Abstract
Despite its efficacy, the use of morphine for the treatment of chronic pain remains limited because of the rapid development of tolerance, dependence and ultimately addiction. These undesired effects are thought to be because of alterations in synaptic transmission and neuroplasticity within the reward circuitry including the striatum. In this study we used subcellular fractionation and quantitative proteomics combined with computational approaches to investigate the morphine-induced protein profile changes at the striatal postsynaptic density. Over 2,600 proteins were identified by mass spectrometry analysis of subcellular fractions enriched in postsynaptic density associated proteins from saline or morphine-treated striata. Among these, the levels of 34 proteins were differentially altered in response to morphine. These include proteins involved in G-protein coupled receptor signaling, regulation of transcription and translation, chaperones, and protein degradation pathways. The altered expression levels of several of these proteins was validated by Western blotting analysis. Using Genes2Fans software suite we connected the differentially expressed proteins with proteins identified within the known background protein-protein interaction network. This led to the generation of a network consisting of 116 proteins with 40 significant intermediates. To validate this, we confirmed the presence of three proteins predicted to be significant intermediates: caspase-3, receptor-interacting serine/threonine protein kinase 3 and NEDD4 (an E3-ubiquitin ligase identified as a neural precursor cell expressed developmentally down-regulated protein 4). Because this morphine-regulated network predicted alterations in proteasomal degradation, we examined the global ubiquitination state of postsynaptic density proteins and found it to be substantially altered. Together, these findings suggest a role for protein degradation and for the ubiquitin/proteasomal system in the etiology of opiate dependence and addiction.
Morphine and other opiates are the drugs of choice for the treatment of both severe and chronic pain. However, the utility of these compounds in the clinical setting is limited because of the rapid development of tolerance, physical dependence and addiction. The underlying cellular and molecular alterations through which chronic opiate exposure results in the persistent behavioral phenomenon of addiction remain poorly understood. However, there is evidence suggesting that the molecular composition of synapses within the reward circuitry of the central nervous system, particularly in the striatum, may be significantly altered (1). Moreover, given the critical involvement of the striatum in translating emotional or rewarding stimuli into motivated behaviors, alterations in this region induced by morphine and other abused drugs could contribute significantly to the pathophysiological responses at the heart of addiction (2). Although the debate surrounding the molecular and cellular mechanisms by which repeated drug administration leads to addiction is still continuing, there is substantial evidence for the involvement of synaptic plasticity within the striatum in this process.
Studies show that morphine administration both reduces the complexity of dendritic branching and spine density of striatal medium spiny neurons (3, 4), and causes structural modifications that are known to lead to alterations in synaptic efficacy (1, 4, 5). In addition to these morphological alterations, morphine also regulates the expression of transcription factors that are involved in mechanisms of learning and synaptic plasticity such as CREB (6), ΔFosB (6, 7), and NF-κ-B (8). Prolonged exposure to morphine and other opiates also results in a number of important molecular and electrophysiological changes at synaptic terminals. Thus morphine exposure reduces transmembrane Ca2+ conductance and neurotransmitter release (9, 10), causes alterations in glutamatergic transmission (11–13), enhances postsynaptic K+ conductance and membrane hyperpolarization (14), and leads to significant up-regulation of μ-δ opiate receptor heteromers in the striatum and other brain regions (15–17). Given the wide-reaching changes induced by chronic morphine administration it is important to develop an understanding of how morphine and other abused drugs generate these neuroplastic changes particularly at the synapse.
Proteomic analysis has been used to elucidate both the in vitro (18, 19) and in vivo (20–26) changes in the protein profiles of synapses that exhibit drug-induced alterations in response to opiate exposure. For example, the use of subcellular fractionation, which allows for the separation and generation of protein fractions selectively enriched in either pre- or postsynaptic proteins (25, 27), in combination with proteomic analysis has revealed that an acute escalating dose of morphine administration produces significant alterations in the postsynaptic density (PSD)1 associated protein network in the hippocampus. This includes significant redistribution of endocytic proteins such as clathrin (20, 22) and glutamatergic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (12, 13). Proteomic analysis coupled with tandem mass spectrometry (MS/MS) followed by the computational analysis that placed the proteomics results within known protein–protein interaction (PPI) networks also revealed a novel function for heat shock proteins and their co-chaperones in the presynaptic active zone of the morphine tolerant striatum. This, in turn, led to the identification of a therapeutic target that, when inhibited, prevented the development of tolerance and dependence while preserving the analgesic properties of morphine (24). This illustrates the power of proteomic analysis in combination with tandem mass spectrometry and data analysis that integrates known PPI networks in detecting and predicting protein changes in a specific subcellular fraction.
In this study we used a quantitative subcellular proteomic approach in order to explore morphine-regulated changes in the PSD fraction of the morphine dependent striatum. For this the striata of animals subjected to chronic escalating doses of morphine (28), were subjected to subcellular fractionation (27) so as to obtain fractions enriched in PSD proteins. In order to reveal relative changes in the abundance of PSD-related proteins, PSD fractions from individual morphine and saline treated animals were labeled with the 8-plex isobaric tags for relative and absolute quantitation (iTRAQ), and then subjected to MS/MS analysis. These experiments identified a total of 2648 proteins, of which 2643 were quantified. Among these proteins, 34 proteins (∼1.4%) exhibited statistically significant regulation in response to chronic morphine treatment and were named the “High Confidence Morphine Regulated Proteins”; 10 of these proteins were significantly down-regulated, whereas the other 24 proteins were significantly up-regulated relative to saline controls. In order to place these differentially expressed proteins within the known biology of the synapse, we used the web-based tool Enrichr (29) for performing protein set enrichment analysis, and Genes2FANs (30) to construct a morphine-regulated subnetwork. Among the proteins identified to be enriched for interactions with the 34 proteins we identified as differentially expressed at the synapse, we selected caspase-3, receptor-interacting serine/threonine protein kinase 3, and NEDD4 for experimental validation by Western blotting. We also found that the global ubiquitination state of the striatal PSD proteins is altered by chronic morphine administration suggesting a broad role for ubiquitination and protein degradation in the development of tolerance to morphine.
EXPERIMENTAL PROCEDURES
Materials
Rabbit-anti-PSD-95 (Catalog# 2507), rabbit-anti-synaptophysin (Catalog# 5467S), rabbit-anti-Gαo (Catalog# 3975S), rabbit-anti-HSP70 (Catalog# 4872), rabbit-anti-GAP43 (D9C8) (Catalog# 8945), rabbit-anti-USP8 (Catalog# 8728), mouse-anti-Ubiquitin (P4D1) (Catalog# 3936), mouse-anti-Caspase-3 (3G2) (Catalog# 9668), rabbit-anti-NEDD4 (C5F5) (Catalog# 3607) were from Cell Signaling Technology, Danvers, MA. Rabbit-anti-Gβ1 (Catalog# NBP1–55307), and rabbit-anti-Annexin 6 (Catalog# NBP1–80514) were from Novus Biologicals, Littleton, Colorado. Rabbit-anti-PPP3R1/Calcineurin B (Catalog# AP09004PU-N) was from Acris Antibodies GmbH, San Diego, CA. Rabbit-anti-RIPK3 (Catalog# 20R-1514) was from Fitzgerald Industries International, Acton, MA.
For extraction of proteins and iTRAQ labeling the following materials and reagents were used: Tris-(2-carboxyethyl) phosphine (TCEP), methyl-methanethiosulfate (MMTS), mobile Phase A (10 mm KH2PO4 with 25% acetonitrile (ACN), pH 3.0), mobile Phase B (500 mm KCl, 10 mm KH2PO4 and 25% ACN, pH 3.0), solvent A (5% ACN with 0.1% trifluoroacetic acid (TFA)), solvent B (95% ACN with 0.1% TFA), MALDI matrix solution (7 mg/mK alpha-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, St. Louis, MO) in 60% ACN, 5 mm ammonium monobasic phosphate and 50 fmol/μl of each of the following internal peptidergic calibrants [Glu-1]-Fibrinopeptide B (Glu-Fib, Sigma-Aldrich) and adrenocorticotropic hormone, fragment 18–39 (ACTH (18–39), Sigma-Aldrich).
Research Animals
Adult morphine-naïve male Sprague-Dawley rats (n = 4 for control and experimental groups respectively) were maintained on a 12 h light/dark cycle with access to food and water ad libitum. All animals were permitted to acclimatize to their environment for approximately 1 week prior to treatment. All experiments were designed and performed in accordance with the recommendations set forth in the Guide for the Care and Use of Laboratory Animals: Eighth Edition (31), and were approved and monitored by the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai (Protocol #LA11–00322).
Morphine Treatment
Morphine and saline control injections were administered essentially as described previously (2, 24, 28, 32). Morphine sulfate (Sigma-Aldrich) was prepared in 0.9% sterile isotonic solution of saline. All injections were administered intraperitoneally (intraperitoneal), and consisted of 0.9% saline (control group) or escalating doses of morphine (experimental group). For morphine treatment, a chronic escalating morphine administration paradigm (2, 24, 28, 32) was used. Briefly, animals received every 12 h escalating doses of morphine ranging from 5 mg/kg on day 1 to 50 mg/kg on the final day (2, 24, 28, 32). All animals were sacrificed 2 h after the final injection, the striata were rapidly dissected on ice and stored at −80 °C until used for subcellular fractionation as described below.
