Abstract
Acanthopanacis cortex has been used in clinical applications for a long time. Considering some historical and geographical factors, Acanthopanacis cortex is easily confused with other herbs in medicine markets, thereby causing potential safety issues. In this study, we used the internal transcribed spacer 2 (ITS2) barcode to identify 69 samples belonging to six species, including Acanthopanacis cortex and its adulterants. The nearest distance, single-nucleotide polymorphisms (SNPs), and neighbor-joining (NJ) tree methods were used to evaluate the identification ability of the ITS2 barcode. According to the kimura-2-parameter model, the intraspecific distance of Eleutherococcus nodiflorus ITS2 sequences ranged from 0 to 0.0132. The minimum interspecific distance between E. nodiflorus and E. giraldii was 0.0221, which was larger than the maximum intraspecific distance of E. nodiflorus. Three stable SNPs in ITS2 can be used to distinguish Acanthopanacis cortex and its closely related species. The NJ tree indicated that the Acanthopanacis cortex samples clustered into one clade, which can be distinguished clearly from the adulterants of this herb. A secondary structure of ITS2 provided another dimensionality to identify species. In conclusion, the ITS2 barcode effectively identifies Acanthopanacis cortex, and DNA barcoding is a convenient tool for medicine market supervision.
Keywords: Acanthopanacis cortex, DNA barcoding, herbal medicine, identification, ITS2
Introduction
Acanthopanacis cortex, a traditional Chinese medicine known as “wujiapi,” is derived from the dried root barks of Eleutherococcus nodiflorus (Dunn) S.Y. Hu (State Pharmacopoeia Committee, 2010). It is commonly used to expel wind-dampness, tonify liver and kidneys, and strengthen muscles and bones, in accordance with Chinese medicine theory (State Pharmacopoeia Committee, 2010). In China, Acanthopanax gracilistylus wine, which is made of Acanthopanacis cortex and other herbs soaked in liquor, is popularly used as a health supplement product to treat rheumatic arthritis because of its clinical effects. To date, Acanthopanax cortex has been reported to exhibit anti-inflammatory and potential antitumor activities, as well as demonstrate treatment effects for bone diseases (Shan et al., 1999, 2000; Zhang et al., 2012). As a candidate therapy, Acanthopanax cortex particularly exerts anti-inflammatory and protective effects on the liver, thereby showing potential treatment for fulminant hepatitis (Zhang et al., 2011). Given the curative effects of Acanthopanax cortex, this herb has been widely used in clinical applications and attracted increasing research attention.
However, Acanthopanacis cortex is easily confused with other herbs in medicine markets, thereby causing potential safety issues (Wu et al., 2014). The original species of Acanthopanacis cortex is E. nodiflorus. The genus Eleutherococcus Maxim. consists of 18 species that are widely distributed throughout China (Flora of China Editorial Committee, 2007). Considering some historical and geographical factors, some closely related species of E. nodiflorus are commonly used as local herbs in various places. For example, E. giraldii (Harms) Nakai, a Tibetan medicine distributed in northwest and southwest China, is mainly used as Acanthopanacis cortex in Sichuan Province (Wang et al., 2005). The root bark from E. sessiliflorus (Rupr. and Maxim.) S.Y. Hu is considered one of the sources of Acanthopanacis cortex in northeast China (Song et al., 2012). The wide application of alternatives may influence the efficacy of clinical medication. Furthermore, Acanthopanacis cortex is frequently adulterated by Periplocae cortex, known as “xiangjiapi,” which is the root cortex of Periploca sepium Bge (Guo et al., 2013; Wu et al., 2014). Both herbs share similar morphological characteristics and name (Figure 1), but the functions and efficacies of these herbs are not quite the same. Periplocae cortex contains toxic components that may cause poisoning and has been implicated in various clinical accidents (Liang et al., 2014). Acanthopanax gracilistylus wine poisoning has been reportedly caused by Periplocae cortex substitution for Acanthopanacis cortex. Therefore, Acanthopanacis cortex should be correctly identified. However, identifying cortex herbs using traditional identification methods is difficult, especially when the cortex is dried and sliced (Sun and Chen, 2013; Huang et al., 2014; Techen et al., 2014). Thus, a good tool is needed to identify Acanthopanacis cortex accurately.
