Abstract
Background. In Kenya, invasive nontyphoidal Salmonella (iNTS) disease causes severe bacteremic illness among adults with human immunodeficiency virus (HIV) and especially among children <5 years of age coinfected with HIV or malaria, or who are compromised by sickle cell disease or severe malnutrition. The incidence of iNTS disease in children ranges from 166 to 568 cases per 100 000 persons per year.
Methods. We review the epidemiology of iNTS disease and genomics of strains causing invasive illness in Kenya. We analyzed a total of 192 NTS isolates (114 Typhimurium, 78 Enteritidis) from blood and stools from pediatric admissions in 2005–2013. Testing for antimicrobial susceptibility to commonly used drugs and whole-genome sequencing were performed to assess prevalence and genetic relatedness of multidrug-resistant iNTS strains, respectively.
Results. A majority (88/114 [77%]) of Salmonella Typhimurium and 30% (24/79) of Salmonella Enteritidis isolates tested were found to be multidrug resistant, whereas a dominant Salmonella Typhimurium pathotype, ST313, was primarily associated with invasive disease and febrile illness. Analysis of the ST313 isolates has identified genome degradation, compared with the ST19 genotype that typically causes diarrhea in humans, especially in industrialized countries, adapting a more host-restricted lifestyle typical of Salmonella Typhi infections.
Conclusions. From 2012, we have observed an emergence of ceftriaxone-resistant strains also showing reduced susceptibility to fluoroquinolones. As most cases present with nonspecific febrile illness with no laboratory-confirmed etiology, empiric treatment of iNTS disease is a major challenge in Kenya. Multidrug resistance, including to ceftriaxone, will pose further difficulty in management of iNTS disease in endemic areas.
Keywords: nontyphoidal Salmonella, epidemiology, genomics, multidrug resistant, Kenya
In industrialized countries, nontyphoidal Salmonella (NTS) infections (ie, infections caused by Salmonella enterica serovars other than Typhi or Paratyphi) are typical zoonotic infections usually associated with self-limiting enterocolitis, commonly acquired from contaminated foods of animal origin [1–4]. In contrast, invasive NTS (iNTS) in sub-Saharan Africa often causes life-threatening bacteremic illness, especially in immunosuppressed individuals [5–11]. Indeed, iNTS, along with Streptococcus pneumoniae, is a leading cause of invasive bacterial disease in children [7, 9, 12–16]. The global burden of iNTS disease is estimated at 3.4 million (range, 2.1–6.5 million) cases annually (overall incidence, 49 cases [range, 30–94] per 100 000 population), with sub-Saharan Africa bearing the biggest burden, especially among children <5 years of age. Disease incidence is estimated at 227 cases [range, 152–341] per 100 000 population, with 1.9 million [range, 1.3–2.9 million] cases and 681 316 deaths (range, 415 164–1 301 520 deaths) annually [17]. The most important risk factors associated with iNTS disease include human immunodeficiency virus (HIV) infection in adults and young children, and severe malnutrition and coinfection with malaria [18, 19] in young children. Currently there is no commercially available vaccine against iNTS disease. Prompt treatment of cases with effective antimicrobials remains the main option for management of the disease. However, the emergence of multidrug resistance in iNTS strains has placed a major challenge in the management of infections, especially in resource-constrained countries where alternative effective antimicrobials are either unavailable or too expensive to be afforded by the public health sector. In this review, we present data on the epidemiology of iNTS disease in Kenya and also discuss the emergence of multidrug-resistant (MDR) iNTS strains and the challenges in management of infection. We explore the emerging data on the genomics of iNTS strains and what these data have revealed concerning the evolution and the geophylogeny of iNTS disease in the country.
EPIDEMIOLOGY OF INVASIVE NTS DISEASE IN KENYA
Disease Burden, Risk Factors, and Implications on Management
In Kenya over the last 2 decades, the 2 main serotypes implicated in iNTS disease in children were Salmonella enterica serotype Typhimurium and Salmonella enterica serotype Enteritidis, which accounted for an average of 87% (range, 79%–90%) of all NTS strains, and these proportions have remained stable and almost equally distributed. Historical data from a referral hospital in Kenya show that iNTS disease caused a high case fatality ratio (CFR) among young children well before the current HIV pandemic emerged in Kenya and the region [20]. This study showed that prevalence of iNTS disease among patients receiving blood cultures was 45% in children <2 years of age and that this fell to 32% in children aged 2–12 years and 23% in those >12 years of age. The CFR among children with iNTS bacteremia was 18% in the youngest age group whereas in those with meningitis, mortality rose to 98%. Malnutrition was cited as a major risk factor for iNTS disease during this period [20].