Subcellular Fractionation and Isolation of Postsynaptic Proteins
The striatum from each control and experimental animal was hemisected, and each half used to generate two fractions: a primary sample for use in experiments, and a backup sample to be used in the event of contamination or loss of the primary sample. Each striatal half was subjected to cell fractionation to obtain PSD fractions as described previously (22, 27, 33) (Fig. 1A). The pellet containing the PSD fraction was re-suspended in 200–250 μl of 1% SDS, the amount of protein determined using the BCA protein estimation kit (Thermo Scientific Pierce, Rockford, IL), and stored at −80 °C until use.
Fig. 1.
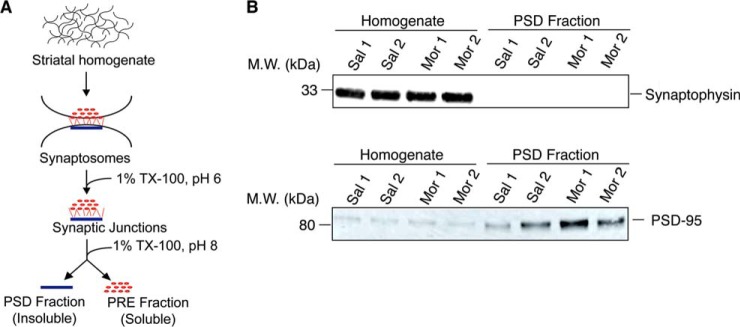
Subcellular fractionation and validation of fractions. A, A schematic of the subcellular fractionation protocol used to generate PSD fractions from the striata of animals treated with either saline or escalating doses of morphine for 5 days (adapted from (27)). TX-100, Triton X-100; PSD, postsynaptic density; PRE, presynaptic density. B. Biochemical validation of the PSD fractions was carried out by Western blot analysis using antibodies to synaptophysin, a presynaptic marker, and to PSD-95, a postsynaptic marker and equal amounts (15 μg) of protein from striatal homogenates and PSD fractions as described under “Experimental Procedures.” A signal for PSD-95 but not for synaptophysin is seen in the PSD fractions.
Immunoblotting
Homogenate or PSD fractions (15 μg protein) from morphine or saline treated animals were resolved by 7.5% SDS-PAGE, and transferred to nitrocellulose membranes (Schleicher & Schuell Bioscience, Keene, NH). Membranes were incubated with primary antibodies for 24 h on an orbital shaker at 4 °C at dilutions recommended by the manufacturer. Following four washes (15 min each) at room temperature with 50 mm Tris-Cl pH 7.4 containing 150 mm NaCl, 1 mm CaCl2 and 0.1% Tween 20 (TBS-T) membranes were incubated in 1:10,000 dilution of either IR800CW- or IR680-labeled goat anti-mouse or anti-rabbit secondary antibodies (LI-COR Biosciences, Lincoln, NE). Membranes were washed four times (15 min each) at room temperature with TBS-T and protein bands were visualized and densitized using the Odyssey infrared imaging system (LI-COR Biosciences).
Protein Extraction and iTRAQ Labeling
PSD fractions were subjected to iTRAQ as described previously (20, 34, 35). Briefly, 100 μg of protein from each saline and morphine treated sample was subjected to SDS-PAGE. The gel was then fixed, stained and rinsed with 25 mm triethylammonium bicarbonate buffer (TEAB) to remove SDS and Tris. The proteins were reduced in the presence of Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), followed by alkylation with methylmethanethiosulfonate (MMTS) (ABSciex, Foster City, CA), and in-gel trypsin digestion (Promega, Madison, WI) overnight at 37 °C. The peptides were extracted with 25 mm TEAB followed by 80% acetonitrile. The peptides were concentrated using a speed-vac and their pH adjusted to 8.5 with 500 mm TEAB. The iTRAQ labeling was performed according to the manufacturer's protocol using the following isobaric iTRAQ tags: 113, 114, 115, and 116 tags for saline control samples, and 117, 118, 119, and 121 tags for morphine treated samples (ABSciex). After incubation with the iTRAQ tags at room temperature for 2 h, the iTRAQ labeled peptides from saline and morphine treated samples were combined and subjected to strong cation exchange liquid chromatography (SCXLC): a polysulfoethyl A strong cation exchange column (4.6 mm × 200 mm, 5 μm, 300Å, Poly LC Inc., Columbia, MD) on a BioCAD SPRINT Perfusion chromatography system (PerSeptive Biosystems Inc., Framingham, MA) that was coupled to an upstream guard column (4.0 mm x 10 mm, Poly LC Inc.) was used. Separation of the different iTRAQ labeled striatal PSD peptides was achieved across a gradient of mobile Phase A (10 mm KH2PO4 in 25% acetonitrile, pH 3) and mobile Phase B (0.6 m KCl and 10 mm KH2PO4 in 25% acetonitrile, pH 3) containing two linear segments: 40 min from 0–50% B, followed by 10 min from 50–100% B at a flow rate of 1 ml/min. Twenty eluted fractions were collected, dried completely with a speed-vac, and desalted using PepClean™ C18 spin columns (Pierce, Rockford, IL).
Tandem Mass Spectrometric (MS/MS) Analysis
Peptides from SCXLC fractions were further analyzed by RPLC-MS/MS on Obitrap Velos mass spectrometer. Briefly, peptides were loaded onto a reversed phase trapping column (0.3 mm × 5.0 mm), and subsequently resolved on a capillary C18 PepMap column (0.1 mm × 150 mm, 3 μm, 100 Å, Dionex). Peptides were eluted using a 70 min gradient of solvent A and solvent B as follows: 5% to 8% B from 0–4 min, 18% B at 34 min, 35% B at 57 min, and 95% B at 64 min. The eluted peptides were introduced into a nano electrospray source on Obitrap Velos MS system with a spray voltage of 2 kV, a capillary temperature of 275 °C and a S-lens voltage of 60%. MS spectra were acquired in a positive ion mode with a resolution of 60,000 full-width at half maximum (FWHM). The higher energy collision dissociation (HCD) MS/MS spectra were acquired in a data-dependent manner. The 10 most abundant ions were selected for HDC fragmentation per MS scan in the Orbitrap at a resolution of 7500 FWHM. The normalized collision energy was set to 40. The lock mass feature was engaged for accurate mass measurements.
Identification and Quantification of Proteins
The MS/MS spectra were searched against UniRef 100 rat database (51,862 entries) using both Mascot (v.2.3) and Sequest search engines via the Proteome Discoverer platform (v 1.3; Thermo Scientific). In conducting this search, the following search parameters were used: (1) fixed modifications included iTRAQ 8plex (K), iTRAQ 8plex (N-terminal), and methylthio (C); (2) variable modifications included iTRAQ 8plex (Y) and oxidation (M); (3) trypsin was selected as the digestive enzyme; (4) a maximum of one missed cleavage site was allowed; (5) the peptide precursor mass tolerance was 10 ppm; and (vi) MS/MS mass tolerance was 0.1 Da. Scaffold (v.3.6.3, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identification. Peptide identifications were accepted if they could be established at ≥ 90% probability by Peptide Prophet (36). A false discovery rate (FDR) was maintained at < 1%. Similarly, protein identifications were accepted if they could be established at ≥ 95% probability and contained at least 1 uniquely identified peptide. Protein probabilities were assigned by the Protein Prophet Algorithm (37). Protein FDR was maintained at < 1%. Homologous protein redundancy was reduced by Scaffold software (V. 3.0) in a minimum. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Quantitative values obtained for each identified protein were determined based only on unique peptides that were detected and assigned to each respective protein. Quantitative ratios for a given protein from each of the saline or morphine treated samples (supplemental Table S1) were calculated as the average of all unique peptide ratios that were associated with a given protein. All quantitative ratios were Log2 normalized for the final quantitative analysis. Proteins with a t test value of p < 0.05 and with a morphine to saline ratio greater than 1.2-fold or less than 0.8-fold were considered as significantly changed.
Enrichment Analysis of Proteomic Data and Generation of Protein-Protein Interaction Networks
The lists of significantly altered proteins were first subjected to gene set enrichment analysis using the tool Enrichr (29). Enrichr uses the Fisher exact test to compute enrichment. In order to generate a PPI network based on proteins identified by proteomics as being modified by morphine treatment we generated a “Morphine Seed List” (53 morphine regulated and altered proteins; 38 up-regulated and 15 down-regulated proteins). This list was created by combining a “High Confidence Morphine Regulated Proteins” data set (34 significantly altered proteins (p < 0.05); 24 up-regulated and 10 down-regulated proteins) (Table I and II) with a “Low Confidence Morphine Regulated Proteins” data set (19 morphine altered proteins (p < 0.1); 14 up-regulated and 5 down-regulated proteins) (supplemental Table S2). The proteins in the Morphine Seed List were connected using the PPI module of Genes2FANs, a web-based software tool (30, 38), that integrates 13 mammalian binary interaction network data sets including HPRD, IntAct, KEGG, MINT, and BioGrid (38). The integration of these data sets results in a background PPI network that contains 11,053 proteins connected through 44,985 direct PPIs. To increase the reliability of the interactions, we only retained interactions arising from sources that contributed five or more PPIs. The proteins from the Morphine Seed List were then connected to each other using the shortest path algorithm with a maximum path length of three. Once the construction of the PPI subnetwork using Genes2 FANs (30) was complete the finalized network was then visualized using Cytoscape v2.8.3 (39).