Figure 1.
Samples of Acanthopanacis cortex and Periplocae cortex collected from medicine markets in China.
DNA barcoding technique uses short DNA sequences to identify species (Hebert et al., 2003), which makes it possible to correctly identify Acanthopanacis cortex. DNA barcoding has been widely used to identify herbs in recent years. Xiang et al. (2013) accurately identified 131 Ophiocordyceps sinensis (Berk.) G. H. Sung samples, as well as 12 common counterfeits and its closely related species by using DNA barcoding. A fast SNP identification based on DNA barcoding was used to identify the genus Panax L. with its closely related species, as well as the mixed powder Panax ginseng C. A. Mey. and P. quinquefolius L. (Chen et al., 2013). Xin et al. (2013) used ITS2 barcode to successfully distinguish Lycium barbarum L., L. chinense Mill., and L. ruthenicum Murray and indicated ITS2 barcode as a powerful tool to trace Goji. Li et al. (2012) used DNA barcode and chemical profiling techniques to successfully identify Baiying (Herba Solani Lyrati) commodity and its toxic substitute Xungufeng (Herba Aristolochiae Mollissimae). Newmaster et al. (2013) used DNA barcoding to detect the North American herbal products, and suggested that the herbal industry should embrace DNA barcoding for authenticating herbal products, which would provide consumers with safe, high quality herbal products. These studies indicated that DNA barcoding is a convenient tool to identify medicinal plants, herbal medicine, and even commercial products. To ensure the clinical safety of Acanthopanacis cortex, the ITS2 barcode was sequenced and used in the present study to identify this herb and its adulterants.
Materials and methods
Plant materials
A total of 69 samples belonging to six species, including E. nodiflorus (7 cortex and 9 leaf samples), P. sepium (12 cortex and 3 leaf samples), E. senticosus Maxim. (10 root, 1 fruit, 1 stem, and 3 leaf samples), E. giraldii (5 cortex and 4 leaf samples), E. sessiliflorus (1 stem and 8 leaf samples), and E. trifoliatus (L.) S. Y. Hu (1 leaf sample and 4 ITS2 sequences downloaded from GenBank), were collected from medicine markets, drug stores, botanical gardens, and fields (Supplementary Table 1). Experimental samples bought from medicine markets and drug stores were already dried in silica gel, and collected from fields. The corresponding voucher samples were deposited in the herbarium of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Beijing, China.
Molecular analyses
The surfaces of the samples were first scraped off then wiped with 75% ethanol. Total genomic DNA was extracted from 20 to 40 mg dry powder of the tissues by using the Plant Genomic DNA Kit (Tiangen Biotech Co.). Polymerase chain reaction (PCR) amplification and sequencing were performed as described in previous study (Chen et al., 2010). Raw trace files were trimmed and assembled using CodonCode Aligner 4.2.7 (CodonCode Co., USA). The ITS2 region was annotated using the Hidden Markov model to remove the 5.8S and 28S sections (Keller et al., 2009). All 69 ITS2 sequences were used to calculate genetic distances and construct a neighbor-joining (NJ) tree. Additionally, 54 ITS2 sequences from E. nodiflorus and its closely related species (E. senticosus, E. giraldii, E. sessiliflorus, and E. trifoliatus) were used to detect single-nucleotide polymorphisms (SNPs). Six haplotypes of Acanthopanacis cortex and its adulterants were used to predict the secondary structure of ITS2. Genetic distances were calculated according to the kimura-2-parameter (K2P) model using MEGA 5.2.2 software (Tamura et al., 2011). SNP sites were detected using Sequencher 5.3 software (Gene Codes Co., USA). An NJ tree was constructed, and bootstrap tests were performed using 1000 replicates to identify Acanthopanacis cortex and its adulterants via MEGA 5.2.2 (Tamura et al., 2011). The ITS2 secondary structure was predicted using the ITS2 Workbench (http://its2.bioapps.biozentrum.uni-wuerzburg.de/) (Koetschan et al., 2012).
