Abstract
Multiciliated epithelial cells protect the upper and lower airways from chronic bacterial infections by moving mucus and debris outward. Congenital disorders of ciliary beating, referred to as primary ciliary dyskinesia (PCD), are characterized by deficient mucociliary clearance and severe, recurrent respiratory infections. Numerous genetic defects, most of which can be detected by transmission electron microscopy (TEM), are so far known to cause different abnormalities of the ciliary axoneme. However, some defects are not regularly discernable by TEM because the ciliary architecture of the axoneme remains preserved. This applies in particular to isolated defects of the nexin links, also known as the nexin-dynein regulatory complex (N-DRC), connecting the peripheral outer microtubular doublets. Immunofluorescence analyses of respiratory cells from PCD-affected individuals detected a N-DRC defect. Genome-wide exome sequence analyses identified recessive loss-of-function mutations in GAS8 encoding DRC4 in three independent PCD-affected families.
Introduction
Primary ciliary dyskinesia (PCD) is a genetically heterogeneous autosomal-recessive disorder characterized by recurrent upper and lower airway infections causing progressive lung damage (MIM: 244400). These chronic infections are triggered by dysfunction of multiple motile cilia lining the respiratory epithelium and resulting in a decreased muco-ciliary clearance. Thus, mucus and pathogens accumulate in the lower airways, leading to chronic inflammation and bronchiectasis.1,2 With an incidence of 1:4,000 to 1:60,000, PCD is a rare heterogeneous genetic disorder.3 During recent years, several distinct genetic variants have been identified.2,4
The architecture of the motile respiratory cilium is highly conserved and shows a 9+2 structure of the axoneme with nine outer doublets surrounding a central pair of two single microtubules (Figure S1A). The outer and inner dynein arms (ODAs and IDAs) are large multimeric protein complexes and generate the force for axonemal bending via ATP hydrolysis. The ODAs are responsible for the main beating force, whereas the IDAs are supposed to coordinate the waveform of the ciliary beating. The dynein arms are attached to the A-tubules of the outer doublets, which are connected to the central pair apparatus by the radial spokes (Figure S1A). Most genetic variants identified so far result in abnormalities of the ODAs and are caused by mutations in genes encoding either structural ODA motor proteins (DNAH5 [MIM: 603335], DNAI1 [MIM: 604366], DNAI2 [MIM: 605483], DNAL1 [MIM: 610062], TXNDC3 [MIM: 607421], DNAH11 [MIM: 603339], CCDC103 [MIM: 614677]),5–12 ODA-docking-complex components (CCDC114 [MIM: 615038], ARMC4 [MIM: 615408], CCDC151 [MIM: 615956]),13–16 or members of the cytoplasmic dynein-arm-assembly machinery (DNAAF1 [KTU] [MIM: 612517], DNAAF2 [LRRC50] [MIM: 613190], DNAAF3 [MIM: 614566], DNAAF4 [DYX1C1] [MIM: 615482], LRRC6 [MIM: 614930], HEATR2 [MIM: 614864], ZMYND10 [MIM: 607070], SPAG1 [MIM: 603395], C21ORF59 [MIM: 615494]).17–26 ODA defects are usually readily identified by transmission electron microscopy (TEM) or immunofluorescence analysis and exhibit severe ciliary beating defects. In addition, PCD variants caused by mutations in genes that result in abnormal radial-spoke (RSPH4A [MIM: 612649], RSPH9 [MIM: 612650], RSPH1 [MIM: 609314], and RSPH3 [MIM: 615876]) or central-pair (HYDIN [MIM: 61081]) composure have been reported.27–30 Detailed summaries of the different PCD variants have been published recently.1,2,4The nexin-dynein regulatory complex (N-DRC), also called the nexin link, is anchored to the A-tubule of ciliary peripheral tubulin doublets and expands toward the B-tubule of the adjacent doublet (Figure S1). The ruler proteins encoded by CCDC39 (MIM: 613798) and CCDC40 (MIM: 613799) (Figure S1B) are important for maintenance of the 9+2 integrity of the axoneme and are responsible for attachment of the N-DRC and IDAs. CCDC39 or CCDC40 mutations cause tubular disorganization and absence of the N-DRC as well as IDA proteins.31,32 We and others recently identified mutations in CCDC164 (MIM: 615294) and CCDC65 [MIM: 611088], encoding the N-DRC proteins DRC1 and DRC2, respectively (Figure S1B).26,33,34 CCDC164 and CCDC65 mutant respiratory cilia show no obvious ultrastructural defects and exhibit only subtle abnormalities of ciliary beating. Here, we report recessive loss-of-function mutations of GAS8 (also originally designated GAS11), encoding DRC4, that are responsible for PCD in three unrelated PCD-affected families. The genetic findings are of particular importance because this PCD variant resembles other N-DRC defects and is hardly detectable by routine diagnostic tests such as high-speed videomicroscopy or TEM. However, we show that this defect is efficiently detected by immunofluorescence analysis and genetic testing.
Material and Methods
PCD-Affected Individuals and Families
We studied DNA of PCD-affected individuals and relatives. Signed and informed consent was obtained from PCD-affected individuals and their family members according to protocols approved by the institutional ethics review boards.
