Abstract
Tooth agenesis is one of the most common developmental anomalies in man. Oligodontia, a severe form of tooth agenesis, occurs both as an isolated anomaly and as a syndromal feature. We performed exome sequencing on 20 unrelated individuals with apparent non-syndromic oligodontia and failed to detect mutations in genes previously associated with oligodontia. In three of the probands, we detected heterozygous variants in LRP6, and sequencing of additional oligodontia-affected individuals yielded one additional mutation in LRP6. Three mutations (c.1144_1145dupAG [p.Ala383Glyfs∗8], c.1779dupT [p.Glu594∗], and c.2224_2225dupTT [p.Leu742Phefs∗7]) are predicted to truncate the protein, whereas the fourth (c.56C>T [p.Ala19Val]) is a missense variant of a conserved residue located at the cleavage site of the protein’s signal peptide. All four affected individuals harboring a LRP6 mutation had a family history of tooth agenesis. LRP6 encodes a transmembrane cell-surface protein that functions as a co-receptor with members from the Frizzled protein family in the canonical Wnt/β-catenin signaling cascade. In this same pathway, WNT10A was recently identified as a major contributor in the etiology of non-syndromic oligodontia. We show that the LRP6 missense variant (c.56C>T) results in altered glycosylation and improper subcellular localization of the protein, resulting in abrogated activation of the Wnt pathway. Our results identify LRP6 variants as contributing to the etiology of non-syndromic autosomal-dominant oligodontia and suggest that this gene is a candidate for screening in DNA diagnostics.
Main Text
Agenesis of one or more permanent teeth is a common developmental anomaly with a prevalence of 5.5% in Europe. Common forms affecting one or a few teeth represent the great majority of cases. More severe phenotypes become increasingly more rare and are observed in approximately 0.14% of the population.1,2 Oligodontia (ICD-10: K00.0), defined as the absence of six or more permanent teeth excluding the third molars, can occur either as an isolated trait (non-syndromic) or as a part of a recognized clinical syndrome and has a heterogeneous dental and dental-facial presentation.3,4
Both environmental and genetic factors, as indicated by familial occurrence, play a role in the etiology of tooth agenesis.5 Inheritance can follow autosomal-dominant, recessive, or sex-linked patterns, and mutations in a number of genes have been associated with non-syndromic oligodontia. The genes currently associated with non-syndromic oligodontia are MSX1 (muscle segment homeobox 1 [MIM: 142983]),6 PAX9 (paired box gene 9 [MIM: 167416]),7 AXIN2 (axis inhibition protein 2 [MIM: 604025]),8 and the ectodermal dysplasia genes EDA (ectodysplasin A [MIM: 300451]), EDAR (ectodysplasin A receptor [MIM: 604095]), and EDARADD (edar-associated death domain [MIM: 606603]).9–12 Recently, mutations in WNT10A (wingless-type MMTV integration site family, member 10A [MIM: 606268]) were found in more than half of 34 unrelated individuals with nonsyndromic oligodontia.13 The objective of the present study was to identify disease-causing variants in genes not known to be involved in the etiology of tooth agenesis by means of whole-exome sequencing (WES) analysis of a cohort of non-syndromic oligodontia cases.
Individuals with apparent isolated oligodontia and visiting the Departments of Oral and Maxillofacial Surgery, Prosthodontics, and Special Dental Care of the University Medical Center Utrecht (UMC Utrecht) and the Antonius Hospital Nieuwegein were referred to the Department of Medical Genetics of UMC Utrecht for syndrome diagnostics and genetic counseling. Tooth agenesis in the affected individuals was assessed by clinical examination by the dentist and on panoramic radiographs. After careful physical examination by a single clinical geneticist, 20 individuals (6 males and 14 females) with six or more missing permanent teeth, excluding the third molars, were classified as displaying non-syndromic oligodontia without ectodermal features. No specific syndrome diagnose could be identified and confirmed. Blood samples were obtained, and after exclusion of mutations in the genes WNT10A, MSX1, PAX9, IRF6, EDA, and AXIN2, WES analysis was performed. Informed consent for WES as a part of the diagnostic process (approved by the medical ethical committee of UMC Utrecht) was obtained for all subjects included in this study. Informed consent for Sanger sequencing analysis was obtained for all subjects included in this study.