In a rural site in western Kenya, incidence of iNTS disease was estimated at 568 per 100 000 per year in 2006–2009 [21], with a 90-day CFR being 7.1% for children and 15.6% for older persons. Earlier studies in adults showed that iNTS disease was associated with HIV infection, with an incidence rate of 21.3% among HIV-infected patients, compared with 3.1% among HIV-uninfected patients (43 of 197 vs 9 of 296; odds ratio, 7.18 [95% confidence interval, 3.58–14.39]), accounting for half of the bacteremic cases, with a CFR ranging between 18.0% and 40.0% [22]. However, the true incidence was thought to be underestimated due to incomplete blood culturing of febrile patients, as many patients with illness in the community never reach the hospital, and blood culturing is a relatively insensitive method due to the low magnitude of Salmonella bacteremia [23].
Socioeconomic status is also a major contributing factor in the prevalence of life-threatening iNTS disease. For instance, a significantly higher proportion of children with iNTS disease came from informal settlements compared with children from the middle-income population and upper socioeconomic classes (n=128) (62 [48.4%] vs 47 [38.2%] and 14 [17.3%], respectively; P < .001); the former had higher prevalence of severe malnutrition, which has been directly associated with high incidence of iNTS disease [9]. In contrast, and as would be expected, a higher proportion of NTS enterocolitis was reported from children from the upper socioeconomic class compared with children from the informal settlements or the middle-income group [14]. Nairobi's informal settlements where these studies were done are characterized by dense population, poor sanitation, and contaminated water supplies. These settings create a perfect environment for rapid spread of enteric and other sanitation-related pathogens through contaminated food and water [24].
Comorbidity with malaria has also been closely associated with increased incidence of iNTS disease in studies in Kenya [25, 26] and elsewhere in Africa including Malawi [27], The Gambia, and Democratic Republic of Congo [28]. In The Gambia, a decline in the prevalence of malaria cases was strongly associated with a decline in incidence of iNTS disease in children, from 60 (during the period 1979–1984) to 10 (during 2003–2005) cases per 100 000 persons per year [29]. However, the association between malaria infection and increased iNTS disease has not been clearly explained.
Sources and Reservoirs of iNTS
Epidemiologic studies of iNTS conducted in parts of Kenya [30] did not find obvious reservoirs among domestic animals. Indeed, the Salmonella Typhimurium isolated from the environment and animals at the homes of index cases were predominantly of different serotypes and often fully susceptible to the antimicrobials tested. In case-control studies in Kilifi and Nairobi, we observed transient carriage and shedding in 6.9% (32/468 individuals) of children and adults from 25 homes of index cases [30]. Asymptomatic carriage of NTS appears to be relatively common [25, 31]. These findings may suggest a role for humans as a symptomatic or asymptomatic reservoir for Salmonella Typhimurium infection, similar to the pattern observed for Salmonella Typhi. It is not known whether stool shedding is relatively transient or whether there is substantial long-term carriage of Salmonella Typhimurium serovars in some individuals.
ANTIMICROBIAL SUSCEPTIBILITY OF INVASIVE NTS STRAINS
In the early 1990s, some 32% of 144 iNTS strains analyzed from cases in urban sites were resistant to ≥3 antimicrobials; the most common resistance phenotype was ampicillin, tetracycline, and trimethoprim-sulfamethoxazole (TMP-SMX) in two-thirds of the isolates and chloramphenicol or ampicillin, tetracycline, and gentamicin in 15% of the isolates. There were no significant differences in prevalence of resistance between the 2 major serotypes, Salmonella Typhimurium and Salmonella Enteritidis. Ciprofloxacin and ceftriaxone were the only antimicrobials to which all NTS were fully susceptible. In subsequent studies in 2006–2010, the proportion of MDR isolates rose to >75% [11, 21]. Although ceftriaxone-resistant Salmonella Typhimurium has previously been reported in other countries in Europe, [32], Asia [33, 34] and the United States [35, 36], only from 2012 did we see the emergence of ceftriaxone-resistant Salmonella Typhimurium sequence type (ST) 313 isolates in Kenya [37]. These isolates came from adults and children who reported to a referral hospital with fever, with or without diarrhea, and were initially treated with ceftriaxone and failed to respond. The spectrum of resistance in these isolates extends beyond the cephalosporins to include tetracyclines, TMP-SMX, chloramphenicol, and aminoglycosides such as streptomycin, but they remained susceptible to azithromycin. Fluoroquinolone resistance is also a growing problem in NTS, with the first report of ciprofloxacin resistance in iNTS disease associated with treatment failure being published in 1990 [32]. The aim of this article was to review existing data on the epidemiology of iNTS disease and to present genomic data on evolving characteristics of subtypes implicated in invasive disease in Kenya.
