Abstract
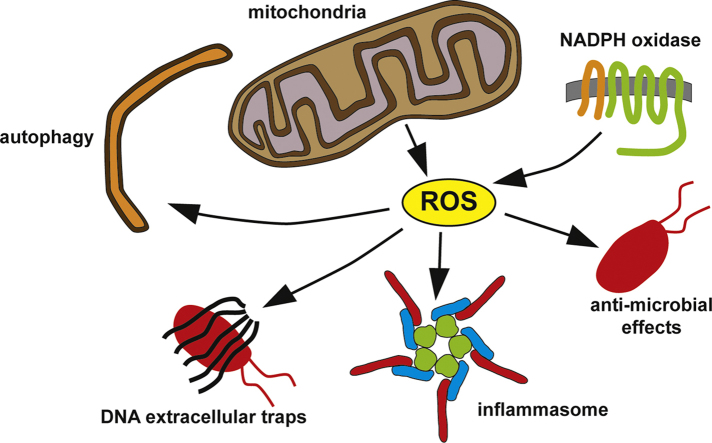
Reactive oxygen species (ROS) are integral components of multiple cellular pathways even though excessive or inappropriately localized ROS damage cells. ROS function as anti-microbial effector molecules and as signaling molecules that regulate such processes as NF-kB transcriptional activity, the production of DNA-based neutrophil extracellular traps (NETs), and autophagy. The main sources of cellular ROS are mitochondria and NADPH oxidases (NOXs). In contrast to NOX-generated ROS, ROS produced in the mitochondria (mtROS) were initially considered to be unwanted by-products of oxidative metabolism. Increasing evidence indicates that mtROS have been incorporated into signaling pathways including those regulating immune responses and autophagy. As metabolic hubs, mitochondria facilitate crosstalk between the metabolic state of the cell with these pathways. Mitochondria and ROS are thus a nexus of multiple pathways that determine the response of cells to disruptions in cellular homeostasis such as infection, sterile damage, and metabolic imbalance. In this review, we discuss the roles of mitochondria in the generation of ROS-derived anti-microbial effectors, the interplay of mitochondria and ROS with autophagy and the formation of DNA extracellular traps, and activation of the NLRP3 inflammasome by ROS and mitochondria.
Abbreviations: ASC, apoptosis-associated speck-like protein containing a caspase recruitment domain; CGD, chronic granulomatous disease; DAMP, damage-associated molecular pattern; Drp1, dynamin-related protein 1; DUSP16/MKP-7, dual specificity protein phosphatase 16/mitogen-activated protein kinase phosphatase-7; ECSIT, evolutionarily conserved signaling intermediate in Toll pathways; eis, enhanced intracellular survival gene; ERRα, estrogen-related receptor α; ETC, electron transport chain; G-CSF, granulocyte colony-stimulating factor; IFN, interferon; IL, interleukin; iNOS, inducible nitric oxide synthase; JNK, c-Jun N-terminal kinase; LPS, lypopolysaccharide; MAVS, mitochondrial antiviral signaling protein; MPO, myeloperoxidase; Mtb, Mycobacterium tuberculosis; mtDAMP, mitochondrial damage-associated molecular pattern; mTOR, target of rapamycin, mammalian homolog; mtROS, ROS produced in the mitochondria; NAC, N-acetyl-l-cysteine; NETs, neutrophil extracellular traps; NF-kB, nuclear factor-kB; NLR, nucleotide binding domain-leucine rich repeat; NLRP3, nucleotide binding domain-leucine rich repeat, pyrin domain-containing 3; NO, nitric oxide; NOX, NADPH oxidase; Nrf2, NF-E2-related factor 2; PAMP, pathogen-associated molecular pattern; PGC-1β, peroxisome proliferator-activated receptor γ coactivator-1β; PI3K-I, class I phosphoinositide 3-kinase; PKC, protein kinase C; PMA, phorbol myristate acetate; PYD, pyrin domain; RAGE, receptor for advanced glycation end-products; RIP1, receptor-interacting serine-threonine kinase 1; RIP3, receptor-interacting serine-threonine kinase 3; ROS, reactive oxygen species, mainly superoxide and hydrogen peroxide; SOD, superoxide dismutase; STAT1, signal transducer and activator of transcription 1; TCA, tricarboxylic acid; TLR, Toll-like receptor; TNF, tumor necrosis factor; TOR, target of rapamycin; TORC1, target of rapamycin complex 1; TRAF6, tumor necrosis factor receptor-associated factor 6; TRX, thioredoxin; TXNIP, thioredoxin-interacting protein; VSV, vesicular stomatitis virus
Keywords: Inflammasome, Mitochondria, Autophagy, Neutrophil extracellular traps, Immunity, Reactive oxygen species
Graphical abstract
Highlights
-
•
Mitochondrial ROS production is regulated by and incorporated into immunity pathways.
-
•
ROS regulate mitophagy and xenophagy and neutrophil extracellular trap formation.
-
•
Mitochondria and mitochondrial ROS regulate NLRP3 inflammasome activation.
1. Introduction
Reactive oxygen species (ROS) such as superoxide and hydrogen peroxide are integral components of multiple cellular pathways even though excessive or inappropriately localized ROS damage cells. In immunity-related pathways ROS can function as anti-microbial effector molecules and as signaling molecules that regulate such processes as nuclear factor-kB (NF-kB) transcriptional activity and the production of DNA-based neutrophil extracellular traps (NETs) [1–3]. ROS are also involved in pathways that regulate autophagy within and outside the context of immunity [4,5].
The main sources of cellular ROS are mitochondria and NADPH oxidases (NOXs). After the identification of NOX as the producer of ROS during the phagocyte oxidative burst, initial studies on immune-related ROS focused on phagocyte NOX. However, increasing evidence indicates that ROS production in the mitochondria, once thought to be an unwanted by-product of oxidative metabolism, is regulated by immune signaling pathways and incorporated into immune responses [6,7]. Mitochondria and potentially mitochondrial ROS are also components of pathways regulating apoptosis [8], stem cell differentiation [9], autophagy [10], and cellular [11,12] and tissue-level [13] inflammation. As hubs of cellular metabolism, mitochondria integrate these pathways with the metabolic state of the cell.
Their involvement in multiple pathways that determine the response of cells to disruptions in cellular physiology such as infection, sterile damage, and metabolic imbalance establish ROS and mitochondria as a nexus of cellular homeostasis. In this review, we discuss the generation of ROS-derived anti-microbial effectors, the interplay of mitochondria and ROS with autophagy and NET formation, and activation of the NLRP3 inflammasome by ROS and mitochondria. While the contributions of NOX-generated ROS to these processes are described, we mainly focus on the emerging roles of ROS produced in the mitochondria (mtROS).
2. Antimicrobial role of ROS
The main role of ROS during bacterial infection has traditionally been described as that of a bactericidal effector during the oxidative burst observed in monocytes after the engulfment of bacterial pathogens and maturation of the resulting phagosome into a phagolysosome [14,15]. The phagocyte NOX (NOX2), a heterodimer of transmembrane proteins Nox2 and p22phox, is recruited and activated by accessory proteins to generate superoxide inside the phagolysosome. After conversion of superoxide to hydrogen peroxide by superoxide dismutase (SOD), an additional step via myeloperoxidase (MPO) is believed to generate hypochlorous acid [16]. This final radical is believed to be the main ROS responsible for bactericidal activity in phagocytes [16]. NOX2 activation has also been linked to pH modulation inside the phagosome [17]. ROS production inside the phagosome leads to an increase in intraphagosomal pH. These changes in pH regulate proteases in the phagosome that can also be key effectors in bacterial killing [18,19].
NOX2 deficiency in humans is the main factor in chronic granulomatous disease (CGD) [20]. In the absence of efficient ROS production through NOX2, CGD patients exhibit a propensity for repeated bacterial and fungal infections that cannot be properly resolved without antibiotics. CGD patients also exhibit chronic inflammation [17]. In contrast, MPO deficiency in humans does not cause a comparable increase in sensitivity to bacterial infection [21]. The more severe phenotype from NOX2 deficiency suggests a role for ROS in innate immunity beyond being substrates for MPO-dependent direct oxidation of pathogens’ molecules.
Although CGD patients do have an increased sensitivity to some bacterial infections, this sensitivity is species-specific and varies depending on the specific mutations responsible for NOX2 deficiency [17]. CGD patients are not completely immunocompromised; thus, it is clear that NOX2-independent mechanisms are involved in efficient bacterial clearance by the innate immune system. Another important source of ROS is the electron transport chain (ETC) of mitochondria. Increasing evidence supports the involvement of mtROS in the immune response.
3. Mitochondrial ROS in the immune response
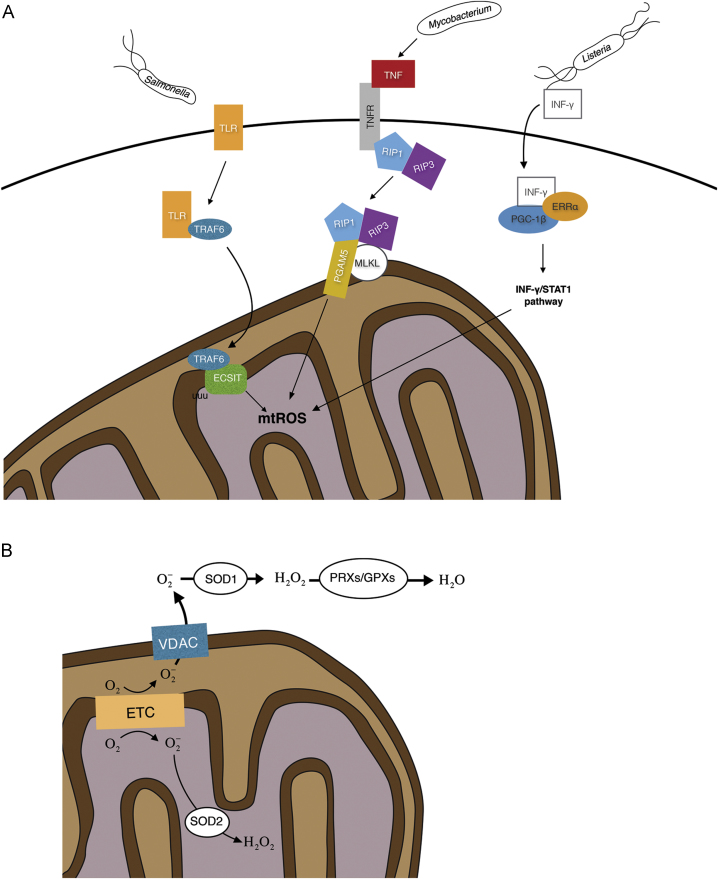
As for phagocyte NOX, mitochondria are recruited to phagosomes to produce ROS during infection. Recruitment and increased mtROS production occur downstream of Toll-like receptor (TLR) signaling during Salmonella infection of macrophages [6]. TLR activation leads to tumor necrosis factor receptor-associated factor 6 (TRAF6) translocation to mitochondria, where it interacts with evolutionarily conserved signaling intermediate in Toll pathways (ECSIT), a protein involved in the assembly of ETC complex I (Fig. 1A). TRAF6-mediated ubiquitination of ECSIT leads to increased mtROS production in macrophages. Without this additional ROS production monocytes in this study were unable to properly clear the bacterial infection, even with an efficient NOX response.