For WES, we prepared DNA libraries of the affected individuals by using Kapa Biosystems reagents and enriched these with Agilent SureSelect All Exon V5 and a custom pooling protocol. In brief, with a Covaris S2 sonicator, we sheared 1 ug of purified gDNA (QIAGEN) into 100–500 bp fragments that we then blunt ended, 5′ phosphorylated, and A-tailed by using KapaBiosystems reagents. Next, we ligated adaptors containing the Illumina barcode sequences to each sample and amplified for seven PCR cycles. Finally, we quantified and equimolarly pooled three samples into one library batch. Barcode blockers for the Illumina adaptor sequences and barcodes were added, and we enriched the library according to the Agilent SureSelect V5 exome protocol and finally PCR amplified the library for ten cycles. Two enriched libraries were then pooled and sequenced on a full-flow cell as a rapid run on the Illumina HiSeq 2500 with 100-bp paired-end reads at the Utrecht Sequencing Facility in the Netherlands.
We performed sequence alignment and variant calling against the reference human genome (UCSC Human Genome Browser hg19) by using the Burrows-Wheeler Aligner (BWA),14 and we processed the data with the Genome Analysis Toolkit (GATK) v.3.1.1,15 according to best practice guidelines.16,17 Mean target coverage was between 66.2× and 110.2× with 94.2%–95.6% of the targets being covered at greater than or equal to 10×.
A multi-sample.vcf file was created and imported into several analysis tools for annotation and interpretation. Cartagenia Bench Lab NGS was used for variant interpretation, as well as Combined Annotation Dependent Depletion (CADD) for variant prioritization. We analyzed the data first with a dominant inheritance model, followed by a recessive model. We prioritized variants on the basis of the population frequency, their presence in clinical variant databases such as the Human Gene Mutation Database (HGMD), and predicted effects at the protein level. Candidate variants were visualized in the integrated genomics viewer (IGV),18 and were validated with Sanger sequencing, followed by segregation analysis in relevant family members. Primer information is available upon request.
For the dominant inheritance model, we excluded variants with a minor allele frequency (MAF) > 1% as seen in the 1000 Genomes Project, NHLBI GO Exome Sequencing Project (ESP, 6500 release), or the Genome of the Netherlands19 variant-frequency databases. Next, we determined putative pathogenic frameshift, stop-loss, stop-gain, start-loss, splice-site, and nonsynonymous variants. Initially, we included nonsynonymous variants on the basis of a majority rule of three functional-effect prediction tools: SIFT (< 0), FATHMM (< 0), and MutationTaster (disease causing). We used the remaining 2,704 variants to rank genes based on recurrence in different individuals. In 349 genes, we found two or more variants. After gene prioritization and relevant literature inspection, we deemed LRP6 (low-density lipoprotein receptor-related protein 6 [MIM: 603507]) the most likely candidate gene to be involved in tooth agenesis. Subsequently, eight additional subjects without causative mutations in oligodontia-associated genes were analyzed by Sanger sequencing. This yielded one additional mutation in LRP6.
In total, we found four heterozygous variants in LRP6 (GenBank: NM_002336.2) in four unrelated individuals. These variants include a nonsense mutation (c.1779dupT [p.Glu594∗]) and two two-nucleotide insertion mutations (c.2224_2225dupTT [p.Leu742Phefs∗7], c.1144_1145dupAG [p.Ala383Glyfs∗8]) resulting in a truncated mRNA product as well as a missense variant (c.56C>T [p.Ala19Val]) located at the cleavage site of the signal peptide of the protein. All four variants are absent in all relevant variation databases, including the Single Nucleotide Polymorphism Database (dbSNP), 1000 Genomes, the NHLBI Exome Project, and the Exome Aggregation Consortium (ExAC; accessed May, 2015). The (c.56C>T) missense variant is rated deleterious by several in-silico-prediction software programs (SIFT score = 0.04; FATHMM score = −3.2; MutationTaster = disease causing; CADD_phred = 19.3).
