang YN, Hou XQ, Li F, Dong SL. Large number of putative chemoreception and pheromone biosynthesis genes revealed by analyzing transcriptome from ovipositor-pheromone glands of Chilo suppressalis . Scientific reports. 2015;5:7888 Epub 2015/01/21. 10.1038/srep07888 srep07888 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li ZQ, Zhang S, Luo JY, Wang CY, Lv LM, Dong SL, et al. Transcriptome comparison of the sex pheromone glands from two sibling Helicoverpa species with opposite sex pheromone components. Scientific reports. 2015;5:9324 Epub 2015/03/21. 10.1038/srep09324 srep09324 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Antony B, Soffan A, Jakse J, Alfaifi S, Sutanto KD, Aldosari SA, et al. Genes involved in sex pheromone biosynthesis of Ephestia cautella, an important food storage pest, are determined by transcriptome sequencing. BMC Genomics. 2015;16:532 10.1186/s12864-015-1710-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta . Proc Natl Acad Sci U S A. 2011;108(18):7449–54. Epub 2011/04/19. 1017963108 [pii] 10.1073/pnas.1017963108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J, Wang B, Dong S, Cao D, Dong J, Walker WB, et al. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta . PLoS One. 2015;10(2):e0117054 Epub 2015/02/07. 10.1371/journal.pone.0117054 PONE-D-14-43348 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poivet E, Gallot A, Montagne N, Glaser N, Legeai F, Jacquin-Joly E. A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis . PLoS One. 2013;8(4):e60263 Epub 2013/04/09. 10.1371/journal.pone.0060263 PONE-D-12-38409 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou JJ, Field LM, He XL. Insect odorant-binding proteins: Do they offer an alternative pest control strategy? Outlooks Pest Manag. 2010;21(1):31–4. [Google Scholar]
- 41. Liu W, Jiao H, O'Connor M, Roelofs WL. Moth desaturase characterized that produces both Z and E isomers of delta 11-tetradecenoic acids. Insect Biochem Mol Biol. 2002;32(11):1489–95. Epub 2003/01/18. S0965174802000693 [pii]. . [DOI] [PubMed] [Google Scholar]
- 42. Hao G, O'Connor M, Liu W, Roelofs WL. Characterization of Z/E11- and Z9-desaturases from the obliquebanded leafroller moth, Choristoneura rosaceana . J Insect Sci. 2002;2:26 Epub 2004/09/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roelofs WL, Liu W, Hao G, Jiao H, Rooney AP, Linn CE Jr. Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci U S A. 2002;99(21):13621–6. Epub 2002/09/19. 10.1073/pnas.152445399 152445399 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu WQ, Zhu JW, Millar J, Lofstedt C. A comparative study of sex pheromone biosynthesis in two strains of the turnip moth, Agrotis segetum, producing different ratios of sex pheromone components. Insect Biochem Mol Biol. 1998;28:895–900. [Google Scholar]
- 45. Liu W, Jiao H, Murray NC, O'Connor M, Roelofs WL. Gene characterized for membrane desaturase that produces (E)-11 isomers of mono- and diunsaturated fatty acids. Proc Natl Acad Sci U S A. 2002;99(2):620–4. Epub 2002/01/24. 99/2/620 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liénard MA, Hagstrom AK, Lassance JM, Löfstedt C. Evolution of multicomponent pheromone signals in small ermine moths involves a single fatty-acyl reductase gene. Proc Natl Acad Sci U S A. 2010;107(24):10955–60. Epub 2010/06/11. 1000823107 [pii] 10.1073/pnas.1000823107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qian S, Fujii T, Ito K, Nakano R, Ishikawa Y. Cloning and functional characterization of a fatty acid transport protein (FATP) from the pheromone gland of a lichen moth, Eilema japonica, which secretes an alkenyl sex pheromone. Insect Biochem Mol Biol. 2011;41(1):22–8. Epub 2010/09/30. S0965-1748(10)00227-4 [pii] 10.1016/j.ibmb.2010.09.008 . [DOI] [PubMed] [Google Scholar]
- 48. Faucheux MJ. Multiporous sensilla on the ovipositor of Monopis crocicapitella Clem. (Lepidoptera: Tineidae) Int J Insect Morphol Embryol. 1988;17:473–75. [Google Scholar]
- 49. Anderson P, Hallberg E. Structure and distribution of tactile and bimodal taste/tactile sensilla on the ovipositor, tarsi and antennae of the flour moth, Ephestia kuehniella (Zeller) (Lepidoptera: Pyralidae). Int J Insect Morphol Embryol 1990;19:13–23. [Google Scholar]