Results
Analysis of PCR amplification and sequence characteristics
The PCR amplification success rate of the ITS2 region was 100%. All purified PCR products were successfully sequenced, and high-quality bidirectional trace files were generated. All the obtained sequences were submitted to GenBank (Supplementary Table 1).
The PCR product sequence lengths of all 69 samples ranged from 420 to 470 bp. After annotation, the ITS2 sequence of 16 E. nodiflorus samples was 230 bp in length, and the average GC content was 62.4%. Four nucleotide variation sites were found in these sample sequences. Four haplotypes of ITS2 sequences were generated. The ITS2 sequence lengths of P. sepium, E. senticosus, E. giraldii, E. sessiliflorus, and E. trifoliatus, which were adulterants of Acanthopanacis cortex, were 235, 230, 230, 230, and 231 bp, respectively, after alignment; the corresponding average GC content of the above adulterants were 55.7, 62.6, 63.4, 62.2, and 63.9%. Only one and three intraspecies variation sites were found in the ITS2 regions of E. giraldii and E. trifoliatus, respectively. The ITS2 regions of P. sepium, E. senticosus, and E. sessiliflorus were highly conservative, and no intraspecies variation sites existed in these regions.
Analysis of SNPs, inter/intra-specific distance, and NJ tree
SNP analysis allowed the successful detection and distinction of specific genetic variations, which were effectively applied in plant and animal identification (Chen et al., 2013; Engdahl et al., 2014). The said method is particularly used to analyze closely related species (Arif et al., 2010; Chen et al., 2013). A total of 54 ITS2 sequences from E. nodiflorus and its closely related species (E. senticosus, E. giraldii, E. sessiliflorus, and E. trifoliatus) were analyzed to detect the SNPs at an interspecific level. Variable sites in the haplotypes of the five Eleutherococcus species are shown in Supplementary Figure 1. Three stable variation sites at the positions of 23, 36, and 170 bp were found (Table 1), which can be used to distinguish Acanthopanacis cortex and its closely related species. All the published ITS2 sequences of E. nodiflorus and its closely related species were downloaded from GenBank. The three SNPs existed stably in these sequences, which further verified the above results.
Table 1.
Three stable SNPs in ITS2 sequences from Eleutherococcus nodiflorus and its closely related species.
| Species name | SNP location | ||
|---|---|---|---|
| 23 bp | 36 bp | 170 bp | |
| E. nodiflorus | A | C | A |
| E. senticosus | C | T | C |
| E. giraldii | C | T | C |
| E. sessiliflorus | C | T | C |
| E. trifoliatus | C | T | C |
The E. nodiflorus sequence (GenBank No. KR080395) is used as reference sequence for locating SNP sites.
All the 69 ITS2 sequences were analyzed to calculate the genetic distances. According to the K2P model, the intraspecific distance of E. nodiflorus varied from 0 to 0.0132, with an average of 0.0078. The maximum intraspecific distances of E. giraldii and E. trifoliatus were 0.0044 and 0.0131, respectively. The average interspecific distance between Acanthopanacis cortex and its adulterants was 0.0510, whereas the maximum interspecific distance was 0.3818, which was attributed to E. nodiflorus and P. sepium. The minimum interspecific distance of 0.0221 between E. nodiflorus and E. giraldii was larger than the maximum intraspecific distance of E. nodiflorus (Table 2). Overall, the results indicated that the ITS2 barcode can correctly identify Acanthopanacis cortex and its adulterants.
Table 2.