Fig. 1.
(A) mtROS in bacterially triggered immune responses. mtROS contribute to Salmonella clearing. TLR/TRAF6 pathway activation leads to poly-ubiquitination of ECSIT in the mitochondria and increase mtROS in macrophages. Mycobacterium infection triggers a TNF response via RIP1/RIP2 that increases mtROS in macrophages, first increasing bactericidal effects but ultimately leading to macrophage death and increased bacterial dissemination. Listeria infection triggers mtROS through an INF-γ response, activating ERRα and PGC-1β and the INF-γ/STAT1 pathway contributing to macrophage clearing of the bacteria. (B) mtROS generation. Superoxide is produced as an intermediate in the electron transport chain (ETC). In the mitochondria matrix excess superoxide is reduced to hydrogen peroxide by SOD2. Superoxide in the inter-membrane space can be exported to the cytoplasm through VDAC. It is then transformed into hydrogen peroxide by SOD1 and finally reduced to water through catalase, peroxiredoxins and glutathione peroxidases.
A recent study revealed that Mycobacterium tuberculosis (Mtb) infection triggers mtROS production via activation of the RIP1/RIP3 pathway by the pro-inflammatory cytokine tumor necrosis factor (TNF) [22]. Increased mtROS correlated with an increased bactericidal effect. However, excessive mtROS levels induced programmed necrosis that released the bacteria from the macrophages. Preventing the ROS-mediated death of the macrophages in this study prolonged the overall bactericidal activity of the innate response.
An mtROS increase in macrophages has been observed to be essential for efficient clearance of Listeria monocytogenes [23]. In this case, interferon (IFN)-γ activates ERRα (estrogen-related receptor α) and PGC-1β (peroxisome proliferator-activated receptor γ coactivator-1β) to promote mtROS production by directly targeting the IFN-γ/signal transducer and activator of transcription 1 (STAT1) signaling pathway. Macrophages lacking either ERRα or PGC-1β did not exhibit increased mtROS and developed a higher L. monocytogenes burden.
It is important to note that, although the bactericidal effect of ROS/mtROS is supported by a plethora of experimental data, the exact mechanisms are not fully understood. Some of our limits in understanding these mechanisms come from the difficulty of quantitating different ROS species in vivo during the immune response. Another challenge is the microorganism-specific sensitivities to different ROS, which prevents the use of a single model organism to study the overall bactericidal effect of ROS. An in-depth description of these limits can be found in a recent review [24] and will not be fully explored here. Recent work has shown the capacity of host-produced hydrogen peroxide during bacterial infection to act as a secondary messenger and decrease the amount of tyrosine phosphorylation of bacterial proteins [25]. The ROS-mediated loss of tyrosine phosphorylation in this study led to a decrease in virulence of Campylobacter jejuni. The direct effects of ROS on bacteria may thus act at different levels with bacterial clearing as the end result.
Another aspect to take into account in these host–pathogen interactions is the selective pressure that immune responses put on pathogens. Indeed, high concentrations of ROS can have a bactericidal effect on Salmonella, but as Burton and colleagues have highlighted, Salmonella has developed the ability to adapt to sub-lethal doses of ROS [26]. It has also been shown that this bacterium can use some of the by-products of ROS chemistry in the lumen of the gastrointestinal tract as electron acceptors to enhance bacterial growth [27].
Salmonella might also manipulate mtROS production via the Wnt signaling pathway. During infection, the secreted Salmonella effector protein AvrA increases expression of Wnt and promotes β-catenin transcriptional activity downstream of Wnt activation [28]. In a separate study, Yoon and colleagues performed a large scale RNA interference screen to identify key proteins that affect mitochondrial metabolism [29]. These authors showed that Wnt signaling significantly upregulates mitochondrial biogenesis and that the corresponding increase in ROS production ultimately leads to oxidative damage. In the Salmonella study, Wnt activation was analyzed in the context of stem cell maintenance in the gut; further analyses are needed to confirm whether this infection-stimulated Wnt activation could also lead to increased mtROS production.
4. Mitochondrial ROS generation and regulation
ROS generation in the mitochondria is due to the oxidation of metabolic intermediates of the ETC and is tightly regulated to prevent the oxidative damage of cellular processes. mtROS is produced in the ETC in the form of superoxide, with complex I often seen as the main source [30]. It is nonetheless difficult to associate mtROS generation in cellulo to a particular complex in the ETC. Quinlan and coworkers have shown that the 2-oxoacid dehydrogenase and the pyruvate dehydrogenase complexes can have a strong contribution to mtROS production [31].
Cytosolic ROS, originating in the mitochondria or produced by NOX, can stimulate additional ROS production via activation of redox-sensing protein kinase C (PKC) isoforms and Src family kinases [32–34]. In neutrophils and endothelial cells, activation of PKC and cSrc by mtROS upon its translocation to the cytosol leads to ROS production by NOX2 [32,34]. In vascular endothelial cells, ROS produced by NOX2 in response to angiotensin II signaling has been reported to cause increased mtROS production downstream of PKCε-mediated activation of the mitochondrial ATP-sensitive K+ channel [32]. However, it is important to note that NOX4, which can be upregulated in response to angiotensin II [35], localizes to mitochondria and can directly affect mtROS production [36].
Major factors in mtROS regulation are ROS scavenging enzymes that eliminate excess ROS. Three SODs facilitate the conversion of superoxide to hydrogen peroxide. SOD1 is found in the intermembrane space as well as in the cytosol [37], SOD2 is targeted to the mitochondrial matrix [38], and SOD3 is anchored to the extracellular matrix [39]. Following conversion of superoxide to hydrogen peroxide, catalase, peroxiredoxins and glutathione peroxidases perform a final reducing step to convert hydrogen peroxide into water (Fig. 1B). In addition to its enzymatic activity, SOD1 has recently been shown to act as a transcription factor in response to a general elevation of cellular ROS. Upon exposure of cells to hydrogen peroxide, SOD1 translocated to the nucleus, bound to promoters, and upregulated the expression of genes involved in oxidative resistance and repair [40].
Changes of either expression or localization of these enzymes are likely to modulate mtROS and thus to regulate various signaling pathways. Factors involved in this spatial segregation include voltage-dependent anion channels [41]. As superoxide is membrane impermeable, it needs either to be produced directly in the cytosol or to diffuse through a pore complex in order to participate in cellular signaling or to have any other effect outside of the mitochondria.
Although mtROS production was once seen as merely an accidental by-product of oxygen metabolism in mitochondria, it is now clear that ROS contributes to various signaling pathways [42]. Depending on the context and triggering stimuli, mtROS production can lead to different cellular responses such as cellular adaptation to hypoxia [43], cellular differentiation [44], autophagy [5], inflammation [45], or an immune response [6]. Overproduction of mtROS can occur in the absence of infection and has been implicated in tumor growth [46] and hypertension [47,48]. While roles for mitochondria and mtROS in immune responses are starting to be well documented, the mechanistic link between immune signaling and mtROS production are not yet clearly understood.
5. Mitochondrial ROS are involved in autophagy
Macroautophagy (herein termed autophagy) is a cellular process conserved in all eukaryotes for degrading dysfunctional organelles or large cytosolic molecules. Different from the single-membraned phagolysosome formation, autophagy employs a double-membraned vesicle, the autophagosome, to progressively engulf cytoplasmic constituents. After the fusion of an autophagosome with single-membraned lysosome(s), an autolysosome is formed, within which the inner autophagosome membrane and its cellular contents are degraded [49]. The key regulator of autophagy induction is target of rapamycin (TOR or mTOR for the mammalian homolog), which forms two distinct complexes. However, only TOR complex 1 (TORC1) functions as the major nutrient and energy sensor of the cell: in nutrient-rich conditions, TORC1 inhibits autophagosome formation while in starvation conditions TORC1 is inactivated to allow induction of autophagy [50]. More than 30 autophagy-related proteins have been identified, and their molecular functions are discussed elsewhere [51,52]. Autophagosomes have been shown to recruit selective receptors that deliver specific cargos, such as cytosolic proteins and organelles, for degradation [53]. Physiological stresses, such as nutrient and energy deprivation can trigger autophagy, which results in the degradation and recycling of nutrients and essential building blocks of the cells for sustaining metabolic functions and survival [54]. Autophagy is also involved in pathological conditions, such as eliminating intracellular pathogens and degrading damaged organelles [55,56].
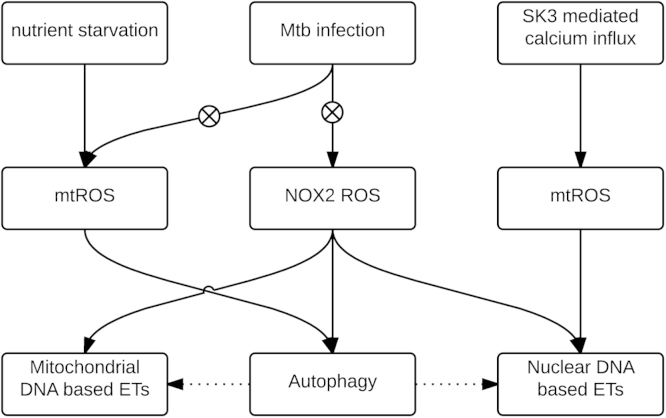
One of the most investigated cargo-specific autophagic pathways is mitophagy, through which damaged mitochondria are degraded. A recent study showed that the homeostasis of mtROS plays an important role in the autophagy pathway. Superoxide has been shown to be the major type of ROS that is responsible for regulating autophagy. Starvation for glucose, L-glutamine, or pyruvate induces superoxide production, while starvation for amino acids induces both superoxide and hydrogen peroxide production, further leading to stimulation of autophagy (Fig. 2). Overexpression of SOD2, which degrades superoxide into hydrogen peroxide in the mitochondria, suppresses autophagy. Catalase, which decreases both superoxide and hydrogen peroxide levels, also suppresses starvation-induced autophagy [10].
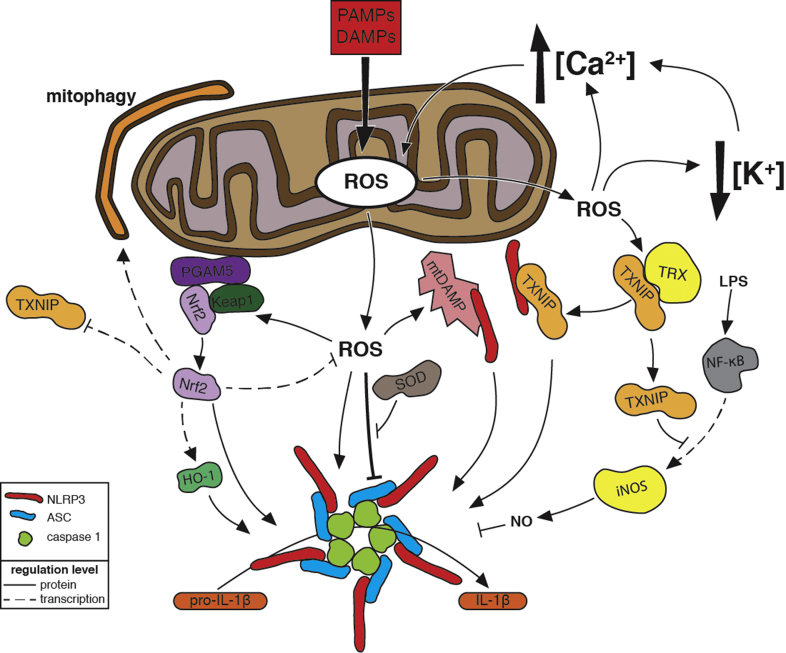
Fig. 3.
