Abstract
We applied whole-genome sequencing to reconstruct the spatial and temporal dynamics underpinning the expansion of Clostridium difficile ribotype 027 in Germany. Based on re-sequencing of genomes from 57 clinical C. difficile isolates, which had been collected from hospitalized patients at 36 locations throughout Germany between 1990 and 2012, we demonstrate that C. difficile genomes have accumulated sequence variation sufficiently fast to document the pathogen's spread at a regional scale. We detected both previously described lineages of fluoroquinolone-resistant C. difficile ribotype 027, FQR1 and FQR2. Using Bayesian phylogeographic analyses, we show that fluoroquinolone-resistant C. difficile 027 was imported into Germany at least four times, that it had been widely disseminated across multiple federal states even before the first outbreak was noted in 2007, and that it has continued to spread since.
Introduction
Clostridium difficile is the most common cause of healthcare-associated diarrhea in Europe and the USA [1, 2]. In 2009, C. difficile infection (CDI) was associated with almost 1% of admissions to US hospitals, resulting in a severe burden of morbidity, mortality, and economic costs [3]. In addition, community-associated CDI has been reported, albeit at a lower rate [4–6].
The increase of CDI incidence observed in North America and Europe during the first decade of this century was accompanied by the emergence of a previously uncommon strain of C. difficile [7, 8], genotyped as PCR ribotype 027, North American pulsotype NAP1, or restriction endonuclease analysis type BI, respectively, depending on the genotyping method used (for simplicity, we will refer to this strain as ribotype 027 throughout the remainder of this paper). This epidemic strain caused large, highly publicized outbreaks in hospitals in Canada [9], the US [10], and the UK [11], which were associated with elevated rates of mortality and caused a change of awareness about CDI severity and epidemiology [12]. While conflicting evidence exists regarding the increased virulence of epidemic ribotype 027 (reviewed in [13]), its high-level resistance to fluoroquinolones may have facilitated its spread in healthcare facilities, where this class of antibiotics is widely used [5, 10].
Population genomic analyses recently showed that, in fact, two distinct ribotype 027 lineages, dubbed FQR1 and FQR2, had emerged in North America, after independently acquiring fluoroquinolone resistance in the 1990s [14]. Evidently, both lineages subsequently had spread into Europe on multiple occasions, and either one of them was also found in Australia and South Korea, respectively [14].
In Germany, cases of CDI with epidemic ribotype 027 were first reported in 2007 from a hospital in Stuttgart [15] and from several hospitals around the city of Trier [16]. A nationwide survey indicated that, by 2008, the dissemination of ribotype 027 was mostly restricted to the southwest of Germany [17]. More recent data suggested that ribotype 027 is among the most predominant C. difficile genotypes in Germany [6] and that its incidence may be increasing [18, 19]. Clostridium difficile 027 was the most frequently isolated ribotype in a recent prospective survey across 20 European countries; however, 43% of ribotype 027 isolates in that study had been found in samples from Germany alone [20]. Ribotype 027 isolates from Germany commonly are highly resistant to fluoroquinolones [6, 17].
It is unclear, at present, to what extent each of the two internationally dispersed fluoroquinolone-resistant lineages, FQR1 and FQR2, contribute to the burden of CDI in Germany, and what the dynamics of their spread among healthcare institutions may be. In the present study, we generated genome sequence data from 57 C. difficile ribotype 027 isolates and analysed this data in a Bayesian phylogeography framework to investigate the temporal dynamics of C. difficile spatial spread in Germany.
