Abstract
Stem cell transplantation is a promising therapeutic strategy to enhance axonal regeneration after spinal cord injury. Unrestricted somatic stem cells (USSC) isolated from human umbilical cord blood is an attractive stem cell population available at GMP grade without any ethical concerns. It has been shown that USSC transplantation into acute injured rat spinal cords leads to axonal regrowth and significant locomotor recovery, yet lacking cell replacement. Instead, USSC secrete trophic factors enhancing neurite growth of primary cortical neurons in vitro. Here, we applied a functional secretome approach characterizing proteins secreted by USSC for the first time and validated candidate neurite growth promoting factors using primary cortical neurons in vitro. By mass spectrometric analysis and exhaustive bioinformatic interrogation we identified 1156 proteins representing the secretome of USSC. Using Gene Ontology we revealed that USSC secretome contains proteins involved in a number of relevant biological processes of nerve regeneration such as cell adhesion, cell motion, blood vessel formation, cytoskeleton organization and extracellular matrix organization. We found for instance that 31 well-known neurite growth promoting factors like, e.g. neuronal growth regulator 1, NDNF, SPARC, and PEDF span the whole abundance range of USSC secretome. By the means of primary cortical neurons in vitro assays we verified SPARC and PEDF as significantly involved in USSC mediated neurite growth and therewith underline their role in improved locomotor recovery after transplantation. From our data we are convinced that USSC are a valuable tool in regenerative medicine as USSC's secretome contains a comprehensive network of trophic factors supporting nerve regeneration not only by a single process but also maintained its regenerative phenotype by a multitude of relevant biological processes.
Injury to the spinal cord leads to a multiple damaging process including axonal contusion and transection with subsequent degeneration, massive apoptosis of oligodendrocytes and break-down of the blood-spinal cord barrier accompanied by invasion of immune cells resulting in sustained motoric and sensory impairments. Glial-fibrotic scarring and the lack of growth promoting factors impair axonal regrowth, which is currently the main target for therapeutic interventions to treat spinal cord injury. In addition, modulation of neuronal survival, remyelination of axons, and the immune reaction could promote functional regeneration (1–3). The inhibition of axonal regeneration might be overcome by exogenous application of growth factors or by transplantation of stem cells directly into the lesion site, which locally release trophic factors and thus support axonal regrowth. For clinical applications, stem cells should be ideally available on a clinical scale without ethical concerns or invasive interventions. Human umbilical cord blood (hUCB)1 is an alternative stem cell source including cells similar to mesenchymal stem cells from bone marrow (BM-MSC) and can be easily expanded as adherent cells in vitro without any risk for the donor. Besides MSC, hUCB contains unrestricted somatic stem cells (USSC) (4), which can be clearly distinguished from BM-MSC as well as from hUCB derived MSC (CB-MSC) by their immunological behavior (5–6), their transcriptome (7), the inability to differentiate into adipocytes (8), and by a specific Hox-gene expression pattern (9). Additionally, USSC exhibit a significant lower senescence rate and possess longer telomeres compared with CB-MSC and BM-MSC. As USSC can be easily isolated at GMP (good manufacturing practice) grade (10) and expanded on a clinical scale, USSC are a promising tool for transplantation studies. In a recent study, we have demonstrated that after transplantation into the acute injured rat spinal cord, USSC induce significant axon regrowth into the lesion side and effectively improve long-term functional locomotor recovery (11). Moreover, USSC transplantation promotes tissue sparing, which might contribute to the locomotor improvement. Although USSC were shown to differentiate into neuronal-like cells under in vitro conditions (4, 12) replacement of endogenous cells in the injured spinal cord was not detected supporting the hypothesis that transplanted stem cells despite their lack of differentiation enhance regeneration by paracrine regulation or direct interactions with endogenous cells. In vitro studies confirmed the potential of USSC to promote neurite growth. Incubation of primary rat dorsal root ganglion neurons or cortical neurons with USSC conditioned medium (USSC-CM) significantly enhanced neurite growth, hence considering USSC-CM is an ideal tool to investigate USSC-derived neurite growth promoting factors. Kögler et al. (13) provided first evidence using cytokine specific antibody arrays that USSC secrete axon growth promoting and neuroprotective factors in vitro such as stromal derived factor-1 (SDF-1) (14–15), leukemia inhibitory factor (LIF) (16), and vascular endothelial growth factor (VEGF) (17–18). In comparison, BM-MSC also secrete trophic factors, which enhance neurite growth in vitro, but the main trophic factors responsible for beneficial effects on neurite growth remain to be elucidated (19). Neurite growth promoting effects of BM-MSC seem to be at least partially mediated by brain derived neurotrophic factor (BDNF) and glial derived neurotrophic factor (GDNF) as demonstrated by neutralization of these neurotrophins using antibodies (20–21). Classical neurotrophic factors, e.g. nerve growth factor (NGF), BDNF, and neurotrophin-3 (NT-3), have not been identified in the USSC secretome yet, and other factors released by USSC, which promote neurite outgrowth, have not been described in detail. Antibody array analysis of the secretome of other somatic stem cell populations of hUCB revealed that the cells release a large panel of cytokines and growth factors (22). Additionally, these cells express genes related to neurogenesis and blood vessel development as determined by gene expression profiling. For the unbiased identification of secreted proteins proteomic approaches including mass spectrometry is the method of choice. For example, CB-MSC secrete typical cartilage-related proteins during chondrogenic differentiation shown by nanoLC MALDI-TOF/TOF (23). Currently, the secretome of several stem cell types has been investigated by mass spectrometry (reviewed by (24–25)). However, a secretome study of USSC derived from hUCB by LC-MS/MS focusing on nerve regeneration has not been performed, yet.
Here, we present the first study aiming at the characterization of the regeneration-supportive phenotype of USSC by detailed secretome analysis using protein mass spectrometry and exhaustive bioinformatic interrogation as well as subsequent functional validation of candidate neurite growth promoting proteins.
EXPERIMENTAL PROCEDURES
Cultivation of USSC
SA 5/73 USSC were isolated from human umbilical cord blood as described by Kögler et al. (4), and have been provided by the Institute for Transplantation Diagnostics and Cell Therapeutics, Heinrich Heine University Medical Center, Düsseldorf, Germany. In brief, cells were separated from fresh umbilical cord blood by Ficoll followed by red blood cell lysis with ammonium chloride. Mononuclear cells were plated in Dulbecco's modified Eagle's medium supplemented with 30% fetal bovine serum (FBS), dexamethasone (10−7 m, Sigma Aldrich, Steinheim, Germany), 2 mm glutamine and penicillin/streptomycin (100 U/ml). Subsequently, USSC clones were picked, expanded in dexamethasone-free medium, and characterized by Hox gene (Hox-negative) (9) and DLK-1 (DLK-positive) (8) expression. For expansion, USSC were cultured in expansion medium containing DMEM (Lonza, Cologne, Germany) supplemented with 30% heat-inactivated FBS (Biochrom, Berlin, Germany), 2 mm glutamine (Invitrogen, Darmstadt, Germany) and penicillin/streptomycin (100 U/ml, Invitrogen) at 37 °C, 5% CO2 and 98% humidity. USSC have been used in passage 6–7 for all experiments.
Preparation of Serum-free Conditioned Medium
To identify secreted proteins USSC were cultured in serum-free medium. USSC were grown to a subconfluency of 80–90% in expansion medium, washed with PBS for three times to remove FBS and then incubated with serum-free N2 medium modified according to Bottenstein and Sato (26) for 48 h. Medium was changed and cells have been incubated for another 48 h. For secretome studies, the second batch of conditioned medium was used to exclude FBS effects. N2 medium is a 1:4 mixture of Dulbecco's modified Eagle's medium with Ham‘s F12, 2 mm glutamine (all Invitrogen), 5 μg/ml insulin, 30 nm sodium selenite, 100 μm putrescine, 20 nm progesterone and 5 μg/ml transferrin (all Sigma Aldrich). The conditioned medium has been collected, centrifuged at 1500 × g for 5 min at 4 °C to remove cell fragments and then stored at −80 °C until application to cortical neurons or secretome analysis.
Protein Concentration
Different strategies have been used to concentrate proteins from USSC conditioned medium. Proteins have been (A) precipitated using a modified method published by Chevallet et al. (27), or (B) concentrated by lyophilization. Briefly, for protein precipitation (method A) 20 ml of conditioned medium was mixed with 10% sarkosyl NL (Sigma Aldrich). After adding trichloroacetic acid (Sigma Aldrich) to a final concentration of 7.5%, samples were incubated on ice for 2h with subsequent centrifugation at 10,000 × g and 4 °C for 10 min. The pellet was resuspended in 2 ml ice cooled tetrahydrofuran (THF, Fluka, Buchs, Switzerland). Afterward, samples were centrifuged again at 10,000 × g and 4 °C for 10 min. The pellet was washed carefully with 2 ml THF and dissolved in 100 μl SDS sample buffer consisting of 600 mm DTT, 30% Glycerin, 12% SDS, 150 mm Tris/HCl, pH 7.0 (all Sigma Aldrich). After SDS-gel-electrophoresis (2 cm, 10 min) and silver staining according to Nesterenko et al. (28), the resulting lane was cut out in one slice. The gel slice was decolorized with a 1:1 mix of 30 mm sodium thiosulfate and 100 mm potassium hexacyanoferrate (III) (both Sigma Aldrich), washed, reduced (10 mm DTT) and alkylated (55 mm iodacetamide). Proteins were digested with 2 μg trypsin (Serva, Heidelberg, Germany) overnight at 37 °C. Resulting peptides were extracted with 50% acetonitrile (Biosolve, Valkenswaard, The Netherlands) and 0.05% TFA. For protein concentration by lyophilisation (method B) 20 ml of conditioned medium were lyophilized, reconstituted in 250 μl SDS sample buffer and 10 μl (2 μg/μl) was run into short SDS-gel, silver stained and subsequently digested and extracted as described above. Furthermore, a SDS-PAGE was performed and 12 lanes have been cut out for further analysis (method C). In supplemental Table S1 the methods are summarized.