Regulation of NLRP3 Inflammasome Activation by Reactive Oxygen Species. In response to pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs), which are stimuli that indicate a disruption in cellular homeostasis, NLRP3, ASC, and caspase-1 assemble into a supramolecular complex, the inflammasome, that processes the inactive form of the pro-inflammatory cytokine interleukin-1β (pro-IL-1β) into its active form (IL-1β) and promotes inflammation. NLRP3 inflammasome stimuli induce mitochondrial reactive oxygen species (ROS) production. ROS might act directly on inflammasome components by oxidizing thiols. Super oxide dismutase (SOD) prevents the accumulation of excess levels of ROS that inhibit inflammasome activation. ROS-mediated release and/or damage of mitochondrial molecules produce mitochondrial DAMPs (mtDAMPs) that bind to NLRP3 (see Fig.4 for more details). ROS promote calcium (Ca2+) influx by activating plasma membrane cation channels. Mitochondrial dysfunction caused by Ca2+ uptake further promotes mitochondrial ROS release. ROS have been implicated in potassium (K+) efflux, which activates the inflammasome via a mechanism that might enhance Ca2+ influx. Oxidation of thioredoxin (TRX) by ROS causes dissociation of thioredoxin-interacting protein (TXNIP) from TRX. Subsequent binding of NLRP3 by TXNIP, possibly at the mitochondria, leads to inflammasome activation. In response to lipopolysaccharide (LPS) detection, NF-κB upregulates expression of inducible nitric oxide synthase (iNOS), which produces nitric oxide (NO) that can inhibit inflammasome activation. TXNIP inhibits the transcriptional activity of NF-κB to attenuate this upregulation and thus prevent inhibition of inflammasome activation. Under no/low oxidative stress conditions, Nrf2 is in a complex comprising Keap1 and the mitochondrial outermembrane protein PGAM5. Oxidation of thiols in Keap1 releases Nrf2 from the complex and leads to Nrf2 association with and activation of the inflammasome. Some Nrf2 translocates to the nucleus and upregulates heme oxygenase-1 (HO-1) expression, which in turn activates the inflammasome. Nrf2 also upregulates expression of anti-oxidant genes, which attenuate ROS levels, and mitophagy-activating genes, which decrease ROS produced by dysfunctional mitochondria. Nrf2 further modulates inflammasome activation by repressing TXNIP expression.
A fast growing area of autophagy research focuses on its role in host defense against bacterial infection, also referred to as xenophagy [57]. As a crucial part of the host innate immune defense, the autophagy pathway is often the target of manipulation by intracellular bacteria. Strategies, such as modification of the bacterial surface, scavenging of ROS, and secretion of virulence factors, enable intracellular bacteria to avoid or circumvent killing by autophagy [58]. Manipulations of autophagy (xenophagy) by various infectious bacteria have been reviewed comprehensively [57,59,60]. Here we focus mainly on findings from recent studies on the roles of ROS in xenophagy. Probably due to the complexity of ROS metabolism and to limitations in current detection methods, a number of the studies did not confirm the source of ROS. However, it is commonly accepted that the principal sources of ROS in the cell during infection are NOX2 and the ETC of mitochondria [61].
During infection by the pathogenic Streptococcus pneumoniae, intracellular ROS produced by host cells induce bactericidal autophagy by inhibiting the PI3K-I/Akt/mTOR signaling pathway [62]. To avoid being digested, Streptococcus suis serotype 2 expresses the superoxide dismutase SodA to inhibit the host xenophagic response by reducing upstream ROS signaling [63].Mtb also bypasses host defense mechanisms by inhibiting autophagy (Fig. 2). The “enhanced intracellular survival” gene (eis) contributes to this inhibition. Infection of macrophages by an eis-deficient Mtb strain induced ROS generation from mitochondria and NOX2 and activated autophagy [64]. A later report identified Mtb eis as an Nɛ-acetyltransferase that participates in acetylating DUSP16/MKP-7, a JNK-specific phosphatase, and negatively regulates autophagy by inhibiting JNK-dependent ROS signaling during Mtb infection [65]. Additionally, the mannosylated glycoprotein phosphoribosyltransferase, a mycobacterial virulence factor located in the cell wall, also assists inhibition of bactericidal autophagy through suppression of host ROS production [66].
Autophagy is the target of some first-line anti-tuberculosis drugs. It has been shown that isoniazid and pyrazinamide stimulate ROS production by NOX2 and mitochondria in Mtb-infected host cells and activate autophagy. Treatment with either a NOX2 inhibitor or mtROS scavenger reduced the antibiotic-induced autophagy in Mtb-infected macrophages [67].
6. The interplay of extracellular traps, autophagy and ROS
As the front-line defense against microbial infections, human neutrophils were previously thought to employ only two strategies to kill invading pathogens: engulfment of microbes and secretion of anti-microbials. A third strategy, formation of Neutrophil Extracellular Traps (NETs), was discovered in 2004 as a phenomenon by which neutrophils kill extracellular pathogens by releasing decondensed chromatin associated with antimicrobial proteins including myeloperoxidase, histones, and proteinases [68]. This DNA-based structure, released from neutrophils during a type of cell death called NETosis, helps to trap bacteria in the blood circulation and inside tissues and to increase the local concentration of anti-microbial enzymes around the trapped bacteria. Thus, NETs increase bacterial killing efficiency while minimizing the damage to neighboring cells by sequestering toxic compounds released from neutrophils into a constrained area [69,70].
It has been shown that NET formation requires both ROS generation and autophagy induction [71]. Phorbol myristate acetate (PMA)-stimulated neutrophils exhibited increased ROS production from NOX2 and an elevated autophagosome formation, leading to NET formation. Inhibition of either NOX2 or autophagy prevents intracellular chromatin decondensation and NET formation [72]. Several other in vivo and in vitro studies also showed that inhibition of autophagy decreases NET formation [73,74]. Autophagy induction is negatively controlled by the metabolic checkpoint kinase mTOR [75]. Itakura and colleagues inhibited mTOR prior to stimulating neutrophils with the bacterial peptide formyl-Met-Leu-Phe and observed increased autophagy and NET formation. This effect was suppressed by inhibiting a NOX2-generated respiratory burst [76].
NETs are associated with cancer. The granulocyte colony-stimulating factor (G-CSF) produced by many tumors also serves as a factor to prime neutrophils to produce NETs [77]. The extracellular chromatin released through NET formation in the blood circulation is a coagulant and promotes the formation of cancer-associated thrombosis [78]. Boone and colleagues found that in pancreatic ductal adenocarcinoma patients and animal models, serum DNA and citrullinated histone H3, the markers of NET formation, increased. Inhibition of autophagy or genetic ablation of receptor for advanced glycation end-products (RAGE), a pattern recognition receptor that activates pro-inflammatory cytokines, lowered the level of NET formation in the tumor microenvironment [79]. However the inhibition mechanism of RAGE on autophagy and NET formation still needs to be further studied.
It has been widely reported that NOX2-generated ROS function as signaling molecules in NET formation [68,70]. Moreover, neutrophils from CGD patients, in contrast to neutrophils with functional NOX2, do not produce NETs upon exposure to Staphylococcus aureus [3]. A NOX2-independent pathway activated by Ca2+ ionophores has also been described [80]. Douda and colleagues recently showed that raising the intracellular level of Ca2+ in neutrophils by adding calcium ionophores significantly activated the small conductance calcium-activated potassium channel SK3 and mitochondrial ROS production, leading to NOX2-independent NET formation (Fig. 2) [81].
The formation of DNA-based extracellular traps has been observed in other types of phagocytic immune cells from vertebrates and invertebrates [82]. Some of the cell types expel mitochondrial DNA instead of chromosomal DNA after bacterial or pharmacological stimulation [83]. There are few reports about the potential interactions of extracellular traps formation, mitochondrial ROS generation and autophagy activation. The detailed study of this topic may expand our current scopes on DNA based host defense mechanism and further enhance our understanding of the mechanisms of immunodeficiency, as well as inflammatory and autoimmune conditions.
7. The NLRP3 inflammasome: a sensor of homeostatic disruptors
Inflammasomes are innate immunity-related signaling complexes that initiate and/or enhance the secretion of pro-inflammatory cytokines in response to pathological disruptions in cell physiology such as infection or the death of neighboring cells. The sensor components of inflammasomes are Nucleotide Binding Domain-Leucine Rich Repeat proteins (NLRs), whose ligands include pathogen-associated molecular patterns (PAMPs) such as flagellin, and AIM2 (Absent In Melanoma 2), which recognizes double-stranded DNA [84]. NLRs also recognize damage-associated molecular patterns (DAMPs) such as mitochondrial DNA [85,86]. Sensing of a PAMP or DAMP promotes the activation of the cysteine protease caspase-1, the subsequent caspase-1-mediated processing of the pro-forms of interleukin (IL)-1β and IL-18, and the secretion of the mature cytokines. Inflammasome signaling can initiate pyroptosis, a mode of induced cell death that releases DAMPs and cytokines to promote inflammation [87]. In contrast to TLRs that monitor for PAMPs and DAMPs at the cell surface and within the endolysosomal system, inflammasome sensors monitor the cytosolic compartment.
NLRP3 (NLR, pyrin domain-containing 3) is the sensor component of the NLRP3 inflammasome. In addition to being activated during infection with viruses [88–90], bacteria [91–96], fungi [97,98], protozoa [99], and helminths [100], the NLRP3 inflammasome responds to DAMPs such as ATP (via the P2×7 receptor) and histones [91,101,102], environmental agents such as silica and asbestos [103–105], and molecules associated with disease states such as uric acid crystals (gout), cholesterol crystals (atherosclerosis), and islet amyloid polypeptide (diabetes) [106–108]. Its activation by wide array of stimuli underscores the central role of the NLRP3 inflammasome in the response to disruptions in cellular homeostasis and its importance in multiple infectious and non-infectious inflammatory diseases.
The structural diversity of NLRP3 inflammasome stimuli suggests that activation is more complex than the binding of PAMPs and DAMPs. Indeed, studies have not yet delineated a central activation pathway upon which all NLRP3 stimuli converge. Shared mechanisms include a drop in intracellular K+ concentration [109,110], increases in Ca2+ concentration [111–114], and lysosomal disruption [103,115,116]. Activation of the NLRP3 inflammasome has been covered in great detail elsewhere [11,84,117–119]. Here we will focus on the mechanisms that involve ROS and mitochondria.