Nasal Nitric Oxide Measurement
While performing an exhalation-against-resistance maneuver, nasal nitric oxide (NO) was measured with chemiluminescence NO analyzers EcoMedics CLD88 (Duernten) or Niox Flex (Aerocrine) as described previously.30
High-Speed Video Analyses of Ciliary Beat Pattern in Human Cells
Ciliary beating was assessed with the SAVA imaging analysis system.35 Nasal brush biopsies were washed in cell culture medium and immediately recorded with a Basler scA640-120 fm digital high-speed video camera (Basler) attached to an inverted phase-contrast microscope (Zeiss Axio Vert. A1; Carl Zeiss) armed with a ×40 and ×63 objective. Digital image sampling was performed at 640 × 480 pixel resolution and 120–150 frames per second. The ciliary beating pattern was evaluated on slow-motion playbacks.
Sequencing
Whole-Exome Sequencing
Exome sequencing of genomic DNA from the two PCD-affected individuals OP-929 and OP-1940 was performed at the Cologne Center for Genomics. For enrichment, the NimbleGen SeqCap EZ Human Exome Library v2.0 was used. Enriched libraries were sequenced on a HiSeq2000 instrument (Illumina) with a paired end 2 × 100 base-pairs protocol. At least 87% (OP-929) and 94% (OP-1940) of target sequence was covered. Sequencing reads that passed quality filtering were mapped to the reference genome sequence (UCSC Genome Browser hg19). Variants were analyzed with the Varbank software.
Sanger Sequencing
Genomic DNA was isolated directly from blood samples by standard methods. Specific primers for all exons, including splice-site regions of GAS8 (GenBank: NM_001481.2), were designed. Each PCR was performed in a volume of 50 μl containing 30 ng DNA, 50 pmol of each primer, 2 mM dNTPs, and 1.0 U GoTaq DNA polymerase (Promega Corporation). Amplifications were carried out by means of an initial denaturation step at 94°C for 3 min and 30 cycles as follows: 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, with a final extension at 72°C for 10 min. PCR products were verified by agarose gel electrophoresis, purified, and sequenced bi-directionally with BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems). Sequence data were evaluated with the CodonCode software (CodonCode Corporation).
TEM
Ciliated respiratory epithelial cells were obtained from the middle turbinate by nasal brush biopsy (Engelbrecht Medicine and Laboratory Technology). The samples were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer at 4°C, washed overnight, and postfixed in 1% osmium tetroxide. After dehydration, the samples were embedded in a mixture of propylene oxide and epoxy resin. After polymerization, several resin sections were cut with an ultramicrotome (60 or 80 nm thin). Sections were picked up onto copper grids. The sections were stained with aqueous 1% uranyl acetate and Reynold’s lead citrate. TEM was performed with Zeiss 10 LEO912AB (zero-loss mode) EFTEM or MORGAGNI 268 (Philips).
High-Resolution Immunofluorescence Microscopy
Respiratory epithelial cells were obtained by nasal brushing and suspended in cell culture medium. Samples were spread onto glass slides, air-dried, and stored at −80°C. Cells were treated with 4% paraformaldehyde (or 100% ice-cold methanol) and 0.2% Triton X-100. The slides used for the anti-GAS8-, anti-CCDC39-, and anti-DNALI1-stainings were blocked overnight with 1% skim milk before incubation with the primary antibodies for about 3 to 4 hr at room temperature the next day. The slides for the anti-LRRC48-staining were blocked with 5% skim milk for 3 hr at room temperature and incubated with the primary antibodies at 4°C overnight. The incubation with the secondary antibodies was performed for 30 min at room temperature. Rabbit polyclonal antibody directed against DNAH5 has previously been reported.36 Rabbit polyclonal anti-CCDC39 (1:300), anti-GAS8 (1:500), and anti-LRRC48 (1:500) were obtained from Atlas Antibodies. The polyclonal anti-DNALI1 antibody (1:250) was offered by Prof. Neesen. Antibody specificity for antibodies directed against CCDC39, LRRC48, and DNALI1 was reported previously.31,33,37
Mouse monoclonal antibody directed against acetylated α-tubulin (1:10,000) was obtained from Sigma. Highly cross-absorbed secondary antibodies, including Alexa-Fluor-488-conjugated goat antibodies to mouse (1:1,000, A11029) and Alexa-Fluor-546-conjugated goat antibodies to rabbit (1:1,000, A11035) were purchased from Molecular Probes (Invitrogen). DNA was stained with Hoechst33342 (1:1,000, 14533-100MG, Sigma). Immunofluorescence images were taken by means of a Zeiss Apotome Axiovert 200 (processed with AxioVision 4.8) or Zeiss LSM880 (processed with ZEN2 software). Figures were prepared with Adobe Creative Suite 4.
Western Blot Analysis
Western blot analysis to demonstrate specificity of the anti-GAS8 antibodies was performed as previously described.21 Human respiratory cell lysates were prepared as previously described.21 Primary antibody was diluted 1:1,000.
Mouse Experiments
Mouse experiments complied with ethical regulations and were approved by local government authorities (AZ 87-51.05.20.11.021, Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen).