- 50. Dani FR, Michelucci E, Francese S, Mastrobuoni G, Cappellozza S, La Marca G, et al. Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori . Chem Senses. 2011;36(4):335–44. Epub 2011/01/12. bjq137 [pii] 10.1093/chemse/bjq137 . [DOI] [PubMed] [Google Scholar]
- 51. Anderson P, Hallberg E. Structure and distribution of tactile and bimodal taste tactile sensilla on the ovipositor, tarsi and antennae of the flour moth, Ephestia Kuehniella (Zeller) (Lepidoptera, Pyralidae). Int J Insect Morphol 1990;19:13–23. [Google Scholar]
- 52. Jacquin-Joly E, Vogt RG, Francois MC, Nagnan-Le Meillour P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae . Chem Senses. 2001;26(7):833–44. Epub 2001/09/14. . [DOI] [PubMed] [Google Scholar]
- 53. Pelosi P, Calvello M, Ban L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem Senses. 2005;30 Suppl 1:i291–2. Epub 2005/03/02. 30/suppl_1/i291 [pii] 10.1093/chemse/bjh229 . [DOI] [PubMed] [Google Scholar]
- 54. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–91. Epub 2012/10/02. 10.1146/annurev-ento-120811-153635 . [DOI] [PubMed] [Google Scholar]
- 55. Zhou JJ. Odorant-binding proteins in insects. Vitam Horm. 2010;83:241–72. Epub 2010/09/14. S0083-6729(10)83010-9 [pii] 10.1016/S0083-6729(10)83010-9 . [DOI] [PubMed] [Google Scholar]
- 56. Zhang YN, Ye ZF, Yang K, Dong SL. Antenna-predominant and male-biased CSP19 of Sesamia inferens is able to bind the female sex pheromones and host plant volatiles. Gene. 2014;536(2):279–86. Epub 2013/12/24. 10.1016/j.gene.2013.12.011 S0378-1119(13)01672-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 57. Pelosi P, Iovinella I, Felicioli A, Dani FR. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 2014;5:320 Epub 2014/09/16. 10.3389/fphys.2014.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–72. Epub 2009/12/19. gr.097261.109 [pii] 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. Epub 2005/08/06. bti610 [pii] 10.1093/bioinformatics/bti610 . [DOI] [PubMed] [Google Scholar]
- 61. Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34(Web Server issue):W293–7. Epub 2006/07/18. 34/suppl_2/W293 [pii] 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. Epub 2008/06/03. nmeth.1226 [pii] 10.1038/nmeth.1226 . [DOI] [PubMed] [Google Scholar]
- 63. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. Epub 2011/10/01. 10.1038/nmeth.1701 nmeth.1701 [pii]. . [DOI] [PubMed] [Google Scholar]
- 64. Legeai F, Malpel S, Montagne N, Monsempes C, Cousserans F, Merlin C, et al. An expressed sequence tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics. 2011;12:86 Epub 2011/02/01. 1471-2164-12-86 [pii] 10.1186/1471-2164-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang YN, Jin JY, Jin R, Xia YH, Zhou JJ, Deng JY, et al. Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS One. 2013;8(7):e69715 10.1371/journal.pone.0069715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics. 2009;10:332 Epub 2009/07/25. 1471-2164-10-332 [pii] 10.1186/1471-2164-10-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Foret S, Wanner KW, Maleszka R. Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol. 2007;37(1):19–28. Epub 2006/12/19. S0965-1748(06)00189-5 [pii] 10.1016/j.ibmb.2006.09.009 . [DOI] [PubMed] [Google Scholar]
- 68. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. Epub 2007/09/12. btm404 [pii] 10.1093/bioinformatics/btm404 . [DOI] [PubMed] [Google Scholar]
- 69. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19(11):1439–40. Epub 2003/07/23. . [DOI] [PubMed] [Google Scholar]
- 71. Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32(6):1372–4, 6,, 8–9. Epub 2002/06/21. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The S. litura translated genes are shown in blue. Accession numbers are given in S2 Table. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values at the nodes are results of bootstrap with 1000 replicates.
(TIF)
The S. litura translated genes are shown in blue. Accession numbers are given in S2 Table. The tree was constructed with MEGA5.0, using the neighbour-joining method. Values at the nodes are results of bootstrap with 1000 replicates.
(TIF)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All genes are available from the NCBI-GenBank database (accession number: KT261647- KT261737).