LC-MS/MS
Peptides were separated using a UltiMate 3000 RSCLnano System (Dionex LC Packings (now Thermo Scientific)) with a C18-Pepman column (2 cm, 75 μm I.D., 3 μm particle size, 100 AA) as precolumn and a C18-Pepmap column (25 cm or 50 cm, 75 μm I.D., 2 μm particle size, 100 AA) as main column. A gradient of 0.1% FA (Fluka) to 0.1% FA/60% acetonitrile over 120–150 min and a constant flow rate of 300 nl/min was used to elute peptides directly via electrospray (voltage 1.2–3.0 keV, capillary temperature 310 °C) into the LTQ Orbitrap Velos or LTQ Orbitrap Elite mass spectrometer (both Thermo Fisher Scientific, Bremen, Germany). All MS spectra were recorded in positive ion mode with a mass range of 300–2000 m/z and a resolution of 35.000 (Orbitrap Velos) or 350–1700 m/z and a resolution of 60.000 (Orbitrap Elite). +2, +3 and higher charged (only Orbitrap Velos) monoisotopic precursors were isolated with a width of 2.0 Da and fragmented using a normalized collision energy of 35% for CID within the linear ion trap. A TOP20-data-dependent acquisition with polysiloxan as a lock mass was applied and dynamic exclusion activated (repeat count 1, duration 30 s on Orbitrap Velos and 45 s on Orbitrap Elite). Seven independent USSC secretomes were analyzed.
MS Data Analysis
Proteins were identified using Proteome Discoverer (Version 1.4, Thermo Fisher Scientific http://www.thermoscientific.com/en/product/proteome-discoverer-software.html) including Mascot search engine (Version 2.4.1, Matrix Science) (29). The UniProtKB/Swiss-Prot database (version from 2014/09, total entries: 546.439) was searched with a mass tolerance of 10 ppm (MS-mode) and 0.4 Da (MS/MS mode), enzyme specificity was set to trypsin and two missed cleavage sites were considered during the search against human subdatabase. Mass range setting was 350–5000 Da. Carbamidomethylation of cysteine was set as fixed modification. Oxidation of methionine was accepted as variable modification. Proteins were assembled by Proteome Discoverer with a Mascot threshold for protein identification set ≥ 50.0 and one unique peptide. For positive identification we considered a FDR of < 1% on peptide level (high peptide confidence, default p < 0.01). For FDR calculation we applied a decoy approach based on reversed protein sequences using Proteome Discoverer. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD002241 (30). Proteins were grouped by Gene ontology (GO) slim term cellular component and biological process.
Bioinformatic Analyses
The prediction of secreted proteins was performed with SecretomeP (31) as well as with SignalP (32). A NN-score > 0.6 (FDR < 0.05) was interpreted as a high probability of secretion via a non-classical pathway using Secretome P. We further applied functional annotation clustering to further characterize the USSC secretome using DAVID database (DAVID Bioinformatics Resources 6.7, (33), http://david.abcc.ncifcrf.gov/home.jsp) including Gene Ontology (GO) terms containing the sub-ontology biological process (defined by the Gene Ontology Consortium) with a false discoverer rate (FDR) < 0.05. Secretion of all UniProt listed proteins were predicted by SecretomeP and supplemented by proteins that are assigned to the UniProt term secreted or extracellular as well as classically secreted proteins with signal peptide predicted by SignalP. The 11.238 proteins served as background list for GO term annotation in DAVID database.
An absolute quantification of protein concentration was performed using Hi-N in Progenesis QI for proteomics 2.0 (Nonlinear Dynamics, Newcastle upon Tyne, see http://www.nonlinear.com/progenesis/qi-for-proteomics/v2.0/faq/how-does-hi-n-work.aspx for further details) based on a publication by Silva et al. (34) using proteins concentrated by method A (TCA precipitation) and B (lyophilisation). Briefly, the Hi-N method considers all peptides for quantification. In the case if more than three peptides are available Hi-N includes the three most abundant unique (non-conflicting) peptides to calculate the absolute concentration. In contrast to top-3 method (34) the Hi-N algorithm applies the average and not the sum of the peptide intensities. Transferrin with a known concentration of 5 μg/ml (64.8 pmol/ml) was applied as single point calibrator and the protein signals were adjusted to transferrin for absolute quantification.
Preparation and Cultivation of Primary Rat Cortical Neurons
To test potential neurite growth promoting factors found in USSC secretome, primary cortical neurons were prepared from E15 Wistar rats as described before (11, 35) and subsequently incubated with serum-free conditioned medium derived from USSC. N2 medium as well as conditioned medium derived from primary cortical astrocytes prepared from p0-p1 Wistar rats (11, 36) served as negative and positive controls, respectively. Embryonic rat cortical neurons were seeded on 96 well plates (Costar, Corning, Wiesbaden, Germany) pre-coated with 1 mg/ml poly-d-lysine and 13 μg/ml laminin (both Sigma Aldrich). A cell density of 40,000/cm2 was proven to be optimal for automated neurite growth analyses. Cells were incubated at 37 °C, 10% CO2 and 98% humidity.
Neutralization and Stimulation Experiments
To influence activity of potential neurite growth promoting factors in USSC-CM, neutralizing antibodies have been incubated for 2 h at 37 °C with USSC-CM prior to incubation of primary cortical neurons. Following antibodies have been used: anti-SPARC (secreted protein acidic and rich in cysteine, also known as Osteonectin, human, monoclonal produced in mouse, sc-33645, Santa Cruz Biotechnology, Heidelberg, Germany), 4 μg/ml; anti-PEDF (pigment epithelium-derived factor, also known as serpinF1, human, polyclonal produced in rabbit, sc-25594, Santa Cruz Biotechnology), 4 μg/ml; anti-CST3 (Cystatin C, human, polyclonal produced in goat, P01034, R&D System, Wiesbaden-Nordenstadt, Germany), 3 μg/ml. Because antibodies have been solved in PBS, USSC-CM with appropriate amounts of PBS has been used as control.
On the other hand, selected recombinant proteins were applied in nonconditioned N2 control medium to stimulate neurite growth. Recombinant proteins have been diluted as follows: SPARC (C388, Novoprotein Scientific, Summit, NJ), 3 μg/ml; PEDF (P3662, Abnova, Taipei, Taiwan), 30 ng/ml.
Immunocytochemical Staining of Cortical Neurons
After 48 h of cultivation, cortical neurons were fixed with 3.7% formaldehyde (Merck, Schwalbach, Germany) for 15 min and carefully washed with PBS. For immunocytochemical analysis, fixed cells were incubated with blocking solution containing 10% normal goat serum (Sigma Aldrich), and 0.03% Triton x-100 (Sigma Aldrich) for 1 h. Staining was performed using a neuron specific anti-β-III-tubulin (Tub) antibody (rabbit, polyclonal, Sigma Aldrich, 1:500 dilution in blocking buffer) over night at 4 °C. After washing with PBS, secondary antibody (goat anti-rabbit, Alexa 488, Invitrogen, 1:500 dilution) was incubated for 4 h at room temperature. DAPI (4′,6-diamidino-2-phenylindole) staining was performed to label all cell nuclei.
Analyses of Neurite Growth
High content screening of cortical neurons was performed in vitro with a Thermo Scientific Cellomics® ArrayScan® VTI HCS Reader and the Neuronal Profiling v3.5 BioApplication software (http://www.thermoscientific.com/en/products/cellular-imaging-analysis.html). Plates were imaged with a 10× objective (25 images per well) and simultaneously analyzed. For quantification, only Tub-positive structures with a DAPI-positive nucleus were included in cell counts. Following parameters were used for quantification: nucleus identification (iso data > −0.119; segment intensity > 80), nucleus validation (size 7–110), cell body identification (iso data > 0; minimum 1 cell body; seed segmentation; cell body segmentation = 2), cell body validation (1 nucleus; cell body size 30–1050; total cell intensity 20,450–521,307), neurite identification (identification modifier = −0.938; length 255; point resolution 2; gap tolerance = 5), neurite validation (neurite length 10–600; neurite intensity 0–4095). Absolute numbers of neurons per well and total neurite length per neuron were used for analyses of different culture conditions. Significance of neurite growth was assessed by paired Student's t test comparing median neurite growth from six wells per condition and experiment with appropriate control condition. Neurite growth was considered to be significantly different at p < 0.05. All data are presented as a mean relative neurite growth (%) of at least three experiments ± S.E.