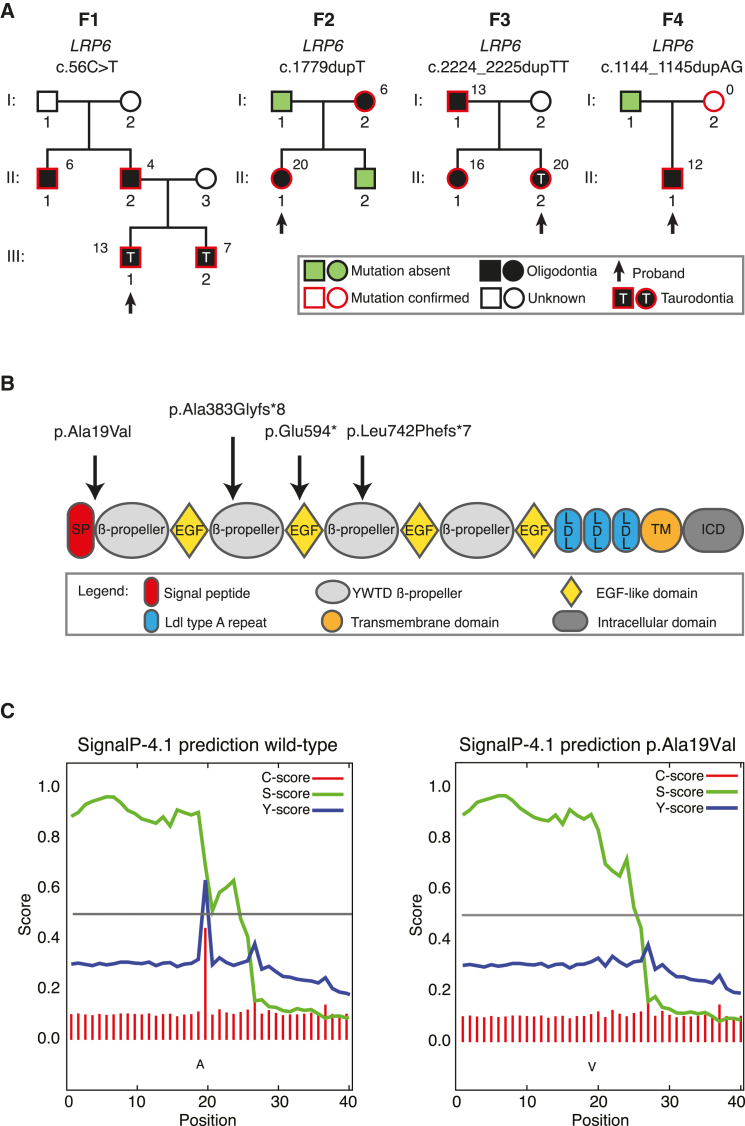
Sanger sequencing confirmed that all four individuals were heterozygous for the mutations. Segregation analysis showed that all had a family history of tooth agenesis. We observed incomplete penetrance for the oligodontia phenotype with the LRP6 variant seen in index-case individual F4-II-1 (Figure 1A). The probands were characterized by a high number of agenetic teeth, whereas the number of missing teeth in the affected family members varied. Furthermore, we found taurodontism in one third of affected individuals (Figure S1). Taurodontism is also associated with WNT10A mutations.20 Dental characteristics for the probands and their family members are described in Table S1. As mentioned, no specific syndrome diagnosis could be confirmed in the probands. However, upon further investigation, all affected members of family F3 showed minor anatomical variation of the ear and underdevelopment of the thumb (Figure S2). Lacrimal glands and ducts showed no abnormalities.
Figure 1.