Table I. Seed list of 25 striatal PSD proteins up-regulated by morphine treatment. Striatal PSD proteins from saline and morphine treated animals (n = 4/group) were subjected to proteomics analysis and quantified as described in Experimental Procedures. Acc. #, accession number; M/S, morphine/saline.
| Full name | Acc. # | Gene I.D. | M.W. (kDa) | Ratio (M/S) | p value |
|---|---|---|---|---|---|
| Similar to cytochrome B-C1 complex, subunit 9 | B2RYX1 | UQCR10 | 7 | 1.4 | 0.001 |
| Claudin-11 | Q99P82 | CLDN11 | 22 | 1.3 | 0.05 |
| Guanine nucleotide-binding protein Gαo | F1LN36 | GNAO1 | 21 | 1.3 | 0.001 |
| 40S ribosomal protein S26 | D3Z8D7 | RPS26 | 13 | 1.2 | 0.001 |
| G-protein β2 subunit (Fragment) | Q91XL4 | GNB2 | 24 | 1.2 | 0.001 |
| 40S ribosomal protein S24 | D4ACJ1 | RPS24 | 15 | 1.2 | 0.01 |
| CD81 antigen | Q62745 | CD81 | 26 | 1.2 | 0.01 |
| Guanine nucleotide-binding protein β1 (Fragment) | Q45QL8 | GNB1 | 28 | 1.2 | 0.01 |
| Heat shock 70 kDa protein 1A/1B | Q07439 | HSPA1A/B | 70 | 1.2 | 0.01 |
| Ras-related protein Rap-1b | Q62636 | RAP1B | 21 | 1.2 | 0.01 |
| Similar to isoform 2 of protein XRP2 | D3ZTJ0 | XRP2 | 43 | 1.2 | 0.02 |
| Similar to leucine zipper protein 1 | D3ZWV9 | LUZP1 | 119 | 1.2 | 0.02 |
| Ubiquitin-like modifier-activating enzyme ATG7 | D3ZP91 | ATG7 | 77 | 1.2 | 0.02 |
| Glutathione S-transferase Mu 5 | Q9Z1B2 | GSTM5 | 27 | 1.2 | 0.03 |
| Protein kinase, AMP-activated, β2 non-catalytic subunit | G3V9X3 | PRKAB2 | 30 | 1.2 | 0.03 |
| Thioredoxin | P11232 | TXN | 12 | 1.2 | 0.03 |
| TNF receptor-associated factor 3 | D3Z9G0 | TRAF3 | 64 | 1.2 | 0.03 |
| 60S ribosomal protein L18a (Fragment) | F1M0K6 | RPL18A | 21 | 1.2 | 0.04 |
| 60S ribosomal protein L21 | D3ZPN7 | RPL21 | 19 | 1.2 | 0.04 |
| Calcineurin subunit B, Type 1 | F1M522 | PPP3R1 | 18 | 1.2 | 0.04 |
| WD repeat-containing protein 41 | B2RYI7 | WDR41 | 51 | 1.2 | 0.04 |
| l-asparaginase | Q8VI04 | ASRGL1 | 34 | 1.2 | 0.05 |
| Neuromodulin | P07936 | GAP43 | 24 | 1.2 | 0.05 |
| Similar to annexin-6 (Mus musculus) | D4ABR6 | ANXA6 | 75 | 1.2 | 0.05 |
The Genes2FANs software prioritizes intermediate proteins that connect the seed list of input proteins using the Binomial proportion test. Predicted intermediates with a Z-score > 3 were deemed “highly significant intermediates,” between 2 and 3 “significant intermediates,” and < 2 “intermediates.” The presence of potential clusters within the generated PPI network was assessed with Cfinder (40). Cfinder utilizes the clique percolation method to localize k-clique percolation clusters within the network.
RESULTS
The present study seeks to elucidate alterations in striatal PSD protein expression in response to prolonged exposure to morphine using a combination of proteomics and network biology. For this animals were treated with either saline or escalating doses of morphine for 5 days (2, 24, 28, 32). The striatum of individual animals was rapidly dissected out and subjected to subcellular fractionation (Fig. 1A) in order to isolate the selectively enriched PSD associated protein fraction (22, 25, 27, 33). The enrichment of PSD associated proteins in this fraction was verified by Western blot analysis using antibodies directed against well-characterized presynaptic (synaptophysin) and postsynaptic (PSD-95) markers. In the case of synaptophysin we detect a strong signal in homogenate fractions from both saline and morphine treated animals but no detectable signal was observed in the PSD fractions of these animals (Fig. 1B). In the case of PSD-95, the intensity of the signal was much stronger in the PSD fractions compared with the homogenate in both saline and morphine treated animals (Fig. 1B). These results indicate that our striatal PSD preparations are enriched in PSD-associated proteins.
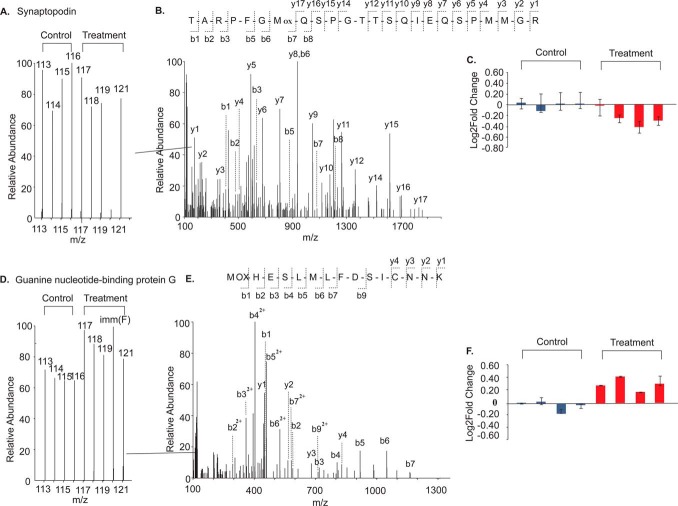
Next, we identified changes in the protein profile of the striatal PSD fractions following morphine treatment. This was achieved by labeling peptide fragments from four saline and four morphine treated animals with eight-plex iTRAQ reagents, mixing the labeled peptides and subjecting the peptide mixture to LC-MS/MS analysis. This led to the identification of 2648 unique proteins in the striatal PSD fractions (supplemental Table S1), of which 2643 were reliably quantified (see “Experimental Procedures”). Representative spectra illustrating the identification and quantification of two distinct proteins detected in this study (synaptopodin which was decreased by morphine treatment and Gαo which was increased) are shown in Fig. 2.
Fig. 2.
Representative Fig. Showing Differential Isotopic Labeling and LC-MS/MS. Representative data for a down-regulated protein, synaptopodin (A) and an up-regulated protein, Gαo is shown (D). (Left panel, A and E) Graph of the normalized intensities of the iTRAQ reporter ions for a peptide fragment; (Middle panel, B and E) the continuous series of the b- and y-ions used for the identification of the peptide fragment; (Right panel, C and F) bar graph showing the ratio as Mean ± S.D. of the iTRAQ labels in relation to iTRAQ-113 signals obtained from all of the peptides derived from the protein.
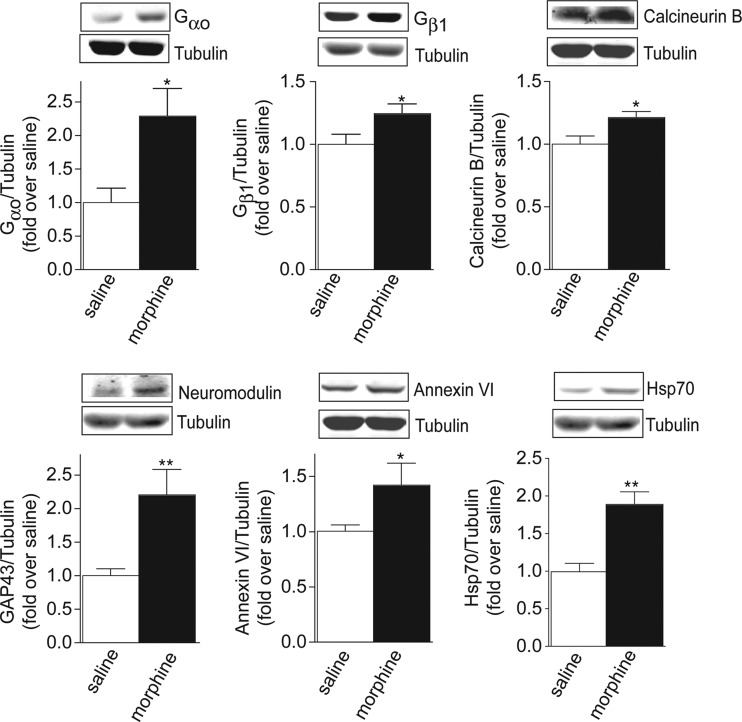
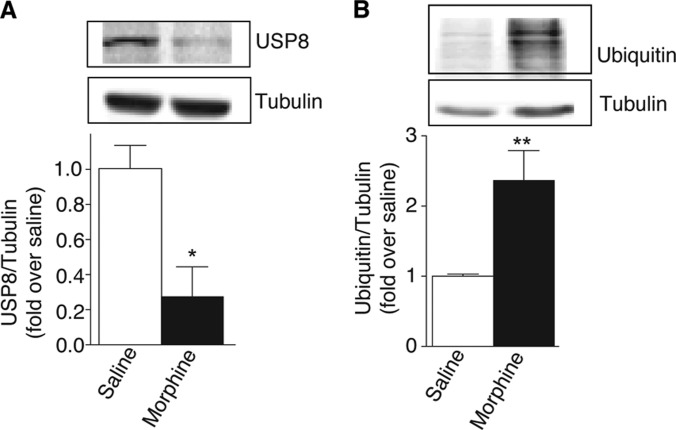
Among the proteins identified by LC-MS/MS analysis, a total of 34 (∼1.4%) exhibited greater than a 20% change (p < 0.05; t test) in the samples from morphine treated animals relative to the saline controls. These proteins, of which 24 were significantly up-regulated and 10 were significantly down-regulated in response to morphine treatment, were labeled as High Probability Morphine Regulated Proteins (Table I and II). Interestingly, among the identified High Probability Morphine Regulated Proteins were a number of signaling molecules, calcium-binding proteins, as well as proteins from the ubiquitin-proteosomal system (UPS). Next, we confirmed by Western blot analysis the changes in the expression levels of several proteins suggested by MS/MS analysis to be significantly altered following morphine treatment. These included up-regulated proteins such as guanine-nucleotide binding protein Gαo, guanine-nucleotide binding protein Gβ1, calcineurin subunit B Type 1, neuromodulin, annexin-VI, and heat shock protein-70 (HSP-70) (Fig. 3) as well as down-regulated proteins such as ubiquitin carboxyl-terminal hydrolase 8 (USP8) (Fig. 4A). We found that the decreased abundance of USP8 in the striatial PSD fractions of morphine treated animals was accompanied by a significant increase in total levels of ubiquitinated proteins (Fig. 4B).