RESULTS
Characterization of USSC Secretome by Mass Spectrometry
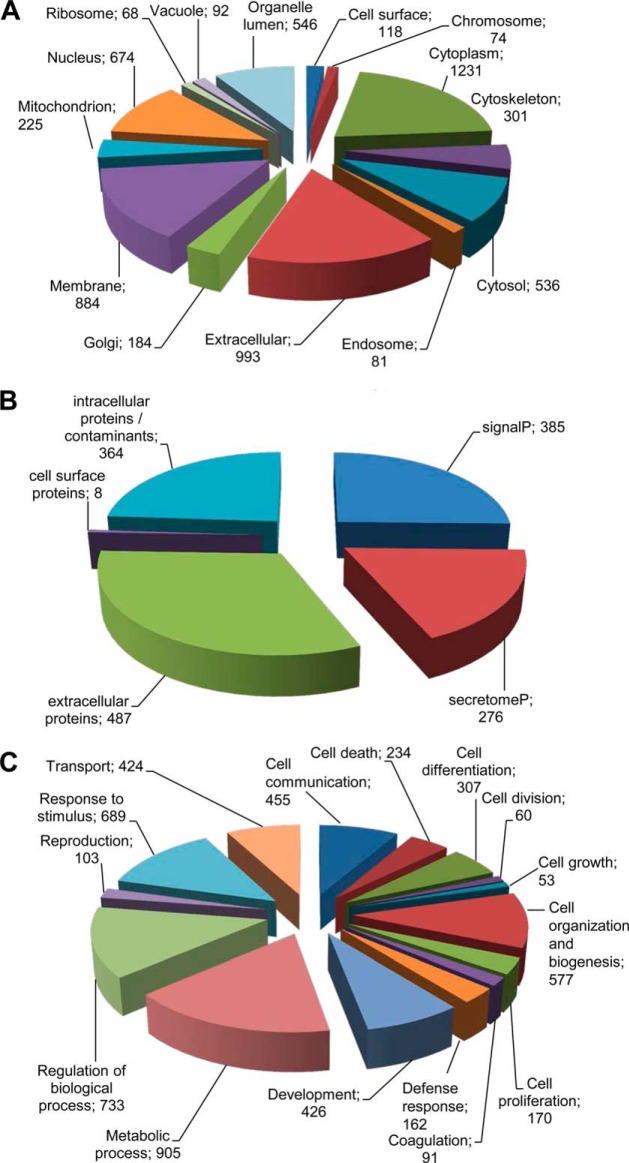
The first evidence that the regenerative potential of USSC is likely to rely on paracrine mechanisms was obtained because we showed that USSC secrete factors that stimulate neurite growth of cultured dorsal root ganglion neurons and cortical neurons (11). As a first approach to test which molecular class mediates the trophic functions of USSC the supernatant was heated to 80 °C for 5 min. The abolished neurite growth potential (data not shown) suggests that temperature-sensitive proteins are likely to enhance neurite growth. Therefore, we set-up a workflow analyzing serum-free supernatants of USSC using nano LC-ESI-MS/MS to identify proteins secreted by USSC. For protein preparation, we applied different methods to obtain a comprehensive protein catalogue of the USSC secretome (supplemental Table S1). Altogether, we identified 1520 proteins in USSC secretome (supplemental Table S2). Because of possible contaminations of conditioned medium by proteins originating from dead cells, not all identified proteins are likely to be secreted or extracellularly localized. Based on UniProt annotation, 993 proteins were classified as being potentially localized extracellularly and 118 proteins to be present at the cell surface (Fig. 1A, multiple protein entries possible). The USSC secretome further includes 884 membrane associated proteins, which could result from shedding. For further identification of secreted proteins, SignalP 4.0 algorithm was applied. We categorized 385 proteins to be classically secreted and—using SecretomeP 2.0 algorithm—276 proteins to be potentially secreted via non-classical pathways, remaining 487 proteins to be localized extracellular predicted by UniProt annotation (supplemental Table S2). Eight proteins, which have been found to be localized at the cell surface as identified by UniProt annotation, were not identified by SignalP and SecretomeP databases (Fig. 1B). Overall, 1156 proteins were determined to be candidate secretory proteins of USSC secretome, whereas 364 proteins were presumably contaminants from dead cells.
Fig. 1.
Categorization of proteins identified in USSC secretome by LC-MS/MS. A, Based on UniProt annotation, 993 proteins were found to be localized extracellularly. The USSC secretome further included 884 membrane associated proteins that might be found in the secretome because of shedding and 118 cell surface proteins (multiple protein entries possible). In total, 1520 proteins were determined. B, SignalP and SecretomeP algorithms revealed that 385 proteins are classically secreted and 276 proteins are nonclassically secreted. GO slim term extracellular included 487 proteins that were not predicted as secreted by SignalP and SecretomeP. Additionally, eight proteins were predicted to be localized at the cell surface. Overall, we identified 1156 proteins predicted to be secreted and 364 contaminants. C, These 1156 were assigned biological processes that in part play important roles during nerve regeneration (multiple protein entries possible) such as cell differentiation, development, cell communication and cell division.
Neurite Growth Promoting Factors Secreted by USSC
Several USSC secreted proteins were found to be associated with neurite growth or axonal regeneration. Comparison of all secreted proteins or proteins predicted to be secreted according to the literature revealed that USSC secrete at least 31 proteins that are known to be involved in neurite growth or axonal regrowth (Table I) including the neurotrophic factors mesencephalic astrocyte-derived neurotrophic factor (MANF) (37), neudesin (38), and neuron-derived neurotrophic factor (NDNF) (39) as well as the growth factors angiopoietin-2 (40), growth/differentiation factor 15 (GDF-15) (41), hepatoma-derived growth factor (HDGF) (42), PDGF (43), and transforming growth factor-beta1/-2 (TGF-β1/TGF-β2) (44–45). Moreover, we identified several matrix- and membrane-associated proteins, e.g. decorin (46), fibulin-1 (47), netrin-4 (48), neuronal growth regulator 1 (49), neuroplastin (50–51), noelin-2 (52), SPARC (53), tenascin (54), and the cytokines colony-stimulating factor 1 (CSF-1) (55), macrophage migration inhibitory factor (MIF) (56) as well as the chemokine stromal cell-derived factor 1 (SDF-1) (15). In addition, the serine protease inhibitors glia-derived nexin (serpinE2) (57), neuroserpin (58), and plasminogen activator inhibitor 1 (serpinE1) (59) as well as the extracellular chaperone clusterin (60–61) were secreted by USSC. In summary, these USSC secreted proteins represent a network of trophic factors that are likely to promote neurite growth in vitro as well as axonal regrowth after transplantation into the injured spinal cord in a synergistic way.
Table I. Neurite growth promoting factors identified in the USSC secretome. Overall, 31 proteins were identified in the USSC secretome (contaminants excluded) which are known to be directly involved in neurite growth.
| ID | Neurite growth promoting factor | Short description | Secretion via/localization | Reference |
|---|---|---|---|---|
| O00468 | Agrin | Heparan sulfate basal lamina glycoprotein | signal peptide | 72 |
| O15123 | Angiopoietin-2 | Growth factor | signal peptide | 40 |
| P10909 | Clusterin (Clu) | Extracellular chaperone | signal peptide | 60–61 |
| P09603 | Colony-stimulating factor 1 (CSF-1) | Cytokine | signal peptide | 55 |
| P07585 | Decorin | Matrix proteoglycan | signal peptide | 46 |
| P23142 | Fibulin-1 | ECM-associated | signal peptide | 47 |
| P06396 | Gelsolin | Actin-binding protein | signal peptide | 80 |
| P07093 | Glia-derived nexin (serpinE2) | Secreted serine protease inhibitor | signal peptide | 57 |
| P28799 | Granulins | Secreted glycosylated peptide, cytokine-like activity | signal peptide | 68–69 |
| Q99988 | Growth/differentiation factor 15 (GDF-15) | Growth factor | signal peptide | 41 |
| P51858 | Hepatoma-derived growth factor (HDGF) | Growth factor | extracellular | 42 |
| P14174 | Macrophage migration inhibitory factor (MIF) | Pro-inflammatory cytokine | non-classical pathway | 56 |
| P55145 | Mesencephalic astrocyte-derived neurotrophic factor (MANF) | Neurotrophic factor | signal peptide | 37 |
| Q9HB63 | Netrin-4 | ECM-associated | signal peptide | 48 |
| Q9UMX5 | Neudesin | Secreted neurotrophic factor | signal peptide | 38 |
| Q8TB73 | Neuron-derived neurotrophic factor (NDNF) | Neurotrophic factor | signal peptide | 39 |
| Q7Z3B1 | Neuronal growth regulator 1 | Membrane-associated | signal peptide | 49 |
| Q9Y639 | Neuroplastin | Type I transmembrane protein | signal peptide | 50–51 |
| Q99574 | Neuroserpin | Secreted serine protease inhibitor | signal peptide | 58 |
| P14543 | Nidogen-1 | Sulfated glycoprotein in basal lamina | signal peptide | 71 |
| O95897 | Noelin-2 | secreted glycoprotein | signal peptide | 52 |
| Q15063 | Periostin | ECM-associated | signal peptide | 67 |