8. ROS and NLRP3 inflammasome activation
8.1. Inflammasome priming
NLRP3 inflammasome activation involves two steps: a priming step that readies the inflammasome components for activation and an activation step that results in inflammasome assembly, processing, and IL-1β secretion [120,121]. In the priming step, the transcription factor NF-κB is activated downstream of TNF, IL-1β, and TLR signaling pathways and subsequently increases the expression of NLRP3 and pro-IL-1β [122–124]. Pretreatment with the general ROS scavenger N-acetyl-l-cysteine (NAC) inhibits NF-κB-dependent priming [125]. This inhibition is presumably due to a requirement for ROS in NF-κB activation [2]. Priming also occurs at the post-translational level [126,127]. Lypopolysaccharide (LPS)-TLR signaling induces mtROS production that might contribute to priming [7]. Mitochondrial complex I inhibitors, which stimulate mtROS production, have been observed to prime in the absence of LPS, although not all reports agree on this [45,110,127,128]. LPS-TLR signaling can prime the NLRP3 inflammasome independently of transcription via deubiquitination of NLRP3, and ROS-scavenging by NAC blocks deubiquitination and subsequent inflammasome activation [127].
8.2. Inflammasome Activation
NLRP3 binds caspase-1 indirectly via the adapter protein ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain) [129,130]. Activation involves the oligomerization of these components. Formation of ASC-containing aggregates, processing of pro-caspase-1, and IL-1β processing and secretion are used to assess NLRP3 inflammasome activation.
ROS have been implicated in the activation step. Initial studies focused on NOX2 as the relevant ROS source: inhibition or knockdown of NOX2 were found to block IL-1β secretion in response to NLRP3 stimuli [105,109,131]. However, cells from CGD patients, which have null mutations in NOX components, still respond to NLRP3 stimuli [132–134]. The inhibition of this NOX-independent activation by ROS scavengers indicates that other sources of ROS can activate the NLRP3 inflammasome [134].
Indeed, mitochondrial ROS are involved in NLRP3 inflammasome activation. Mitochondrial complex I inhibitors increase the production of mtROS and induce IL-1β secretion via NLRP3 activation, and this activation is blocked by ROS scavengers [45,128]. Inflammasome activation by ATP also involves an increase in mtROS production [113,135–138] and is blocked by mitochondria-specific ROS scavengers [90,138]. Ca2+ appears to mediate induction of mtROS production by ATP. Murakami and colleagues observed that inhibiting cytosolic Ca2+ mobilization blocked mtROS production and inflammasome activation by ATP and proposed a model in which mitochondrial uptake of increased cytosolic Ca2+ causes mitochondrial damage and mtROS production [113]. NLRP3 inflammasome activation by liposomes and silica crystals requires an mtROS-dependent influx of Ca2+ via the TRPM2 cation channel [139]. This Ca2+ influx could promote additional mtROS production.
Defects in mitophagy, an autophagy pathway that degrades malfunctioning mitochondria, activate NLRP3 inflammasome partially due to an increase in mtROS [45,138,140,141]. Increased mtROS downstream of NLRP3 activation may act as a positive feedback mechanism that sustains activation [138,140,141].
9. Links between ROS and NLRP3 inflammasome activation
Although numerous studies indicate a role for ROS in activation of the NLRP3 inflammasome, the actual mechanisms remain poorly defined. ROS have been implicated in Ca2+ influx, K+ efflux, the exacerbation of mitochondrial damage, modification of DAMPS sensed by NLRP3, and post-translational modification of inflammasome components [86,89,127,138,139,142]. Blocking cellular antioxidant responses to ROS can inhibit NLRP3 inflammasome activation [143–145]. This observation is partially due to the fact that excess ROS levels can negatively affect protein function. In response to ATP, sod1−/− mice were observed to secrete less IL-1β than wild type controls due to overproduction of ROS and a consequential oxidation of caspase-1 [146]. Nrf2 and TXNIP, proteins involved in the response to oxidative stress, contribute to NLRP3 inflammasome activation independently of regulating caspase-1 oxidation.
9.1. Activation by thioredoxin-interacting protein
Thioredoxin-interacting protein (TXNIP), which binds the antioxidant thioredoxin (TRX) under no/low oxidative stress conditions and is thus considered to be pro-oxidant, is released upon oxidative stress [147,148]. Zhou and colleagues observed ROS- and TXNIP-dependent caspase-1 processing and IL-1β secretion upon stimulation with ATP, alum, uric acid crystals, or silica [149]. Co-precipitation experiments indicate an interaction between TXNIP and NLRP3 [149,150], and immunostaining showed an ROS-dependent recruitment of TXNIP and NLRP3 to mitochondria and or mitochondria-associated membranes in a subsequent study [45]. The authors proposed a model in which the ROS induced by NLRP3 stimuli cause dissociation of TXNIP from TRX. TXNIP then binds to NLRP3 to activate the inflammasome. TXNIP plays a role in insulin resistance and is upregulated in response to glucose; these data may explain the implication of IL-1β and inflammation in type 2 diabetes [149]. Inflammasome activation mediated by binding of NLRP3 by TXNIP was also observed in studies on homocysteine-induced kidney damage and on liver damage induced by a combination of D-galactosamine and LPS [151,152]. In contrast to the above results, another study on inflammasome activation within the context of diabetes observed ROS-dependent and TXNIP-independent activation of NLRP3 by islet amyloid polypeptide. Wild-type and txnip−/− macrophages secreted similar levels of IL-1β in response to IAPP and also when stimulated with silica, ATP, uric acid crystals [108].
Park et al. observed no requirement for TXNIP in NLRP3 activation by ATP, nigericin, or uric acid crystals but did observe that macrophages from txnip−/− mice secreted less IL-1β than wild-type cells in response to LPS or to E. coli in the absence of additional stimuli [153]. LPS treatment induced higher levels of inducible nitric oxide synthase (iNOS) expression in txnip−/− macrophages than wild-type cells; this increased expression correlated with increased binding of the iNOS promoter by NF-κB. The resulting elevated NO levels caused an increase in S-nitrosylation of NLRP3 and caspase-1, which was previously demonstrated to inhibit NLRP3 inflammasome activation after prolonged exposure to LPS [135]. Treatment with an iNOS inhibitor restored IL-1β secretion to wild-type levels [153]. The authors propose a model in which TXNIP negatively regulates induction of iNOS by LPS-induced activation of NF-κB. In the absence of TXNIP, enhanced induction of iNOS by NF-kB leads to levels of NO sufficient to inactivate NLRP3 (Fig.3).
Fig. 4.
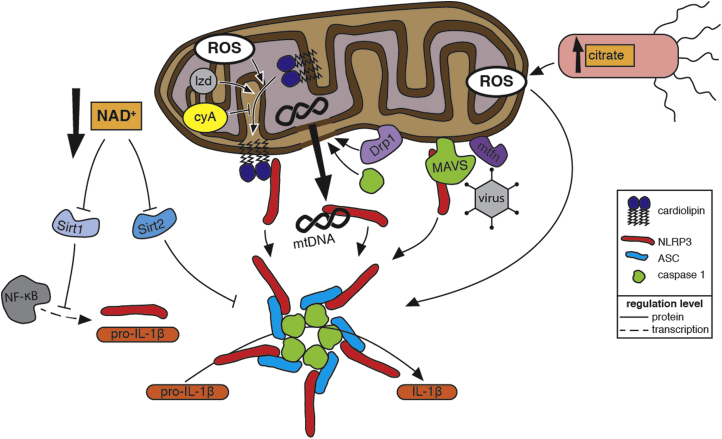
Mitochondrial Regulation of NLRP3 Inflammasome Activation.Mitochondria activate the NLRP3 inflammasome, a supramolecular signalling complex, comprising NLRP3, ASC, and caspase-1, that processes the inactive form of the pro-inflammatory cytokine interleukin-1β (pro-IL-1β) into its active form (IL-1β) to promote inflammation. Indicators of dysfunctional mitochondria contribute to inflammasome activation. Within eukaryotes, molecules such as cardiolipin and mitochondrial DNA (mtDNA), which are unique to the proteobacterially derived mitochondria, are sequestered inside healthy mitochondria. Damaged mitochondria expose these molecules to the cytosol, where they are recognized as mitochondrial damage-associated molecular patterns (mtDAMPs). Damaged mitochondrial also produce reactive oxygen species (ROS) that promote inflammasome activation (see Fig. 3 for more details). Binding of ROS-oxidized mtDNA to NLRP3 promotes inflammasome activation. Cardiolipin is translocated to the mitochondrial outermembrane, where it binds NLRP3, in response to increased mtROS or in response to the antibiotic linezolid (lzd) via a ROS-independent mechanism. This translocation is blocked by cyclosporin A (cyA), which inhibits mitochondrial permeability transition. Caspase-1 activated by NLRP3 further damages mitochondria, and the resulting release of mtROS and mtDAMPs enhances inflammasome activation. Dynamin-related protein 1(Drp1), which promotes mitochondrial fragmentation and potentially the release of mtDAMPs, contributes to inflammasome activation during viral infection. A decrease in NAD+ levels caused by disruptions in the electron transport chain of damaged mitochondria inhibits the activity of the NAD+-dependent deacetylases Sirt1 and Sirt2. Inactive Sirt2 leads to inflammasome activation via a mechanism that involves the trafficking of mitochondria along acetylated microtubules to be in close proximity to the ER. Deacetylation by Sirt1 inhibits the transcriptional activity of NF-κB. Decreased Sirt1 activity allows increased expression of NLRP3 and pro-IL-1β. Salmonella mutants that produce excessive levels of the tricarboxylic acid cycle intermediate citrate activate the inflammasome via a mechanism that requires mitochondrial ROS; a build-up of mitochondrially produced citrate in the cytosol might also promote inflammasome activation. Prior to inflammasome assembly during viral infection, NLRP3 is recruited to the mitochondria by mitochondrial anti-viral signalling protein (MAVS), and this association is enhanced by mitofusins 1 and 2 (mtfn).
S-nitrosylation of NLRP3 appears to be a mechanism by which the adaptive immune response minimizes inflammation-induced pathology during M. tuberculosis infection [154]. The crystal structure of NLRP3 identified a disulfide bond between a cysteine in the pyrin domain (PYD) and a cysteine in a loop connecting to the nucleotide-binding domain [155]. PYD is in the C-terminus of NLRP3 and is required for interaction with ASC; oxidation of the thiols into a disulfide bond might be a mechanism by which ROS contribute to inflammasome activation. The C-terminus of NLRP3 is more sensitive to S-nitrosylation than the N-terminus [135]. Disruption of this disulfide bridge by NO could inhibit NLRP3 association with ASC and thereby prevent NLRP3 inflammasome activation.
The two models for TXNIP regulation of the NLRP3 inflammasome are not mutually exclusive and might intersect. For example, TXNIP released from TRX due to oxidative stress could inhibit NF-kB signaling to dampen the inflammatory response and production of additional reactive oxygen and nitrogen species. Interestingly, the transient repression of TXNIP mRNA levels by LPS stimulation might enable additional modulation of the inflammatory response [108,153,156].