Fig. 3.
Biochemical validation of proteins shown to be up-regulated by quantitative proteomics. PSD fractions (15 μg protein) from morphine or saline treated animals were subjected to Western blot analysis using antibodies to either Gαo, Gβ1, calcineurin B, neuromodulin (GAP43), annexin VI or Hsp70 as described under “Experimental Procedures.” Representative blot is shown in the figure. Data represent Mean ± S.E. of 4 independent animals. *p < 0.05; **p < 0.01; t test.
Fig. 4.
Biochemical validation of USP8, a protein shown to be down-regulated by quantitative proteomics. PSD fractions (15 μg protein) from morphine or saline treated animals were subjected to Western blot analysis using antibodies to USP8 (A) as described under “Experimental Procedures.” The total level of ubiquitinated proteins (B) was assessed using anti-ubiquitin antibodies. Representative blot is shown in the figure. Data represent Mean ± S.E. of 4 independent animals. *p < 0.05; **p < 0.01; t test.
Next we performed a protein set enrichment analysis using the tool Enrichr (29), and relevant gene set libraries such as mammalian phenotypes for knockout mice, Reactome pathways, Human Gene Atlas, ENCODE and TargetScan. The results show that both the up-regulated and down-regulated proteins are enriched for neuronal defect phenotypes (Fig. 5). Reactome enrichment analysis suggest an increase in opioid signaling components which are likely to be an adaptive mechanism (Fig. 5). Interestingly, many of the up-regulated proteins are targets of two microRNA family members, whereas the down-regulated proteins are enriched in genes highly expressed in the pineal gland and regulated by the transcription factor CTCF (Fig. 5).
Fig. 5.
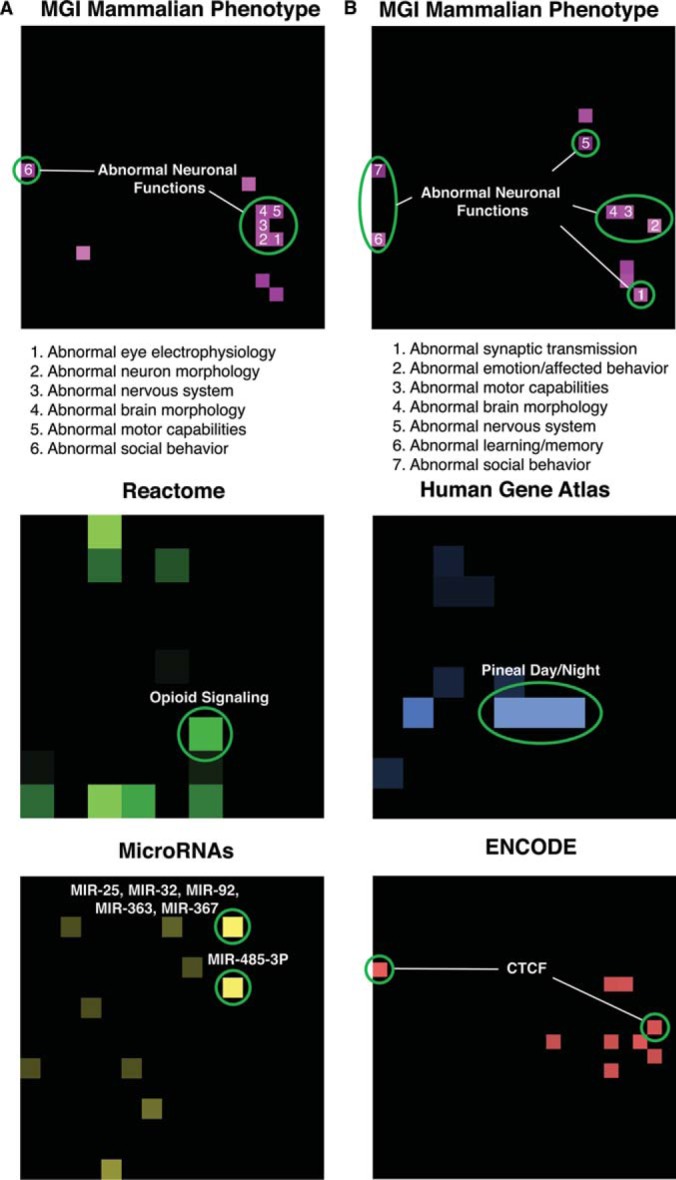
Enrichment analysis of significantly up-regulated and down-regulated proteins. Enrichr (29) was used to perform enrichment analysis on the significantly up-regulated (A) and down-regulated (B) proteins (24 were up-regulated and 10 were down-regulated in morphine treated samples) as described under “Experimental Procedures”. The proteins were first mapped to gene symbols and then used as input for Enrichr. The figure shows the canvas representation of the results.
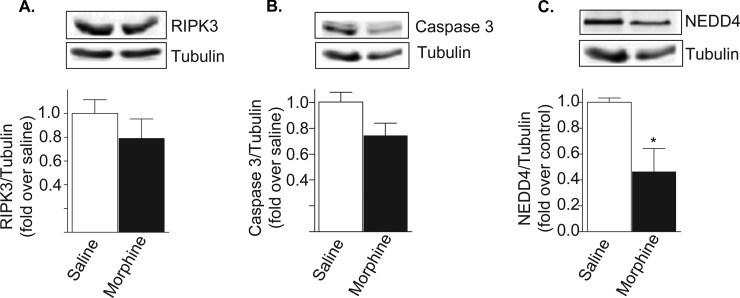
Next we generated a PPI network based on proteins modified by morphine treatment using the Genes2Fans software suite (30). For this we generated a Morphine Regulated Protein Seed List (further referred to simply as the “seed list”) by combining the observed High Probability Morphine Regulated Proteins (Table I and II) with a set of proteins that exhibited lower levels of statistical significance (0.05 < p < 0.10), defined as the Low Probability Morphine Regulated Proteins (supplemental Table S2). We used Genes2Fans to connect as many pairs of seed list proteins as possible using intermediate proteins interactions identified within the known background PPI network. This allowed us to obtain a PPI subnetwork that comprised a total of 116 proteins, 17 from the seed list and 93 from the background PPI network (Table III shows the list of Significant intermediates). These proteins were connected via 223 identified interactions (Fig. 6). Genes2Fans applied a binomial proportions test to prioritize the additional proteins that connect our seed list and found 40 significant intermediate proteins (Z-score > 2.0). Among these 40 proteins, 28 were found to be highly significant intermediates (Z-score > 3) whereas 12 were identified as significant intermediates (2.0 ≤ Z-score ≤ 3.0). We also carried out cluster analysis using CFinder (v.2.0.5) (40, 41). Cluster analyses showed that our network exhibited a higher clustering coefficient (0.169, p < 0.01) compared with 100 scrambled networks generated from the background data set that had identical network topology (clustering coefficient of 0.086) which suggests a degree clustering that is distinctly non-random. Interestingly, our network predicts a number of receptors including opioid receptors, signaling molecules involved in calcium signaling as well as molecules whose function at the PSD is not as yet known as significant intermediates. Next, we validated the predicted significant intermediate proteins by Western blot analysis. For this we selected three of the most unique proteins identified by Genes2FANs as none of them were known to localize to, or function at, mature synapses prior to this investigation: caspase-3 (CASP3), receptor interacting serine/threonine protein kinase 3 (RIPK3), and the E3-ubiquitin ligase neural precursor cell expressed developmentally down-regulated protein 4 (or NEDD4). Western blot analysis using synaptosomal fractions showed that the expression levels of RIPK3 and CASP3 tended to decrease in morphine treated animals whereas levels of NEDD4 were significantly decreased compared with saline treated controls (Fig. 7).