Trachea of wild-type and lnks/lnks mice (The lnks-mutant mouse line was established as part of the Sloan-Kettering Institute Mouse Project R37-HD035455)32 were dissected in 1× PBS, embedded in Cryomatrix (Thermo Scientific), frozen on dry ice, and cut into 25-μm sections with a cryostat (CM 3050s, Leica). For immunofluorescent analysis on embryos, staged embryos were dissected at embryonic day E7.75 in 1× PBS. The sections and dissected embryos were fixed in 4% paraformaldehyde in 1× PBS for 15 min. After three washing steps with 0.1% Triton X-100 in 1× PBS for 5 min each, the sections were blocked in 1% BSA in 1× PBS containing 0.1%Triton-X for 2 hr (overnight for embryos) and incubated with primary antibodies overnight. The following primary antibodies were used: mouse monoclonal antibody to acetylated α-tubulin (T7451, Sigma, 1:4,000 dilution) and rabbit polyclonal anti-GAS8 (HPA041311, Atlas Antibodies, 1:500 dilution). After being washed three times in 0.1% Triton-X in 1× PBS, tissues were incubated with appropriate dye-conjugated secondary antibodies (anti-mouse Alexa Flour 488 [A-21200, Molecular Probes], anti-rabbit Rhodamine Red-X [711-295-152, Dianova]) at a dilution of 1:200 for 1 hr. Secondary antibodies alone were used as a control. Samples were washed three times in 1× PBS containing DAPI (1:10,000 dilution) for counterstaining of nuclei. Stained sections were rinsed with 1× PBS, and coverslips were applied with Fluorescence Mounting Medium (Dako). Staged embryos were mounted on StarFrost slides (Knittel Glas), and coverslips were applied with Fluorescence Mounting Medium (Dako). Images were acquired on a Zeiss LMS880 laser scanning microscope and processed with ZEN2 Software and Adobe Creative Suite 4.
Results
Identification of PCD-Affected Individuals with GAS8-Deficient Respiratory Cilia
Our laboratory routinely uses antibodies targeting the ODAs (anti-DNAH5) and the N-DRCs (anti-GAS8) for diagnostic purposes in the work-up of individuals suspected to suffer from PCD. The used polyclonal anti-GAS8 antibodies specifically recognize two of the known isoforms of GAS8 (Figure S2). To identify individuals with abnormal N-DRCs, we searched our database for PCD-affected individuals with abnormal ciliary composition of GAS8 (human DRC4) and normal axonemal localization of the ODA protein DNAH5 as documented by immunofluorescence microscopy.26,33 To exclude N-DRC defects due to abnormal function of the ruler proteins CCDC39 or CCDC40, we also restricted our studies to individuals with normal axonemal localization of CCDC39.31,32 We initially identified two individuals (OP-929, OP-1940) with this staining pattern in our database. During the study, we detected a third individual, OP-1627, with the same staining pattern. The absence of the N-DRC protein GAS8 from the whole ciliary axoneme as well as the normal axonemal localization of the ODA protein DNAH5 are shown in Figure 1. The normal localization of the ruler protein CCDC39 is presented in Figure S3.
Figure 1.
Immunofluorescence Analyses of Ciliated Respiratory Cells Reveal GAS8 Deficiency in Individuals OP-929, OP-1940, and OP-1627
In control respiratory epithelial cells, the ODA protein DNAH5 (green) and the N-DRC protein GAS8 (red) co-localize within the axonemes. The respiratory cilia of the PCD-affected individuals demonstrate normal axonemal DNAH5 localization, whereas GAS8 is absent from the ciliary axonemes in the PCD-affected individuals OP-929, OP-1940, and OP-1627. Nuclei were stained with Hoechst 33342 (blue). Scale bars represent 10 μm.
Mutation Analyses
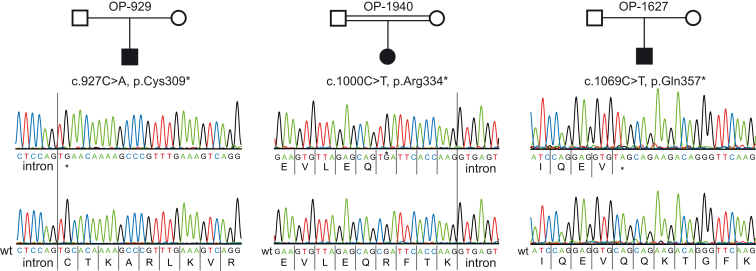
We have previously demonstrated that recessive mutations of CCDC164 and CCDC65 encoding DRC1 and DRC2, respectively, can result in PCD characterized by isolated defects of the N-DRC (Figure S1B).26,33 Therefore, we first performed Sanger sequencing of all coding exons from CCDC164 and CCDC65 in the DNA samples from OP-929 and OP-1940.26,33 Interestingly, we found no evidence for CCDC164 or CCDC65 mutations, indicating that mutations in another gene are responsible for the observed phenotype in those PCD-affected individuals. To identify the underlying genetic defect, we performed whole-exome sequencing and prioritized our analyses for the presence of mutations in genes encoding other DRC proteins (summarized in Table S1). With this approach, we found two different homozygous nonsense mutations (OP-929: c.927C>A [p.Cys309∗]; OP-1940: c.1000C>T [p.Arg334∗]) within exon eight of GAS8 (GenBank: NM_001481.2). We verified these mutations by Sanger sequencing. During the progress of the study, we identified an additional PCD-affected individual with an identical defect of the N-DRC. Therefore, we also analyzed this DNA for the presence of GAS8 mutations and identified a homozygous nonsense mutation (c.1069C>T [p.Gln357∗]) (Figure 2). This mutation is annotated in the Single Nucleotide Polymorphism database (dbSNP) with a minor allele frequency, whereas the other two mutations are absent from the 1000 Genomes38 and Exome Aggregation Consortium (ExAC) databases. Consistent with homozygous mutations due to a common ancestor, consanguinity was reported in OP-1940. Consanguinity is also probably responsible for homozygous mutations in OP-1627 because both parents originate from the same small village in a remote area of Sri Lanka. Consanguinity in the Danish family OP-929 has not been reported, thus we cannot rule out the possibility of a heterozygous deletion on one allele. Parental DNA was not available to answer that point. However, in any case, the detected GAS8 mutations represent classical loss-of-function mutations. Consistently, GAS8 was not detectable in GAS8-mutant respiratory cells (Figure 1).