Results and Discussion
Phylogeny and population structure
Applying Illumina technology, we re-sequenced the genomes from 57 C. difficile ribotype 027 isolates, 56 of which had been collected from hospitalized patients at 35 locations in Germany between 2007 and 2012, and one isolate had been isolated already in 1990 (S1 Table; sequences were submitted to the European Nucleotide Archive, accession number PRJEB9067). In addition, we included 11 previously published genome sequences representing an international context [14]. Sequencing reads were mapped onto the reference genome sequence from isolate R20291 [14], which had been isolated during a large outbreak in the Stoke Mandeville hospital, UK, in 2005 [11]. After masking variation in mobile genetic elements and repetitive regions, we identified 268 single nucleotide polymorphisms (SNP) in the 3,770,610 basepair core genome (S2 Table). Only a small fraction of mutations (i. e., 13 out of 268 SNPs) were detected in close proximity (≤300 bp) to each other. After removal of these SNPs, the level of homoplasy was very low (homoplasy index, 0.019), suggesting that sequence variation had been generated primarily through mutations and that homologous recombination was rare. The resulting set of 255 SNPs provided the basis for phylogenetic analyses.
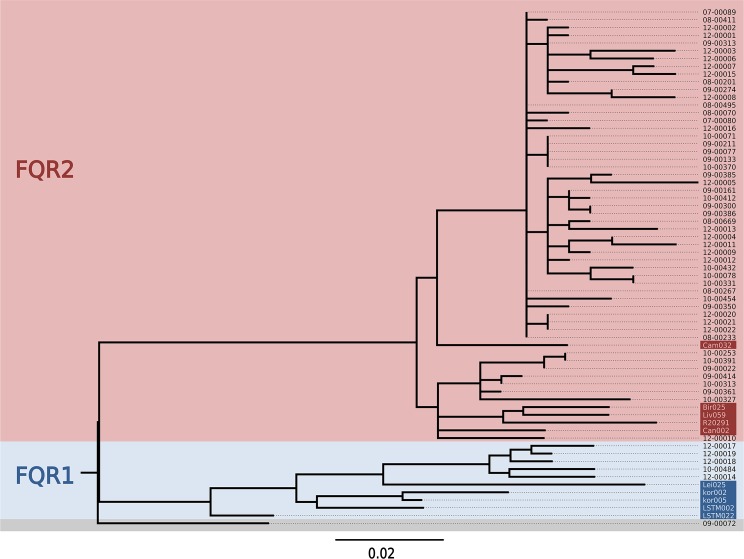
A maximum-likelihood phylogenetic tree revealed two major clades (Fig 1). These clades were identified as corresponding to the previously described strains of fluoroquinolone-resistant C. difficile 027, FQR1 and FQR2, by comparison to several genome sequences from that previous study (Fig 1) [14]. This result demonstrated that both, FQR1 and FQR2, were present in Germany. The majority of isolates in our sample (i. e., 51 out of 57) were related to FQR2, however (Fig 1). As reported previously [14], all FQR1 and FQR2 isolates carried a missense mutation in their DNA gyrase subunit A gene that rendered them fluoroquinolone resistant (S2 Table). In addition, one isolate from Germany (09–00072), which had been sampled in 1990 and was not resistant to fluoroquinolones, sat at a basal position in the tree and represented the ancestral ribotype 027 population, from which FQR1 and FQR2 have emerged (Fig 1, S1 Table) [14]. Bayesian phylogenetic analysis estimated that point mutations had accumulated in the core genome of C. difficile 027 at an average rate of 0.17 mutations per 106 basepairs per year (95% confidence intervals, 0.12 to 0.22 mutations per 106 basepairs per year), which is similar to previous estimates [14, 21]. Based on this calibration, the emergence of FQR1 was estimated to 1998 (95% confidence intervals, 1988 to 2001) and the emergence of FQR2 to 1997 (95% confidence intervals, 1987 to 2000), also in accordance with previous estimates [14].
Fig 1. Maximum likelihood phylogenetic tree.
The phylogeny of C. difficile ribotype 027 was reconstructed based on 255 core-genome SNPs. Previously published genome sequences (indicated by shaded isolate names; [14]) were included to enable identification of clades FQR1 and FQR2. The tree was rooted by using the distantly related BI 3 genome [14] as an outgroup.