Table III. Significant intermediates from the background data set that link seed list proteins. A binomial proportions test (z-score) was used to identify the intermediates that had higher preference to interact with proteins from the seed list as compared to the background PPI network. Proteins with a z-score > 2.5 were designated as significant intermediates and with a z-score < 2.5 as other intermediates.
| Intermediate nodes | Protein name | Intermediate Nodes | Protein name |
|---|---|---|---|
| ABL1 | Tyrosine protein kinase ABL1 | MAPK1 | Mitogen-activated protein kinase 1 |
| ACTN4 | α-actinin 4 | MAPK3 | Mitogen-activated protein kinase 3 |
| ADRA2A | α2A adrenergic receptor | MAPT | Microtubule-associated protein tau |
| ADRBK1 | β-adrenergic receptor kinase 1 | MATR3 | Matrin 3 |
| AKT1 | RAC-α serine/threonine protein kinase | NEDD4 | E3 ubiquitin protein ligase NEDD4 |
| APLP2 | Amyloid-like protein 2 | NFKB2 | Nuclear facto NF-κ-B p100 subunit |
| ARRB1 | β-arrestin 1 | NSF | Vesicle-fusing ATPase |
| ARRB2 | β-arrestin 2 | OPRD1 | δ type opioid receptor |
| CACNA1A | Voltage-dependent P/Q type calcium channel subunit α1A | OPRM1 | μ type opioid receptor |
| CACNA1C | Voltage-dependent L type calcium channel subunit α1C | PA2G4 | Proliferation-associated protein 2G4 |
| CALM1 | Calmodulin | PACSIN1 | Protein kinase C and casein kinase substrate in neurons protein 1 |
| CAND1 | Cullin-associated NEDD8-dissociated protein 1 | PARP1 | Poly [ADP-ribose] polymerase 1 |
| CASP3 | Caspase-3 | PLEC | Plectin |
| CCT7 | T-complex protein 1 subunit η | PPP3CA | Serine/threonine-protein phosphatase 2B catalytic subunit α isoform |
| COPS6 | COP9 signalosome complex subunit 6 | PPP3CB | Serine/threonine-protein phosphatase 2B catalytic subunit β isoform |
| CSNK1A1 | Casein kinase I isoform α | PRKAA1 | 5′-AMP-activated protein kinase catalytic subunit α1 |
| CTBP1 | C-terminal binding protein 1 | PRKCA | Protein kinase C α type |
| CTNNB1 | Catenin β1 | PRKCD | Protein kinase C δ type |
| CUL2 | Cullin-2 | PRKCE | Protein kinase C ϵ type |
| CUL5 | Cullin-5 | PRNP | Major prion protein |
| DLG4 | Disks large homolog 4 | RAC1 | Ras-related C3 botulinum toxin substrate 1 |
| EWSR1 | RNA-binding protein EWS | RAP1GAP | Rap1 GTPase-activating protein 1 |
| FLNA | Filamin-A | RASA1 | Ras GTPase-activating protein 1 |
| FMR1 | Fragile X mental retardation protein1 | RASGRF1 | Ras-specific guanine nucleotide-releasing factor 1 |
| GABBR1 | γ-aminobutyric acid type B receptor subunit 1 | RB1CC1 | RB1-inducible coiled-coil protein 1 |
| GNAQ | Guanine nucleotide-binding protein Gαq | RGS6 | Regulator of G-protein signaling 6 |
| GNG2 | Guanine nucleotide-binding protein Gi/Gs/Go subunit γ2 | RNF128 | E3 ubiquitin protein ligase RNF128 |
| GOLGA2 | Golgin subfamily A member 2 | RNF41 | E3 ubiquitin protein ligase NRDP1 |
| GPX1 | Glutathione peroxidase 1 | RPS24 | 40S ribosomal protein S24 |
| GRB2 | Growth factor receptor-bound protein 2 | RTN4 | Reticulon-4 |
| GRIN1 | Glutamate receptor ionotropic, NMDA1 | SPTAN1 | Spectrin α chain, non-erythrocytic 1 |
| GRIN2A | Glutamate receptor ionotropic, NMDA 2A | SPTBN1 | Spectrin β chain, non-erythrocytic 1 |
| GRIN2B | Glutamate receptor ionotropic, NMDA 2B | SQSTM1 | Sequestosome-1 |
| GRM7 | Metabotropic glutamate receptor 7 | SRC | Proto-oncogene tyrosine protein kinase Src |
| GSK3B | Glycogen synthase kinase 3β | SRR | Serine racemase |
| GSN | Gelsolin | STAU1 | Double-stranded RNA-binding protein Staufen homolog 1 |
| HDAC2 | Histone deacetylase 2 | SYN1 | Synapsin-1 |
| HDAC4 | Histone deacetylase 4 | TBK1 | Serine/threonine protein kinase TBK1 |
| HDAC5 | Histone deacetylase 5 | TJP1 | Tight junction protein ZO-1 |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | TP53 | Cellular tumor antigen p53 |
| HSP90AA1 | Heat shock protein HSP 90α | TSPAN4 | Tetraspanin-4 |
| HSPA5 | 78 kDa glucose-regulated protein | TSSC1 | Protein TSSC1 |
| HTT | Sodium-dependent serotonin transpoter | UBC | E2 Ubiquitin-conjugating enzyme |
| IKBKG | NF-κ-B essential modulator | ULK1 | Serine/threonine protein kinase ULK1 |
| ITPR1 | Inositol 1,4,5-triphosphate receptor type 1 | UNC119 | Protein unc-119 homolog A |
| KRT10 | Keratin, type I cytoskeletal 10 | VCL | Vinculin |
| LRRC4 | Leucine-rich repeat-containing protein 4 | VIM | Vimentin |
| MAP2K4 | Dual specificity mitogen-activated protein kinase kinase 4 | YWHAB | 14–3-3 protein β/α |
| MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | YWHAQ | 14–3-3 protein θ |
| MAP3K3 | Mitogen-activated protein kinase kinase kinase 3 | YWHAZ | 14–3-3 protein ζ/δ |
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 |
Fig. 6.
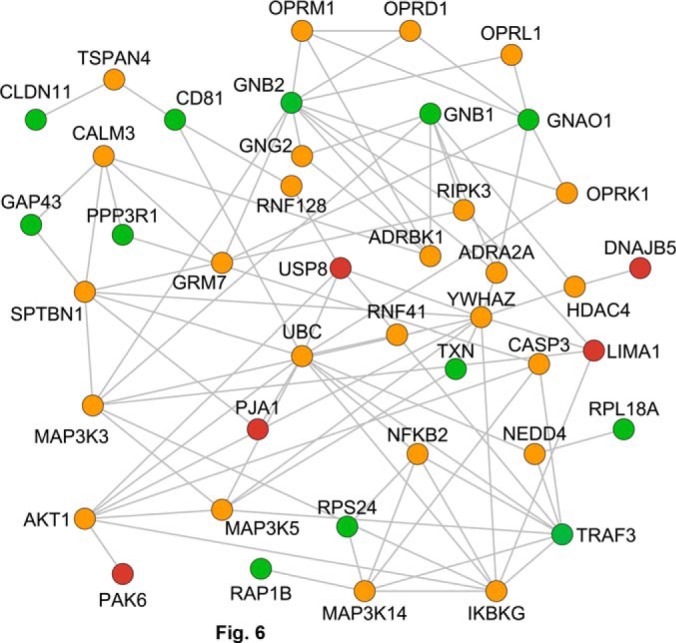
Network representation of proteins altered by morphine treatment generated using intermediates from a background data set. Genes2FANS (30, 38) was used to connect the up-regulated proteins (green) with the down-regulated proteins (red) from the seed list using a maximum of two intermediates from the background literature-based PPI network. The network contains a total of 99 proteins and 180 interactions. Significant intermediates are shown in orange (z-score > 2.5). Up-regulated proteins (green): CD81, CD81 protein; CLDN11, Claudin-11; GAP43, Neuromodulin; GNAO1, Guanine nucleotide-binding protein G(o) subunit alpha; GNB1, Guanine nucleotide-binding protein G(i)/G(s)/G(t) subunit beta-1; GNB2, Guanine nucleotide-binding protein G(i)/G(s)/G(t) subunit beta-2; PPP3R1, Calcineurin subunit B type 1; RAP1B, Ras-related protein Rap-1b; RPL18A, 60S ribosomal protein L18a; RPS24, 40S ribosomal protein S24; TNX, Thioredoxin. Down-regulated proteins (red): DNAJB5, DnaJ homolog subfamily B member 5; LIMA1, LIM domain and actin-binding protein 1; PAK6, Serine/threonine-protein kinase PAK6; PJA1, E3 ubiquitin-protein ligase Praja-1; TRAF3, TNF receptor-associated factor 3; USP8, Ubiquitin carboxyl-terminal hydrolase 8. Significant intermediates (orange): ADRA2A, Alpha-2A adrenergic receptor; ADRBK1, Beta-adrenergic receptor kinase 1; AKT1, Protein kinase B alpha; CALM3, Calmodulin-3; CASP3, Caspase-3; GNG2, Guanine nucleotide-binding protein G(i)/G(s)/G(o) subunit gamma-2; GRM7, Metabotropic glutamate receptor 7; HD4C4, Histone deacetylase 4; IKBKG, NF-kappa B essential modulator (NEMO); MAP3K14, Mitogen-activated protein kinase kinase kinase 14; MAP3K3, Mitogen-activated protein kinase kinase kinase 3; MAP3K5, Mitogen-activated protein kinase kinase kinase 5; NEDD4, E3 ubiquitin-protein ligase NEDD4; NFKB2, Nuclear factor NF-kappa B p100 subunit; OPRD1, Delta type opioid receptor; OPRk1, Kappa type opioid receptor; OPRL1, Nociceptin receptor; OPRM1, Mu type opioid receptor; RIPK3, Receptor-interacting serine/threonine protein kinase 3; RNF128, E3 ubiquitin protein ligase RNF128; RNF41, E3 ubiquitin protein ligase NRDP1; SPTBN1, Spectrin beta chain, non-erythrocytic 1; TSPAN4, Tetraspanin-4; UBC, Ubiquitin-conjugating enzyme; YWHAZ, 14–3-3 protein zeta/delta.