MATERIALS AND METHODS
Bacterial Isolates and Antimicrobial Susceptibility Testing
A total of 192 NTS isolates (114 Salmonella Typhimurium, 78 Salmonella Enteritidis) from blood and stools from pediatric admissions at 4 main hospitals in Kenya during 2000–2013 were studied. NTS isolates were tested for susceptibility to antimicrobials initially by a controlled disk diffusion technique on Mueller-Hinton agar (Oxoid Ltd, Basingstoke, United Kingdom) plates. The antibiotic disks (all from Oxoid) contained ampicillin (10 µg), ceftriaxone (30 µg), chloramphenicol (30 µg), ciprofloxacin (1 µg), nalidixic acid (30 µg), tetracycline (10 µg), TMP (2.5 µg), TMP-SMX 1:19 (25 µg), and nitrofurantoin (300 µg). Escherichia coli ATCC 25922 was included to control for disk potency and quality of the agar on each test occasion. Susceptibility tests were interpreted using Clinical and Laboratory Standards Institute (CLSI) guidelines [23]. Annual current CLSI updates on the interpretation of ciprofloxacin susceptibility cutoffs on Salmonella species were utilized when reporting susceptibility test results of blood culture isolates to clinicians for management of patients.
Genomic DNA Preparation
Bacteria for genomic analysis were first grown on Luria-Bertani medium (Oxoid). Single, pure isolated colonies were inoculated in Luria-Bertani broth and incubated overnight at 37°C. The growth was pelleted by centrifugation and whole-genome DNA was extracted using the Wizard Genomic DNA kit (Promega, Southampton, United Kingdom) according to the manufacturer's instructions. Aliquots of 20–50 ng/μL of DNA from each isolate were submitted for whole-genome sequencing.
Library Preparation and DNA Sequencing
Multiplex libraries with 108 base-pair (bp) reads and a mean insert size of 272 bp were prepared as previously described [38]. The cluster formation, primer hybridization, and sequencing reactions were done using the Illumina Hiseq sequencer (LGC, Middlesex, United Kingdom). An average of 91.7% of each strain was mapped using SMALT with a mean depth of 118.6-fold in mapped regions across all isolates as previously described [39].
Phylogenetic Analysis
Maximum-likelihood phylogenetic trees were constructed using RAxML version 7.0.4 and an alignment of all the concatenated variant sites from the nonrepetitive and nonrecombinant chromosome of each of the isolates were included in the analyses as previously described [39]. The maximum-likelihood ratios were then calculated using the general time-reversible model with a γ correction for site variation as the nucleotide substitution model. The likelihood test ratios were determined as previously described [40]. The support for nodes on the trees were checked using 100 random bootstrap replicates. Resulting phylogenetic trees were visualized using the Fig tree package version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
RESULTS AND DISCUSSION
Antimicrobial Resistance
Overall, serovar Salmonella Typhimurium had a higher proportion of isolates exhibiting resistance, with 97% (111) of the isolates showing resistance to at least 1 antimicrobial, compared to 92% (73) of the Salmonella Enteritidis isolates that are resistant to at least 1 antimicrobial (Table 1). The majority (77%) of the Salmonella Typhimurium isolates were resistant to at least 3 antimicrobials, whereas only 3% were fully susceptible to all the antimicrobials tested. The most common antimicrobial resistance phenotype among Salmonella Typhimurium isolates was ampicillin/chloramphenicol/nalidixic acid/TMP/TMP-SMX, with 20% [24] of Salmonella Typhimurium isolates having this phenotype. Of the 79 Salmonella Enteritidis isolates, 30% were resistant to at least 3 antibiotics, whereas only 8% were fully susceptible to all the antimicrobials tested; the most common resistance phenotype was ampicillin/chloramphenicol/tetracycline/TMP/TMP-SMX (Table 2).
Table 1.