Intraspecific and interspecific genetic distances between Acanthopanacis cortex and its adulterants.
| K2P genetic distances | Range of genetic distances (mean) |
|---|---|
| Intraspecific distances of Eleutherococcus nodiflorus | 0–0.0132 (0.0078) |
| Intraspecific distances of E. giraldii | 0–0.0044 (0.0017) |
| Intraspecific distances of E. trifoliatus | 0–0.0131 (0.0061) |
| Interspecific distances between Acanthopanacis cortex and its adulterants | 0.0221–0.3818 (0.0510) |
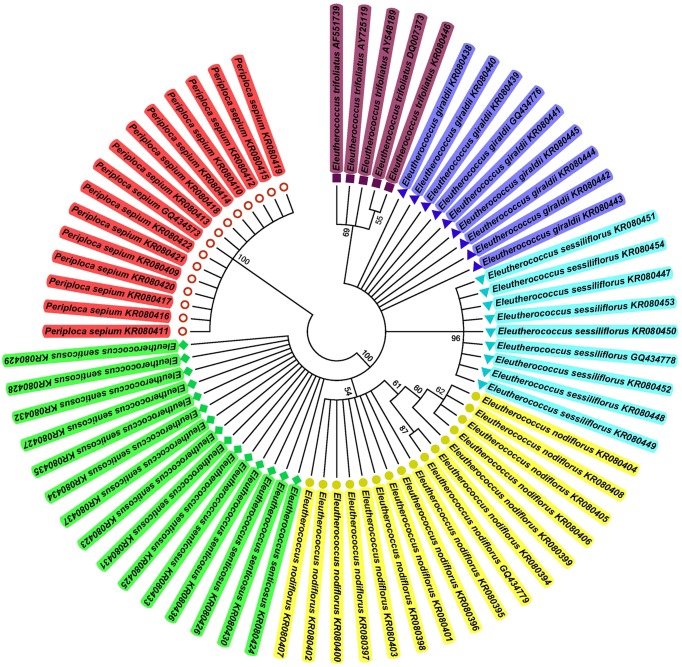
All the 69 ITS2 sequences were used to construct the NJ tree. The findings showed that E. nodiflorus formed into one clade, whereas the adulterants clustered into other clades. Thus, the NJ tree can successfully distinguish Acanthopanacis cortex from its adulterants (Figure 2).
Figure 2.
NJ tree of Acanthopanacis cortex and its adulterants constructed with ITS2 sequences. The bootstrap scores (1000 replicates) are shown (≥50%) for each branch.
Analysis of secondary structure
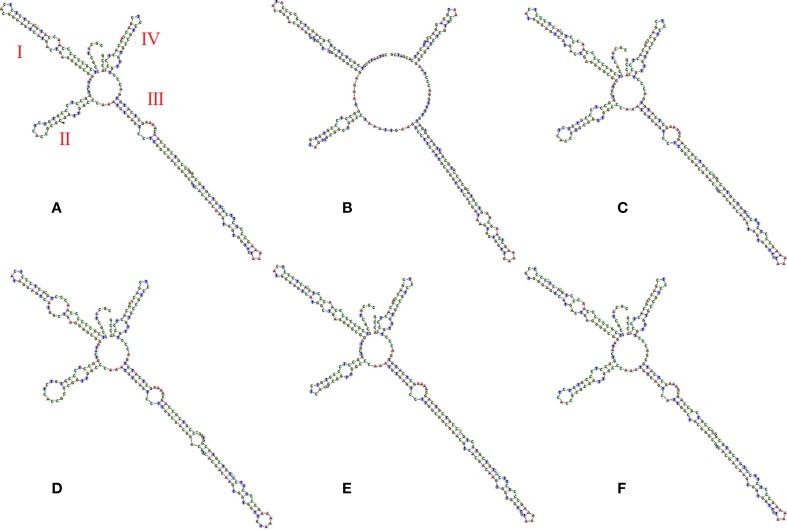
Six haplotypes of Acanthopanacis cortex and its adulterants were used to predict the secondary structure of ITS2. All six ITS2 secondary structures exhibited four similar helices: Helix I, II, III, and IV. The secondary structures between E. nodiflorus and P. sepium showed significant differences on Helix I, II, III, and IV. The secondary structures between E. nodiflorus and its related species were relatively conservative, but the position, size, and number of Loops in Helix I, II, and III showed differences, which can be used as molecular morphological characteristics to identify species (Figure 3). Thus, the ITS2 secondary structure provided an auxiliary method to distinguish Acanthopanacis cortex and its adulterants.