Figure 2.
GAS8 Mutations in Three PCD-Affected Individuals with Absent GAS8/DRC4 in Respiratory Cells
All affected individuals carry GAS8 homozygous nonsense mutations, which are predicted to cause a premature stop of translation. The mutations are located in exon eight (OP-929: c.927C>A [p.Cys309∗]; OP-1940: c.1000C>T [p.Arg334∗]) and exon nine (OP-1627: c.1069C>T [p.Gln357∗]). Consanguinity was reported for OP-1940.
Clinical Data
The parents of the PCD-affected individual OP-1627 are not aware of consanguinity but originate from the same remote village in Sri Lanka. PCD diagnosis was established at the age of 6 years in individual OP-1627. This individual has a history of chronic rhinosinusitis, chronic wet cough since early childhood, recurrent bronchitis and pneumonia, and recurrent otitis media with effusion. After insertion of grommets, he had suffered from purulent otorrhea for several months. Audiometry revealed conductive hearing loss without signs of inner ear impairment. Computed tomography of the lungs at the age of 7 years demonstrated consolidation of the middle and lingual lobes with early bronchiectasis. His nasal NO production rate was low (100 nl/min), consistent with PCD.39
OP-1940 is a 21-year-old female from Libya who attended our PCD clinics for a diagnostic work-up; she has a history of chronic cough since early childhood, chronic rhinosinusitis, recurrent chest infections, and marked bronchiectatic lung disease. Her parents are first-degree cousins. Two of her seven siblings also suffer from chronic cough and bronchiectasis but were not available for further genetic analyses. Her nasal NO production rate was low (31.4 nl/min), consistent with PCD.
OP-929 is a 27-year-old Danish male born to non-consanguineous parents. PCD was diagnosed in 2000 due to classical clinical symptoms. He has a history of recurrent pneumonia since birth, chronic rhinitis, recurrent otitis media with effusion leading to several grommet insertions, and reduced conductive hearing. Middle lobe atelectasis has been demonstrated by chest X-ray. His nasal NO production rate was also low (36.6 nl/min), consistent with PCD.
High-Speed Videomicroscopy
We were able to perform nasal brushing biopsies in all affected persons. The analysis of the respiratory cilia did not show any significant anomalies. The frequency of the ciliary beat was within the normal range of 5–8 Hz at room temperature (Supplemental Movies S1 and S2). High-speed videomicroscopy analyses were initially rated as normal. Careful analyses of the ciliary beat pattern revealed a subtle reduction of the beating amplitude compared to control cilia (Supplemental Movie S3). Because GAS8 mutations resulted in very subtle beating defects, diagnosis of PCD in the three PCD-affected individuals presented here relied on detection of low nasal NO levels as well as demonstration of abnormal motor protein composition by immunofluorescence analyses.
TEM
TEM images of cross sections of respiratory cilia were available from PCD-affected individuals OP-929 and OP-1627 with GAS8 mutations. We analyzed the ultrastructure of a total of 151 cilia cross sections (OP-929 [n = 60]; OP-1627 [n = 91]). As expected, careful ultrastructural analyses did not identify a defect of the ODAs, which is consistent with the demonstration of normal axonemal localization of the ODA chain DNAH5 by immunofluorescence analyses. Overall, we found a normal ultrastructural 9+2 composition of the cilia. We also did not observe any defects of the central single tubules or gross tubular disorganization comparable to those associated with CCDC39 or CCDC40 mutations. However, careful analyses of the cross sections for subtle changes revealed an increased frequency of misaligned outer doublets in GAS8-mutant cilia (Figure 3). In approximately 26% (40 of 151) of the analyzed cross sections, outer doublets were not correctly aligned. For comparison, cilia cross sections from two healthy control individuals did not show this extent of misaligned outer doublets. Here, we found a range from 4.5% (2 of 44 cross sections) to 11% (6 of 55 cross sections). However, because altered positioning of outer doublets can also occur as a result of inflammation due to other causes, diagnosis of the GAS8 defect, like other N-DRC defects, cannot be determined solely by TEM.26,33,34
Figure 3.
TEM of GAS8-Mutant Cilia Does Not Exhibit Gross Ultrastructural Abnormalities
The analyses of cross sections from OP-1627 and OP-929 predominantly showed a normal ultrastructure with a 9+2 architecture. Abnormalities of the ODAs, as well as of central tubules, were not detected. Only the frequency of misaligned outer doublets was slightly increased in comparison to that of control cilia (25% versus 5%–10%). Misaligned outer doublets are marked with a white ellipse. Scale bars represent 100 nm.