Fig. 7.
Biochemical validation of predicted proteins from network analysis. Synaptosomal fractions (15 μg protein) from morphine or saline treated animals were subjected to Western blot analysis using antibodies to either RIPK3 (A), caspase-3 (B) or NEDD4 (C) as described under “Experimental Procedures.”
DISCUSSION
The present study utilized a combination of subcellular fractionation (22, 25, 27, 33), quantitative proteomic approaches and network analysis in an effort to examine alterations in the postsynaptic protein profile within the striatum as a consequence of prolonged exposure to morphine. Of the 2648 proteins that were identified in this study, 34 (or ∼1.4% of the proteins identified) exhibited significantly altered expression following the morphine treatment paradigm. Among these 34 morphine-regulated proteins, 24 exhibited significant up-regulation (Table I), whereas the remaining 10 were significantly down-regulated (Table II). Among the proteins up-regulated following morphine treatment were proteins involved in G-protein signaling, including GαO, Gβ1, and Gβ2. These results are in agreement with previous studies that showed increased expression of G-protein subunits particularly GαO and Gαi during opiate addiction (42, 43) in a number of neuroanatomical regions including the prefrontal cortex, locus coeruleus and nucleus accumbens (44). This effect of morphine could extend to the molecular level, because an increased synthesis of GαO and a decrease in the synthesis of GαS was observed in the rodent hippocampus subsequent to chronic morphine exposure (45). Interestingly, proteomic studies using hippocampal PSD fractions of mice treated with escalating doses of morphine reported a slight (0.92-fold) decrease in the abundance of a closely related G-protein subunit, GαO2, compared with saline treated controls (22).
Table II. Seed list of 13 striatal PSD proteins down-regulated by morphine treatment. Striatal PSD proteins from saline and morphine treated animals (n = 4/group) were subjected to proteomics analysis and quantified as described in Experimental Procedures. Acc. #, accession number; M/S, morphine/saline.
| Full name | Acc. # | Gene I.D. | M.W. (kDa) | Ratio (M/S) | p value |
|---|---|---|---|---|---|
| EST Domain-containing transcription factor ERF | D3ZJW0 | ERF | 59 | 0.8 | 0.01 |
| FTS and Hook interacting protein | D4A7B7 | FAM160A2 | 99 | 0.8 | 0.02 |
| Opioid growth factor receptor | D4ABV6 | OGFR | 63 | 0.8 | 0.02 |
| Similar to E3 ubiquitin-protein ligase Praja-1 | Q66HF7 | PJA1 | 45 | 0.8 | 0.03 |
| Synaptopodin | Q9Z327 | SYNPO | 100 | 0.8 | 0.03 |
| Ubiquitin carboxyl-terminal hydrolase | D3ZN39 | USP8 | 124 | 0.8 | 0.03 |
| Serine/threonine protein kinase PAK6 | D3ZQ51 | PAK6 | 75 | 0.8 | 0.03 |
| Protein Lima1 | F1LR10 | LIMA1 | 83 | 0.3 | 0.03 |
| CST complex subunit STN1 | Q6AYD2 | OBFC1 | 47 | 0.8 | 0.05 |
| Dnajb5 protein (Fragment) | B2GV48 | DNAJB5 | 44 | 0.8 | 0.05 |
In this study we find that among the striatal postsynaptic proteins altered by morphine administration are several that are associated with the Ubiquitin-Proteasome System (UPS) such as PRAJA-1, FAM160A2, ubiquitin carboxyl-terminal hydrolase 8 (USP8), and ATG7. Of these Praja-1, FAM160A2, and USP8 were down-regulated while ATG7 was up-regulated in morphine treated animals. Interestingly a previous proteomics study investigating changes in phosphotyrosinated proteins in the frontal cortex of morphine dependent animals also detected alterations in a number of UPS related proteins (43). To date very little is known about the role of these proteins in the central nervous system. PRAJA-1 is a E2-dependent E3 ubiquitin ligase that is abundantly expressed in the brain (46), and has as its putative substrates the postsynaptic proteins Homer2, CAMK1G, EPHB3 and SEPT1 (47). PRAJA-1 has been implicated in learning and memory associated with fear conditioning (48). FAM160A2 is a FHIP protein that is a component of the FTS and Hook-interacting protein/FHIP (FHF) complex (49). The FTS component of the complex is an E2 ubiquitin-conjugating enzyme (49) and the complex as a whole has been implicated in sorting to early endosomes (50). ATG7 has been shown to be involved in autophagy, a process that can be recruited to clear aggregated ubiquitin-tagged proteins following USP disruption (51). USP8, also known as USPy or HUMORF8, is a deubiquitinating hydrolase that regulates protein turnover by removing ubiquitin from proteins targeted for degradation (52). USP8 has been shown to regulate the surface expression, endocytosis and degradation of a number of channels and receptors including the delta opioid receptor (53–57). Interestingly, in this study we find that morphine treatment leads to a decrease in the expression of USP8 in striatal PSD fractions and this is accompanied by a significant increase in the total levels of ubiquitinated proteins. This would suggest an UPS involvement in the development of tolerance and dependence to morphine. Thus, further studies are needed to evaluate the role of ubiquitinating and deubiquitinating enzymes in drug addiction.
The PPI network generated by Genes2FANs predicts caspase-3, RIPK3 and NEDD4 as significant intermediates in morphine induced changes at striatal PSDs; however, very little is known about the role of these proteins at the synapse during addiction to morphine. Caspase-3 (CASP3), which we find in this study to be decreased in synaptosomal fractions of morphine treated animals, is best known for its role during apoptosis as an executioner caspase that leads to neuronal cell death (58, 59). However, recent studies have revealed not only developmental functions but also a functional role for this enzyme in the regulation of synaptic plasticity, learning, and memory (60). Studies have demonstrated the presence of caspase-3 in both dendrites and PSD fractions (61), and have shown that it can modulate synaptic transmission by interacting with and cleaving specific AMPA subunits at the PSD (62), and by regulating the internalization of synaptic AMPA receptors (63). In this context, it is interesting to note that both caspase-3 and NEDD4 (another significant intermediate protein predicted by Genes2FANs) have been shown to modulate AMPA receptor surface expression through their interactions with the GluR1 subunit (63). In addition, caspase-3 and NEDD4 have been shown to modulate the activity of E2- and E3-ubiquitin ligases of the ubiquitin-proteasome system (61, 64), some of which were found to be altered in this study following morphine administration. Therefore caspase-3 may play an important role in neurological disorders. This is supported by studies that implicate interactions between caspase-3 and glutamate receptors in Alzheimer's disease (65), as well as studies showing that mutations in leucine-rich repeat kinase2 (LRRK2) render dopaminergic neurons more sensitive to caspase-3 activation in Parkinson's disease (66). Although the current study indicates that chronic morphine treatment causes a decrease in the levels of caspase-3, further studies are needed to elucidate the role on this enzyme both at the protein level as well as with regards to its enzymatic activity in the development of tolerance and addiction to drugs of abuse.
Another protein predicted to have a significant involvement in morphine-mediated changes at the striatal PSD is the receptor interacting protein kinase 3 (RIPK3) (Fig. 6 and 7). RIPK3 and the closely related protein RIPK1, are important regulatory proteins involved in the process of necroptosis, a process associated with programmed necrotic cell death (67). Necroptosis is negatively regulated by caspase-8 (67); thus decreased levels of caspase-8 can lead to either RIPK1-dependent or independent mechanisms of recruitment of RIPK3 (67, 68). To date very little information is available about the role of necroptosis in the central nervous system with one study reporting that 5-aminolevulinic acid-based photodynamic therapy stimulates necroptosis in glioblastomas by increasing the formation of necrosomes enriched in RIPK3 and RIPK1, but lacking other common proteinacious components such as FADD and caspase-8 (69). Thus further studies are needed to elucidate the role of RIPK3 in the brain particularly during the development of tolerance to drugs of abuse.