Figure 3.
Secondary structure of ITS2 in Acanthopanacis cortex and its adulterants. (A) Eleutherococcus nodiflorus (GQ434779), (B) Periploca sepium (KR080415), (C) E. senticosus (KR080436), (D) E. giraldii (KR080439), (E) E. sessiliflorus (GQ434778), and (F) E. trifoliatus (KR080446).
Discussion
Why we selected ITS2 barcode to identify acanthopanacis cortex
The establishment of a standard DNA barcode is an important premise of DNA barcoding applications. Chen et al. (2010) analyzed more than 6600 medicinal plants and their closely related species and evaluated the identification ability of the genomic regions psbA-trnH, matK, rbcL, rpoC1, ycf5, internal transcribed spacer (ITS), and ITS2; they determined that the rate of successful identification with ITS2 barcode was 92.7% at the species level and suggested that the ITS2 DNA barcode represents the most suitable region to identify medicinal plants. The identification efficiency of the ITS2 barcode has been validated in several families and genera of medicinal plants (Gao et al., 2010; Pang et al., 2011; Gu et al., 2013). The China Plant BOL Group (2011) confirmed that the ITS/ITS2 regions should be incorporated into the core barcode for seed plants. Furthermore, the stability and accuracy of the ITS2 barcode have been verified in herbal medicine (Chen et al., 2013; Hou et al., 2013; Xiang et al., 2013; Xin et al., 2013). A DNA barcoding system for herbal medicine, which is based on ITS2 as the core DNA barcode, has been established. An online DNA barcoding database has also been constructed (http://www.tcmbarcode.cn) (Chen et al., 2014). The principle for the DNA barcoding of traditional Chinese medicine has been added to Supplement 3 of the Chinese Pharmacopeia (2010 edition). Therefore, the ITS2 barcode provides a suitable tool to identify traditional Chinese medicine and address problems regarding substitutes and adulterants.
ITS2 barcode effectively identifies acanthopanacis cortex
Acanthopanacis cortex is increasingly becoming popular in clinical applications. However, several closely related species of E. nodiflorus, such as E. giraldii, E. senticosus, and E. sessiliflorus, are locally used as Acanthopanacis cortex in several places (Wang et al., 2005; Song et al., 2012). Moreover, Acanthopanacis cortex is seriously mistaken for Periplocae cortex when used in Chinese patent medicines or health supplement products (Guo et al., 2013). Existing toxic adulterants are important factors causing safety issues. Thus, correct identification of Acanthopanacis cortex is absolutely essential to ensure clinical safety. Traditional identification methods cannot easily authenticate naturally dried, sliced, shredded, or simply processed herbal medicine. Morphological authentication approach largely depends on taxonomists and becomes infeasible because of the absence of identifying features. The ITS2 barcode offers the potential to resolve this problem. Liu et al. (2012) indicated that ITS2 can be easily used for PCR amplification and sequencing and is a powerful barcode for Araliaceae identification. Sun and Chen (2013)used ITS2 to identify cortex herbs listed in the Chinese Pharmacopeia; they found that genuine cortex herbs can be discriminated from false ones and also concluded that the ITS2 barcode is suitable to identify cortex herbs in the Chinese Pharmacopeia. The materials in the aforementioned studies comprised leaf tissues and sequences downloaded from GenBank. In the present research, the studied materials mainly included cortexes from medicine markets and drug stores. Adulterants of Acanthopanacis cortex were also analyzed. Genomic DNA from the cortex tissues was successfully acquired. The results of SNPs, nearest distance, and NJ tree methods indicated that the ITS2 barcode can successfully identify Acanthopanacis cortex and its adulterants. Among the adulterants, E. senticosus and E. sessiliflorus were distributed in the same areas and easily confused (Zhu et al., 2011). Our research demonstrated that ITS2 can also distinguish these two species from one another (Figure 2).