Dominant LRP6 Loss-of-Function Mutations Co-segregate with the Oligodontia Phenotype
(A) Black squares and circles represent male and female individuals affected by oligodontia, respectively. An arrow indicates the index individual in each family. Numbers in italics indicate the number of missing teeth for that particular family member. T indicates that taurodontia is observed. Colors indicate presence (red) or absence (green) of the LRP6 variant as found in the index case. All affected family members carry LRP6 variants. No variants have been found in tested unaffected family members.
(B) Representation of LRP6 with the depicted domains. Black arrows indicate the locations of variants found in this study. The missense variant (p.Ala19Val) is found at the penultimate position of the signal peptide domain. The predicted protein truncating variants (p.Glu594∗ and p.Leu742Phefs∗7) are located in the third EGF-like and β-propeller domains, respectively.
(C) SignalP predictions for the WT and p.Ala19Val LRP6 signal peptide sequences. The S score is a general signal peptide score and this score is mostly unaltered between the p.Ala19Val and WT LRP6 signal peptide sequences. However, the C-score (raw cleavage-site score) and Y-score (combined cleavage-site score) predict a complete loss of a signal peptide cleavage site after amino acid 20 in the altered amino acid sequence.
LRP6 encodes a transmembrane cell-surface protein that, together with members of the seven-pass transmembrane receptors of the Frizzled family, functions as a co-receptor in the canonical Wnt/β-catenin signaling cascade. LRP6 contains a large extracellular domain (ECD) to which Wnt-proteins are known to bind. It has a modular structure of four β-propeller domains interconnected by an EGF-like domain followed by three LDLR type A ligand-binding domains.21 Figure 1B shows a schematic representation of the various protein domains and the location of the three mutations identified in this study.
Both the nonsense mutation (c.1779dupT) and the insertion mutations (c.2224_2225dupTT, c.1144_1145dupAG) result in a premature stop codon in the middle of the putative protein product, and the resulting transcripts are predicted to undergo nonsense-mediated mRNA decay. The missense variant (c.56C>T) is found at the penultimate amino acid position of the signal peptide sequence. Interestingly, the SignalP 4.1 server predicts the altered amino acid sequence for this variant to have lost its signal peptide cleavage site, (Figure 1C).22