Another interesting protein predicted to be a significant intermediate in this study is neural precursor cell expressed, developmentally down-regulated 4 (NEDD4). NEDD4 was first characterized as a developmentally regulated protein with peak expression during early neurogenesis, followed by substantially decreased expression throughout postnatal development (70). NEDD4 is also known to function as an E3 ubiquitin ligase (71) and has recently been reported to contribute to several mechanisms associated with synaptic plasticity in the nervous system. For example, NEDD4-dependent ubiquitination of AMPA receptors bearing GluR1 subunits has been shown to regulate receptor localization, stability, endocytosis and trafficking to lysosomes (72, 73). Interestingly, studies have demonstrated the involvement of AMPA receptors in the persistent synaptic changes following repeated morphine administration (13) as well as increased internalization of AMPA receptors in a clathrin-dependent manner following morphine administration (22). Thus further studies are needed to evaluate the role of NEDD4 in the development of tolerance and dependence to drugs of abuse.
Taken together, this study demonstrates how a combination of proteomics in combination with network analysis can be a powerful tool to detect changes at the striatal synapse following chronic morphine administration and help predict novel proteins that could be potential therapeutic targets to diminish the side-effects associated with chronic morphine use such as the development of tolerance, dependence, and addiction.
Supplementary Material
Footnotes
Author contributions: A.M., J.A.M., H.L., and L.A.D. designed research; S.D.S., I.G., T.L., C.M., L.H., and M.R.J. performed research; S.D.S., I.G., T.L., L.H., M.R.J., A.M., J.A.M., H.L., and L.A.D. analyzed data; I.G., T.L., A.M., and L.A.D. wrote the paper.
* This work was supported in part by NIH grants NS026880 and DA019521 to L.A.D., R01GM098316, U54HG008230 and U54CA189201 to A.M., DA027460 and DA036826 to J.A.M., and NS046593 to H.L.
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
1 The abbreviations used are:
- PSD
- postsynaptic density
- ACN
- acetonitrile
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- BCA
- bicinchoninic acid
- CASP3
- caspase-3
- CREB
- cAMP response element-binding protein
- CTCF
- CCCTC-binding factor
- i.p.
- intraperitoneal
- FDR
- false discovery rate
- FWHM
- full-width at half maximum
- HCD
- higher energy collision dissociation
- HSP-70
- heat shock protein-70
- iTRAQ
- isobaric tag for relative and absolute quantitation
- LRRK2
- leucine-rich repeat kinase2
- MALDI
- matrix-assisted laser desorption/ionization
- MMTS
- methyl-methanethiosulfate
- MS
- mass spectrometry
- NEDD4
- neural precursor cell expressed developmentally down-regulated protein 4
- PAGE
- polyacrylamide gel electrophoresis
- PPI
- protein-protein interaction
- PRE
- presynaptic density
- RIPK
- receptor interacting serine/threonine protein kinase
- RPLC
- reverse phase liquid chromatography
- SCXLC
- strong cation exchange liquid chromatography
- SDS
- sodium dodecyl sulfate
- TCEP
- Tris-(2-carboxyethyl)phosphine
- TEAB
- triethylammonium bicarbonate
- TFA
- trifluoroacetic acid
- TX-100
- Triton X-100
- UPS
- ubiquitin-proteasomal system
- USP8
- ubiquitin carboxyl terminal hydrolase 8.
REFERENCES
- 1. Russo S. J., Dietz D. M., Dumitriu D., Morrison J. H., Malenka R. C., Nestler E. J. (2010) The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 33, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang N. K., Tseng C. J., Wong C. S., Tung C. S. (1997) Effects of acute and chronic morphine on DOPAC and glutamate at subcortical DA terminals in awake rats. Pharmacol. Biochem. Behav. 56, 363–371 [DOI] [PubMed] [Google Scholar]
- 3. Robinson T. E., Kolb B. (1999) Morphine alters the structure of neurons in the nucleus accumbens and neocortex of rats. Synapse 33, 160–162 [DOI] [PubMed] [Google Scholar]
- 4. Grueter B. A., Rothwell P. E., Malenka R. C. (2012) Integrating synaptic plasticity and striatal circuit function in addiction. Curr. Opin. Neurobiol. 22, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lüscher C., Malenka R. C. (2011) Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron 69, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chao J., Nestler E. J. (2004) Molecular neurobiology of drug addiction. Annu. Rev. Med. 55, 113–132 [DOI] [PubMed] [Google Scholar]
- 7. Perrotti L. I., Weaver R. R., Robison B., Renthal W., Maze I., Yazdani S., Elmore R. G., Knapp D. J., Selley D. E., Martin B. R., Sim-Selley L., Bachtell R. K., Self D. W., Nestler E. J. (2008) Distinct patterns of DeltaFosB induction in brain by drugs of abuse. Synapse 62, 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nestler E. J. (2001) Molecular basis of long-term plasticity underlying addiction. Nat Rev. Neurosci. 2, 119–128 [DOI] [PubMed] [Google Scholar]
- 9. Lovinger D. M., Partridge J. G., Tang K. C. (2003) Plastic control of striatal glutamatergic transmission by ensemble actions of several neurotransmitters and targets for drugs of abuse. Ann. N.Y. Acad. Sci. 1003, 226–240 [DOI] [PubMed] [Google Scholar]
- 10. Lovinger D. M. (2010) Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology 58, 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu N. J., Bao L., Fan H. P., Bao G. B., Pu L., Lu Y. J., Wu C. F., Zhang X., Pei G. (2003) Morphine withdrawal increases glutamate uptake and surface expression of glutamate transporter GLT1 at hippocampal synapses. J. Neurosci. 23, 4775–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Billa S. K., Liu J., Bjorklund N. L., Sinha N., Fu Y., Shinnick-Gallagher P., Morón J. A. (2010) Increased insertion of glutamate receptor 2-lacking alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors at hippocampal synapses upon repeated morphine administration. Mol. Pharmacol. 77, 874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xia Y., Portugal G. S., Fakira A. K., Melyan Z., Neve R., Lee H. T., Russo S. J., Liu J., Morón J. A. (2011) Hippocampal GluA1-containing AMPA receptors mediate context-dependent sensitization to morphine. J. Neurosci. 31, 16279–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faber E. S., Sah P. (2004) Opioids inhibit lateral amygdala pyramidal neurons by enhancing a dendritic potassium current. J. Neurosci. 24, 3031–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gupta A., Mulder J., Gomes I., Rozenfeld R., Bushlin I., Ong E., Lim M., Maillet E., Junek M., Cahill C. M., Harkany T., Devi L. A. (2010) Increased abundance of opioid receptor heteromers after chronic morphine administration. Sci. Signal. 3, ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stockton S. D. Jr., Devi L. A. (2012) Functional relevance of mu-delta opioid receptor heteromerization: a role in novel signaling and implications for the treatment of addiction disorders: from a symposium on new concepts in mu-opioid pharmacology. Drug Alcohol Depend. 121, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costantino C. M., Gomes I., Stockton S. D., Lim M. P., Devi L. A. (2012) Opioid receptor heteromers in analgesia. Expert Rev. Mol. Med. 14, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodzon-Kulakowska A., Suder P., Mak P., Bierczynska-Krzysik A., Lubec G., Walczak B., Kotlinska J., Silberring J. (2009) Proteomic analysis of striatal neuronal cell cultures after morphine administration. J. Sep. Sci. 32, 1200–1210 [DOI] [PubMed] [Google Scholar]
- 19. Suder P., Bodzon-Kulakowska A., Mak P., Bierczynska-Krzysik A., Daszykowski M., Walczak B., Lubec G., Kotlinska J. H., Silberring J. (2009) The proteomic analysis of primary cortical astrocyte cell culture after morphine administration. J. Proteome Res. 8, 4633–4640 [DOI] [PubMed] [Google Scholar]
- 20. Prokai L., Zharikova A. D., Stevens S. M. Jr. (2005) Effect of chronic morphine exposure on the synaptic plasma-membrane subproteome of rats: a quantitative protein profiling study based on isotope-coded affinity tags and liquid chromatography/mass spectrometry. J. Mass Spectrom. 40, 169–175 [DOI] [PubMed] [Google Scholar]
- 21. Abul-Husn N. S., Devi L. A. (2006) Neuroproteomics of the synapse and drug addiction. J. Pharmacol. Exp. Ther. 318, 461–468 [DOI] [PubMed] [Google Scholar]
- 22. Morón J. A., Abul-Husn N. S., Rozenfeld R., Dolios G., Wang R., Devi L. A. (2007) Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol. Cell. Proteomics 6, 29–42 [DOI] [PubMed] [Google Scholar]
- 23. Li K. W., Jimenez C. R., van der Schors R. C., Hornshaw M. P., Schoffelmeer A. N., Smit A. B. (2006) Intermittent administration of morphine alters protein expression in rat nucleus accumbens. Proteomics 6, 2003–2008 [DOI] [PubMed] [Google Scholar]
- 24. Abul-Husn N. S., Annangudi S. P., Ma'ayan A., Ramos-Ortolaza D. L., Stockton S. D. Jr., Gomes I., Sweedler J. V., Devi L. A. (2011) Chronic morphine alters the presynaptic protein profile: identification of novel molecular targets using proteomics and network analysis. PLoS ONE 6, e25535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bu Q., Yang Y., Yan G., Hu Z., Hu C., Duan J., Lv L., Zhou J., Zhao J., Shao X., Deng Y., Li Y., Li H., Zhu R., Zhao Y., Cen X. (2012) Proteomic analysis of the nucleus accumbens in rhesus monkeys of morphine dependence and withdrawal intervention. J. Proteomics 75, 1330–1342 [DOI] [PubMed] [Google Scholar]