| P36955 | Pigment epithelium-derived factor (PEDF) | Secreted neurotrophic factor | signal peptide | 84–86 |
| P05121 | Plasminogen activator inhibitor 1 (serpinE1) | Secreted serine protease inhibitor | signal peptide | 59 |
| P04085 | Platelet-derived growth factor (PDGFA) | Growth factor | signal peptide | 43 |
| P09486 | Secreted protein, acidic, cysteine-rich (SPARC) | Matrix-asscociated | signal peptide | 53 |
| P48061 | Stromal cell-derived factor 1 (SDF-1) | Chemokine | signal peptide | 14–15 |
| P24821 | Tenascin | ECM-associated | signal peptide | 54 |
| P07996 | Thrombospondin-1 (TSP-1) | Adhesive glycoprotein | signal peptide | 63 |
| P01137 | Transforming growth factor, beta-1 (TGF-b1) | Growth factor | signal peptide | 45 |
| P61812 | Transforming growth factor, beta-2 (TGF-b2) | Growth factor | signal peptide | 44 |
Clustering of USSC Secreted Proteins into Biological Processes
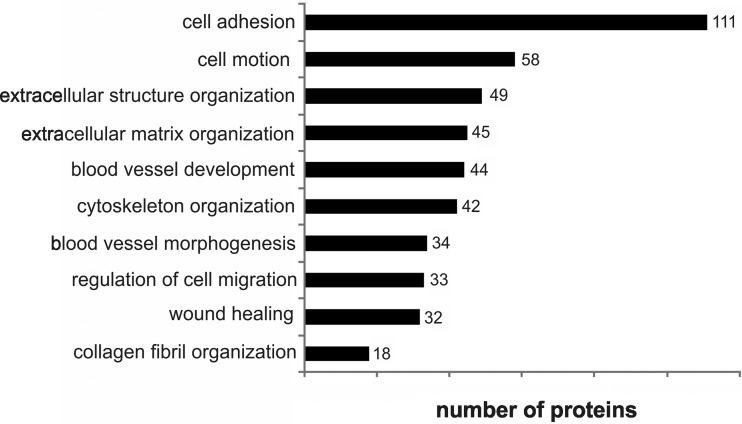
In order to gain an understanding of the complex composition of the USSC secretome, we assigned all proteins (1156 proteins) to biological processes by UniProt annotation resulting in biological processes associated with regeneration after spinal cord injury. USSC secreted proteins were assigned to cell communication (455 proteins), development (426 proteins), cell differentiation (307 proteins), cell proliferation (170 proteins), and cell growth (53 proteins) suggesting that USSC transplantation affects these biological processes relevant for nerve regeneration (Fig. 1C, multiple protein entries possible). Next, we performed enrichment analysis using DAVID bioinformatics database (DAVID Bioinformatics Resources 6.7, (33)) to reveal biological processes overrepresented in USSC secretome. Several biological processes were identified to be significantly enriched that are associated with neurite growth, neuronal differentiation and survival as well as nerve regeneration, e.g. cell adhesion (111 proteins), cell motion (58 proteins), cytoskeleton organization (42 proteins), and extracellular matrix organization (45 proteins) (Fig. 2 and supplemental Table S3). Numerous proteins involved in cell adhesion expressed by USSC including cadherins, neuronal growth regulator 1, neuropilin 1, neuroplastin, roundabout axon guidance receptor and tenascin C are known to have a strong impact on nerve regeneration. Moreover, the USSC secretome included proteins associated with the functional category blood vessel development (44 proteins). The latter is known to be important for regeneration after neuronal trauma, containing angiopoietin 2, SDF-1, matrix metallopeptidases MMP-2, MMP-14, MMP-19, platelet-derived growth factor A (PDGFA), and TGF-β2. In addition, the biological process collagen fibril organization (18 proteins) included proteins associated with extracellular matrix, e.g. lumican as well as aggrecan and TGF-β2.
Fig. 2.
Subset of biological processes associated with neurite growth, neuron differentiation, survival and regeneration enriched in comparison to all secreted proteins. All proteins identified in the USSC secretome by LC-MS/MS excluding contaminants were included (multiple protein entries possible). GO terms were evaluated with DAVID database with a false discoverer rate (FDR) p < 0.05.
Quantification of USSC Secreted Proteins
Further we were interested to determine the concentration of identified proteins involved in relevant biological processes of neuronal regeneration. Therefore, we determined the protein concentrations based on a publication by Silva et al. (34) which enabled us to quantify 1020 proteins. For the USSC secretome we found that the protein concentrations varied in the range of 2–3 orders of magnitude with fibronectin as the highest abundant (88 pmol/ml) and quinone oxidoreductase (2 fmol/ml) as the lowest abundant protein detected by MS (supplemental Table S4). Quantified proteins were ranked into three classes based on their abundance. Enrichment analysis of the three abundance classes by DAVID bioinformatics database thereby using all secreted proteins as background revealed that the high abundant proteins (class I) are linked to extracellular matrix organization and ectoderm development (Fig. 3). Class II included proteins likewise associated with extracellular matrix organization but also with cytoskeleton organization and actin filament-based processes whereas class III proteins are linked to biological adhesion and cell adhesion. Analysis further revealed that 26 of the 31 identified neurite growth promoting factors were quantifiable and evenly distributed in class II (71–2457 fmol/ml) and class III (2–71 fmol/ml). Class II included growth factors and extracellular matrix (ECM) associated proteins whereas class III mainly included growth factors and small cytokine-like proteins. In class I, four neurite growth promoting proteins (2–88 pmol/ml) have been found that are all associated with ECM.
Fig. 3.
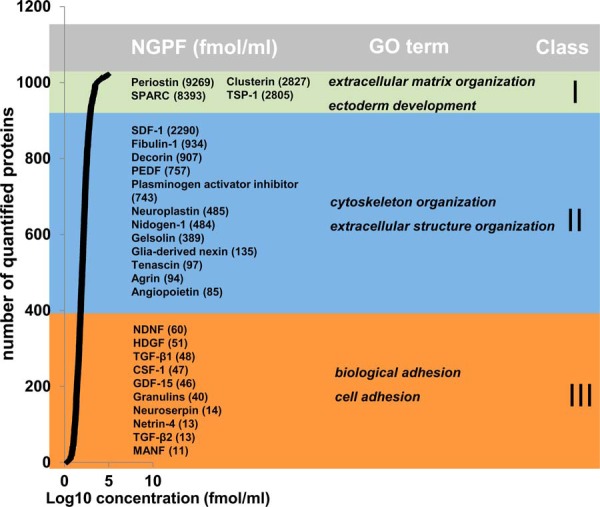
Concentration range of neurite growth promoting factors within USSC secretome. Protein concentrations (Log10 fmol/ml) of USSC secretome were ranked into three classes. For each class neurite growth promoting factors (NGPF) with corresponding concentration (fmol/ml) were assigned. Enrichment analysis was applied for each abundance class. GO terms important for regeneration are indicated.
Validation of Potential Neurite Growth Promoting Factors in a Neurite Outgrowth Assay
Neutralization of Candidate Neurite Growth Promoting Factors
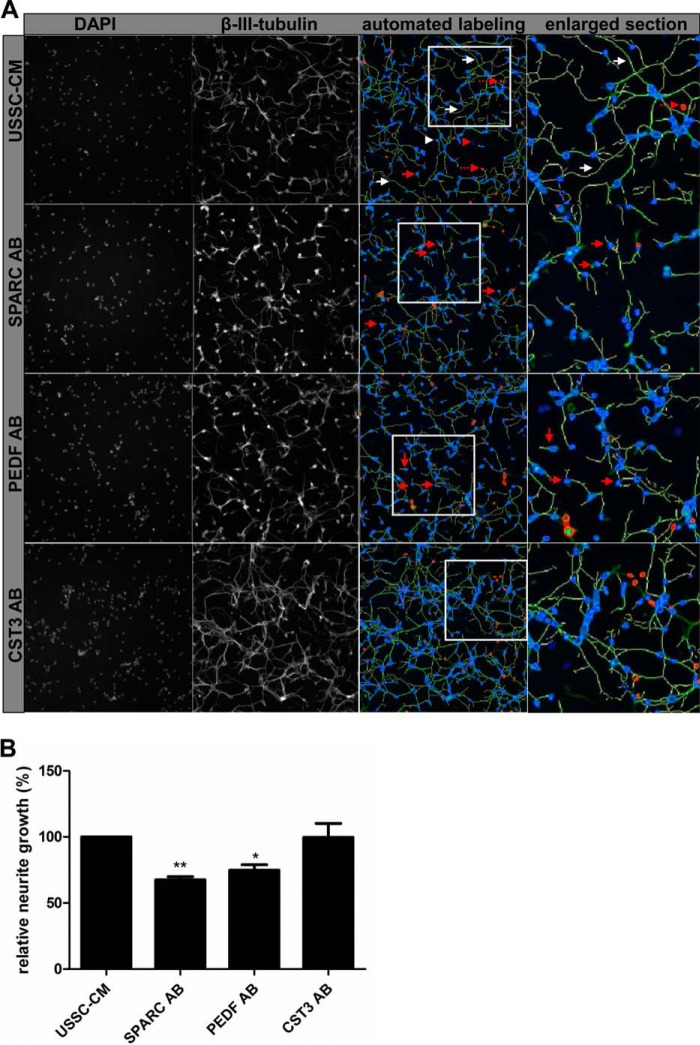
With intend to validate neurite growth promoting function of potential candidates we investigated selected proteins assigned to first two abundance classes in a neurite outgrowth assay in vitro. Therefore, we inhibited functionality of candidate proteins by neutralizing antibodies applied to USSC-CM 2 h before application to primary rat cortical neurons. Cortical neurons were incubated for 48 h, fixed and stained with a neuron-specific antibody (β-III-tubulin) to visualize cell bodies and neurites and analyzed by automated scanning with a Cellomics® ArrayScan® VTI HCS Reader. Astrocyte conditioned medium known to enhance neuronal survival and neurite growth in vitro (11, 36, 62) served as positive control for culture preparations in each experiment (data not shown). Our data indicate that neutralization of SPARC as well as PEDF (also known as serpinF1) in USSC-CM was effective with 4 μg/ml neutralizing antibody each resulting in significantly decreased neurite growth compared with control cultures treated with USSC-CM (Fig. 4). Neurites were significantly shorter (67.6% ± 2.3 as well as 74.8% ± 4.1 of control) because of neutralization of SPARC and PEDF, respectively. Neutralizing antibodies against cystatin C (CST3) (Fig. 4) had no effect on neurite growth, which also indicates that application of immunoglobulin does not in general suppress neurite growth. Cell survival was unchanged upon neutralization of proteins (data not shown).