Molecular Characterization of the GAS8 Defect
To further understand the functional role of GAS8 (DRC4) for the N-DRC, we analyzed the ciliary composition of GAS8-deficient respiratory cilia by high-resolution immunofluorescence analysis. We found that GAS8-deficient cilia of the three PCD-affected individuals completely lacked LRRC48 (DRC3) (Figure 4). Thus, GAS8 (DRC4) function is essential for axonemal localization of LRRC48. We previously found that CCDC164 (DRC1) and CCDC65 (DRC2) mutant cilia cannot assemble LRRC48 in ciliary axonemes. Therefore, we speculate that the multimeric N-DRC protein complex cannot properly assemble when one of the DRC proteins CCDC164 (DRC1), CCDC65 (DRC2), or GAS8 (DRC4) is mutated (Figure S1).
Figure 4.
GAS8-Mutant Respiratory Cilia Are Deficient for the N-DRC Protein LRRC48
In control respiratory epithelial cells, the N-DRC protein LRRC48 (red) and the axonemal marker acetylated α-tubulin (green) co-localize within the axonemes. Immunofluorescence analyses in respiratory cells from OP-929, OP-1940, and OP-1627 show absence of the N-DRC protein LRRC48 from the ciliary axonemes. Nuclei were stained with Hoechst 33342 (blue). Scale bars represent 10 μm
We previously demonstrated that mutations in genes encoding the ruler proteins CCDC39 and CCDC40 cause absent axonemal localization of GAS8 (DRC4) in human samples. Absent localization of GAS8 was confirmed in tracheal sections of mice homozygously carrying the lnks mutation of CCDC40 (Figure S4), indicating a conserved relevance of CCDC40 for the proper localization of GAS8. In addition, mutations in CCDC39 and CCDC40 cause reduced axonemal localization of the IDA protein DNALI1. Therefore, we also checked DNALI1 localization in GAS8-mutant respiratory cells. Immunofluorescence analyses of GAS8-deficient cilia demonstrated a normal axonemal DNALI1 composition (Figure S5) identical to findings obtained in CCDC164 (DRC1) mutant respiratory cilia reported previously.33 This finding indicates that the ruler proteins CCDC39 and CCDC40 can affect assembly of the IDA light chain DNALI1 through a mechanism independent from GAS8 (DRC4) and CCDC164 (DRC1).
Discussion
The Chlamydomonas orthologs of CCDC39 (FAP59) and CCDC40 (FAP172) are ruler proteins determining the axonemal 96-nm repeat length.40 CCDC39 and CCDC40 form a dimer, closely attached to the protofilaments of the outer doublets. The IDA complexes, as well as the N-DRCs, cannot bind to the tubules by themselves and attach to specific positions along the CCDC39-CCDC40 heterodimer.41 We recently identified recessive loss-of-function mutations in the genes CCDC39 and CCDC40 in PCD-affected individuals.31,32 We demonstrated that mutations in those two genes result in severe axonemal disorganization easily detectable by TEM and axonemal absence of GAS8 (DRC4) documented by immunofluorescence analyses. This was the first report that the N-DRC protein GAS8 (DRC4) plays a role in human disease. We also showed that PCD defects due to CCDC39 or CCDC40 mutations can be detected by demonstration of the absence of CCDC39 from the ciliary axonemes by immunofluorescence analysis.
We also recently reported recessive loss-of-function mutations in the genes CCDC164 and CCDC65 encoding the N-DRC components DRC1 and DRC2 in PCD individuals with normal ultrastructure and subtle ciliary beating abnormalities (Figure S1B).26,33 On the basis of these results, we speculated that PCD might also be caused by mutations in genes encoding other N-DRC components such as GAS8 (DRC4) (Table S1). Therefore, we systematically searched for PCD-affected individuals with abnormal axonemal GAS8 localization and normal CCDC39 localization without evidence of CCDC164 and CCDC65 mutations (Figure 1; Figure S2).
With this strategy, we identified two PCD-affected individuals matching our criteria and performed high-throughput whole-exome sequence analyses. Within the dataset, we focused our search for mutations in genes encoding N-DRC subunits (Table S1) and subsequently identified homozygous GAS8 nonsense mutations in two individuals, which we confirmed by Sanger sequencing (Figure 2). During the study, we found an additional PCD-affected individual matching our criteria and subsequently also identified a distinct homozygous GAS8 nonsense mutation. Overall, we found three distinct GAS8 nonsense mutations (Figure 2) in three PCD-affected individuals originating from Asia and Europe. Absence of GAS8 in GAS8-mutant respiratory cells was consistent with our genetic analyses (Figure 1).