Antimicrobial Susceptibility Patterns Among Invasive Nontyphoidal Salmonella Isolates, 2010–2012
| No. of Antibiotics Exhibiting Resistance | Proportion, % (Frequency) |
|
|---|---|---|
| Salmonella enterica Serovar Typhimurium (n = 114) | Salmonella enterica Serovar Enteritidis (n = 79) | |
| Fully susceptible | 3 (3) | 8 (6) |
| 1 | 13 (15) | 32 (25) |
| 2 | 7 (8) | 30 (24) |
| 3 | 12 (14) | 1 (1) |
| 4 | 30 (34) | 1 (1) |
| 5 | 27 (31) | 24 (19) |
| 6 | 5 (6) | 4 (3) |
| 7 | 2 (2) | 0 (0) |
| 8 | 0 (0) | 0 (0) |
| 9 | 1 (1) | 0 (0) |
Table 2.
Antimicrobial Resistance Phenotype Distribution Among Nontyphoidal Salmonella Isolates
| Antimicrobial Resistance Phenotypes | Proportion, % (Frequency) |
|
|---|---|---|
| Salmonella enterica Serovar Typhimurium (n = 114) | Salmonella enterica Serovar Enteritidis (n = 79) | |
| AMP/C/NA/W/TMP-SMX | 20 (23) | 0 (0) |
| AMP/C/TE/W/TMP-SMX | 0 (0) | 24 (19) |
| AMP/C/W/TMP-SMX | 17 (19) | 0 (0) |
| AMP/W/TMP-SMX | 12 (14) | 1 (1) |
| AMP/W/TMP-SMX/F | 5 (6) | 0 (0) |
| AMP/TE/W/TMP-SMX | 5 (6) | 0 (0) |
| AMP/CRO/C/TE/W/TMP-SMX | 4 (5) | 1 (1) |
| AMP/C/TE/W/TMP-SMX/F | 1 (1) | 3 (2) |
| AMP/TE/W/TMP-SMX | 3 (3) | 0 (0) |
| AMP/CRO/C/CIP/TE/W/TMP-SMX | 2 (2) | 0 (0) |
| AMP/NA/W/TMP-SMX | 2 (2) | 0 (0) |
| AMP/NA/W/TMP-SMX/F | 2 (2) | 0 (0) |
| AMP/C/W/TMP-SMX/F | 1 (1) | 0 (0) |
| AMP/CRO/W/TMP-SMX | 1 (1) | 0 (0) |
| AMP/CRO/W/TMP-SMX/F | 1 (1) | 0 (0) |
| AMP/NA/TE/W/TMP-SMX | 1 (1) | 0 (0) |
| CRO/C/NA/F | 0 (0) | 1 (1) |
| AMP/CRO/C/CIP/NA/TE/W/TMP-SMX/F | 1 (1) | 0 (0) |
Abbreviations: AMP, ampicillin (10 µg); C, chloramphenicol (30 µg); CIP, ciprofloxacin (1 µg); CRO, ceftriaxone (30 µg); F, nitrofurantoin (300 µg); NA, nalidixic acid (30 µg); TE, tetracycline (10 µg); TMP-SMX, trimethoprim-sulfamethoxazole 1:19 (25 µg); W, trimethoprim (2.5 µg).
A number of studies have suggested that the development of resistance stems from the use of antimicrobials in human medicine, animal husbandry, and agricultural and aquaculture practices. In animal husbandry practices, antimicrobial agents are used for treatment and prevention of diseases as well as for growth promotion [41]. These practices contribute to emergence of antimicrobial resistant bacteria that subsequently can be transferred from animals to humans through the food chain. In many industrialized countries, most antimicrobial-resistant Salmonella infections are acquired from eating contaminated foods of animal origin [42]. In Kenya, we were unable to show any significant association between NTS isolates from humans and those from animals living in close contact. In these settings, it is likely that humans are more important reservoirs of MDR isolates than are food animals [14], with contaminated water and foods playing important roles as vehicles in communities with poor hygiene and sanitation [43].
It is noteworthy that in Kenya the availability of commonly used antimicrobials over the counter and without prescription for self-treatment of suspected infection in humans may have played a major role in the high prevalence of the multidrug resistance. In addition, the availability of cheaper generic drugs of variable quality for treatment of bacterial infections may also have contributed to the increased levels of resistance [44].