Fig. 4.
Reduced neurite growth after immune neutralization of neurite growth promoting factors. A, Neutralization of SPARC and PEDF leads to reduced neurite growth of cortical neurons compared with USSC-CM. Neutralization of CST3 had no significant effect on neurite growth. Red arrow: neuron with short neurite; white arrow: long neurite; red arrowhead: nuclei but no neuron, excluded from quantification; white arrowhead: counted cell body of a neuron; interrupted red arrow: artifact excluded from quantification. Neurites labeled by the Neuronal Profiling v3.5 BioApplication software (yellow labeled neurites) were included into quantification. See also enlarged sections for further details. B, Quantification of relative neurite length (%) compared with USSC-CM control. Results derived from at least three independent experiments are presented as mean values ± SEM. * p < 0.05, ** p < 0.01 (Student′s t test).
Stimulation of Neurite Growth by Application of Candidate Neurite Growth Promoting Factors
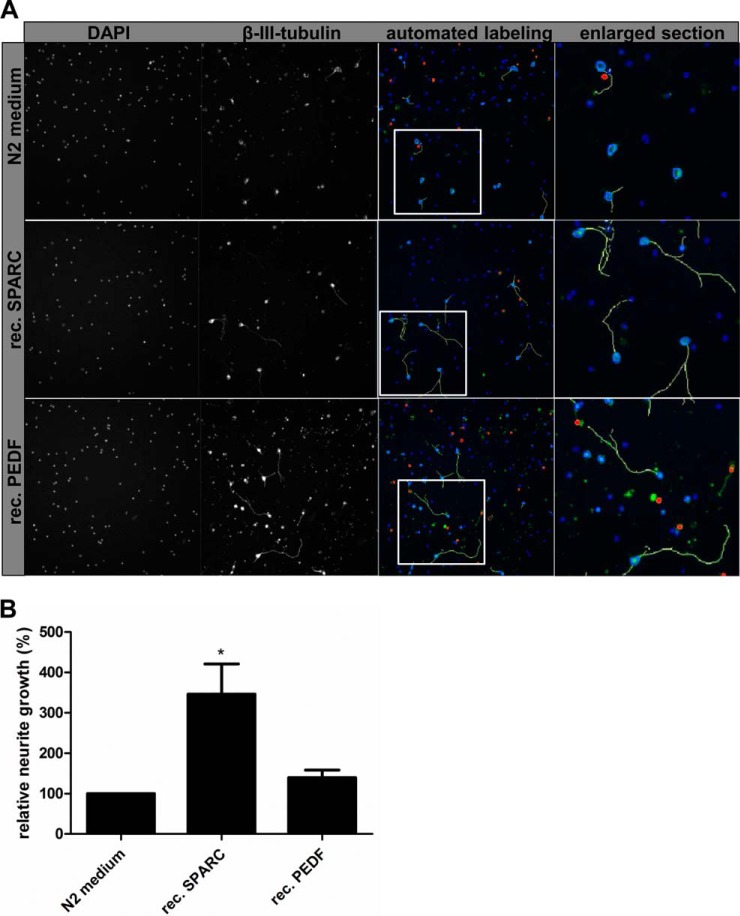
To further validate candidate proteins, we applied recombinant proteins in N2 (negative) control medium on primary cortical neurons and analyzed neurite growth after 48 h of incubation. Recombinant SPARC led to significantly enhanced neurite growth after application on cortical neurons up to 364.6% ± 74.4 (Fig. 5). PEDF application alone had a small effect on neurite growth (139.7% ± 18.6, p = 0.07), which did not meet our significance criteria. Again, cell survival was not influenced by application of recombinant proteins (data not shown). In summary, neutralization of SPARC and PEDF in USSC-CM resulted in significantly decreased neurite growth. However, the decline in neurite growth did not reach the low basal levels of N2 control medium. We demonstrated further that application of recombinant SPARC to N2 medium enhanced neurite growth, but less strong than USSC conditioned medium.
Fig. 5.
Enhanced neurite growth by application of recombinant proteins. A, Application of recombinant SPARC leads to enhanced neurite growth of cortical neurons compared with N2 control medium. Application of recombinant PEDF has only minor effects on neurite growth. Neurites labeled by the Neuronal Profiling v3.5 BioApplication software (yellow labeled neurites) were included into quantification. See also enlarged sections for further details. B, Quantification of relative neurite length (%) compared with N2 control medium. Results derived from at least three independent experiments are presented as mean values ± SEM. * p < 0.05 (Student′s t test).
DISCUSSION
USSC are a promising tool to support regeneration after spinal cord injury (SCI), because this stem cell type was previously shown to improve axonal regeneration and tissue sparing after transplantation into an acute rat SCI model (11). Beside axonal regeneration, support of cell survival and modulation of the ECM as well as angiogenesis are main targets for therapeutic interventions after SCI. As USSC secrete factors enhancing neurite growth and neuronal survival in vitro, we were interested to identify and characterize these factors in the USSC secretome to gain insight into potential paracrine mechanisms leading to enhanced regeneration after USSC transplantation. In the present study we established a USSC secretome comprising 1156 proteins and revealed that USSC release at least 31 well-established neurite growth promoting factors. In addition, USSC release proteins that are related to several biological processes involved in nerve regeneration. We were able to functionally validate two of the candidate neurite growth promoting factors by neurite outgrowth assays in vitro validating our proteomic approach and confirming a paracrine mechanism of regeneration support for transplanted USSC in SCI.
Secretion of Proteins Involved in Neurite Growth or Axonal Regeneration
By means of secretome approach applying LC-MS/MS we determined 1156 proteins, whereof at least 31 proteins in the USSC secretome are already known to be associated with neurite growth or axonal regeneration. The group of 31 proteins include SPARC and thrombospondin-1 (TSP-1), which together with tenascin C modulate interactions with ECM molecules (63). We further identified netrin-4 known to attract or repel growing axons depending on neuron type (48). PDGF and TGF-β1, both secreted by USSC, are known to interact with SPARC (64–66), indicating a solid network between the secreted factors. Moreover, USSC secrete MIF, which directs the initial neurite outgrowth from the statoacoustic ganglion (SAG) to the developing inner ear (56), and NDNF, which could together with MANF, HDGF, and neudesin stimulate neurite growth as well as migration, cell growth, and survival (37–39, 42). Recently, periostin, which is normally expressed in bone and muscles, has been shown to play an essential role in neurite growth and axonal regeneration after SCI to overcome the inhibitory effect of scar-associated molecules by signaling through focal adhesion kinase and Akt (67). Progranulin has similar characteristics like neurotrophins as it is cleaved into mature granulins by extracellular proteases and has been shown to stimulate neurite growth (68–69). TGF-β2, which is an immunomodulatory factor, normally facilitating ECM synthesis resulting in enhanced scar formation has also been shown to stimulate neurite growth in cultured dorsal root ganglion neurons (44). Using Hi-N method we showed that the identified proteins are distributed over a concentration range from fmol/ml - pmol/ml. As expected, growth factors were present at low concentration (fmol/ml range), whereas ECM associated proteins were highly abundant (pmol/ml range) in the USSC secretome. However, it has been shown that this kind of quantification method is associated with large quantification errors of small proteins (70), which likely depend on limited number of peptides as well as protein-to-protein variation. As our experiments were not designed for protein quantification in the first instance we took prior advantage of Hi-N method allowing quantification of hundreds of proteins in parallel to get a first impression about the distribution of proteins' concentration in USSC secretome. Although, this global approach gave acceptable results with mean standard deviation from 49–115% concentrations of specific proteins need confirmation by complementary techniques.
Altogether, these different neurite growth promoting factors, which were expressed by USSC, either alone or in combination, are likely to participate in neurite growth stimulation of cultured primary cortical neurons as well as to support axonal regrowth observed after USSC transplantation into the injured spinal cord. The combined secretion of neurite growth promoting factors as well as inhibitory proteins led to enhanced neurite growth as well as to axonal regeneration indicating an over-all supportive balance of USSC secreted proteins for regeneration associated processes.
Native USSC cultured in serum-containing medium were shown to release several cytokines and growth factors (13). In the present study, expression of CSF and SDF-1α, which are known to be directly or indirectly involved in neurite growth or axonal regeneration (14–15, 55), was confirmed under serum-free conditions. However, expression of the other proteins identified in the former work of Kögler et al. (13) was not detected in our experiments, suggesting changes in the USSC expression profile under different culture conditions as well as the different detection modalities of the applied analytical platforms.
USSC secrete proteins involved in multiple regeneration associated processes
To establish the USSC secretome we performed rigorous bioinformatics data analysis. We considered accessible knowledge about proven extracellular localization and further we took advantage of the two bioinformatics tools SignalP and SecretomeP allowing prediction of classical and non-classical protein secretion, respectively. From the 1520 proteins identified by LC-MS/MS 1156 are known or were predicted to be secreted including 385 classically secreted proteins and 276 proteins secreted via nonclassical pathways. Three hundred and sixty-four proteins did not match our criteria and were removed from our candidate list. We are confident that our approach of combining experimental data with bioinformatics prediction tools is straightforward to establish USSC secretome. This holds true especially for classical protein secretion by signal peptide that is well studied and prediction algorithms like e.g. SignalP were successfully proven. In contrast, SecretomeP, which, based on simplified assumption that extracellular proteins share the same characteristics, have to be taken with caution and should only be applied in combination with experimental data.