The N-DRC subunit DRC4, orthologous to human GAS8, is conserved across diverse phyla, and the functional role has been studied in several organisms, including the green unicellular alga Chlamydomonas, the African protozoan Trypanosoma, and the zebrafish Danio rerio.42–45 Genetic analyses in Chlamydomonas found that the wild-type PF2 gene mutated in pf2 mutants encodes DRC4.42 pf2 mutant alga display both reduced swimming velocities and aberrant waveforms with reduced beating amplitudes of the flagella as a result of dysfunction of the N-DRC.42,46 In DRC4-deficient pf2 mutant alga, five different members (DRC3–DRC7) of the N-DRC complex fail to assemble in the flagella, indicating that DRC4 function is important for the functional integrity of the N-DRC multi-protein complex.47–49 Structural labeling with the biotin-streptavidin system and cryo-electron tomography enabled the precise localization of DRC1, DRC2, and DRC4, which have elongated conformations and span the length of the N-DRC in the unicellular Chlamydomonas alga (Figure S1B). This indicates that the N-DRC scaffold is made of bundled coiled-coil proteins rather than clustering globular proteins.40 Further Chlamydomonas in situ localization studies utilizing the SNAP system to visualize DRC3 and DRC4 by cryo-electron microscopy found that DRC3 is located within the L1 projection of the nexin linker close to DRC4 (Figure S1B).50 Here, we demonstrate that GAS8-mutant respiratory cells harboring homozygous nonsense mutations are deficient for GAS8 (DRC4) (Figure 1) and lack axonemal LRRC48 (DRC3) localization (Figure 4). GAS8-deficient cilia display a reduced beating amplitude, indicating that the functional role of DRC4 for N-DRC integrity has been evolutionary conserved from the unicellular algae to man. Consistent with an important function for DRC4 for axonemal motility, sRNAi knockdown of Trypanin (DRC4) expression in Trypanosoma resulted in defective flagellar beat coordination and reduced cell motility.43,44 Analyses in gas8 morphant zebrafish also confirmed an evolutionarily conserved role for cilia beating in vertebrates. Gas8 morphants exhibited phenotypes such as hydrocephalus, left-right axis defects, and abnormal otolith composition of the inner ear, which are all typical defects encountered in zebrafish when ciliary beating is disrupted.45
In contrast, in PCD-affected individuals, the hallmark of the disease phenotype is a chronic destructive respiratory disorder caused by insufficient mucociliary clearance of the airways. All three GAS8-mutant PCD-affected individuals had recurrent infections of the upper and lower airways and had evidence of bronchiectasis and chronic atelectasis. Conductive hearing impairment is a common finding in PCD and was also reported in the GAS8-mutant individuals; it is caused by abnormal clearance of the middle ears due to defective ciliary motility. Interestingly, the three PCD-affected individuals did not display any deficits of inner ear hearing, indicating that GAS8 function–in contrast to fish–is not essential for inner ear function in humans.45 Randomization of left-right body asymmetry is caused by dysmotility of rotating motile monocilia at the node during early embryogenesis.1 Deficient ciliary motility at the node results in absence of a fluid flow important for down-stream signaling events that subsequently determine the left-right body composition. Therefore, most molecular PCD defects also result in randomization of left-right body asymmetry, and approximately half of PCD-affected individuals display situs inversus (mirror image of the body composition). Interestingly, the three GAS8-mutant PCD-affected individuals did not display situs inversus, which might have occurred by chance. However, the six reported PCD-affected individuals carrying either CCDC164 or CCDC65 mutations encoding N-DRC proteins26,33,34 also had normal body composition. The likelihood that this occurred by chance is 1:512. Thus, it is possible that in the human system, mutations in genes encoding N-DRC subunits cause such subtle beating defects that during early embryogenesis a fluid flow at the ventral node can still be established in most cases.
In conclusion, we identified recessive loss-of-function GAS8 mutations as a cause of PCD with very subtle abnormalities of ciliary beating (Supplemental Movies S1 and S2) and ultrastructure (Figure 3). Consistently, PCD variants due to mutations in genes (CCDC164, CCDC65, GAS8) encoding N-DRC proteins are not readily identified by high-speed videomicroscopy evaluation of the ciliary beating or TEM analysis.26,33 Therefore, the diagnosis of this rare PCD variant is greatly aided by high-resolution immunofluorescence microscopy, which can demonstrate absence of GAS8 from the ciliary axonemes, as well as by genetic analyses. We expect that PCD might also be caused by mutations in genes encoding other N-DRC proteins such LRRC48 (DRC3) (Figure S1; Table S1).
Acknowledgments
We are grateful to the PCD-affected individuals and their family members whose cooperation made this study possible, and we thank all referring physicians. We thank Prof. J. Neesen (Medical University of Vienna) for providing us anti-DNALI1 antibodies. We thank A. Wolter for help in immunofluorescence analysis. We thank D. Ernst, M. Herting, B. Lechtape, L. Overkamp, F.J. Seesing, and C. Westermann for excellent technical work. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. This work was supported by a fellowship of the Nordrhein-Westfalen (NRW) Research School, “Cell Dynamics and Disease, CEDAD,” to H. Omran, by the Deutsche Forschungsgemeinschaft grants OM 6/4, OM 6/7, and OM6/8 to H. Omran and OL450/1 to H. Olbrich, by the Interdisziplinären Zentrum für Klinische Forschung (IZKF) Muenster grant to H. Omran (Om2/009/12), by the European Commission (Seventh Framework Programme [FP7] 2007–2013) grant agreement no. 262055 (H. Omran) as a Transnational Access project of the European Sequencing and Genotyping Infrastructure (ESGI), and by the European Union seventh FP under grant agreement no. 241955, project SYSCILIA (H. Omran) and grant agreement no. 305404, project BESTCILIA (K.G.N. and H. Omran).