SALMONELLA TYPHIMURIUM PHYLOGENY IN KENYA IS DEFINED BY MDR PHENOTYPE
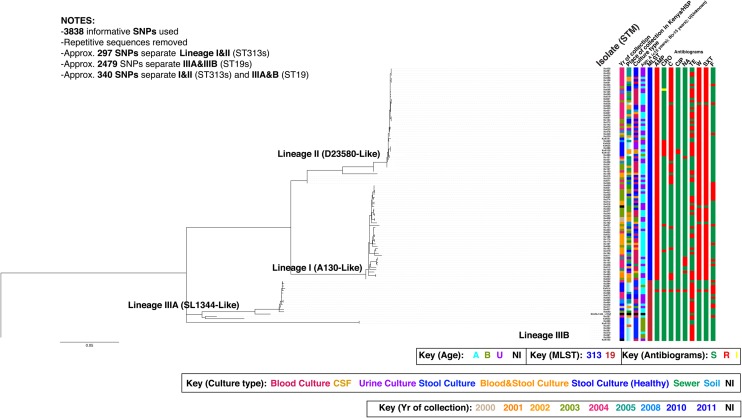
Whole-genome sequencing showed that the majority (80% [91/114]) of the Salmonella Typhimurium isolates belonged to ST313 [1, 45], whereas 23 (20%) of the isolates belonged to ST19 (an ST common in Europe and the United States). Further detailed analysis showed that the isolates fell into 4 lineages that were designated as I, II, IIIA, and IIIB (Figure 1). The isolates in lineage I (invasive-associated A130-like) and II (invasive-associated D23580-like) all belong to ST313, whereas isolates in lineage IIIA (enterocolitis-associated SL1344-like) and IIIB all belong to ST19. These data agree with previous findings [45], but ST19 strains are not known to be a common cause of enterocolitis in Kenya and the region. Thus, we defined 2 new ST19 lineages circulating within Kenya. An average of 297 single-nucleotide polymorphisms (SNPs) separates ST313 lineage I from ST313 lineage II, whereas an average of 2479 SNPs separates ST19 lineage IIIA from ST19 lineage IIIB, implying that ST19 lineage isolates are more diverse than ST313 lineage isolates.
Figure 1.
The single-nucleotide polymorphism (SNP)–based phylogenetic structure of 114 clinical and nonclinical Salmonella Typhimurium isolates implicated in invasive and gastrointestinal Salmonella disease in humans in Kenya between 2000 and 2011. This is an unrooted maximum-likelihood tree showing the relationship between isolates associated with invasive disease and gastroenteritis. The identified lineages for the studied isolates are designated as I, II, IIIA, and IIIB. The extrapolative phylogenetic positioning in regard to the previously characterized invasive Salmonella Typhimurium A130 and D23580 [1] and enterocolitis-associated SL1344 are based on the previous findings by Okoro et al [45]. Branch lengths are indicative of the estimated substitution rate per variable site. Scale bar = 0.07 substitutions per variable site. Colors in legend highlight isolate information (place, year, and patient age), culture type, multilocus sequence type, and antimicrobial resistance profiles. Abbreviations: AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; CSF, cerebrospinal fluid; F, nitrofurantoin; HSP, Hospital; MLST, multilocus sequence type; NA, nalidixic acid; NI, no information available; STM, Salmonella Typhimurium; SXT, trimethoprim-sulfamethoxazole; TE, tetracycline; W, trimethoprim.
The clinical metadata showed that the majority (61%) of the Salmonella Typhimurium isolates within lineages I and II were isolated from invasive-associated tissues (blood, cerebrospinal fluid, urine), strongly indicating that the Salmonella Typhimurium isolates in lineages I and II are associated with iNTS disease. This is consistent with the previous findings that the majority of Salmonella Typhimurium isolates implicated in invasive disease in sub-Saharan Africa mainly comprised 2 closely related ST313 lineages that were more closely related to each other than to other Salmonella Typhimurium [45]. Interestingly, isolates from blood of 6 patients belonged to ST19 and mapped within lineages IIIA and IIIB. Although additional clinical data for 4 patients were not available, 2 patients had cough, fever, respiratory distress, and fever, with general body weakness. This supports a premise that other Salmonella Typhimurium lineages are potentially capable of establishing opportunistic invasive disease in immunocompromised individuals, as reported in studies conducted outside the sub-Saharan Africa region [45]. The antimicrobial susceptibility profiles of the iNTS isolates were found to be generally congruent with their phylogenetic positioning (Figure 1). Again, consistent with previous reports, nearly all (99% [91/92]) of the ST313 isolates in lineages I and II were MDR [45]. Conversely, the majority (68% [17/25]) of ST19 isolates in lineage IIIA and B were susceptible to most antimicrobials in the testing panel. Comparative whole-genome analysis of these and previous Salmonella Typhimurium strains revealed distinct genomic signatures that suggested that the ST313 isolates may be undergoing adaptation to a host-restricted pathogenic lifestyle [1, 45], similar to findings in the other human-adapted Salmonella pathogens, including Salmonella Typhi and S. Paratyphi A [1].