The observed evolutionary rate of C. difficile is about 10-fold lower than for Staphylococcus aureus [22–24] and some other bacteria [25], which limits the discriminatory power of C. difficile genome sequencing for molecular epidemiology analyses. Accordingly, and due to the stochastic nature of spontaneous mutations, several groups of our isolates had core genome sequences with no detectable differences, despite their temporal or spatial distances. For example, we found identical genomes in five isolates which had been sampled over 19 months (November 2008 to June 2010) in four different hospitals in Southern Germany (Stuttgart, Sindelfingen, Esslingen, Augsburg) (Fig 1, see 09–00077 and related isolates).
Phylogeographic analyses
For Bayesian phylogeographic inference, we analysed 66 FQR1 and FQR2 genomes, 56 of which had been sampled from 35 geographic locations within Germany (S1 Table), and ten of which originated from other countries (including the UK, USA, Canada, Switzerland, Korea, [14]); the latter were included to place the C. difficile 027 population in Germany into an international context. Not unexpectedly, estimates of rates of spatial spread between pairs of these 35 locations yielded insufficient statistical support, because very few data points were available for each individual estimate. Accordingly, discrete Bayes factor tests failed to verify transmissions determined by the Markov chain Monte Carlo analysis (Bayes factors <3 for all individual transmissions, not shown). Therefore, to increase the statistical power of Bayesian phylogeographic analysis, we grouped neighboring locations into regions based on their straight-line distances (S1 Fig). When the number of locations was reduced to 17 regions in Germany, Bayes factors were ≥20 for individual transmissions within Germany (S5 Table), which commonly is considered a significant level of support [26]. Further reduction to 11 regions in Germany yielded Bayes factors >290 for individual transmissions (S5 Table), indicating strong support [26].
Dynamics of spatial spread
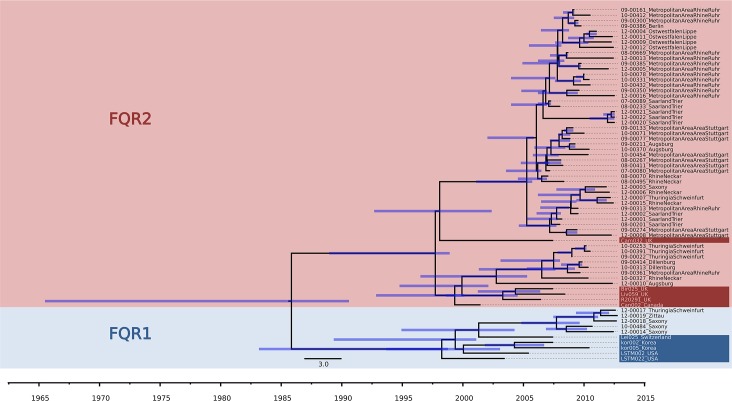
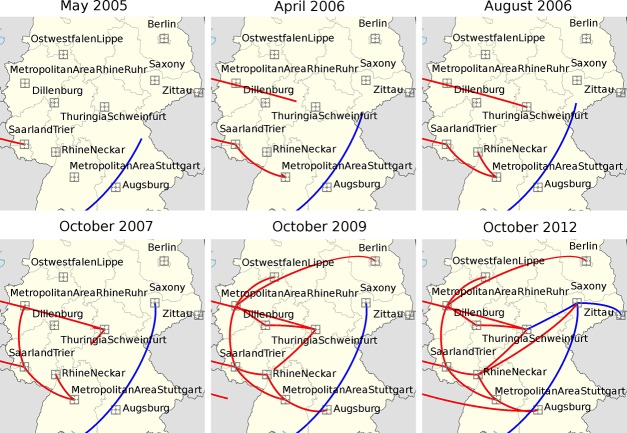
Independent from the level of parameter reduction, our phylogeographic analyses consistently indicated four imports of fluoroquinolone-resistant C. difficile 027 into Germany (Fig 2, Fig 3, S2 Fig and S3 Fig). These four introduction events were inferred from a Bayesian, time-calibrated phylogeny reconstruction (Fig 2) and their directionality was confirmed by Bayesian stochastic search variable selection (BSSVS) (S5 Table) [27, 28]. Lineage FQR2 was indicated to have been introduced into Southwestern Germany during the first half of 2005 (95% confidence interval, 2001 to end of 2005) (Fig 2, Fig 3), hence some years before it caused a major outbreak affecting several hospitals in the area around the city of Trier [16]. Our analysis considering 11 regions indicated the Saarland/Trier region as the first entry point for FQR2 (Fig 3), and subsequent spread from there into the Rhein/Ruhr region, the Stuttgart region, and Thuringia (Fig 3). In contrast, our analysis considering 17 regions (i. e., with moderate parameter reduction) suggested the Stuttgart region as the first entry point (S2 Fig), hence this was not clearly resolved. Interestingly, routine surveillance had detected fluoroquinolone-resistant C. difficile 027 in a hospital in Stuttgart in January 2007 [15], simultaneously with the Trier outbreak [16]. In