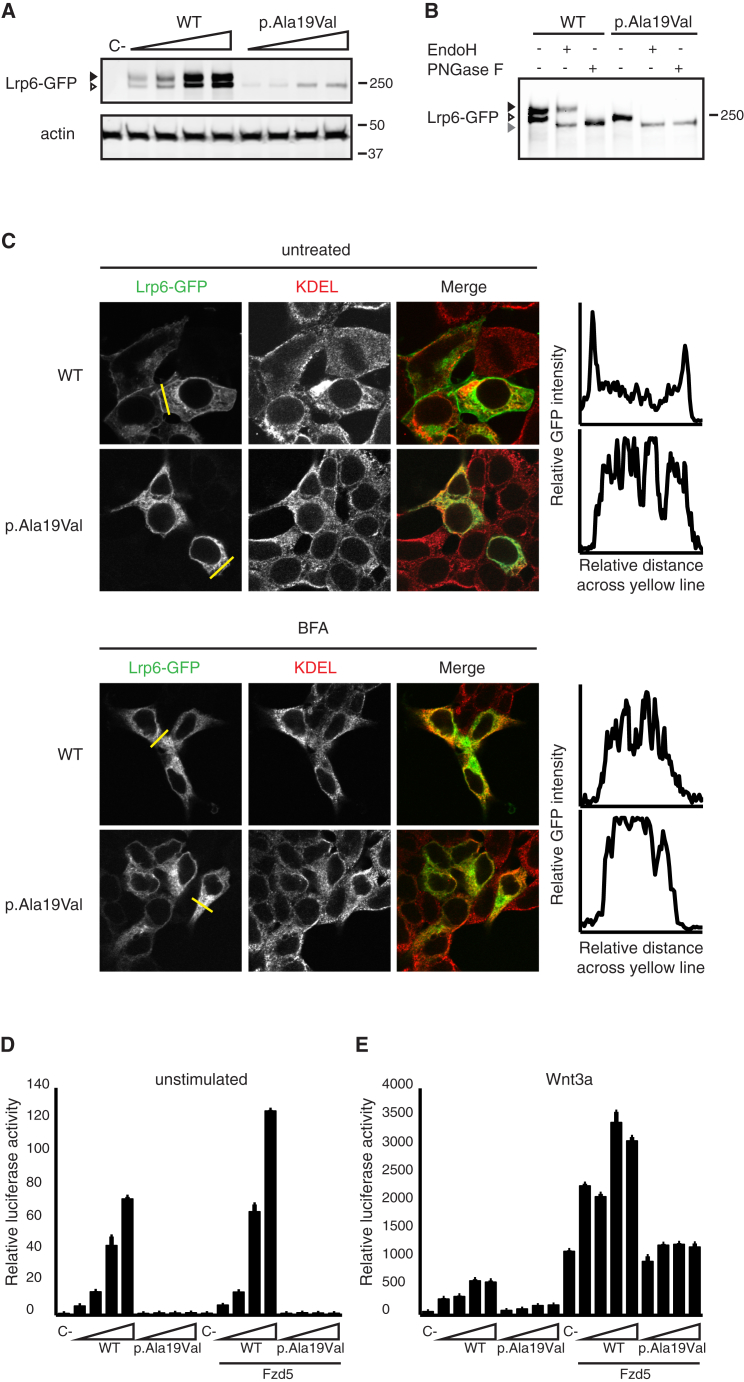
To gain insights into the molecular mechanism by which the p.Ala19Val substitution in LRP6 affects its biological activity, we compared the protein amounts of mouse wild-type (WT) Lrp6 (Lrp6-GFP) and the variant counterpart, generated by site-directed mutagenesis (Lrp6 p.Ala19Val-GFP). HEK293T cells grown in 24-well plates were transfected with increasing amounts of Lrp6-GFP or Lrp6 p.Ala19Val-GFP (1.25 ng, 2 ng, 5 ng, and 10 ng), lysed after 24 hr, and analyzed by immunoblotting with mouse anti-GFP (Roche) and mouse anti-actin (C4; MP Biomedicals). Whereas WT Lrp6-GFP was processed to the fully glycosylated mature form, Lrp6 p.Ala19Val-GFP only displayed a single protein band of lower molecular weight (MW), most likely representing the immature high-mannose precursor that is generated in the endoplasmic reticulum (ER) during the early steps of biogenesis (Figure 2A). Furthermore, the protein amount of Lrp6 p.Ala19Val-GFP was reproducibly lower as compared to that of WT Lrp6, suggesting impaired biogenesis of the altered protein or enhanced turnover.
Figure 2.
The Defective Signaling Capacity of Lrp6 p.Ala19Val Is Due to ER Retention of the Altered Protein
(A) Lrp6 p.Ala19Val fails to acquire Golgi-dependent complex glycosylation. Immunoblot analysis of increasing amounts of WT Lrp6-GFP and Lrp6 p.Ala19Val-GFP transfected in HEK293T cells is shown. Actin loading controls are shown. The immature and fully glycosylated mature forms of Lrp6 are indicated by a white and a black arrowhead, respectively.
(B) Lrp6 p.Ala19Val is trapped in the ER. HEK293T cells were transfected with WT Lrp6-GFP or Lrp6 p.Ala19Val-GFP and cell lysates were treated with the indicated glycosidases. Gray, white, and black arrowheads indicate deglycosylated, immature ER-resident (high-mannose), and mature complex glycosylated Lrp6-GFP forms, respectively.
(C) Lrp6 p.Ala19Val fails to reach the cell surface. Cellular localization of WT and altered Lrp6-GFP transfected in HEK293T cells in the presence or absence of Brefeldin A (BFA) treatment, as analyzed by confocal immunofluorescence microscopy. The ER was stained with anti-KDEL antibody. Graphs indicate relative intensity of GFP fluorescence across the yellow line.