- 26. Freeman W. M., Hemby S. E. (2004) Proteomics for protein expression profiling in neuroscience. Neurochem. Res. 29, 1065–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phillips G. R., Huang J. K., Wang Y., Tanaka H., Shapiro L., Zhang W., Shan W. S., Arndt K., Frank M., Gordon R. E., Gawinowicz M. A., Zhao Y., Colman D. R. (2001) The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 32, 63–77 [DOI] [PubMed] [Google Scholar]
- 28. Trang T., Sutak M., Quirion R., Jhamandas K. (2003) Spinal administration of lipoxygenase inhibitors suppresses behavioural and neurochemical manifestations of naloxone-precipitated opioid withdrawal. Br. J. Pharmacol. 140, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dannenfelser R., Clark N. R., Ma'ayan A. (2012) Genes2FANs: connecting genes through functional association networks. BMC Bioinformatics 13, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Council N. R. (2011) Guide fo the Care and Use of Laboratory Animals: Eight Edition, Tha National Academies Press, Washington, DC. [Google Scholar]
- 32. Thollander M., Hellström P. M., Svensson T. H. (1989) Suppression of small intestinal motility and morphine withdrawal diarrhoea by clonidine: peripheral site of action. Acta Physiol Scand. 137, 385–392 [DOI] [PubMed] [Google Scholar]
- 33. Jordan B. A., Fernholz B. D., Boussac M., Xu C., Grigorean G., Ziff E. B., Neubert T. A. (2004) Identification and verification of novel rodent postsynaptic density proteins. Mol. Cell. Proteomics 3, 857–871 [DOI] [PubMed] [Google Scholar]
- 34. Tyler W. A., Jain M. R., Cifelli S. E., Li Q., Ku L., Feng Y., Li H., Wood T. L. (2011) Proteomic identification of novel targets regulated by the mammalian target of rapamycin pathway during oligodendrocyte differentiation. Glia 59, 1754–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jain M. R., Li Q., Liu T., Rinaggio J., Ketkar A., Tournier V., Madura K., Elkabes S., Li H. (2012) Proteomic identification of immunoproteasome accumulation in formalin-fixed rodent spinal cords with experimental autoimmune encephalomyelitis. J. Proteome Res. 11, 1791–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 37. Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 38. Berger S. I., Posner J. M., Ma'ayan A. (2007) Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics 8, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adamcsek B., Palla G., Farkas I. J., Derényi I., Vicsek T. (2006) CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics 22, 1021–1023 [DOI] [PubMed] [Google Scholar]
- 41. Derényi I., Palla G., Vicsek T. (2005) Clique percolation in random networks. Phys. Rev. Lett. 94, 160202. [DOI] [PubMed] [Google Scholar]
- 42. Terwilliger R. Z., Beitner-Johnson D., Sevarino K. A., Crain S. M., Nestler E. J. (1991) A general role for adaptations in G-proteins and the cyclic AMP system in mediating the chronic actions of morphine and cocaine on neuronal function. Brain Res. 548, 100–110 [DOI] [PubMed] [Google Scholar]
- 43. Kim S. Y., Chudapongse N., Lee S. M., Levin M. C., Oh J. T., Park H. J., Ho I. K. (2005) Proteomic analysis of phosphotyrosyl proteins in morphine-dependent rat brains. Brain Res. Mol. Brain Res. 133, 58–70 [DOI] [PubMed] [Google Scholar]
- 44. Abul-Husn N. S., Bushlin I., Morón J. A., Jenkins S. L., Dolios G., Wang R., Iyengar R., Ma'ayan A., Devi L. A. (2009) Systems approach to explore components and interactions in the presynapse. Proteomics 9, 3303–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Przewlocka B., Lasoń W., Przewlocki R. (1994) The effect of chronic morphine and cocaine administration on the Gs and Go protein messenger RNA levels in the rat hippocampus. Neuroscience 63, 1111–1116 [DOI] [PubMed] [Google Scholar]
- 46. Yu P., Chen Y., Tagle D. A., Cai T. (2002) PJA1, encoding a RING-H2 finger ubiquitin ligase, is a novel human X chromosome gene abundantly expressed in brain. Genomics 79, 869–874 [DOI] [PubMed] [Google Scholar]
- 47. Loch C. M., Eddins M. J., Strickler J. E. (2011) Protein microarrays for the identification of praja1 e3 ubiquitin ligase substrates. Cell Biochem. Biophys. 60, 127–135 [DOI] [PubMed] [Google Scholar]
- 48. Stork O., Stork S., Pape H. C., Obata K. (2001) Identification of genes expressed in the amygdala during the formation of fear memory. Learn Mem. 8, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu L., Sowa M. E., Chen J., Li X., Gygi S. P., Harper J. W. (2008) An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol. Biol. Cell 19, 5059–5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Richardson S. C., Winistorfer S. C., Poupon V., Luzio J. P., Piper R. C. (2004) Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell 15, 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng Q., Li J., Wang X. (2009) Interplay between the ubiquitin-proteasome system and autophagy in proteinopathies. Int. J. Physiol. Pathophysiol. Pharmacol. 1, 127–142 [PMC free article] [PubMed] [Google Scholar]
- 52. Naviglio S., Mattecucci C., Matoskova B., Nagase T., Nomura N., Di Fiore P. P., Draetta G. F. (1998) UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J. 17, 3241–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balut C. M., Loch C. M., Devor D. C. (2011) Role of ubiquitylation and USP8-dependent deubiquitylation in the endocytosis and lysosomal targeting of plasma membrane KCa3.1. FASEB J. 25, 3938–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niendorf S., Oksche A., Kisser A., Löhler J., Prinz M., Schorle H., Feller S., Lewitzky M., Horak I., Knobeloch K. P. (2007) Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell. Biol. 27, 5029–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Berlin I., Higginbotham K. M., Dise R. S., Sierra M. I., Nash P. D. (2010) The deubiquitinating enzyme USP8 promotes trafficking and degradation of the chemokine receptor 4 at the sorting endosome. J. Biol. Chem. 285, 37895–37908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hasdemir B., Murphy J. E., Cottrell G. S., Bunnett N. W. (2009) Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J. Biol. Chem. 284, 28453–28466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hislop J. N., Henry A. G., Marchese A., von Zastrow M. (2009) Ubiquitination regulates proteolytic processing of G protein-coupled receptors after their sorting to lysosomes. J. Biol. Chem. 284, 19361–19370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hengartner M. O. (2000) The biochemistry of apoptosis. Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 59. Kumar S. (2007) Caspase function in programmed cell death. Cell Death Differ. 14, 32–43 [DOI] [PubMed] [Google Scholar]
- 60. Snigdha S., Smith E. D., Prieto G. A., Cotman C. W. (2012) Caspase-3 activation as a bifurcation point between plasticity and cell death. Neurosci. Bull. 28, 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Williams D. W., Kondo S., Krzyzanowska A., Hiromi Y., Truman J. W. (2006) Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234–1236 [DOI] [PubMed] [Google Scholar]
- 62. Lu C., Fu W., Salvesen G. S., Mattson M. P. (2002) Direct cleavage of AMPA receptor subunit GluR1 and suppression of AMPA currents by caspase-3: implications for synaptic plasticity and excitotoxic neuronal death. Neuromol. Med. 1, 69–79 [DOI] [PubMed] [Google Scholar]
- 63. Li Z., Jo J., Jia J. M., Lo S. C., Whitcomb D. J., Jiao S., Cho K., Sheng M. (2010) Caspase-3 Activation via Mitochondria Is Required for Long-Term Depression and AMPA Receptor Internalization. Cell 141, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harvey K. F., Kumar S. (1999) Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 9, 166–169 [DOI] [PubMed] [Google Scholar]
- 65. Hu N. W., Ondrejcak T., Rowan M. J. (2012) Glutamate receptors in preclinical research on Alzheimer's disease: update on recent advances. Pharmacol. Biochem. Behav. 100, 855–862 [DOI] [PubMed] [Google Scholar]
- 66. Byers B., Lee H. L., Pera R. R. (2012) Modeling Parkinson's disease using induced pluripotent stem cells. Curr. Neurol. Neurosci. Rep. 12, 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaczmarek A., Vandenabeele P., Krysko D. V. (2013) Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 38, 209–223 [DOI] [PubMed] [Google Scholar]
- 68. Moujalled D. M., Cook W. D., Okamoto T., Murphy J., Lawlor K. E., Vince J. E., Vaux D. L. (2013) TNF can activate RIPK3 and cause programmed necrosis in the absence of RIPK1. Cell Death Disease 4, e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Coupienne I., Fettweis G., Rubio N., Agostinis P., Piette J. (2011) 5-ALA-PDT induces RIP3-dependent necrosis in glioblastoma. Photochem. Photobiol. Sci. 10, 1868–1878 [DOI] [PubMed] [Google Scholar]
- 70. Kumar S., Tomooka Y., Noda M. (1992) Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 185, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 71. Ingham R. J., Gish G., Pawson T. (2004) The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 72. Schwarz L. A., Hall B. J., Patrick G. N. (2010) Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J. Neurosci. 30, 16718–16729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lin A., Hou Q., Jarzylo L., Amato S., Gilbert J., Shang F., Man H. Y. (2011) Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J. Neurochem. 119, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.