contrast to previous epidemiological analyses, however, our present analyses consistently showed that, by the beginning of 2007, FQR2 had already been widespread in Germany, affecting four regions in at least four different federal states (Fig 3). Phylogeographic analysis also indicated that the FQR2 lineage was imported two more times, around 2006 and 2012, apparently into the Thuringia and Augsburg regions, respectively (Fig 3). Interestingly, spread of FQR2 was restricted mostly to the West of Germany for several years and did not reach Berlin and Saxony prior to 2009 (Fig 3). This finding is in concordance with the distribution of C. difficile 027 in German hospitals prior to 2009 as detected through epidemiological surveillance [17]. In addition, FQR1 was imported around 2007 (95% HPD, 2004 to 2009), yet its current distribution appears restricted to the East of Germany (i. e., we detected it in the federal states of Sachsen and Thueringen; Fig 3, S1 Table). While available data is limited, it appears from our results and from those of He et al. [14] that FQR1 is less prevalent than FQR2, both in Germany and globally, even though they both emerged in North America around the same time. Possibly, FQR1 is less proliferative than FQR2, but the reasons for this difference have not been resolved.
Fig 2. Maximum clade credibility tree based on BEAST analysis of C. difficile genome sequences.
Tips of the tree were constrained by sampling dates, the time scale is shown at the bottom. Blue bars indicate 95% Bayesian credibility intervals of bacterial divergence dates (node heights).
Fig 3. Bayesian reconstruction of the spread of C. difficile 027 in Germany.
Squares indicate the centroids of 11 regions and lines indicate the inferred spread of FQR1 (blue) and FQR2 (red). Note that, in this discrete analysis, arrival at a region centroid indicates arrival at that region, but trajectories do not represent precise transport routes.
Limitations
Even though our set of C. difficile isolates is the most comprehensive collection of C. difficile ribotype 027 isolates from Germany studied to date, the number of isolates investigated (n = 57) is very small compared to the number of hospitals or to annual numbers of CDI cases [17, 20, 29]. Therefore, the scenario of C. difficile dissemination and spread we describe may underestimate the complexity of the real situation. Because data available from isolates collected internationally are even more limited [14], we did not attempt to identify sources for introduction into Germany.
Conclusions
Genome-based phylogenetic analysis indicated that fluoroquinolone-resistant C. difficile ribotype 027 was imported into Germany at least four times independently. Despite the limited number of isolates included and the comparatively low evolutionary rate of C. difficile, genome re-sequencing provided conclusive information for Bayesian inference of spread between regions within Germany based on a discrete phylogeographic model. Our analyses indicated that one of the previously described fluoroquinolone-resistant variants of C. difficile 027 (i. e., FQR2) has spread within Germany for more than a decade, and the other variant (FQR1) had been introduced few years later. At the time when the first outbreak of C. difficile 027 was noticed in Germany in 2007 [16], these newly emerged strains had already been disseminated across hospitals in several federal states, and they have continued to spread since then.