DNA barcoding is a convenient tool for medicine market supervision
With the development of DNA barcoding, its application in identifying Chinese medicine has also expanded. In some studies, DNA barcoding was applied to supervise medicine markets. For instance, Xin et al. (2015) constructed a DNA barcode database with 82 voucher samples from 10 Rhodiola species and used this database to identify 100 Rhodiolae Crenulatae Radix et Rhizoma decoction samples from hospitals and drug stores; their results indicated that only 36 samples (40%) were authentic. Wu et al. (2015) established efficient method with DNA barcoding to identify Aristolochiaceous herbs; this method can protect consumers from health risks caused by aristolochic acids, which can lead to aristolochic acid nephropathy. In the current research, DNA barcoding was precisely verified to supervise medicine markets. We detected five adulterants by using the ITS2 barcode from all nine Acanthopanacis cortex samples purchased from drug stores and medicine markets (Supplementary Table 2). One of the five adulterants was derived from E. giraldii, also known as “hongmao-wujiapi,” which was reportedly used as Acanthopanacis cortex in several places (Wang et al., 2005). Previous studies have reported that P. sepium, also known as “Periplocae cortex,” was easily confused with Acanthopanacis cortex (Guo et al., 2013; Wu et al., 2014). In the present study, four out of the five adulterants were derived from P. sepium (Supplementary Table 2). We concluded that Periplocae cortex is a common adulterant of Acanthopanacis cortex in medicine markets and drug stores. Two main reasons can explain why the adulterant Periplocae cortex is common in medicine markets. First, Acanthopanacis cortex and Periplocae cortex are similar in morphological characteristics and difficult to distinguish from each other (Figure 1). Second, Acanthopanacis cortex and Periplocae cortex always use the names “Nanwujiapi” and “Beiwujiapi” in medicine markets, respectively. These two names sound similar and can be easily confused. Hence, we suggest that the ITS2 barcode should be used to monitor Acanthopanacis cortex, and the name “Xiangjiapi” should be used instead of “Beiwujiapi” in medicine markets.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Professor Yulin Lin (the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences) for identification of experimental samples. This research was supported by grants from the Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2014ZX09304307001) and “National Science and Technology Infrastructure Program” (No. 2014FY130400).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00840
Plant materials used in this study.
Authenticity of commercial samples of Acanthopanacis cortex collected from medicine markets and drug stores.
Variable sites in haplotypes of the five Eleutherococcus species.
References
- Arif I. A., Bakir M. A., Khan H. A., Al Farhan A. H., Al Homaidan A. A., Bahkali A. H., et al. (2010). A brief review of molecular techniques to assess plant diversity. Int. J. Mol. Sci. 11, 2079–2096. 10.3390/ijms11052079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L., Pang X. H., Song J. Y., Shi L. C., Yao H., Han J. P., et al. (2014). A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol. Adv. 32, 1237–1244. 10.1016/j.biotechadv.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Chen S. L., Yao H., Han J. P., Liu C., Song J. Y., Shi L. C., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5:e8613. 10.1371/journal.pone.0008613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. C., Liao B. S., Song J. Y., Pang X. H., Han J. P., Chen S. L. (2013). A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene 530, 39–43. 10.1016/j.gene.2013.07.097 [DOI] [PubMed] [Google Scholar]