9.2. Activation by Nrf2
The transcription factor NF-E2-related factor 2 (Nrf2) links ROS to NLRP3 activation via the antioxidant response. In no/low stress conditions, Nrf2 is bound to the cullin E3 ubiquitin ligase adapter Keap1, which targets Nrf2 for ubiquitination and subsequent proteasomal degradation [157–159]. Modifications of cysteine thiols in Keap1 by oxidative stress promotes Nrf2 stabilization by disrupting the Keap1–Nrf2 interaction [160].
The initial evidence demonstrating a role for Nrf2 in NLRP3 inflammasome activation is from a study on inflammation within the context of atherosclerosis. Cholesterol crystals, abundant in atherosclerotic plaques, were found to activate the NLRP3 inflammasome via an Nrf2-dependent mechanism in macrophages [107]. Later studies demonstrated that macrophages from nrf2−/− mice secreted less IL-1β than wild-type cells in response to the established NLRP3 activators alum, silica, uric acid crystals, ATP, and nigericin and that knockdown of Nrf2 inhibited the response to uric acid crystals in THP-1 macrophages [128,150].
Nrf2 protein but not transcript levels increase in response to LPS exposure, and pretreatment of cells with ROS scavengers block this accumulation [128]. Although the timing of this increase in Nrf2 levels corresponds to the priming step of inflammasome signaling, Nrf2 does not appear to have a direct role in priming: absence of Nrf2 had no effect on IL-1β, NLRP3, and ASC expression [107,128]. In its resting state, a fraction of Nrf2 associates with the mitochondria in a ternary complex with Keap1 and PGAM5, a mitochondrial outer membrane resident phosphatase that has been implicated in programmed cell death [161,162]. ROS produced downstream of NLRP3 stimuli cause dissociation of Nrf2 from Keap1 and translocation to the cytosol, where it associates with ASC aggregates [128]. This mitochondrial localization places Nrf2 in a prime position to sense mtROS. Indeed, increased mtROS due to mitochondrial complex I inhibitors activate Nrf2- and NLRP3-dependent IL-1β secretion [128]. However, treatment with ROS scavengers after LPS-induced increase in Nrf2 protein did not block IL-1β secretion in response to silica [128]. It is possible that the localization of Nrf2 at the mitochondrial outer membrane renders preventing its interaction with mtROS difficult. Determining whether this activation in the presence of ROS scavengers is due to a ROS-independent activation of Nrf2, a unique aspect of activation by silica, or an insufficient sequestration of mtROS from Nrf2 requires further examination.
Defects in autophagy, which have been linked to activation of NLRP3 inflammasome via the accumulation of defective mitochondria and a concomitant increase in mtROS [45,138], also increase Nrf2 levels. Accumulated p62/SQSTM1 binds Keap1 and causes its dissociation from Nrf2 [163,164]. Transcription factor activity of Nrf2 also contributes to NLRP3 activation. Nrf2 upregulates heme oxygenase-1 (HO-1) in response to uric acid crystals stimulation, and inhibition of HO-1 reduced IL-1β secretion in response to uric acid crystals [150]. A role for HO-1 in NLRP3 inflammasome activation during bacterial infection has also been described [165]. Nrf2 downregulates expression of TXNIP and upregulates genes involved in the antioxidant response, in autophagy, and in mitochondrial biogenesis [164,166,167]. Thus, Nrf2 appears to contribute to modulation of NLRP3 as it also activates it (Fig. 3).
10. Additional roles for mitochondria and NLRP3 activation
10.1. Activation by mitochondrial DAMPs
Mitochondrial DAMPs (mtDAMPs), indicators of mitochondrial damage/dysfunction, can activate the inflammasome in addition to or in lieu of mtROS (Fig. 4). Shimada et al. reported that infection with Salmonella or Chlamydia or treatment with NLRP3 stimulators such as ATP caused mitochondrial dysfunction and release of mitochondrial DNA (mtDNA). Upon oxidation, presumably due to the observed increased mtROS levels produced under the same conditions, oxidized mtDNA bound to and activated NLRP3 [86]. Nakahira et al. observed release of mtDNA downstream of mtROS-dependent NLRP3 activation that enhanced IL-1β secretion, but direct binding of mtDNA by NLRP3 was not assessed [138].
Fig 2.
The Roles of ROS in autophagy and extracellular trap formation. Autophagy can be induced by reactive oxygen species (ROS) produced by mitochondria and phagocyte NADPH oxidase (NOX2) in response to nutrient starvation and bacterial infection. Bacteria such as Mycobacterium tuberculosis inhibit autophagy by decreasing mitochondrial and NOX2-generated ROS. Autophagy, mitochondrial ROS, and NOX2-generated ROS have also been found to be important for extracellular trap (ET) formation. The mechanisms of how these three factors are involved and interact in ET formation, especially in mitochondrial DNA-based ET formation, are not clear.
Cardiolipin, a phospholipid of bacteria and the inner mitochondrial membrane, activates the NLRP3 inflammasome downstream of mitochondria dysfunction [142]. Iyer et al. found that the antiobiotic linezolid, which is used to treat infections with multi-drug resistant bacteria and can cause myelosuppression, activates the NLRP3 inflammasome via a mechanism that is independent of ROS, dependent on K+ efflux, and involves recruitment of NLRP3 to the mitochondria and its binding to cardiolipin [142]. Cardiolipin was also required for NLRP3 activation downstream of mtROS-dependent stimuli such as ATP, silica, and nigericin [142]. Cyclosporin A, which stabilizes mitochondria by inhibiting mitochondrial permeability transition, and knockdown of cardiolipin synthase inhibited IL-1β secretion and caspase-1 processing by linezolid, ATP, and silica [142]. These data suggest that ROS-dependent and -independent pathways converge on mitochondrial damage and identify cardiolipin as an mtDAMP. Moreover, these results implicate bacterial cardiolipin as a potential NLRP3 activator.
The GTPase dynamin-related protein 1 (Drp1), which promotes fragmentation of mitochondria, is required for activation of NLRP3 during infection with vesicular stomatitis virus (VSV) [88]. NLRP3 activation involved recruitment of Drp1 to the mitochondria where it caused fragmentation and aggregation of mitochondria and production of mtROS. Knockdown of Drp1 inhibited mitochondrial damage and IL-1β secretion in response to VSV but not uric acid crystals, ATP, or nigericin [88]. The link between mitochondrial damage and NLRP3 activation might be mtROS or mtDAMPs.
10.2. Activation by mitochondrial metabolites
Mitochondrial metabolites related to the tricarboxylic acid (TCA) cycle and the ETC can also contribute to NLRP3 activation. Misawa et al. observed a role for NAD+, which decreases when the ETC is inhibited. NLRP3 activators such as ATP and uric acid crystals decreased NAD+ levels, which, via decreased activity of the NAD+ dependent Sirt2 deacetylase, caused accumulation of acetylated tubulin and subsequent trafficking of mitochondria to the perinuclear region in close apposition to the ER [168]. Interactions between the ER and mitochondria contribute to NLRP3 inflammasome activation [45]. Colchicine, an inhibitor of microtubule polymerization, disrupted this trafficking and reduced IL-1β secretion [168]. The NAD+-dependent deacetylase Sirt1 inactivates NF-κB and Nrf2 [169,170]. Given the role of NF-κB in inflammasome priming, a drop in NAD+ might inhibit Sirt1 and contribute to increased expression of NLRP3 and pro-IL-1β and further inflammasome activation [124]. The effects of the Nrf2 acetylation state on its activation of NLRP3 activation are not known.
The TCA cycle intermediate citrate accumulates in the cytosol of LPS-stimulated macrophages, likely due to an NF-κB-dependent increase in expression of the mitochondrial citrate carrier, and contributes to ROS and NO production [171,172]. An intriguing recent study demonstrated that Salmonella strains with TCA cycle mutations that caused citrate buildup activate the NLRP3 inflammasome via an mtROS-dependent mechanism [95]. Citrate of bacterial origin should be indistinguishable from that of mitochondrial origin. Thus, it is possible that the cell senses the citrate as an indicator of mitochondrial damage or altered metabolism and that this activates the inflammasome within the context of infection. How the bacterial citrate gains access to the host cytosol and how it activates NLRP3 await elucidation.
10.3. Activation by the mitochondrial antiviral signaling protein
The mitochondrial antiviral signaling protein (MAVS) was originally identified as a mitochondrial membrane protein that activates the interferon response upon infection with viruses [173]. Recent evidence suggests that MAVS also activates the NLRP3 inflammasome during viral infection and in response to non-particulate NLRP3 stimuli [89,174–176]. MAVS-mediated activation involved transient recruitment of NLRP3 to the mitochondria prior to NLRP3 association with ASC [174,175]. It also involved a ROS-dependent increase in plasma membrane permeability and subsequent drop in intracellular K+ [89]. It should be noted that the data do not agree on whether non-viral stimuli activate NLRP3 via MAVS and that the involvement of ROS was not addressed in all of the studies. Further work is needed to determine the mechanism(s) of NLRP3 activation by MAVS and whether it is unique to viral infection.
Mitofusins are outer mitochondrial membrane GTPases that are proposed to regulate mitochondrial fusion. Ichinohe and colleagues observed that mitofusin 1 and 2 are required for activation of NLRP3 during viral infection [90]. Activation required recruitment of NLRP3 to mitochondria, was independent of ROS production, and enhanced the association of NLRP3 with MAVS [90].
11. A Unified model of NLRP3 inflammasome activation?
The NLRP3 inflammasome is activated by a wide array of stimuli and conditions whose only common feature appears to be that they are indicators of disruptions in cellular homeostasis. This diversity in stimuli is reflected by the numerous molecules, proteins, and events that have been implicated in its activation pathways. Whether these pathways converge upstream of, at, or downstream of NLRP3 itself (i.e., no commonalities other than NLRP3-dependent IL-1β secretion) remains a matter of debate. Though consensus has certainly not been achieved, most proposed pathways involve ROS/mtROS and/or mitochondria.
Mitochondria may serve in non-mutually exclusive capacities as a signaling platform and as a source of DAMPs. Recruitment of NLRP3 to the mitochondria in response to infection and endogenous stimuli places it in close proximity to stimuli (mtROS), ligands (mtDNA and cardiolipin), and activating/adaptor proteins (Nrf2, mitofusins, MAVS). Mitochondria are thus a focal point of NLRP3 activation. Even so, different mechanisms of NLRP3 activation are possible. For example, cardiolipin and mtDNA may not be required for NLRP3 activation during viral infection.
ROS have multiple points of entry: the priming step, initiation of an antioxidant response (dissociation of Nrf2 and TXNIP), induction and/or indication of mitochondrial damage, oxidation of NLRP3 itself or its ligands, and possibly K+ efflux. Some of the disagreements over the role of ROS might be explained by mitochondrial DAMPs or the fact that high ROS levels can inhibit caspase-1. However, other discrepancies, such as whether ROS are dispensable for priming and whether stimuli such as ATP and uric acid crystals require ROS are more difficult to resolve [135–137].
Based on observations that the antioxidant response is important for NLRP3 activation [107,144,146,149], it has been proposed that the overall redox state of the cell may account for some of the contradictory data [143,177,178]. Cells in tissue culture are exposed to higher levels of oxygen than their normal environments and thus upregulate their antioxidant capacity. Consequently, high levels of ROS production are necessary to induce an antioxidant response sufficient for NLRP3 activation. In contrast, freshly prepared primary cells have a low antioxidant capacity. In these cells, the low level of ROS induced by LPS is sufficient to activate NLRP3 in the absence of a second signal [143]. More research is required. As the field continues to identify additional aspects of NLRP3 inflammasome signaling and regulation and the extent to which these mechanisms are impacted by the redox state of the cell, it will get closer to resolving the current contradictions.