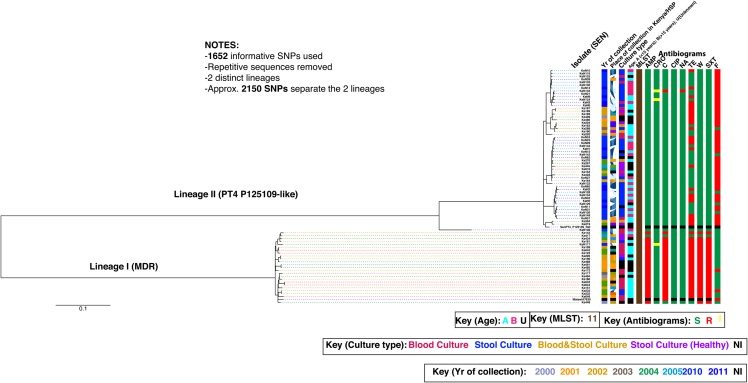
SALMONELLA ENTERITIDIS PHYLOGENY IN KENYA SHOWS A CORRELATION WITH MDR PHENOTYPE
All the Salmonella Enteritidis isolates tested belonged to ST11. Interestingly, the Salmonella Enteritidis isolates fell into 2 distinct and separate lineages that were designated I and II (PT4 P125109-like) (Figure 2). The lineages are separated by approximately 2150 SNPs. Similar to what was observed in Salmonella Typhimurium phylogeny, clinical metadata showed that 46% (11/24) of the Salmonella Enteritidis isolates within lineage I were from bacteremic patients compared with 29% (16/55) of lineage II, thus suggesting that the Salmonella Enteritidis isolates in lineages I could be more closely associated with iNTS disease. Likewise, a strong association was observed between antimicrobial susceptibility and the population structure of Salmonella Enteritidis isolates (Figure 2). The majority (96% [23/24]) of isolates in lineage I were MDR, compared with 45% (25/55) in lineage II (PT4 P125109-like), which are MDR. Conversely, the majority (55% [30/55]) of isolates in lineage II (PT4 P125109-like) are susceptible to most antimicrobials in the testing panel. The high proportion of MDR among lineage I isolates typifies the reported high antimicrobial resistance against NTS isolates in sub-Saharan Africa [44]. So far we have not observed genomic degradation in Salmonella Enteriditis as observed in Salmonella Typhimurium. We hypothesize that Salmonella Enteritidis may have different reservoir status in human hosts and may be undergoing a different evolutionary pathway compared with Salmonella Typhimurium.
Figure 2.
The predicted single-nucleotide polymorphism (SNP)–based population structure for 79 clinical and nonclinical Salmonella Enteritidis isolates implicated in invasive and enterocolitis disease in humans in Kenya between 2000 and 2011. This is an unrooted maximum-likelihood tree showing the relationship between isolates associated with invasive disease and enterocolitis. The identified lineages for the studied isolates are designated as I and II. The phylogenetic position of the European reference Salmonella Enteritidis strain PT4P125109 [46] is indicated. Branch lengths are indicative of the estimated substitution rate per variable site. Scale bar = 0.07 substitutions per variable site. Colors in legend highlight isolate information (place, year, and patient age), culture type, multilocus sequence type, and antibiotic resistance profiles. Abbreviations: AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; F, nitrofurantoin; HSP, Hospital; MDR, multidrug resistant; MLST, multilocus sequence type; NA, nalidixic acid; NI, no information available; SEN, Salmonella Enteritidis; SXT, trimethoprim-sulfamethoxazole; TE, tetracycline; W, trimethoprim.
CONCLUSIONS
Invasive NTS disease in Kenya is largely driven by 2 serovars, Salmonella Typhimurium and Salmonella Enteritidis, and causes most illness among children <5 years of age and in HIV-infected adults. Although whole-genome sequencing has helped to gain insights into the epidemiology and phylogenetics of iNTS disease, there are still important gaps in knowledge, such as establishing reservoirs of iNTS strains in the community and mapping modes and routes of transmission that require to be filled in order to devise effective management and control strategies. The increasing prevalence of MDR strains causing iNTS disease poses a major challenge for public health management and control of iNTS disease in Kenya, especially as alternative effective antimicrobials are either too expensive or simply unavailable.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. S. K. is supported by the National Institute of Allergy and Infectious Diseases of the NIH (award number 1R01AI099525) and the Wellcome Trust.