Methods and Tools
Isolates
Clostridium difficile ribotype 027 isolates (S1 Table) were selected based on results from classical ribotyping [17] or surface layer protein A sequence typing [30], respectively, and based on their geographic origin, in order to achieve maximum representation of the pathogen's spatial distribution in Germany. As a result, we included 57 C. difficile ribotype 027 isolates, collected from hospitalized patients at 36 hospitals in Germany (S1 Table).
Ethics statement
A formal ethical review process and approval was not required in accordance with article 25, section 1 of the German Infection Protection Act of 2001. All data were analyzed anonymously.
Raw data
The genomes from C. difficile isolates were sequenced on HiScan and MiSeq systems (Illumina), producing paired-end reads with lengths of 100 or 250 bases, respectively, to an average >35-fold coverage. Sequencing data were submitted to the European Nucleotide Archive (ENA) and assigned study accession number PRJEB9067. To enable comparisons to previously published genome data [14], sequencing reads from representative FQR1 and FQR2 isolates (Fig 1) were downloaded from ENA and included in our analyses. Consensus sequences for individual genomes were determined by applying a read mapping approach combining BWA-SW version 0.7.3a [31] for mapping to a reference genome sequence (acc. no. NC_013316 [32]), SAMtools [33] to handle SAM files (Sequence Alignment/Map) and VarScan (version 2.3) [34] for consensus calling in a customized pipeline framework (VarScan parameters: minimum coverage, 10; minimum average base quality, 20; minimum variant frequency, 0.8; p-value threshold, 0.01). The high-level interpreted language GNU Octave [35] was used to format output files, to analyse the SNP content, and to assemble an alignment of SNPs. Previously published sequencing reads from 11 C. difficile genomes were included for reference [14](Fig 1).
Mobile genetic elements and repetitive sequences
Mobile genetic elements may evolve at a different mode and rate than the remainder of the genome, and it is therefore wise to exclude them from phylogenetic analyses. To assort a list of mobile genetic elements in the reference genome, the annotation was screened for keywords and the web server based application PHAST [36] were used to identify mobile genetic elements [32, 37, 38], which were then excluded from phylogenetic analyses (S3 Table). We also excluded CRISPR (S3 Table) and other repetitive DNA sequences, as they tend to create ambiguities in read alignments [39]. Repetitive DNA was detected with the pattern matching engine available in KODON software (Applied Math).
Phylogenetic analyses
Phylogeny reconstruction is based on the assumption of tree-like evolution, which is violated by homologous recombination that creates mosaic sequences [40]. Therefore we screened our sequences for clustered variation and masked all mutations that had a distance of ≤300 bp from the next mutation. Based on an alignment of core-genome SNPs, PAUP 4b10 (http://paup.csit.fsu.edu/) was used to calculate the homoplasy index, and PhyML implemented in Seaview 4 was used to calculate a maximum-likelihood phylogenetic tree.
Bayesian inference with BEAST 2.0
To incorporate spatial and temporal components into the phylogeny, the BEAST 2.0 [41] cross-platform was used. It implements a Bayesian framework focused on using strict or relaxed molecular clock models for inference of time-measured phylogenies. As a total re-implementation of the BEAST 1.x software package it provides the option to extend the system with new models via the package system. BEAST 2.0 provides a full Bayesian framework for phylogeographic analysis [27]. The model setup was done following the tutorial "Ancestral reconstruction/discrete phylogeography with BEAST 2.0 (available at http://www.beast2.org/wiki/index.php/Tutorials) with the BEAUti2 xml editor as a part of the BEAST 2.0 framework. We used the beast-classic (BEAST_CLASSIC) add-on and the BEASTii add-on as described in the tutorial. Each sequence was labeled with the year and month of isolate sampling and with ETRS89 (European Terrestrial Reference System 1989) coordinates of sampling locations. Using the HKY model of nucleotide substitution, a strict clock and an uncorrelated relaxed lognormal clock model were tested. The initial clock rate was set to 2 x 10−7 substitutions per nucleotide site and per year. On the prior for the nonzero rates (Poisson distribution), the lambda (expected value, variance) was set to 100,693. To test the influence of several tree priors, analyses with different coalescent priors were applied in several runs, under the assumption of a strict clock and an uncorrelated lognormal relaxed clock (S4 Table). For the combined FQR1/FQR2 set without 09–00072 and BI–3 (66 sequences total), 6 x 107 generations was enough to verify proper mixing. Effective sample sizes (ESS) were >200. Trees from every 6,000 generations were sampled. To verify proper priors and to insure that the results are significantly informed by the data, several runs with sampling from priors only were performed and analysed. Samples from three independent Markov chain Monte Carlo runs were combined by applying the LogCombiner software with an exclusion of 15% burn-in [41]. For model comparison, Bayes factors were calculated based on marginal likelihood estimated by using the smoothed harmonic mean estimator [42, 43], as implemented in the Tracer application version v1.6.0.