Secretion of Proteins Involved in Extracellular Matrix Organization
Further data interrogation by annotation clustering revealed that USSC secrete proteins are involved in differentiation and developmental processes. For more detailed characterization of the USSC secretome, proteins were assigned GO terms by DAVID bioinformatics database revealing that USSC secrete proteins associated with biological processes involved in regeneration after SCI.
Several proteins of the USSC secretome were assigned to cell adhesion, extracellular structure organization, and extracellular matrix organization, e.g. the chondroitin sulfate proteoglycan (CSPG) aggrecan is known to inhibit axonal regrowth but also the growth promoting decorin, which antagonizes scar formation (46). USSC further secrete laminin subunits, which might promote neural differentiation and migration of neural precursor cells after CNS lesion and together with nidogen-1 could guide growth cone turning (71) as well as the heparan sulfate proteoglycan agrin, which is involved in synaptogenesis during regeneration (72). Thus, our results suggest that USSC strongly interact with the local environment after transplantation into the injured spinal cord via modifications of ECM structures as well as cell-cell-contact. Previous studies described that even in the presence of inhibitory molecules successful axon growth after injury were achieved when a prevalence of growth permissive signals is present (73–74). Thus, USSC release ECM proteins that seem to shift the balance toward an axon growth permissive microenvironment thereby enhancing axonal regrowth in vivo.
Secretion of Proteins Involved in Angiogenesis
Beside axonal regrowth, regulation of cell adhesion and ECM modulation, angiogenesis is essential for proper regeneration as neurons have high metabolic requirements. Despite massive blood vessel sprouting, new blood vessels exhibit abnormal permeability after SCI suggesting limited functionality (75). USSC are likely to promote angiogenesis because these cells secrete several proteins assigned to blood vessel development or blood vessel morphogenesis, e.g. angiopoietin-2, angiopoietin-like 4, MMP-2, −14, and −19, PDGF, and TGF-β, which together with guidance molecules such as netrin-4, semaphorin-7 and slit-3, could enhance blood vessel formation during regeneration.
USSC further secrete proteins grouped into GO terms cell motion and regulation of cell migration indicating that USSC might influence migration of endogenous neural precursor cells and endothelial as well as immune cells. Enhanced migration of precursor cells derived from the ependymal zone of the central canal could led to better recovery because these cells have been shown to differentiate into oligodendroglial cells (76). Because the SDF-1/CXCR4 axis is suggested to play a key role in migration and differentiation of adult endogenous stem cells and oligodendrocyte precursor cells after SCI (77–79), USSC expressed SDF-1 could influence precursor cell properties. In addition, secreted CSF-1, laminin subtypes, neuropilin-1, PDGF, migration and invasion enhancer 1 (MIEN-1) and TGF-β could regulate migration of precursor cells, astrocytes, fibroblasts, and immune cells, which contribute to proper regeneration.
Secretion of Proteins Involved in Cytoskeletal Organization
Most of the above mentioned proteins are known to be secreted via the classical pathway. In addition, the USSC secretome includes several proteins predicted to be either secreted by nonclassical pathways or annotated as extracellular by UniProt database. These proteins were found to be enriched in biological processes cytoskeleton organization, e.g. capping proteins, actin-related protein complexes, and gelsolin, which is known to play a role in neurite growth (80). Together with syntaxin, synaptotagmin, and kinesins, which were also found in the USSC secretome these proteins potentially result from exocytosis processes as these proteins are known to be involved in exocytosis. Furthermore, cytoskeleton proteins could originate from shedding because of their localization at the cell membrane, in particular ezrin and moesin both identified in the USSC secretome that link cell surface receptors and the actin cytoskeleton. It is unclear whether these proteins were secreted under certain conditions (25), but we cannot exclude that these proteins partially originate from dead cells. However, the biological processes related to the cytoskeleton included classically secreted proteins normally directly or indirectly regulating the dynamic remodeling of actin cytoskeleton (PDGF, SDF-1) indicating that USSC could also influence cytoskeleton dynamics of regenerating axons. By protein quantification we revealed that biological processes like, e.g. extracellular matrix organization and ectoderm development are enriched among highly abundant proteins, whereas proteins involved in cell adhesion are enriched within the group of low abundant proteins. We only can speculate if the abundance of a protein or a protein family is a measure for relevance of the associated biological process because both low abundant proteins like neurotrophins and high abundant ECM proteins are known to be critical for regeneration.
Functional Analysis of Candidate Proteins
SPARC is one of the most abundant proteins identified in the USSC secretome. It is a multifunctional glycoprotein that binds to collagens and vitronectins and modulates the activity of PDGF, FGF-2 and VEGF. It was further demonstrated that SPARC stimulates Schwann cells to facilitate axonal regeneration via laminin-1 and TGF-β1 signaling (53). Schwann cells also express SPARC, which have been shown to promote both neurite growth and survival of purified retinal ganglion cells (81). SPARC is discussed to have synergistic effects on classical neurotrophins enhancing neurite outgrowth of retinal ganglion cells (82–83). USSC-CM has been shown to stimulate neurite growth of primary cortical neurons (11). In the present study, we demonstrated that neurite growth is significantly decreased when a SPARC neutralizing antibody is applied to USSC-CM. On cortical neurons, recombinant SPARC directly enhances neurite growth. Therefore, we conclude that cortical neurons respond to SPARC signaling and that SPARC is one of the major neurite growth promoting factors secreted by USSC.
Recombinant PEDF (serpinF1) has been applied at low concentration representing the rather low amounts identified in USSC-CM, resulting only in a slightly increased neurite growth. On the other hand, neutralization resulted in a significant decrease in neurite growth, similar in range as SPARC antibody, indicating that PEDF may act in combination with other USSC secreted factors to stimulate cortical neurite growth. PEDF has been shown to enhance neuronal survival as well as neurite growth of retinoblastoma cells (84), cerebellar granule cells (85) and spinal motor neurons (86). Under our culture conditions, SPARC and PEDF did not affect neuronal survival of cortical neurons, indicating that other USSC secreted proteins promote neuron survival of cultured cortical neurons. In vivo, both proteins are likely to stimulate axonal regrowth directly but could also have indirect effects on regeneration via stimulation of glial cells or cell survival.
CONCLUSIONS
The lack of regeneration after SCI might be overcome by transplantation of stem cells directly into the lesion site, which locally release trophic factors and thus support axonal regrowth. The present study demonstrates that the regenerative phenotype of USSC secretome bases on a complex network of proteins associated with several biological processes that lead to enhanced neurite growth in vitro and therefore suggest a relevant role on axonal regeneration in vivo. The complex network of trophic factors secreted by USSC seems to act synergistically on several levels of nerve regeneration. With this observation it appears that transplantation of somatic stem cells instead or in combination with defined factors has great potential in regenerative medicine.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Ellen Fritsche (IUF - Leibniz-Institut für Umweltmedizinische Forschung, Düsseldorf) for providing the Thermo Scientific Cellomics® ArrayScan® VTI HCS Reader, Eva Bruns for technical support and Gereon Poschmann for bioinformatical support.
Footnotes
Author contributions: J.S., H.F., H.W.M., and K.S. designed research; J.S., H.F., and M.H. performed research; G.K. and H.E.M. contributed new reagents or analytic tools; J.S., H.F., and D.M.W. analyzed data; J.S. and K.S. wrote the paper; H.W.M. and K.S. supervised the study.
* The study was supported by German Research Council Deutsche Forschungsgemeinschaft (DFG) (grant number MU 630/10–1), Christiane and Claudia Hempel Foundation for Clinical Stem Cell Research and Forschungskommission of Heinrich Heine University Düsseldorf.
 This article contains supplemental Tables S1 to S4.
This article contains supplemental Tables S1 to S4.
1 The abbreviations used are:
- hUCB
- human umbilical cord blood
- BDNF
- brain derived neurotrophic factor
- CSPG
- chondroitin sulfate; proteoglycan
- CST3
- Cystatin C
- CSF-1
- colony-stimulating factor 1
- CV
- coefficient of variation
- DAPI
- 4′,6-diamidino-2-phenylindole
- ECM
- extracellular matrix
- FDR
- false discoverer rate
- FBS
- fetal bovine serum
- FGF-2
- fibroblast growth factor-2
- GO
- gene ontology
- GDNF
- glial derived neurotrophic factor
- GMP
- good manufacturing practice
- GDF-15
- growth/differentiation factor 15
- HDGF
- hepatoma-derived growth factor
- CB-MSC
- derived MSC
- LIF
- leukemia inhibitory factor
- MIF
- macrophage migration inhibitory factor
- MMP
- matrix metallopeptidases
- MANF
- mesencephalic astrocyte-derived neurotrophic factor
- BM-MSC
- mesenchymal stem cells from bone marrow
- MIEN-1
- migration and invasion enhancer 1
- NGF
- nerve growth factor
- NGPF
- neurite growth promoting factor
- NDNF
- neuron-derived neurotrophic factor
- NT-3
- neurotrophin-3
- PEDF
- pigment epithelium-derived factor
- PDGFA
- platelet-derived growth factor A
- p0
- postnatal day 0
- rec
- recombinant
- SPARC
- secreted protein acidic and rich in cysteine
- SCI
- spinal cord injury
- SDF-1
- stromal derived factor-1
- TSP-1
- thrombospondin-1
- TGF-β
- transforming growth factor-beta
- USSC
- unrestricted somatic stem cells
- USSC-CM
- USSC conditioned medium
- VEGF
- vascular endothelial growth factor.