Supplement sponsorship. This article appeared as part of the supplement “Invasive Salmonella Disease in Africa,” sponsored by the University of Otago.
Potential conflict of interest. Both authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kingsley RA, Msefula CL, Thomson NR et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 2009; 19:2279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laupland KB, Schonheyder HC, Kennedy KJ et al. Salmonella enterica bacteraemia: a multi-national population-based cohort study. BMC Infect Dis 2010; 10:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laufer AS, Grass J, Holt K, Whichard JM, Griffin PM, Gould LH. Outbreaks of Salmonella infections attributed to beef—United States, 1973–2011. Epidemiol Infect 2015; 143:2003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DE Knegt LV, Pires SM, Hald T. Attributing foodborne salmonellosis in humans to animal reservoirs in the European Union using a multi-country stochastic model. Epidemiol Infect 2015; 143:1175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis 2010; 10:417–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon MA, Graham SM, Walsh AL et al. Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis 2008; 46:963–9. [DOI] [PubMed] [Google Scholar]
- 7.Sigauque B, Roca A, Mandomando I et al. Community-acquired bacteremia among children admitted to a rural hospital in Mozambique. Pediatr Infect Dis J 2009; 28:108–13. [DOI] [PubMed] [Google Scholar]
- 8.Gilks CF, Brindle RJ, Otieno LS et al. Life-threatening bacteraemia in HIV-1 seropositive adults admitted to hospital in Nairobi, Kenya. Lancet 1990; 336:545–9. [DOI] [PubMed] [Google Scholar]
- 9.Berkley JA, Lowe BS, Mwangi I et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med 2005; 352:39–47. [DOI] [PubMed] [Google Scholar]
- 10.Berkley J, Mwangi I, Griffiths K et al. Assessment of severe malnutrition among hospitalized children in rural Kenya: comparison of weight for height and mid upper arm circumference. JAMA 2005; 294:591–7. [DOI] [PubMed] [Google Scholar]
- 11.Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 2015; 33(suppl 3):C21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahwere P, Levy J, Hennart P et al. Community-acquired bacteremia among hospitalized children in rural central Africa. Int J Infect Dis 2001; 5:180–8. [DOI] [PubMed] [Google Scholar]
- 13.Green SD, Cheesbrough JS. Salmonella bacteraemia among young children at a rural hospital in western Zaire. Ann Trop Paediatr 1993; 13:45–53. [DOI] [PubMed] [Google Scholar]
- 14.Kariuki S, Revathi G, Kariuki N, Kiiru J, Mwituria J, Hart CA. Characterisation of community acquired non-typhoidal Salmonella from bacteraemia and diarrhoeal infections in children admitted to hospital in Nairobi, Kenya. BMC Microbiol 2006; 6:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassa-Kelembho E, Mbolidi CD, Service YB, Morvan J, Minssart P. Bacteremia in adults admitted to the Department of Medicine of Bangui Community Hospital (Central African Republic). Acta Trop 2003; 89:67–72. [DOI] [PubMed] [Google Scholar]
- 16.Berkley JA, Ross A, Mwangi I et al. Prognostic indicators of early and late death in children admitted to district hospital in Kenya: cohort study. BMJ 2003; 326:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg Infect Dis 2015; 21:941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feasey N, Wansbrough-Jones M, Mabey DC, Solomon AW. Neglected tropical diseases. Br Med Bull 2010; 93:179–200. [DOI] [PubMed] [Google Scholar]
- 19.Williams TN, Uyoga S, Macharia A et al. Bacteraemia in Kenyan children with sickle-cell anaemia: a retrospective cohort and case-control study. Lancet 2009; 374:1364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wamola IA, Mirza NB. Problems of Salmonella infections in a hospital in Kenya. East Afr Med J 1981; 58:677–83. [PubMed] [Google Scholar]
- 21.Tabu C, Breiman RF, Ochieng B et al. Differing burden and epidemiology of non-Typhi Salmonella bacteremia in rural and urban Kenya, 2006–2009. PLoS One 2012; 7:e31237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arthur G, Nduba VN, Kariuki SM, Kimari J, Bhatt SM, Gilks CF. Trends in bloodstream infections among human immunodeficiency virus-infected adults admitted to a hospital in Nairobi, Kenya, during the last decade. Clin Infect Dis 2001; 33:248–56. [DOI] [PubMed] [Google Scholar]
- 23.Clinical Laboratory and Standards Institute. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. Document M100-S24. Wayne, PA: CLSI, 2014. [Google Scholar]