Bayesian phylogeography
For discrete phylogeographic analysis, ETRS89 coordinates of sampling locations were incorporated into the model as an additional discrete location trait, applying the BEAST-classic add-on during the model setup with BEAUti 2.0 as described by Lemey et al [27]. The Markov chain Monte Carlo analysis was performed as described above, assuming an uncorrelated lognormal relaxed clock and a coalescent constant tree prior. Effective sample sizes (ESS) were >200. Tree files from three independent runs were combined and then processed with the SPREAD 1.0.6 software (Spatial Phylogenetic Reconstruction of Evolutionary Dynamics) [28] to determine the spatial spread in Germany. To identify well-supported transition rates, discrete Bayes factor tests were performed on a BEAST log file with rate indicators (Bayesian stochastic search variable selection (BSSVS) procedure; cutoff, 3) [27]. To increase the statistical power of Bayesian phylogeographic inference, we grouped neighboring locations into regions by using a hierarchical cluster analysis based on straight-line-distances and using the centroid method assuming Euclidean metric (S1 Fig). Setting the level of hierarchy to reduce the number of locations is a compromise to maintain informative spatial resolution and ensure robust statistics at the same time. This way, 11 or 17 regions, respectively, were defined to represent the total of 35 locations in Germany from which isolates had originated, where each region was represented by the coordinates of the centroid of the locations [44]. With these adjustments to the priors, Markov chain Monte Carlo analysis was re-run, and the combined sample data from three independent runs underwent discrete Bayes factor tests to verify the inferred transition rates S5 Table. The visualization of the proliferation dynamics of C. difficile in time and space was derived from models assuming a relaxed clock.
Supporting Information
(TIF)
Discrete phylogeographic analysis was performed with BEAST 2.0 and SPREAD 1.0.6.
(TIF)
Discrete phylogeographic analysis was performed with BEAST 2.0 and SPREAD 1.0.6.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Clock rates and Bayes factor tests.
(XLSX)
Level of statistical support for individual spreading events.
(XLSX)
Acknowledgments
We thank Wiebke Streckel and Anna Nimmesgern for technical assistance and Boyke Bunk and Adam Podstawka for bioinformatics support. Remco Bouckaert provided expert advice on usage of BEAST 2.0 which we greatly appreciated.
Data Availability
All sequence data files are available from the European Nucleotide Archive database (accession number PRJEB9067).