REFERENCES
- 1. Blight A. R. (1985) Delayed demyelination and macrophage invasion: a candidate for secondary cell damage in spinal cord injury. Central Nervous Syst. Trauma 2, 299–315 [DOI] [PubMed] [Google Scholar]
- 2. Popovich P. G., Guan Z., Wei P., Huitinga I., van Rooijen N., Stokes B. T. (1999) Depletion of hematogenous macrophages promotes partial hindlimb recovery and neuroanatomical repair after experimental spinal cord injury. Exp. Neurol. 158, 351–365 [DOI] [PubMed] [Google Scholar]
- 3. Hauben E., Nevo U., Yoles E., Moalem G., Agranov E., Mor F., Akselrod S., Neeman M., Cohen I. R., Schwartz M. (2000) Autoimmune T cells as potential neuroprotective therapy for spinal cord injury. Lancet 355, 286–287 [DOI] [PubMed] [Google Scholar]
- 4. Kogler G., Sensken S., Airey J. A., Trapp T., Muschen M., Feldhahn N., Liedtke S., Sorg R. V., Fischer J., Rosenbaum C., Greschat S., Knipper A., Bender J., Degistirici O., Gao J., Caplan A. I., Colletti E. J., Almeida-Porada G., Muller H. W., Zanjani E., Wernet P. (2004) A new human somatic stem cell from placental cord blood with intrinsic pluripotent differentiation potential. J. Exp. Med. 200, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van den Berk L. C., Jansen B. J., Siebers-Vermeulen K. G., Netea M. G., Latuhihin T., Bergevoet S., Raymakers R. A., Kogler G., Figdor C. C., Adema G. J., Torensma R. (2009) Toll-like receptor triggering in cord blood mesenchymal stem cells. J. Cell Mol. Med. 13, 3415–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winter M., Wang X. N., Daubener W., Eyking A., Rae M., Dickinson A. M., Wernet P., Kogler G., Sorg R. V. (2009) Suppression of cellular immunity by cord blood-derived unrestricted somatic stem cells is cytokine-dependent. J. Cell Mol. Med. 13, 2465–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jansen B. J., Gilissen C., Roelofs H., Schaap-Oziemlak A., Veltman J. A., Raymakers R. A., Jansen J. H., Kogler G., Figdor C. G., Torensma R., Adema G. J. (2010) Functional differences between mesenchymal stem cell populations are reflected by their transcriptome. Stem Cells Dev. 19, 481–490 [DOI] [PubMed] [Google Scholar]
- 8. Kluth S. M., Buchheiser A., Houben A. P., Geyh S., Krenz T., Radke T. F., Wiek C., Hanenberg H., Reinecke P., Wernet P., Kogler G. (2010) DLK-1 as a marker to distinguish unrestricted somatic stem cells and mesenchymal stromal cells in cord blood. Stem Cells Dev. 19, 1471–1483 [DOI] [PubMed] [Google Scholar]
- 9. Liedtke S., Buchheiser A., Bosch J., Bosse F., Kruse F., Zhao X., Santourlidis S., Kogler G. (2010) The HOX Code as a “biological fingerprint” to distinguish functionally distinct stem cell populations derived from cord blood. Stem Cell Res. 5, 40–50 [DOI] [PubMed] [Google Scholar]
- 10. Aktas M., Buchheiser A., Houben A., Reimann V., Radke T., Jeltsch K., Maier P., Zeller W. J., Kogler G. (2010) Good manufacturing practice-grade production of unrestricted somatic stem cell from fresh cord blood. Cytotherapy 12, 338–348 [DOI] [PubMed] [Google Scholar]
- 11. Schira J., Gasis M., Estrada V., Hendricks M., Schmitz C., Trapp T., Kruse F., Kogler G., Wernet P., Hartung H. P., Muller H. W. (2012) Significant clinical, neuropathological and behavioural recovery from acute spinal cord trauma by transplantation of a well-defined somatic stem cell from human umbilical cord blood. Brain 135, 431–446 [DOI] [PubMed] [Google Scholar]
- 12. Greschat S., Schira J., Kury P., Rosenbaum C., de Souza Silva M. A., Kogler G., Wernet P., Muller H. W. (2008) Unrestricted somatic stem cells from human umbilical cord blood can be differentiated into neurons with a dopaminergic phenotype. Stem Cells Dev. 17, 221–232 [DOI] [PubMed] [Google Scholar]
- 13. Kogler G., Radke T. F., Lefort A., Sensken S., Fischer J., Sorg R. V., Wernet P. (2005) Cytokine production and hematopoiesis supporting activity of cord blood-derived unrestricted somatic stem cells. Exp. Hematol. 33, 573–583 [DOI] [PubMed] [Google Scholar]
- 14. Heskamp A., Leibinger M., Andreadaki A., Gobrecht P., Diekmann H., Fischer D. (2013) CXCL12/SDF-1 facilitates optic nerve regeneration. Neurobiol. Dis. 55, 76–86 [DOI] [PubMed] [Google Scholar]
- 15. Opatz J., Kury P., Schiwy N., Jarve A., Estrada V., Brazda N., Bosse F., Muller H. W. (2009) SDF-1 stimulates neurite growth on inhibitory CNS myelin. Mol. Cell. Neurosci. 40, 293–300 [DOI] [PubMed] [Google Scholar]
- 16. Blesch A., Uy H. S., Grill R. J., Cheng J. G., Patterson P. H., Tuszynski M. H. (1999) Leukemia inhibitory factor augments neurotrophin expression and corticospinal axon growth after adult CNS injury. J. Neurosci. 19, 3556–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khaibullina A. A., Rosenstein J. M., Krum J. M. (2004) Vascular endothelial growth factor promotes neurite maturation in primary CNS neuronal cultures. Brain Res. Dev. Brain Res. 148, 59–68 [DOI] [PubMed] [Google Scholar]
- 18. Kim H. M., Hwang D. H., Lee J. E., Kim S. U., Kim B. G. (2009) Ex vivo VEGF delivery by neural stem cells enhances proliferation of glial progenitors, angiogenesis, and tissue sparing after spinal cord injury. PLoS ONE 4, e4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakano N., Nakai Y., Seo T. B., Yamada Y., Ohno T., Yamanaka A., Nagai Y., Fukushima M., Suzuki Y., Nakatani T., Ide C. (2010) Characterization of conditioned medium of cultured bone marrow stromal cells. Neurosci. Lett. 483, 57–61 [DOI] [PubMed] [Google Scholar]
- 20. Crigler L., Robey R. C., Asawachaicharn A., Gaupp D., Phinney D. G. (2006) Human mesenchymal stem cell subpopulations express a variety of neuro-regulatory molecules and promote neuronal cell survival and neuritogenesis. Exp. Neurol. 198, 54–64 [DOI] [PubMed] [Google Scholar]
- 21. Gu W., Zhang F., Xue Q., Ma Z., Lu P., Yu B. (2012) Bone mesenchymal stromal cells stimulate neurite outgrowth of spinal neurons by secreting neurotrophic factors. Neurol. Res. 34, 172–180 [DOI] [PubMed] [Google Scholar]
- 22. Liu C. H., Hwang S. M. (2005) Cytokine interactions in mesenchymal stem cells from cord blood. Cytokine 32, 270–279 [DOI] [PubMed] [Google Scholar]
- 23. Arufe M. C., De la Fuente A., Mateos J., Fuentes I., De Toro F. J., Blanco F. J. (2011) Analysis of the chondrogenic potential and secretome of mesenchymal stem cells derived from human umbilical cord stroma. Stem Cells Dev. 20, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 24. Makridakis M., Roubelakis M. G., Vlahou A. (2013) Stem cells: insights into the secretome. Biochim. Biophys. Acta 1834, 2380–2384 [DOI] [PubMed] [Google Scholar]
- 25. Skalnikova H., Motlik J., Gadher S. J., Kovarova H. (2011) Mapping of the secretome of primary isolates of mammalian cells, stem cells and derived cell lines. Proteomics 11, 691–708 [DOI] [PubMed] [Google Scholar]
- 26. Bottenstein J. E., Sato G. H. (1979) Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc. Natl. Acad. Sci. U.S.A. 76, 514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chevallet M., Diemer H., Van Dorssealer A., Villiers C., Rabilloud T. (2007) Toward a better analysis of secreted proteins: the example of the myeloid cells secretome. Proteomics 7, 1757–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nesterenko M. V., Tilley M., Upton S. J. (1994) A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Meth. 28, 239–242 [DOI] [PubMed] [Google Scholar]
- 29. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 30. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bendtsen J. D., Kiemer L., Fausboll A., Brunak S. (2005) Non-classical protein secretion in bacteria. BMC Microbiol. 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Meth. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 33. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 34. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 35. Kappler J., Junghans U., Koops A., Stichel C. C., Hausser H. J., Kresse H., Muller H. W. (1997) Chondroitin/dermatan sulphate promotes the survival of neurons from rat embryonic neocortex. Eur. J. Neurosci. 9, 306–318 [DOI] [PubMed] [Google Scholar]
- 36. Schmalenbach C., Muller H. W. (1993) Astroglia-neuron interactions that promote long-term neuronal survival. J. Chem. Neuroanat. 6, 229–237 [DOI] [PubMed] [Google Scholar]