- China Plant BOL Group Li, D. Z., Gao L. M., Li H. T., Wang H., Ge X. J., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora of China Editorial Committee (2007). Flora of China. Beijing; St. Louis: Science Press/Missouri Botanical Garden Press. [Google Scholar]
- Engdahl C., Larsson P., Näslund J., Bravo M., Evander M., Lundström J. O., et al. (2014). Identification of Swedish mosquitoes based on molecular barcoding of the COI gene and SNP analysis. Mol. Ecol. Resour. 14, 478–488. 10.1111/1755-0998.12202 [DOI] [PubMed] [Google Scholar]
- Gao T., Yao H., Song J. Y., Zhu Y. J., Liu C., Chen S. L. (2010). Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol. Biol. 10:324. 10.1186/1471-2148-10-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Song J. Y., Cao Y., Sun Q. W., Yao H., Wu Q. N., et al. (2013). Application of the ITS2 region for barcoding medicinal plants of Selaginellaceae in Pteridophyta. PLoS ONE 8:e67818. 10.1371/journal.pone.0067818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Mao H. P., Pan G. X., Zhang H., Fan G. W., Li W. W., et al. (2013). Antagonism of Cortex Periplocae extract-induced catecholamines secretion by Panax notoginseng saponins in cultured bovine adrenal medullary cells by drug combinations. J. Ethnopharmacol. 147, 447–455. 10.1016/j.jep.2013.03.036 [DOI] [PubMed] [Google Scholar]
- Hebert P. D., Ratnasingham S., Dewaard J. R. (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 270 (Suppl. 1), S96–S99. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou D. Y., Song J. Y., Shi L. C., Ma X. C., Xin T. Y., Han J. P., et al. (2013). Stability and accuracy assessment of identification of traditional Chinese materia medica using DNA barcoding: a case study on Flos Lonicerae Japonicae. Biomed Res. Int. 2013:549037. 10.1155/2013/549037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Liang Z., Chen H., Zhao Z., Li P. (2014). Identification of Chinese herbal medicines by fluorescence microscopy: fluorescent characteristics of medicinal bark. J. Microsc. 256, 6–22. 10.1111/jmi.12153 [DOI] [PubMed] [Google Scholar]
- Keller A., Schleicher T., Schultz J., Muller T., Dandekar T., Wolf M. (2009). 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430, 50–57. 10.1016/j.gene.2008.10.012 [DOI] [PubMed] [Google Scholar]
- Koetschan C., Hackl T., Müller T., Wolf M., Förster F., Schultz J. (2012). ITS2 database IV: interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Mol. Phylogenet. Evol. 63, 585–588. 10.1016/j.ympev.2012.01.026 [DOI] [PubMed] [Google Scholar]
- Li M., Au K. Y., Lam H., Cheng L., Jiang R. W., But P. P. H., et al. (2012). Identification of Baiying (Herba Solani Lyrati) commodity and its toxic substitute Xungufeng (Herba Aristolochiae Mollissimae) using DNA barcoding and chemical profiling techniques. Food Chem. 135, 1653–1658. 10.1016/j.foodchem.2012.06.049 [DOI] [PubMed] [Google Scholar]
- Liang S., Deng F. C., Xing H. Y., Wen H., Shi X. Y., Martey O. N., et al. (2014). P-glycoprotein- and organic anion-transporting polypeptide-mediated transport of periplocin may lead to drug-herb/drug-drug interactions. Drug Des. Devel. Ther. 8, 475–483. 10.2147/DDDT.S61024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. H., Zeng X., Yang D., Chu G. Y., Yuan Z. R., Chen S. L. (2012). Applying DNA barcodes for identification of plant species in the family Araliaceae. Gene 499, 76–80. 10.1016/j.gene.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Newmaster S. G., Grguric M., Shanmughanandhan D., Ramalingam S., Ragupathy S. (2013). DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 11, 222. 10.1186/1741-7015-11-222 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pang X. H., Song J. Y., Zhu Y. J., Xu H. X., Huang L. F., Chen S. L. (2011). Applying plant DNA barcodes for Rosaceae species identification. Cladistics 27, 165–170. 10.1111/j.1096-0031.2010.00328.x [DOI] [PubMed] [Google Scholar]
- State Pharmacopoeia Committee (2010). Chinese Pharmacopoeia. Beijing: Medical Science and Technology Press. [Google Scholar]