Funding Statement
The National Consultant Laboratory for Clostridium difficile at the University of Saarland Medical Centre in Homburg was partially funded by a grant from the Robert Koch Institute (to MH). The work of MS was funded through a PhD stipend from the Robert Koch Institute (to UN). This work was partially funded by the European Union, project PathNgenTrace (FP7-278864-2). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Suetens C, Hopkins S, Kolman J, Högberg LD. Point prevalence survey of healthcareassociated infections and antimicrobial use in European acute care hospitals 2011–2012: European Centre for Disease Prevention and Control; 2013. Available from: www.ecdc.europa.eu. [Google Scholar]
- 2. Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, et al. Multistate point-prevalence survey of health care-associated infections. The New England journal of medicine. 2014;370(13):1198–208. 10.1056/NEJMoa1306801 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pacheco SM, Johnson S. Important clinical advances in the understanding of Clostridium difficile infection. Current opinion in gastroenterology. 2013;29(1):42–8. 10.1097/MOG.0b013e32835a68d4 . [DOI] [PubMed] [Google Scholar]
- 4. Bassetti M, Villa G, Pecori D, Arzese A, Wilcox M. Epidemiology, diagnosis and treatment of Clostridium difficile infection. Expert review of anti-infective therapy. 2012;10(12):1405–23. 10.1586/eri.12.135 . [DOI] [PubMed] [Google Scholar]
- 5. Lessa FC, Gould CV, McDonald LC. Current status of Clostridium difficile infection epidemiology. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;55 Suppl 2:S65–70. 10.1093/cid/cis319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. von Müller L, Haffmann A, Herrmann M. Aktuelle Daten und Trends zur Antibiotikaresistenzentwicklung von Clostridium difficile . Bundesgesundheitsblatt. 2012;55:1410–7. [DOI] [PubMed] [Google Scholar]
- 7. McDonald LC, Killgore GE, Thompson A, Owens RC Jr., Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile . The New England journal of medicine. 2005;353(23):2433–41. . [DOI] [PubMed] [Google Scholar]
- 8. Kuijper EJ, Coignard B, Tüll P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2006;12 Suppl 6:2–18. . [DOI] [PubMed] [Google Scholar]
- 9. Pepin J, Valiquette L, Cossette B. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ. 2005;173(9):1037–42. Epub 2005/09/24. cmaj.050978 [pii] 10.1503/cmaj.050978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muto CA, Pokrywka M, Shutt K, Mendelsohn AB, Nouri K, Posey K, et al. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infection control and hospital epidemiology: the official journal of the Society of Hospital Epidemiologists of America. 2005;26(3):273–80. 10.1086/502539 . [DOI] [PubMed] [Google Scholar]
- 11.Anonymous. Investigation into outbreaks of Clostridium difficile at Stoke Mandeville Hospital, Buckinghamshire Hospitals NHS Trust: Healthcare Commission Report; available at http://www.buckinghamshirehospitals.nhs.uk/healthcarecommision/HCC-Investigation-into-the-Outbreak-of-Clostridium-Difficile.pdf2006.
- 12. Hunt JJ, Ballard JD. Variations in virulence and molecular biology among emerging strains of Clostridium difficile . Microbiology and molecular biology reviews: MMBR. 2013;77(4):567–81. 10.1128/MMBR.00017-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerding DN, Johnson S. Does infection with specific Clostridium difficile strains or clades influence clinical outcome? Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2013;56(11):1601–3. 10.1093/cid/cit133 . [DOI] [PubMed] [Google Scholar]
- 14. He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile . Nature genetics. 2013;45(1):109–13. Epub 2012/12/12. 10.1038/ng.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaiß NH, Weile J, Ackermann G, Kuijper E, Witte W, Nübel U. A case of Clostridium difficile-associated disease due to the highly virulent clone of Clostridium difficile PCR-ribotype 027, March 2007 in Germany. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2007;12(11):E0711151. . [DOI] [PubMed] [Google Scholar]
- 16. Kleinkauf N, Weiss B, Jansen A, Eckmanns T, Bornhofen B, Kuehnen E, et al. Confirmed cases and report of clusters of severe infections due to Clostridium difficile PCR ribotype 027 in Germany. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2007;12(11):E0711152. Epub 2007/11/17. 2307 [pii]. . [DOI] [PubMed] [Google Scholar]