(D and E) The capacity of Lrp6 p.Ala19Val to induce Wnt signaling is severely compromised. Wnt pathway activation induced by increasing amounts of Lrp6-GFP and Lrp6 p.Ala19Val-GFP was measured with a luciferase gene reporter assay in HEK293T cells. Activities were compared in the absence and presence of Wnt3a and co-transfection of Frizzled-5 (Fzd5), as indicated. Empty vector-transfected cells (C-) serve as an internal control. Results represent duplicate experiments. Luciferase activity was normalized against unstimulated controls; error bars depict SDs.
To confirm that Lrp6 p.Ala19Val is retained in the ER in its immature high-mannose form, we determined the sensitivity of the altered protein to Endoglycosidase H (EndoH) activity. EndoH cleaves high-mannose glycans that are added to nascent proteins in the ER. When proteins enter the Golgi, additional glycan modifications occur, rendering the resulting complex glycan chains resistant to EndoH digestion. In brief, HEK293T cells grown in six-well plates were transfected with 200 ng of Lrp6-GFP or Lrp6 p.Ala19Val-GFP and lysed after 24 hr. Cell lysates were incubated with either EndoH (Roche) or Peptide-N-Glycosidase F (PNGase F; New England Biolabs) for 5 hr at 37°C and analyzed by immunoblotting. As shown in Figure 2B, the low MW protein bands of both WT and altered Lrp6 were sensitive to EndoH treatment, as suggested by a shift to a lower MW deglycosylated form, whereas the complex glycosylated mature form of WT Lrp6 remained unaffected. As expected, both protein bands were sensitive to PNGase F, which hydrolyses all N-linked glycans. These results argue that the p.Ala19Val variant causes retention of the nascent Lrp6 in the ER, potentially due to a failure in the cleavage of the signal peptide.
To substantiate these findings, we investigated by confocal microscopy whether Lrp6 p.Ala19Val could be transported to the plasma membrane. To this end, HEK293T cells grown on laminin-coated glass coverslips in 24-well plates were transfected with 100 ng of either Lrp6-GFP or Lrp6 p.Ala19Val-GFP. The next day, cells were fixed in 4% paraformaldehyde or incubated with 10 μg/ml Brefeldin A (BFA) for 5 hr before fixation. Anti-KDEL antibody (Abcam) was used as an ER marker. Whereas the WT protein is mainly present at the cell surface, Lrp6 p.Ala19Val colocalizes with KDEL and does not reach the plasma membrane (Figure 2C). Importantly, upon treatment with the ER-Golgi trafficking inhibitor BFA, WT Lrp6 displayed a similar perinuclear ER-like localization, whereas no major differences were detected for Lrp6 p.Ala19Val, suggesting that the mutation phenocopies the BFA effect.
Given that Lrp6 is a major component of the Wnt receptor complex in the canonical Wnt pathway, we sought to investigate the signaling capacity of the altered protein by using the previously described TOPFlash luciferase reporter assay, which provides a highly specific and quantitative readout for Wnt pathway activity.23 Increasing amounts of WT Lrp6-GFP in HEK293T cells induced a dose-dependent activation of β-catenin-mediated transcription (Figure 2D). Strikingly, under the same conditions, altered Lrp6 p.Ala19Val-GFP failed to trigger β-catenin activation, indicating its inability to activate the Wnt pathway. In Wnt stimulated cells, the Lrp6 receptor relays the Wnt signal together with the seven span Frizzled (Fzd) receptor, forming a trimeric complex (Wnt-Fzd-Lrp6) at the cell surface.24 We next addressed the signaling capacity of the Lrp6 p.Ala19Val variant in the presence of high Fzd5 amounts. To this end, cells were co-transfected with increasing amounts of either WT or altered Lrp6 along with 30 ng Fzd5.25 Similar to the results above, Lrp6 p.Ala19Val-GFP was not able to enhance Wnt signaling induced by Fzd5 expression, irrespective of the amount of transfected altered Lrp6 DNA (Figure 2D). By comparison, combined expression of both WT Lrp6 and Fzd5 strongly enhanced reporter activity. Moreover, Lrp6 p.Ala19Val-GFP expressing cells failed to induce enhanced Wnt pathway activation upon stimulation for 16 hr with exogenous Wnt3a-conditioned medium derived from stably transfected murine L cells (Figure 2E). Together, these findings show that the Lrp6 p.Ala19Val variant confers loss-of-function effects upon the Lrp6 receptor as a result of ER retention, thereby rendering the protein invisible for incoming Wnt ligands at the cell surface.