Published: September 17, 2015
Footnotes
Supplemental Data include five figures, one table, and three movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.08.012.
Contributor Information
Heike Olbrich, Email: heike.olbrich@ukmuenster.de.
Heymut Omran, Email: heymut.omran@ukmuenster.de.
Web Resources
The URLs for data presented herein are as follows:
ExAC Browser, http://exac.broadinstitute.org/
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Varbank analysis software, https://varbank.ccg.uni-koeln.de/
Supplemental Data
References
- 1.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 2.Werner C., Onnebrink J.G., Omran H. Diagnosis and management of primary ciliary dyskinesia. Cilia. 2015;4:2. doi: 10.1186/s13630-014-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuehni C.E., Frischer T., Strippoli M.P., Maurer E., Bush A., Nielsen K.G., Escribano A., Lucas J.S., Yiallouros P., Omran H., ERS Task Force on Primary Ciliary Dyskinesia in Children Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur. Respir. J. 2010;36:1248–1258. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 4.Zariwala M.A., Knowles M.R., Omran H. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 5.Olbrich H., Häffner K., Kispert A., Völkel A., Volz A., Sasmaz G., Reinhardt R., Hennig S., Lehrach H., Konietzko N. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 6.Pennarun G., Escudier E., Chapelin C., Bridoux A.M., Cacheux V., Roger G., Clément A., Goossens M., Amselem S., Duriez B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 1999;65:1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loges N.T., Olbrich H., Fenske L., Mussaffi H., Horvath J., Fliegauf M., Kuhl H., Baktai G., Peterffy E., Chodhari R. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am. J. Hum. Genet. 2008;83:547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazor M., Alkrinawi S., Chalifa-Caspi V., Manor E., Sheffield V.C., Aviram M., Parvari R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 2011;88:599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duriez B., Duquesnoy P., Escudier E., Bridoux A.M., Escalier D., Rayet I., Marcos E., Vojtek A.M., Bercher J.F., Amselem S. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartoloni L., Blouin J.L., Pan Y., Gehrig C., Maiti A.K., Scamuffa N., Rossier C., Jorissen M., Armengot M., Meeks M. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA. 2002;99:10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwabe G.C., Hoffmann K., Loges N.T., Birker D., Rossier C., de Santi M.M., Olbrich H., Fliegauf M., Failly M., Liebers U. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat. 2008;29:289–298. doi: 10.1002/humu.20656. [DOI] [PubMed] [Google Scholar]
- 12.Panizzi J.R., Becker-Heck A., Castleman V.H., Al-Mutairi D.A., Liu Y., Loges N.T., Pathak N., Austin-Tse C., Sheridan E., Schmidts M. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onoufriadis A., Paff T., Antony D., Shoemark A., Micha D., Kuyt B., Schmidts M., Petridi S., Dankert-Roelse J.E., Haarman E.G., UK10K Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles M.R., Leigh M.W., Ostrowski L.E., Huang L., Carson J.L., Hazucha M.J., Yin W., Berg J.S., Davis S.D., Dell S.D., Genetic Disorders of Mucociliary Clearance Consortium Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjeij R., Onoufriadis A., Watson C.M., Slagle C.E., Klena N.T., Dougherty G.W., Kurkowiak M., Loges N.T., Diggle C.P., Morante N.F.C., UK10K Consortium CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N.T., Hagiwara H., Zhang Q., Leblond G., O’Toole E., Hara C. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loges N.T., Olbrich H., Becker-Heck A., Häffner K., Heer A., Reinhard C., Schmidts M., Kispert A., Zariwala M.A., Leigh M.W. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A.M., Coste A., Deschildre A., de Blic J., Legendre M., Montantin G. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchison H.M., Schmidts M., Loges N.T., Freshour J., Dritsoula A., Hirst R.A., O’Callaghan C., Blau H., Al Dabbagh M., Olbrich H. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012;44:381–389. doi: 10.1038/ng.1106. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarkar A., Loges N.T., Slagle C.E., Francis R., Dougherty G.W., Tamayo J.V., Shook B., Cantino M., Schwartz D., Jahnke C., UK10K DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013;45:995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kott E., Duquesnoy P., Copin B., Legendre M., Dastot-Le Moal F., Montantin G., Jeanson L., Tamalet A., Papon J.-F., Siffroi J.-P. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horani A., Druley T.E., Zariwala M.A., Patel A.C., Levinson B.T., Van Arendonk L.G., Thornton K.C., Giacalone J.C., Albee A.J., Wilson K.S. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zariwala M.A., Gee H.Y., Kurkowiak M., Al-Mutairi D.A., Leigh M.W., Hurd T.W., Hjeij R., Dell S.D., Chaki M., Dougherty G.W. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 2013;93:336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles M.R., Ostrowski L.E., Loges N.T., Hurd T., Leigh M.W., Huang L., Wolf W.E., Carson J.L., Hazucha M.J., Yin W. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am. J. Hum. Genet. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin-Tse C., Halbritter J., Zariwala M.A., Gilberti R.M., Gee H.Y., Hellman N., Pathak N., Liu Y., Panizzi J.R., Patel-King R.S. Zebrafish Ciliopathy Screen Plus Human Mutational Analysis Identifies C21orf59 and CCDC65 Defects as Causing Primary Ciliary Dyskinesia. Am. J. Hum. Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castleman V.H., Romio L., Chodhari R., Hirst R.A., de Castro S.C., Parker K.A., Ybot-Gonzalez P., Emes R.D., Wilson S.W., Wallis C. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kott E., Legendre M., Copin B., Papon J.-F., Dastot-Le Moal F., Montantin G., Duquesnoy P., Piterboth W., Amram D., Bassinet L. Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 2013;93:561–570. doi: 10.1016/j.ajhg.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeanson L., Copin B., Papon J.-F., Dastot-Le Moal F., Duquesnoy P., Montantin G., Cadranel J., Corvol H., Coste A., Désir J. RSPH3 Mutations Cause Primary Ciliary Dyskinesia with Central-Complex Defects and a Near Absence of Radial Spokes. Am. J. Hum. Genet. 2015;97:153–162. doi: 10.1016/j.ajhg.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olbrich H., Schmidts M., Werner C., Onoufriadis A., Loges N.T., Raidt J., Banki N.F., Shoemark A., Burgoyne T., Al Turki S., UK10K Consortium Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merveille A.C., Davis E.E., Becker-Heck A., Legendre M., Amirav I., Bataille G., Belmont J., Beydon N., Billen F., Clément A. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2011;43:72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker-Heck A., Zohn I.E., Okabe N., Pollock A., Lenhart K.B., Sullivan-Brown J., McSheene J., Loges N.T., Olbrich H., Haeffner K. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wirschell M., Olbrich H., Werner C., Tritschler D., Bower R., Sale W.S., Loges N.T., Pennekamp P., Lindberg S., Stenram U. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat. Genet. 2013;45:262–268. doi: 10.1038/ng.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horani A., Brody S.L., Ferkol T.W., Shoseyov D., Wasserman M.G., Ta-shma A., Wilson K.S., Bayly P.V., Amirav I., Cohen-Cymberknoh M. CCDC65 mutation causes primary ciliary dyskinesia with normal ultrastructure and hyperkinetic cilia. PLoS ONE. 2013;8:e72299. doi: 10.1371/journal.pone.0072299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sisson J.H., Stoner J.A., Ammons B.A., Wyatt T.A. All-digital image capture and whole-field analysis of ciliary beat frequency. J. Microsc. 2003;211:103–111. doi: 10.1046/j.1365-2818.2003.01209.x. [DOI] [PubMed] [Google Scholar]
- 36.Fliegauf M., Olbrich H., Horvath J., Wildhaber J.H., Zariwala M.A., Kennedy M., Knowles M.R., Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashid S., Breckle R., Hupe M., Geisler S., Doerwald N., Neesen J. The murine Dnali1 gene encodes a flagellar protein that interacts with the cytoplasmic dynein heavy chain 1. Mol. Reprod. Dev. 2006;73:784–794. doi: 10.1002/mrd.20475. [DOI] [PubMed] [Google Scholar]
- 38.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1092 human genomes. Nature. 2014;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins S.A., Gove K., Walker W., Lucas J.S. Nasal nitric oxide screening for primary ciliary dyskinesia: systematic review and meta-analysis. Eur. Respir. J. 2014;44:1589–1599. doi: 10.1183/09031936.00088614. [DOI] [PubMed] [Google Scholar]
- 40.Oda T., Yanagisawa H., Kamiya R., Kikkawa M. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science. 2014;346:857–860. doi: 10.1126/science.1260214. [DOI] [PubMed] [Google Scholar]
- 41.Oda T., Yanagisawa H., Kikkawa M. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol. Biol. Cell. 2015;26:294–304. doi: 10.1091/mbc.E14-09-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rupp G., Porter M.E. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J. Cell Biol. 2003;162:47–57. doi: 10.1083/jcb.200303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutchings N.R., Donelson J.E., Hill K.L. Trypanin is a cytoskeletal linker protein and is required for cell motility in African trypanosomes. J. Cell Biol. 2002;156:867–877. doi: 10.1083/jcb.200201036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabututu Z.P., Thayer M., Melehani J.H., Hill K.L. CMF70 is a subunit of the dynein regulatory complex. J. Cell Sci. 2010;123:3587–3595. doi: 10.1242/jcs.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colantonio J.R., Vermot J., Wu D., Langenbacher A.D., Fraser S., Chen J.N., Hill K.L. The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature. 2009;457:205–209. doi: 10.1038/nature07520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brokaw C.J., Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- 47.Piperno G., Mead K., LeDizet M., Moscatelli A. Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell Biol. 1994;125:1109–1117. doi: 10.1083/jcb.125.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J., Tritschler D., Song K., Barber C.F., Cobb J.S., Porter M.E., Nicastro D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bower R., Tritschler D., Vanderwaal K., Perrone C.A., Mueller J., Fox L., Sale W.S., Porter M.E. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol. Biol. Cell. 2013;24:1134–1152. doi: 10.1091/mbc.E12-11-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song K., Awata J., Tritschler D., Bower R., Witman G.B., Porter M.E., Nicastro D. In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography. J. Biol. Chem. 2015;290:5341–5353. doi: 10.1074/jbc.M114.626556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.