12. Final remarks
The acquisition of the endosymbiont protobacterium that evolved into mitochondria gave early eukaryotes a more efficient means of ATP production but also a source of potentially dangerous ROS. It was thus advantageous to cells to develop and/or enhance antioxidant mechanisms to counteract increased ROS from damaged mitochondria and to evolve mechanisms such as autophagy to degrade damaged mitochondria in response to ROS produced by dysfunctional mitochondria. These countermeasures mitigate the toxic potential of ROS. As a consequence, ROS have been co-opted as not just danger signals but also as actively generated signaling molecules.
Similarly, the mechanisms regulating mitochondria function and sensing mitochondrial damage have been co-opted by cellular immune response pathways. Mitochondria are used as a source of ROS to produce anti-microbial effectors. Autophagy is used to eliminate bacteria from the cytosol. ROS have roles in activating autophagy and in the formation of extracellular DNA traps. Pathways that regulate the metabolic homeostasis of cells intersect with immune pathways via the mitochondria. This intersection is exemplified by the NLRP3 inflammasome. Mitochondrial dysfunction, sensed by the exposure of mtDAMPs to the cytosol, alterations in metabolite levels, and/or increased mtROS, activates the NLRP3 inflammasome. The dysfunction can result from the activation of pathways that sense the presence of microbes or from the direct targeting of mitochondria by pathogens. It can also result from non-infectious diseases such as diabetes, gout, and atherosclerosis.
Many questions regarding the central roles of mitochondria and ROS in regulating cellular homeostasis remain. Do mtROS have direct anti-microbial effects, or do they only function as signaling molecules? If mtROS are anti-microbial effectors, then how do they gain access to phagosome-bound pathogens? What are the actual mechanisms that mediate ROS signaling? Is it a direct modification of proteins? Is it indirectly via activation of antioxidant proteins? Which species of ROS are important in which pathways? How does the redox state of the cell impact ROS signaling? Answering such questions will increase our understanding of cellular regulatory mechanisms and provide potential points for therapeutic intervention in a range of infectious and non-infectious diseases.
Acknowledgments
We thank Igor Brodsky (University of Pennsylvania, Philadelphia, PA, USA) for comments on portions of the manuscript. All authors of this review were supported by the European Cooperation in Science and Technology (COST) Action BM1203/EU‐ROS. Additional support came from the Swiss SEFRI-COST N. C13.0137 (J.D.D), the Swiss National Science Foundation (X.Z.) (SNSF grant: PDFMP3 127302/ 1), Science Foundation Ireland (L.A.J.A.) (10/ IN.1/B2988), and the Children’s Medical and Research Foundation Ireland (L.A.J.A.) (K12/1).
Contributor Information
Joe Dan Dunn, Email: joedan.dunn@unige.ch.
Luis AJ Alvarez, Email: luis.alvarez@ucd.ie.
Xuezhi Zhang, Email: Xuezhi.Zhang@unige.ch.
Thierry Soldati, Email: Thierry.Soldati@unige.ch.
References
- 1.Nauseef W.M. Myeloperoxidase in human neutrophil host defence. Cell Microbiol. 2014;16:1146–1155. doi: 10.1111/cmi.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan M.J., Liu Z.-G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs T.A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam G.Y., Huang J., Brumell J.H. The many roles of NOX2 NADPH oxidase-derived ROS in immunity. Semin Immunopathol. 2010;32:415–430. doi: 10.1007/s00281-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 5.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulua A.C., Simon A., Maddipati R., Pelletier M., Park H., Kim K.-Y. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) J. Exp. Med. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender T., Martinou J.-C. Where killers meet—permeabilization of the outer mitochondrial membrane during apoptosis. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a011106. a011106–a011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maryanovich M., Gross A. A ROS rheostat for cell fate regulation. Trends Cell Biol. 2013;23:129–134. doi: 10.1016/j.tcb.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 11.Gurung P., Lukens J.R., Kanneganti T.-D. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol.r Med. 2015;21:193–201. doi: 10.1016/j.molmed.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills E., O’Neill L.A.J. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014;24:313–320. doi: 10.1016/j.tcb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Mittal M., Siddiqui M.R., Tran K., Reddy S.P., Malik A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014;20:1126–1167. doi: 10.1089/ars.2012.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rada B., Leto T.L. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib. Microbiol. 2008;15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soldati T., Neyrolles O. Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic. 2012;13:1042–1052. doi: 10.1111/j.1600-0854.2012.01358.x. [DOI] [PubMed] [Google Scholar]
- 16.Al Ghouleh I., Khoo N.K.H., Knaus U.G., Griendling K.K., Touyz R.M., Thannickal V.J. Oxidases and peroxidases in cardiovascular and lung disease: new concepts in reactive oxygen species signaling. Free Radic. Biol. Med. 2011;51:1271–1288. doi: 10.1016/j.freeradbiomed.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinauer M.C. Chronic granulomatous disease and other disorders of phagocyte function. Hematology / the Education Program of the American Society of Hematology. American Society of Hematology. Education Program. 2005:89–95. doi: 10.1182/asheducation-2005.1.89. [DOI] [PubMed] [Google Scholar]
- 18.Mantegazza A.R., Savina A., Vermeulen M., Pérez L., Geffner J., Hermine O. NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood. 2008;112:4712–4722. doi: 10.1182/blood-2008-01-134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rybicka J.M., Balce D.R., Khan M.F., Krohn R.M., Yates R.M. NADPH oxidase activity controls phagosomal proteolysis in macrophages through modulation of the lumenal redox environment of phagosomes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10496–10501. doi: 10.1073/pnas.0914867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stasia M.J., Li X.J. Genetics and immunopathology of chronic granulomatous disease. Semin. Immunopathol. 2008;30:209–235. doi: 10.1007/s00281-008-0121-8. [DOI] [PubMed] [Google Scholar]
- 21.Lanza F. Clinical manifestation of myeloperoxidase deficiency. J. Mol. Med. 1998;76:676–681. doi: 10.1007/s001090050267. [DOI] [PubMed] [Google Scholar]
- 22.Roca F.J., Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell. 2013;153:521–534. doi: 10.1016/j.cell.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda J., Laganiere J., Mehl I.R., Barish G.D., Chong L.W., Li X. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Devt. 2007;21:1909–1920. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupre-Crochet S., Erard M., Nüße O. ROS production in phagocytes: why, when, and where? J. Leukoc. Biol. 2013;94:657–670. doi: 10.1189/jlb.1012544. [DOI] [PubMed] [Google Scholar]
- 25.Corcionivoschi N., Alvarez L.A.J., Sharp T.H., Strengert M., Alemka A., Mantell J. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe. 2012;12:47–59. doi: 10.1016/j.chom.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton N.A., Schurmann N., Casse O., Steeb A.K., Claudi B., Zankl J. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbé. 2014;15:72–83. doi: 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Winter S.E., Thiennimitr P., Winter M.G., Butler B.P., Huseby D.L., Crawford R.W. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., Lu R., Wu S., Sun J. Salmonella regulation of intestinal stem cells through the Wnt/beta-catenin pathway. FEBS Lett. 2010;584:911–916. doi: 10.1016/j.febslet.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon J.C., Ng A., Kim B.H., Bianco A., Xavier R.J., Elledge S.J. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabharwal S.S., Schumacker P.T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan C.L., Goncalves R.L., Hey-Mogensen M., Yadava N., Bunik V.I., Brand M.D. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dikalov S.I., Nazarewicz R.R., Bikineyeva A., Hilenski L., Lassègue B., Griendling K.K. Nox2-induced production of mitochondrial superoxide in angiotensin II-mediated endothelial oxidative stress and hypertension. Antioxid. Redox Signal. 2014;20:281–294. doi: 10.1089/ars.2012.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazarewicz R.R., Dikalova A.E., Bikineyeva A., Dikalov S.I. Nox2 as a potential target of mitochondrial superoxide and its role in endothelial oxidative stress. Am. J. Physiol. Heart Circ. Physiol. 2013;305 doi: 10.1152/ajpheart.00063.2013. H1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kröller-Schön S., Steven S., Kossmann S., Scholz A., Daub S., Oelze M. Molecular mechanisms of the crosstalk between mitochondria and NADPH oxidase through reactive oxygen species-studies in white blood cells and in animal models. Antioxid. Redox Signal. 2014;20:247–266. doi: 10.1089/ars.2012.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D.-Y., Wauquier F., Eid A.A., Roman L.J., Ghosh-Choudhury G., Khazim K. Nox4 NADPH oxidase mediates peroxynitrite-dependent uncoupling of endothelial nitric-oxide synthase and fibronectin expression in response to angiotensin II: role of mitochondrial reactive oxygen species. J. Biol. Chem. 2013;288:28668–28686. doi: 10.1074/jbc.M113.470971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozieł R., Pircher H., Kratochwil M., Lener B., Hermann M., Dencher N.A. Mitochondrial respiratory chain complex I is inactivated by NADPH oxidase Nox4. Biochem. J. 2013;452:231–239. doi: 10.1042/BJ20121778. [DOI] [PubMed] [Google Scholar]
- 37.Kawamata H., Manfredi G. Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid. Redox Signal. 2010;13:1375–1384. doi: 10.1089/ars.2010.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karnati S., Luers G., Pfreimer S., Baumgart-Vogt E. Mammalian SOD2 is exclusively located in mitochondria and not present in peroxisomes. Histochem Cell Biol. 2013;140:105–117. doi: 10.1007/s00418-013-1099-4. [DOI] [PubMed] [Google Scholar]
- 39.Gottfredsen R.H., Goldstrohm D.A., Hartney J.M., Larsen U.G., Bowler R.P., Petersen S.V. The cellular distribution of extracellular superoxide dismutase in macrophages is altered by cellular activation but unaffected by the naturally occurring R213G substitution. Free Radic. Biol. Med. 2014;69:348–356. doi: 10.1016/j.freeradbiomed.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang C.K., Liu Y., Thomas J., Zhang Y., Zheng X.F. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014;5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han D., Antunes F., Canali R., Rettori D., Cadenas E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 2003;278:5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- 42.Collins Y., Chouchani E.T., James A.M., Menger K.E., Cocheme H.M., Murphy M.P. Mitochondrial redox signalling at a glance. J. Cell Sci. 2012;125:801–806. doi: 10.1242/jcs.098475. [DOI] [PubMed] [Google Scholar]
- 43.Guzy R.D., Schumacker P.T. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp. Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 44.Mandal S., Lindgren A.G., Srivastava A.S., Clark A.T., Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells. 2011;29:486–495. doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou R., Yazdi A.S., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 46.Bell E.L., Emerling B.M., Ricoult S.J., Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura S., Zhang G.-X., Nishiyama A., Shokoji T., Yao L., Fan Y.-Y. Mitochondria-derived reactive oxygen species and vascular MAP kinases: comparison of angiotensin II and diazoxide. Hypertension. 2005;45:438–444. doi: 10.1161/01.HYP.0000157169.27818.ae. [DOI] [PubMed] [Google Scholar]