- 24.Olack B, Feikin DR, Cosmas LO et al. Mortality trends observed in population-based surveillance of an urban slum settlement, Kibera, Kenya, 2007–2010. PLoS One 2014; 9:e85913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brent AJ, Oundo JO, Mwangi I, Ochola L, Lowe B, Berkley JA. Salmonella bacteremia in Kenyan children. Pediatr Infect Dis J 2006; 25:230–6. [DOI] [PubMed] [Google Scholar]
- 26.Berkley JA, Bejon P, Mwangi T et al. HIV infection, malnutrition, and invasive bacterial infection among children with severe malaria. Clin Infect Dis 2009; 49:336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bronzan RN, Taylor TE, Mwenechanya J et al. Bacteremia in Malawian children with severe malaria: prevalence, etiology, HIV coinfection, and outcome. J Infect Dis 2007; 195:895–904. [DOI] [PubMed] [Google Scholar]
- 28.Phoba MF, Lunguya O, Mayimon DV et al. Multidrug-resistant Salmonella enterica, Democratic Republic of the Congo. Emerg Infect Dis 2012; 18:1692–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackenzie G, Ceesay SJ, Hill PC et al. A decline in the incidence of invasive non-typhoidal Salmonella infection in The Gambia temporally associated with a decline in malaria infection. PLoS One 2010; 5:e10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kariuki S, Revathi G, Kariuki N et al. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: zoonotic or anthroponotic transmission? J Med Microbiol 2006; 55(Pt 5):585–91. [DOI] [PubMed] [Google Scholar]
- 31.Kariuki S, Revathi G, Kariuki N et al. Increasing prevalence of multidrug-resistant non-typhoidal salmonellae, Kenya, 1994–2003. Int J Antimicrob Agents 2005; 25:38–43. [DOI] [PubMed] [Google Scholar]
- 32.Majtan J, Majtanova L, Majtan V. Increasing trend of resistance to nalidixic acid and emerging ceftriaxone and ciprofloxacin resistance in Salmonella enterica serovar Typhimurium in Slovakia, 2005 to 2009. Diagn Microbiol Infect Dis 2010; 68:86–8. [DOI] [PubMed] [Google Scholar]
- 33.Wong MH, Yan M, Chan EW, Biao K, Chen S. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother 2014; 58:3752–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B, Wang Q, Cui S et al. Characterization of extended-spectrum beta-lactamases-producing Salmonella strains isolated from retail foods in Shaanxi and Henan Province, China. Food Microbiol 2014; 42:14–8. [DOI] [PubMed] [Google Scholar]
- 35.Crump JA, Medalla FM, Joyce KW et al. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring system, 1996 to 2007. Antimicrob Agents Chemother 2011; 55:1148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medalla F, Hoekstra RM, Whichard JM et al. Increase in resistance to ceftriaxone and nonsusceptibility to ciprofloxacin and decrease in multidrug resistance among Salmonella strains, United States, 1996–2009. Foodborne Pathog Dis 2013; 10:302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kariuki S, Okoro C, Kiiru J et al. Ceftriaxone resistant Salmonella Typhimurium sequence type 313 from Kenyan patients is associated with blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob Agents Chemother 2015; 59:3133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris SR, Feil EJ, Holden MT et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 2010; 327:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688–90. [DOI] [PubMed] [Google Scholar]
- 40.Okoro CK, Kingsley RA, Quail MA et al. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin Infect Dis 2012; 54:955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham JP, Boland JJ, Silbergeld E. Growth promoting antibiotics in food animal production: an economic analysis. Public Health Rep 2007; 122:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White DG, Zhao S, Singh R, McDermott PF. Antimicrobial resistance among gram-negative foodborne bacterial pathogens associated with foods of animal origin. Foodborne Pathog Dis 2004; 1:137–52. [DOI] [PubMed] [Google Scholar]
- 43.Gasem MH, Dolmans WM, Keuter MM, Djokomoeljanto RR. Poor food hygiene and housing as risk factors for typhoid fever in Semarang, Indonesia. Trop Med Int Health 2001; 6:484–90. [DOI] [PubMed] [Google Scholar]
- 44.Kariuki S, Dougan G. Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann N Y Acad Sci 2014; 1323:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okoro CK, Kingsley RA, Connor TR et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet 2012; 44:1215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomson NR, Clayton DJ, Windhorst D et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res 2008; 18:1624–37. [DOI] [PMC free article] [PubMed] [Google Scholar]