- 17. Zaiß NH, Witte W, Nübel U. Fluoroquinolone resistance and Clostridium difficile, Germany. Emerg Infect Dis. 2010;16(4):675–7. Epub 2010/03/31. 10.3201/eid1604.090859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arvand M, Vollandt D, Bettge-Weller G, Harmanus C, Kuijper EJ, Clostridium difficile study group H. Increased incidence of Clostridium difficile PCR ribotype 027 in Hesse, Germany, 2011 to 2013. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2014;19(10). . [DOI] [PubMed] [Google Scholar]
- 19. Anonymous. Schwer verlaufende Clostridium-difficile-Infektionen: IfSG-Surveillancedaten von 2013. Epidemiologisches Bulletin. 2014;(27):233–7. [Google Scholar]
- 20. Davies KA, Longshaw CM, Davis GL, Bouza E, Barbut F, Barna Z, et al. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID). The Lancet infectious diseases. 2014;14(12):1208–19. 10.1016/S1473-3099(14)70991-0 . [DOI] [PubMed] [Google Scholar]
- 21. Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, et al. Diverse sources of Clostrdium difficile infection identified on whole-genome sequencing. The New England journal of medicine. 2013;369(13):1195–205. 10.1056/NEJMoa1216064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nübel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, et al. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus . PLoS pathogens. 2010;6(4):e1000855 10.1371/journal.ppat.1000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–74. Epub 2010/01/23. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, et al. A genomic portrait of the emergence, evolution and global spread of a methicillin resistant Staphylococcus aureus pandemic. Genome research. 2013;23:653–64. Epub 2013/01/10. 10.1101/gr.147710.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–4. Epub 2011/01/29. 10.1126/science.1198545 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kass RE, Raftery AE. Bayes factors Journal of the American Statistical Association. 1995;90:773–95. [Google Scholar]
- 27. Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS computational biology. 2009;5(9):e1000520 Epub 2009/09/26. 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27(20):2910–2. 10.1093/bioinformatics/btr481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geffers C, Gastmeier P. Nosokomiale Infektionen und multiresistente Erreger in Deutschland. Deutsches Ärzteblatt. 2011;108(6):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joost I, Speck K, Herrmann M, von Muller L. Characterisation of Clostridium difficile isolates by slpA and tcdC gene sequencing. International journal of antimicrobial agents. 2009;33 Suppl 1:S13–8. 10.1016/S0924-8579(09)70010-X . [DOI] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome biology. 2009;10(9):R102 Epub 2009/09/29. gb-2009-10-9-r102 [pii] 10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22(3):568–76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton JW, Bateman D, Hauberg S, Wehbring R. GNU Octave version 3.8.1 manual: a high-level interactive language for numerical computations. CreateSpace Independent Publishing Platform2014.
- 36. Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic acids research. 2011;39(Web Server issue):W347–52. Epub 2011/06/16. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One. 2011;6(8):e23014 10.1371/journal.pone.0023014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. Journal of clinical microbiology. 1998;36(8):2240–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature reviews Genetics. 2012;13(1):36–46. 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172(4):2665–81. 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS computational biology. 2014;10(4):e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Newton MA, Raftery AE. Approximate Bayesian inference with the weighted likelihood bootstrap (with discussion). Journal of the Royal Statistical Society Series B. 1994;56:3–48. [Google Scholar]
- 43. Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Molecular biology and evolution. 2001;18(6):1001–13. . [DOI] [PubMed] [Google Scholar]
- 44. Gray RR, Tatem AJ, Johnson JA, Alekseyenko AV, Pybus OG, Suchard MA, et al. Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a bayesian framework. Molecular biology and evolution. 2011;28(5):1593–603. Epub 2010/11/30. 10.1093/molbev/msq319 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Discrete phylogeographic analysis was performed with BEAST 2.0 and SPREAD 1.0.6.
(TIF)
Discrete phylogeographic analysis was performed with BEAST 2.0 and SPREAD 1.0.6.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Clock rates and Bayes factor tests.
(XLSX)
Level of statistical support for individual spreading events.
(XLSX)
Data Availability Statement
All sequence data files are available from the European Nucleotide Archive database (accession number PRJEB9067).