- 48.Dikalova A.E., Bikineyeva A.T., Budzyn K., Nazarewicz R.R., McCann L., Lewis W. Therapeutic targeting of mitochondrial superoxide in hypertension. Circ. Res. 2010;107:106–116. doi: 10.1161/CIRCRESAHA.109.214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klionsky D.J., Eskelinen E.L., Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes wait, I’m confused. Autophagy. 2014;10:549–551. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boya P., Reggiori F., Codogno P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge L., Baskaran S., Schekman R., Hurley J.H. The protein-vesicle network of autophagy. Curr. Opin. Cell Biol. 2014;29:18–24. doi: 10.1016/j.ceb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Weidberg H., Shvets E., Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 54.Romanov J., Walczak M., Ibiricu I., Schuchner S., Ogris E., Kraft C. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knodler L.A., Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol. 2011;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baxt L.A., Garza-Mayers A.C., Goldberg M.B. Bacterial subversion of host innate immune pathways. Science. 2013;340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 59.Deretic V. Autophagy in infection. Curr. Opin. Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li P., Shi J., He Q., Hu Q., Wang Y.Y., Zhang L.J. Streptococcus pneumoniae induces autophagy through the inhibition of the PI3K-I/Akt/mTOR pathway and ROS hypergeneration in A549 cells. PLoS ONE. 2015;10:e0122753. doi: 10.1371/journal.pone.0122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang L., Shen H., Tang Y., Fang W. Superoxide dismutase of Streptococcus suis serotype 2 plays a role in anti-autophagic response by scavenging reactive oxygen species in infected macrophages. Vet. Microbiol. 2015;176:328–336. doi: 10.1016/j.vetmic.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Shin D.-M., Jeon B.-Y., Lee H.-M., Jin H.S., Yuk J.-M., Song C.-H. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K.H., An D.R., Song J., Yoon J.Y., Kim H.S., Yoon H.J. Mycobacterium tuberculosis eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7729–7734. doi: 10.1073/pnas.1120251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohanty S., Jagannathan L., Ganguli G., Padhi A., Roy D., Alaridah N. A mycobacterial phosphoribosyltransferase promotes bacillary survival by inhibiting oxidative stress and autophagy pathways in macrophages and zebrafish. J. Biol. Chem. 2015;290:13321–13343. doi: 10.1074/jbc.M114.598482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J.J., Lee H.M., Shin D.M., Kim W., Yuk J.M., Jin H.S. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11:457–468. doi: 10.1016/j.chom.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 69.Brinkmann V., Zychlinsky A. Beneficial suicide: why neutrophils die to make NETs. Nat. Rev. Microbiol. 2007;5:577–582. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 70.Wartha F., Henriques-Normark B. ETosis: a novel cell death pathway. Sci. Signal. 2008;1:pe25. doi: 10.1126/stke.121pe25. [DOI] [PubMed] [Google Scholar]
- 71.Remijsen Q., Kuijpers T.W., Wirawan E., Lippens S., Vandenabeele P., Vanden Berghe T. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ. 2011;18:581–588. doi: 10.1038/cdd.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Remijsen Q., Vanden Berghe T., Wirawan E., Asselbergh B., Parthoens E., De Rycke R. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chrysanthopoulou A., Mitroulis I., Apostolidou E., Arelaki S., Mikroulis D., Konstantinidis T. Neutrophil extracellular traps promote differentiation and function of fibroblasts. J. Pathol. 2014;233:294–307. doi: 10.1002/path.4359. [DOI] [PubMed] [Google Scholar]
- 74.Tang S., Zhang Y., Yin S.W., Gao X.J., Shi W.W., Wang Y. Neutrophil extracellular trap formation is associated with autophagy-related signalling in ANCA-associated vasculitis. Clin. Exp. Immunol. 2015 doi: 10.1111/cei.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tattoli I., Sorbara M.T., Vuckovic D., Ling A., Soares F., Carneiro L.A. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 76.Itakura A., McCarty O.J. Pivotal role for the mTOR pathway in the formation of neutrophil extracellular traps via regulation of autophagy. Am. J. Physiol. Cell Physiol. 2013;305 doi: 10.1152/ajpcell.00108.2013. C348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Demers M., Wagner D.D. Neutrophil extracellular traps: A new link to cancer-associated thrombosis and potential implications for tumor progression. Oncoimmunology. 2013;2:e22946. doi: 10.4161/onci.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demers M., Krause D.S., Schatzberg D., Martinod K., Voorhees J.R., Fuchs T.A. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boone B.A., Orlichenko L., Schapiro N.E., Loughran P., Gianfrate G.C., Ellis J.T. The receptor for advanced glycation end products (RAGE) enhances autophagy and neutrophil extracellular traps in pancreatic cancer. Cancer Gene Ther. 2015 doi: 10.1038/cgt.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parker H., Dragunow M., Hampton M.B., Kettle A.J., Winterbourn C.C. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J. Leukoc. Biol. 2012;92:841–849. doi: 10.1189/jlb.1211601. [DOI] [PubMed] [Google Scholar]
- 81.Douda D.N., Khan M.A., Grasemann H., Palaniyar N. SK3 channel and mitochondrial ROS mediate NADPH oxidase-independent NETosis induced by calcium influx. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2817–2822. doi: 10.1073/pnas.1414055112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goldmann O., Medina E. The expanding world of extracellular traps: not only neutrophils but much more. Front. Immunol. 2012;3:420. doi: 10.3389/fimmu.2012.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yousefi S., Gold J.A., Andina N., Lee J.J., Kelly A.M., Kozlowski E. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 84.Abais J.M., Xia M., Zhang Y., Boini K.M., Li P.-L. Redox Regulation of NLRP3 inflammasomes: ROS as trigger or effector? Antioxid. Redox Signal. 2015;22:1111–1129. doi: 10.1089/ars.2014.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jabir M.S., Hopkins L., Ritchie N.D., Ullah I., Bayes H.K., Li D. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy. 2015;11:166–182. doi: 10.4161/15548627.2014.981915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shimada K., Crother T.R., Karlin J., Dagvadorj J., Chiba N., Chen S. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fink S.L., Bergsbaken T., Cookson B.T. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang X., Jiang W., Yan Y., Gong T., Han J., Tian Z. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol. 2014;15:1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- 89.Franchi L., Eigenbrod T., Muñoz-Planillo R., Ozkurede U., Kim Y.-G., Chakrabarti A. Cytosolic double-stranded RNA activates the NLRP3 inflammasome via MAVS-induced membrane permeabilization and K+ efflux. J. Immunol. 2014;193:4214–4222. doi: 10.4049/jimmunol.1400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ichinohe T., Yamazaki T., Koshiba T., Yanagi Y. Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17963–17968. doi: 10.1073/pnas.1312571110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mariathasan S., Weiss D.S., Newton K., McBride J., O’Rourke K., Roose-Girma M. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 92.Toma C., Higa N., Koizumi Y., Nakasone N., Ogura Y., McCoy A.J. Pathogenic Vibrio activate NLRP3 inflammasome via cytotoxins and TLR/nucleotide-binding oligomerization domain-mediated NF-kappa B signaling. J. Immunol. 2010;184:5287–5297. doi: 10.4049/jimmunol.0903536. [DOI] [PubMed] [Google Scholar]
- 93.Semper R.P., Mejías-Luque R., Groß C., Anderl F., Müller A., Vieth M. Helicobacter pylori-induced IL-1β secretion in innate immune cells is regulated by the NLRP3 inflammasome and requires the cag pathogenicity island. J. Immunol. 2014;193:3566–3576. doi: 10.4049/jimmunol.1400362. [DOI] [PubMed] [Google Scholar]
- 94.Zwack E.E., Snyder A.G., Wynosky-Dolfi M.A., Ruthel G., Philip N.H., Marketon M.M. nflammasome activation in response to the Yersinia type III secretion system requires hyperinjection of translocon proteins YopB and YopD. MBio. 2015;6 doi: 10.1128/mBio.02095-14. e02095–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wynosky-Dolfi M.A., Snyder A.G., Philip N.H., Doonan P.J., Poffenberger M.C., Avizonis D. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J. Exp. Med. 2014;211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mishra B.B., Moura-Alves P., Sonawane A., Hacohen N., Griffiths G., Moita L.F. Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell Microbiol. 2010;12:1046–1063. doi: 10.1111/j.1462-5822.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- 97.Lamkanfi M., Malireddi R.K.S., Kanneganti T.-D. Fungal zymosan and mannan activate the cryopyrin inflammasome. J. Biol. Chem. 2009;284:20574–20581. doi: 10.1074/jbc.M109.023689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Groß O., Poeck H., Bscheider M., Dostert C., Hannesschläger N., Endres S. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 99.Gorfu G., Cirelli K.M., Melo M.B., Mayer-Barber K., Crown D., Koller B.H. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. MBio. 2014;5 doi: 10.1128/mBio.01117-13. e01117–13–e01117–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ritter M., Groß O., Kays S., Ruland J., Nimmerjahn F., Saijo S. Schistosoma mansoni triggers Dectin-2, which activates the Nlrp3 inflammasome and alters adaptive immune responses. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20459–20464. doi: 10.1073/pnas.1010337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sutterwala F.S., Ogura Y., Szczepanik M., Lara-Tejero M., Lichtenberger G.S., Grant E.P. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 102.Allam R., Darisipudi M.N., Rupanagudi K.V., Lichtnekert J., Tschopp J., Anders H.-J. Cutting edge: cyclic polypeptide and aminoglycoside antibiotics trigger IL-1β secretion by activating the NLRP3 inflammasome. J. Immunol. 2011;186:2714–2718. doi: 10.4049/jimmunol.1002657. [DOI] [PubMed] [Google Scholar]
- 103.Hornung V., Bauernfeind F., Halle A., Samstad E.O., Kono H., Rock K.L. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cassel S.L., Eisenbarth S.C., Iyer S.S., Sadler J.J., Colegio O.R., Tephly L.A. The Nalp3 inflammasome is essential for the development of silicosis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B.T., Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 107.Freigang S., Ampenberger F., Spohn G., Heer S., Shamshiev A.T., Kisielow J. Nrf2 is essential for cholesterol crystal-induced inflammasome activation and exacerbation of atherosclerosis. Eur. J. Immunol. 2011;41:2040–2051. doi: 10.1002/eji.201041316. [DOI] [PubMed] [Google Scholar]
- 108.Masters S.L., Dunne A., Subramanian S.L., Hull R.L., Tannahill G.M., Sharp F.A. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 2010;11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 110.Muñoz-Planillo R., Kuffa P., Martínez-Colón G., Smith B.L., Rajendiran T.M., Núñez G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Triantafilou K., Hughes T.R., Triantafilou M., Morgan B.P. The complement membrane attack complex triggers intracellular Ca2+ fluxes leading to NLRP3 inflammasome activation. J. Cell Sci. 2013;126:2903–2913. doi: 10.1242/jcs.124388. [DOI] [PubMed] [Google Scholar]