Missense variants in LRP6 have been associated with coronary artery disease (CAD)26,27 and metabolic syndrome;28 however, these phenotypes were not observed in our affected individuals. In contrast, no nonsense, frameshift, or splice-site variants have been reported thus far. Most likely, the loss-of-function variants reported here represent a different functional class of mutations.
Mouse embryos homozygous for an Lrp6 insertion mutation have been shown to die at birth and exhibit a variety of severe developmental abnormalities that are a striking composite of developmental abnormalities caused by mutations in individual Wnt genes. Also, the phenotype of a classical Wnt-3a hypomorphic allele, vestigial tail (vt), is enhanced upon elimination of one functional copy of Lrp6. This observation is believed to provide genetic evidence that Lrp6 and Wnt-3a function in the same pathway.29 Subsequently, it has been shown that 10% of mice heterozygous for this Lrp6 insertion mutation show a tail-kink phenotype, indicating that haploinsufficiency of Lrp6 is sufficient to generate a phenotype in the mouse.30 However, tooth anomalies have not been reported in these animals.
In this study, we identified four heterozygous LRP6 loss-of-function mutations in four independent families affected by severe tooth agenesis. Segregation in these families follows an autosomal-dominant inheritance pattern with reduced penetrance and variable expression; this observation is in accordance with familial segregation and twin studies.2 Given that LRP6 is a key co-receptor in the canonical Wnt/β-catenin pathway, our results strengthen the notion that disruption of important developmental pathways such as the Wnt-pathway is of importance in the etiology of tooth agenesis.
In conclusion, we here provide genetic and functional evidence that mutations in LRP6 are a cause of non-syndromic tooth agenesis, and our results suggest that this gene is a candidate for screening in DNA diagnostics.
Acknowledgments
We gratefully acknowledge the affected individuals and their families whose participation made this research possible. We kindly thank Hans Clevers (Hubrecht Institute, Utrecht, the Netherlands) for providing us the mouse Lrp6 plasmid (pcDNA3). This work was supported by Utrecht University (seed grant to M.M.M.), the European Union (FP7 Marie-Curie ITN 608180 “WntsApp” to M.M.M.), and the Netherlands Organization for Scientific Research NWO (VICI grant to M.M.M.).
Published: September 17, 2015
Footnotes
Supplemental Data include two figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.08.014.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Cartagenia, http://www.cartagenia.com/
ExAC Browser, http://exac.broadinstitute.org/
FATHMM, http://fathmm.biocompute.org.uk/
MutationTaster, http://www.mutationtaster.org/
NHLBI GO Exome Sequencing Project, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org
SignalP 4.1, http://www.cbs.dtu.dk/services/SignalP/
SIFT, http://sift.jcvi.org
Supplemental Data
References
- 1.Polder B.J., Van’t Hof M.A., Van der Linden F.P., Kuijpers-Jagtman A.M. A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent. Oral Epidemiol. 2004;32:217–226. doi: 10.1111/j.1600-0528.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- 2.Nieminen P. Genetic basis of tooth agenesis. J. Exp. Zoolog. B Mol. Dev. Evol. 2009;312B:320–342. doi: 10.1002/jez.b.21277. [DOI] [PubMed] [Google Scholar]
- 3.Créton M.A., Cune M.S., Verhoeven W., Meijer G.J. Patterns of missing teeth in a population of oligodontia patients. Int. J. Prosthodont. 2007;20:409–413. [PubMed] [Google Scholar]
- 4.Créton M., Cune M.S., de Putter C., Ruijter J.M., Kuijpers-Jagtman A.M. Dentofacial characteristics of patients with hypodontia. Clin. Oral Investig. 2010;14:467–477. doi: 10.1007/s00784-009-0308-y. [DOI] [PubMed] [Google Scholar]