- Shan B. E., Yoshita Y., Sugiura T., Yamashita U. (1999). Suppressive effect of Chinese medicinal herb, Acanthopanax gracilistylus, extract on human lymphocytes in vitro. Clin. Exp. Immunol. 118, 41–48. 10.1046/j.1365-2249.1999.01031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B. E., Zeki K., Sugiura T., Yoshida Y., Yamashita U. (2000). Chinese medicinal herb, Acanthopanax gracilistylus, extract induces cell cycle arrest of human tumor cells in vitro. Jpn. J. Cancer Res. 91, 383–389. 10.1111/j.1349-7006.2000.tb00956.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Deng Y., Huang D. X., Wen J., Liu Z. G., Li F. M. (2012). LC-MS/MS determination and pharmacokinetic study of four lignan components in rat plasma after oral administration of Acanthopanax sessiliflorus extract. J. Ethnopharmacol. 141, 957–963. 10.1016/j.jep.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Sun Z. Y., Chen S. L. (2013). Identification of cortex herbs using the DNA barcode nrITS2. J. Nat. Med. 67, 296–302. 10.1007/s11418-012-0681-8 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techen N., Parveen I., Pan Z., Khan I. A. (2014). DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotech. 25, 103–110. 10.1016/j.copbio.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Wang Z. W., Zhang L. X., Sun Y. Q. (2005). Semipreparative separation and determination of eleutheroside E in Acanthopanax giraldii Harms by high-performance liquid chromatography. J. Chromatogr. Sci. 43, 249–252. 10.1093/chromsci/43.5.249 [DOI] [PubMed] [Google Scholar]
- Wu L., Sun W., Wang B., Zhao H. Y., Li Y. L., Cai S. Q., et al. (2015). An integrated system for identifying the hidden assassins intraditional medicines containing aristolochic acids. Sci. Rep. 5:11318. 10.1038/srep11318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. Y., Zhang Y. B., Zhu K. K., Luo C., Zhang J. X., Cheng C. R., et al. (2014). Anti-inflammatory diterpenoids from the root bark of Acanthopanax gracilistylus. J. Nat. Prod. 77, 2342–2351. 10.1021/np500125x [DOI] [PubMed] [Google Scholar]
- Xiang L., Song J. Y., Xin T. Y., Zhu Y. J., Shi L. C., Xu X. L., et al. (2013). DNA barcoding the commercial Chinese caterpillar fungus. FEMS Microbiol. Lett. 347, 156–162. 10.1111/1574-6968.12233 [DOI] [PubMed] [Google Scholar]
- Xin T. Y., Li X. J., Yao H., Lin Y. L., Ma X. C., Cheng R. Y., et al. (2015). Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci. Rep. 5:8337. 10.1038/srep08337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin T. Y., Yao H., Gao H. H., Zhou X. Z., Ma X. C., Xu C. Q., et al. (2013). Super food Lycium barbarum (Solanaceae) traceability via an internal transcribed spacer 2 barcode. Food Res. Int. 54, 1699–1704. 10.1016/j.foodres.2013.10.007 [DOI] [Google Scholar]
- Zhang B. X., Li N., Zhang Z. P., Liu H. B., Zhou R. R., Zhong B. Y., et al. (2011). Protective effect of Acanthopanax gracilistylus-extracted Acankoreanogenin A on mice with fulminant hepatitis. Int. Immunopharmacol. 11, 1018–1023. 10.1016/j.intimp.2011.02.019 [DOI] [PubMed] [Google Scholar]
- Zhang Z. G., Dong J. Z., Liu M. J., Li Y., Pan J. H., Liu H., et al. (2012). Therapeutic effects of Cortex acanthopanacis aqueous extract on bone metabolism of ovariectomized rats. Evid Based Compl. Alt. 2012:492627. 10.1155/2012/492627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Bai Y. J., Oya M., Tanaka K., Komatsu K., Maruyama T., et al. (2011). Genetic and chemical diversity of Eleutherococcus senticosus and molecular identification of Siberian ginseng by PCR-RFLP analysis based on chloroplast trnK intron sequence. Food Chem. 129, 1844–1850. 10.1016/j.foodchem.2011.05.128 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plant materials used in this study.
Authenticity of commercial samples of Acanthopanacis cortex collected from medicine markets and drug stores.
Variable sites in haplotypes of the five Eleutherococcus species.