- 112.Lee G.-S., Subramanian N., Kim A.I., Aksentijevich I., Goldbach-Mansky R., Sacks D.B. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murakami T., Ockinger J., Yu J., Byles V., McColl A., Hofer A.M. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rossol M., Pierer M., Raulien N., Quandt D., Meusch U., Rothe K. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nat. Commun. 2012;3:1329. doi: 10.1038/ncomms2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weber K., Schilling J.D. Lysosomes integrate metabolic-inflammatory cross-talk in primary macrophage inflammasome activation. J. Biol. Chem. 2014;289:9158–9171. doi: 10.1074/jbc.M113.531202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fubini B., Hubbard A. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) generation by silica in inflammation and fibrosis. Free Radic. Biol. Med. 2003;34:1507–1516. doi: 10.1016/s0891-5849(03)00149-7. [DOI] [PubMed] [Google Scholar]
- 117.Elliott E.I., Sutterwala F.S. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harijith A., Ebenezer D.L., Natarajan V. Reactive oxygen species at the crossroads of inflammasome and inflammation. Front. Physiol. 2014;5:352. doi: 10.3389/fphys.2014.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abderrazak A., Syrovets T., Couchie D., El Hadri K., Friguet B., Simmet T. NLRP3 inflammasome: from a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biol. 2015;4:296–307. doi: 10.1016/j.redox.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Franchi L., Eigenbrod T., Muñoz-Planillo R., Núñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Faustin B., Lartigue L., Bruey J.-M., Luciano F., Sergienko E., Bailly-Maitre B. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 122.O’Connor W., Harton J.A., Zhu X., Linhoff M.W., Ting J.P.-Y. Cutting edge: CIAS1/cryopyrin/PYPAF1/NALP3/CATERPILLER 1.1 is an inducible inflammatory mediator with NF-kappa B suppressive properties. J. Immunol. 2003;171:6329–6333. doi: 10.4049/jimmunol.171.12.6329. [DOI] [PubMed] [Google Scholar]
- 123.Franchi L., Eigenbrod T., Núñez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bauernfeind F.G., Horvath G., Stutz A., Alnemri E.S., MacDonald K., Speert D. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bauernfeind F., Bartok E., Rieger A., Franchi L., Núñez G., Hornung V. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol. 2011;187:613–617. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schroder K., Sagulenko V., Zamoshnikova A., Richards A.A., Cridland J.A., Irvine K.M. Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology. 2012;217:1325–1329. doi: 10.1016/j.imbio.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 127.Juliana C., Fernandes-Alnemri T., Kang S., Farias A., Qin F., Alnemri E.S. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao C., Gillette D.D., Li X., Zhang Z., Wen H. Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J. Biol. Chem. 2014;289:17020–17029. doi: 10.1074/jbc.M114.563114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Agostini L., Martinon F., Burns K., McDermott M.F., Hawkins P.N., Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 130.Stehlik C., Lee S.H., Dorfleutner A., Stassinopoulos A., Sagara J., Reed J.C. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. J. Immunol. 2003;171:6154–6163. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- 131.Cruz C.M., Rinna A., Forman H.J., Ventura A.L.M., Persechini P.M., Ojcius D.M. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J. Biol. Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meissner F., Seger R.A., Moshous D., Fischer A., Reichenbach J., Zychlinsky A. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood. 2010;116:1570–1573. doi: 10.1182/blood-2010-01-264218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van de Veerdonk F.L., Smeekens S.P., Joosten L.A.B., Kullberg B.J., Dinarello C.A., van der Meer J.W.M. Reactive oxygen species-independent activation of the IL-1beta inflammasome in cells from patients with chronic granulomatous disease. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3030–3033. doi: 10.1073/pnas.0914795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Van Bruggen R., Köker M.Y., Jansen M., van Houdt M., Roos D., Kuijpers T.W. Human NLRP3 inflammasome activation is Nox1-4 independent. Blood. 2010;115:5398–5400. doi: 10.1182/blood-2009-10-250803. [DOI] [PubMed] [Google Scholar]
- 135.Hernandez-Cuellar E., Tsuchiya K., Hara H., Fang R., Sakai S., Kawamura I. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J Immunol. 2012;189:5113–5117. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- 136.Mao K., Chen S., Chen M., Ma Y., Wang Y., Huang B. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 2013;23:201–212. doi: 10.1038/cr.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hua K.-F., Chou J.-C., Ka S.-M., Tasi Y.-L., Chen A., Wu S.-H. Cyclooxygenase-2 regulates NLRP3 inflammasome-derived IL-1β production. J. Cell Physiol. 2015;230:863–874. doi: 10.1002/jcp.24815. [DOI] [PubMed] [Google Scholar]
- 138.Nakahira K., Haspel J.A., Rathinam V.A.K., Lee S.-J., Dolinay T., Lam H.C. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhong Z., Zhai Y., Liang S., Mori Y., Han R., Sutterwala F.S. TRPM2 links oxidative stress to NLRP3 inflammasome activation. Nat. Commun. 2013;4:1611. doi: 10.1038/ncomms2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu J., Nagasu H., Murakami T., Hoang H., Broderick L., Hoffman H.M. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15514–15519. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lupfer C., Thomas P.G., Anand P.K., Vogel P., Milasta S., Martinez J. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Iyer S.S., He Q., Janczy J.R., Elliott E.I., Zhong Z., Olivier A.K. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carta S., Tassi S., Pettinati I., Delfino L., Dinarello C.A., Rubartelli A. The rate of interleukin-1beta secretion in different myeloid cells varies with the extent of redox response to Toll-like receptor triggering. J. Biol. Chem. 2011;286:27069–27080. doi: 10.1074/jbc.M110.203398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tassi S., Carta S., Vené R., Delfino L., Ciriolo M.R., Rubartelli A. Pathogen-induced interleukin-1beta processing and secretion is regulated by a biphasic redox response. J. Immunol. 2009;183:1456–1462. doi: 10.4049/jimmunol.0900578. [DOI] [PubMed] [Google Scholar]
- 145.Tassi S., Carta S., Delfino L., Caorsi R., Martini A., Gattorno M. Altered redox state of monocytes from cryopyrin-associated periodic syndromes causes accelerated IL-1beta secretion. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9789–9794. doi: 10.1073/pnas.1000779107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meissner F., Molawi K., Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat. Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 147.Junn E., Han S.H., Im J.Y., Yang Y., Cho E.W., Um H.D. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J. Immunol. 2000;164:6287–6295. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- 148.Nishiyama A., Matsui M., Iwata S., Hirota K., Masutani H., Nakamura H. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 149.Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2009;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 150.Jhang J.-J., Cheng Y.-T., Ho C.-Y., Yen G.-C. Monosodium urate crystals trigger Nrf2- and heme oxygenase-1-dependent inflammation in THP-1 cells. Cell. Mol. Immunol. 2014 doi: 10.1038/cmi.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Abais J.M., Xia M., Li G., Chen Y., Conley S.M., Gehr T.W.B. Nod-like receptor protein 3 (NLRP3) inflammasome activation and podocyte injury via thioredoxin-interacting protein (TXNIP) during hyperhomocysteinemia. J. Biol. Chem. 2014;289:27159–27168. doi: 10.1074/jbc.M114.567537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim S.-J., Lee S.-M. NLRP3 inflammasome activation in D-galactosamine and lipopolysaccharide-induced acute liver failure: role of heme oxygenase-1. Free Radic. Biol. Med. 2013;65:997–1004. doi: 10.1016/j.freeradbiomed.2013.08.178. [DOI] [PubMed] [Google Scholar]
- 153.Park Y.-J., Yoon S.-J., Suh H.-W., Kim D.O., Park J.-R., Jung H. TXNIP deficiency exacerbates endotoxic shock via the induction of excessive nitric oxide synthesis. PLoS Pathog. 2013;9:e1003646. doi: 10.1371/journal.ppat.1003646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mishra B.B., Rathinam V.A.K., Martens G.W., Martinot A.J., Kornfeld H., Fitzgerald K.A. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat. Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bae J.Y., Park H.H. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J. Biol. Chem. 2011;286:39528–39536. doi: 10.1074/jbc.M111.278812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kanari Y., Sato Y., Aoyama S., Muta T. Thioredoxin-interacting protein gene expression via MondoA is rapidly and transiently suppressed during inflammatory responses. PLoS One. 2013;8:e59026. doi: 10.1371/journal.pone.0059026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang D.D., Lo S.-C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kobayashi A., Kang M.-I., Okawa H., Ohtsuji M., Zenke Y., Chiba T. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cullinan S.B., Gordan J.D., Jin J., Harper J.W., Diehl J.A. Vol. 24. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase; pp. 8477–8486. (Mol. Cell. Biol.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lo S.-C., Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Plafker K.S., Plafker S.M. The ubiquitin-conjugating enzyme UBE2E3 and its import receptor importin-11 regulate the localization and activity of the antioxidant transcription factor NRF2. Mol. Biol. Cell. 2015;26:327–338. doi: 10.1091/mbc.E14-06-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 164.Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wegiel B., Larsen R., Gallo D., Chin B.Y., Harris C., Mannam P. Macrophages sense and kill bacteria through carbon monoxide-dependent inflammasome activation. J. Clin. Invest. 2014;124:4926–4940. doi: 10.1172/JCI72853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.He X., Ma Q. Redox regulation by nuclear factor erythroid 2-related factor 2: gatekeeping for the basal and diabetes-induced expression of thioredoxin-interacting protein. Mol. Pharmacol. 2012;82:887–897. doi: 10.1124/mol.112.081133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Piantadosi C.A., Withers C.M., Bartz R.R., MacGarvey N.C., Fu P., Sweeney T.E. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. J. Biol. Chem. 2011;286:16374–16385. doi: 10.1074/jbc.M110.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Misawa T., Takahama M., Kozaki T., Lee H., Zou J., Saitoh T. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 169.Yeung F., Hoberg J.E., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kawai Y., Garduño L., Theodore M., Yang J., Arinze I.J. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem. 2011;286:7629–7640. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Infantino V., Convertini P., Cucci L., Panaro M.A., Di Noia M.A., Calvello R. The mitochondrial citrate carrier: a new player in inflammation. Biochem. J. 2011;438:433–436. doi: 10.1042/BJ20111275. [DOI] [PubMed] [Google Scholar]
- 173.Seth R.B., Sun L., Ea C.-K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 174.Subramanian N., Natarajan K., Clatworthy M.R., Wang Z., Germain R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chu C.T., Bayır H., Kagan V.E. LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: implications for Parkinson disease. Autophagy. 2014;10:376–378. doi: 10.4161/auto.27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chakrabarti A., Banerjee S., Franchi L., Loo Y.-M., Gale M., Núñez G. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe. 2015;17:466–477. doi: 10.1016/j.chom.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rubartelli A., Gattorno M., Netea M.G., Dinarello C.A. Interplay between redox status and inflammasome activation. Trends Immunol. 2011;32:559–566. doi: 10.1016/j.it.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 178.Rubartelli A. Redox control of NLRP3 inflammasome activation in health and disease. J. Leukoc. Biol. 2012;92:951–958. doi: 10.1189/jlb.0512265. [DOI] [PubMed] [Google Scholar]