- 5.Brook A.H. Multilevel complex interactions between genetic, epigenetic and environmental factors in the aetiology of anomalies of dental development. Arch. Oral Biol. 2009;54:S3–S17. doi: 10.1016/j.archoralbio.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vastardis H., Karimbux N., Guthua S.W., Seidman J.G., Seidman C.E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 7.Stockton D.W., Das P., Goldenberg M., D’Souza R.N., Patel P.I. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- 8.Lammi L., Arte S., Somer M., Jarvinen H., Lahermo P., Thesleff I., Pirinen S., Nieminen P. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am. J. Hum. Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han D., Gong Y., Wu H., Zhang X., Yan M., Wang X., Qu H., Feng H., Song S. Novel EDA mutation resulting in X-linked non-syndromic hypodontia and the pattern of EDA-associated isolated tooth agenesis. Eur. J. Med. Genet. 2008;51:536–546. doi: 10.1016/j.ejmg.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Song S., Han D., Qu H., Gong Y., Wu H., Zhang X., Zhong N., Feng H. EDA gene mutations underlie non-syndromic oligodontia. J. Dent. Res. 2009;88:126–131. doi: 10.1177/0022034508328627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arte S., Parmanen S., Pirinen S., Alaluusua S., Nieminen P. Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS ONE. 2013;8:e73705. doi: 10.1371/journal.pone.0073705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergendal B., Klar J., Stecksén-Blicks C., Norderyd J., Dahl N. Isolated oligodontia associated with mutations in EDARADD, AXIN2, MSX1, and PAX9 genes. Am. J. Med. Genet. A. 2011;155A:1616–1622. doi: 10.1002/ajmg.a.34045. [DOI] [PubMed] [Google Scholar]
- 13.van den Boogaard M.J., Créton M., Bronkhorst Y., van der Hout A., Hennekam E., Lindhout D., Cune M., Ploos van Amstel H.K. Mutations in WNT10A are present in more than half of isolated hypodontia cases. J. Med. Genet. 2012;49:327–331. doi: 10.1136/jmedgenet-2012-100750. [DOI] [PubMed] [Google Scholar]
- 14.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;11 doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 20.Yang J., Wang S.K., Choi M., Reid B.M., Hu Y., Lee Y.L., Herzog C.R., Kim-Berman H., Lee M., Benke P.J. Taurodontism, variations in tooth number, and misshapened crowns in Wnt10a null mice and human kindreds. Mol. Genet. Genomic Med. 2015;3:40–58. doi: 10.1002/mgg3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joiner D.M., Ke J., Zhong Z., Xu H.E., Williams B.O. LRP5 and LRP6 in development and disease. Trends Endocrinol. Metab. 2013;24:31–39. doi: 10.1016/j.tem.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 23.Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 24.Clevers H., Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Tauriello D.V., Haegebarth A., Kuper I., Edelmann M.J., Henraat M., Canninga-van Dijk M.R., Kessler B.M., Clevers H., Maurice M.M. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol. Cell. 2010;37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y., Gong W., Peng J., Wang H., Huang J., Ding H., Wang D.W. Functional analysis LRP6 novel mutations in patients with coronary artery disease. PLoS ONE. 2014;9:e84345. doi: 10.1371/journal.pone.0084345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mani A., Radhakrishnan J., Wang H., Mani A., Mani M.A., Nelson-Williams C., Carew K.S., Mane S., Najmabadi H., Wu D., Lifton R.P. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R., Smith E., Fathzadeh M., Liu W., Go G.W., Subrahmanyan L., Faramarzi S., McKenna W., Mani A. Rare nonconservative LRP6 mutations are associated with metabolic syndrome. Hum. Mutat. 2013;34:1221–1225. doi: 10.1002/humu.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinson K.I., Brennan J., Monkley S., Avery B.J., Skarnes W.C. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 30.Kelly O.G., Pinson K.I., Skarnes W.C. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. 2004;131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.