- 37. Voutilainen M. H., Back S., Porsti E., Toppinen L., Lindgren L., Lindholm P., Peranen J., Saarma M., Tuominen R. K. (2009) Mesencephalic astrocyte-derived neurotrophic factor is neurorestorative in rat model of Parkinson's disease. J. Neurosci. 29, 9651–9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura I., Konishi M., Miyake A., Fujimoto M., Itoh N. (2006) Neudesin, a secreted factor, promotes neural cell proliferation and neuronal differentiation in mouse neural precursor cells. J. Neurosci. Res. 83, 1415–1424 [DOI] [PubMed] [Google Scholar]
- 39. Kuang X. L., Zhao X. M., Xu H. F., Shi Y. Y., Deng J. B., Sun G. T. (2010) Spatio-temporal expression of a novel neuron-derived neurotrophic factor (NDNF) in mouse brains during development. BMC Neurosci. 11, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosa A. I., Goncalves J., Cortes L., Bernardino L., Malva J. O., Agasse F. (2010) The angiogenic factor angiopoietin-1 is a proneurogenic peptide on subventricular zone stem/progenitor cells. J. Neurosci. 30, 4573–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mensching L., Borger A. K., Wang X., Charalambous P., Unsicker K., Haastert-Talini K. (2012) Local substitution of GDF-15 improves axonal and sensory recovery after peripheral nerve injury. Cell Tissue Res. 350, 225–238 [DOI] [PubMed] [Google Scholar]
- 42. Marubuchi S., Okuda T., Tagawa K., Enokido Y., Horiuchi D., Shimokawa R., Tamura T., Qi M. L., Eishi Y., Watabe K., Shibata M., Nakagawa M., Okazawa H. (2006) Hepatoma-derived growth factor, a new trophic factor for motor neurons, is up-regulated in the spinal cord of PQBP-1 transgenic mice before onset of degeneration. J. Neurochem. 99, 70–83 [DOI] [PubMed] [Google Scholar]
- 43. Othberg A., Odin P., Ballagi A., Ahgren A., Funa K., Lindvall O. (1995) Specific effects of platelet derived growth factor (PDGF) on fetal rat and human dopaminergic neurons in vitro. Exp. Brain Res. 105, 111–122 [DOI] [PubMed] [Google Scholar]
- 44. Roet K. C., Franssen E. H., de Bree F. M., Essing A. H., Zijlstra S. J., Fagoe N. D., Eggink H. M., Eggers R., Smit A. B., van Kesteren R. E., Verhaagen J. (2013) A multilevel screening strategy defines a molecular fingerprint of proregenerative olfactory ensheathing cells and identifies SCARB2, a protein that improves regenerative sprouting of injured sensory spinal axons. J. Neurosci. 33, 11116–11135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walshe T. E., Leach L. L., D'Amore P. A. (2011) TGF-beta signaling is required for maintenance of retinal ganglion cell differentiation and survival. Neuroscience 189, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Minor K., Tang X., Kahrilas G., Archibald S. J., Davies J. E., Davies S. J. (2008) Decorin promotes robust axon growth on inhibitory CSPGs and myelin via a direct effect on neurons. Neurobiol. Dis. 32, 88–95 [DOI] [PubMed] [Google Scholar]
- 47. Cooley M. A., Kern C. B., Fresco V. M., Wessels A., Thompson R. P., McQuinn T. C., Twal W. O., Mjaatvedt C. H., Drake C. J., Argraves W. S. (2008) Fibulin-1 is required for morphogenesis of neural crest-derived structures. Dev. Biol. 319, 336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayano Y., Sasaki K., Ohmura N., Takemoto M., Maeda Y., Yamashita T., Hata Y., Kitada K., Yamamoto N. (2014) Netrin-4 regulates thalamocortical axon branching in an activity-dependent fashion. Proc. Natl. Acad. Sci. U.S.A. 111, 15226–15231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pischedda F., Szczurkowska J., Cirnaru M. D., Giesert F., Vezzoli E., Ueffing M., Sala C., Francolini M., Hauck S. M., Cancedda L., Piccoli G. (2014) A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization. Mol. Cell. Proteomics 13, 733–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Owczarek S., Kiryushko D., Larsen M. H., Kastrup J. S., Gajhede M., Sandi C., Berezin V., Bock E., Soroka V. (2010) Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor. FASEB J. 24, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 51. Owczarek S., Soroka V., Kiryushko D., Larsen M. H., Yuan Q., Sandi C., Berezin V., Bock E. (2011) Neuroplastin-65 and a mimetic peptide derived from its homophilic binding site modulate neuritogenesis and neuronal plasticity. J. Neurochem. 117, 984–994 [DOI] [PubMed] [Google Scholar]
- 52. Lee J. A., Anholt R. R., Cole G. J. (2008) Olfactomedin-2 mediates development of the anterior central nervous system and head structures in zebrafish. Mech. Dev. 125, 167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Au E., Richter M. W., Vincent A. J., Tetzlaff W., Aebersold R., Sage E. H., Roskams A. J. (2007) SPARC from olfactory ensheathing cells stimulates Schwann cells to promote neurite outgrowth and enhances spinal cord repair. J. Neurosci. 27, 7208–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gotz B., Scholze A., Clement A., Joester A., Schutte K., Wigger F., Frank R., Spiess E., Ekblom P., Faissner A. (1996) Tenascin-C contains distinct adhesive, anti-adhesive, and neurite outgrowth promoting sites for neurons. J. Cell Biol. 132, 681–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Su Y., Cui L., Piao C., Li B., Zhao L. R. (2013) The effects of hematopoietic growth factors on neurite outgrowth. PLoS ONE 8, e75562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bank L. M., Bianchi L. M., Ebisu F., Lerman-Sinkoff D., Smiley E. C., Shen Y. C., Ramamurthy P., Thompson D. L., Roth T. M., Beck C. R., Flynn M., Teller R. S., Feng L., Llewellyn G. N., Holmes B., Sharples C., Coutinho-Budd J., Linn S. A., Chervenak A. P., Dolan D. F., Benson J., Kanicki A., Martin C. A., Altschuler R., Koch A. E., Jewett E. M., Germiller J. A., Barald K. F. (2012) Macrophage migration inhibitory factor acts as a neurotrophin in the developing inner ear. Development 139, 4666–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Farmer L., Sommer J., Monard D. (1990) Glia-derived nexin potentiates neurite extension in hippocampal pyramidal cells in vitro. Develop. Neurosci. 12, 73–80 [DOI] [PubMed] [Google Scholar]
- 58. Parmar P. K., Coates L. C., Pearson J. F., Hill R. M., Birch N. P. (2002) Neuroserpin regulates neurite outgrowth in nerve growth factor-treated PC12 cells. J. Neurochem. 82, 1406–1415 [DOI] [PubMed] [Google Scholar]
- 59. Simon D., Martin-Bermejo M. J., Gallego-Hernandez M. T., Pastrana E., Garcia-Escudero V., Garcia-Gomez A., Lim F., Diaz-Nido J., Avila J., Moreno-Flores M. T. (2011) Expression of plasminogen activator inhibitor-1 by olfactory ensheathing glia promotes axonal regeneration. Glia 59, 1458–1471 [DOI] [PubMed] [Google Scholar]
- 60. Wicher G., Fex-Svenningsen A., Velsecchi I., Charnay Y., Aldskogius H. (2008) Extracellular clusterin promotes neuronal network complexity in vitro. Neuroreport 19, 1487–1491 [DOI] [PubMed] [Google Scholar]
- 61. Wright M. C., Mi R., Connor E., Reed N., Vyas A., Alspalter M., Coppola G., Geschwind D. H., Brushart T. M., Hoke A. (2014) Novel roles for osteopontin and clusterin in peripheral motor and sensory axon regeneration. J. Neurosci. 34, 1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muller H. W., Beckh S., Seifert W. (1984) Neurotrophic factor for central neurons. Proc. Natl. Acad. Sci. U.S.A. 81, 1248–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brekken R. A., Sage E. H. (2001) SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 19, 816–827 [DOI] [PubMed] [Google Scholar]
- 64. Francki A., Bradshaw A. D., Bassuk J. A., Howe C. C., Couser W. G., Sage E. H. (1999) SPARC regulates the expression of collagen type I and transforming growth factor-beta1 in mesangial cells. J. Biol. Chem. 274, 32145–32152 [DOI] [PubMed] [Google Scholar]
- 65. Raines E. W., Lane T. F., Iruela-Arispe M. L., Ross R., Sage E. H. (1992) The extracellular glycoprotein SPARC interacts with platelet-derived growth factor (PDGF)-AB and -BB and inhibits the binding of PDGF to its receptors. Proc. Natl. Acad. Sci. U.S.A. 89, 1281–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reed M. J., Vernon R. B., Abrass I. B., Sage E. H. (1994) TGF-beta 1 induces the expression of type I collagen and SPARC, and enhances contraction of collagen gels, by fibroblasts from young and aged donors. J. Cell. Physiol. 158, 169–179 [DOI] [PubMed] [Google Scholar]
- 67. Shih C. H., Lacagnina M., Leuer-Bisciotti K., Proschel C. (2014) Astroglial-derived periostin promotes axonal regeneration after spinal cord injury. J. Neurosci. 34, 2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gass J., Lee W. C., Cook C., Finch N., Stetler C., Jansen-West K., Lewis J., Link C. D., Rademakers R., Nykjaer A., Petrucelli L. (2012) Progranulin regulates neuronal outgrowth independent of sortilin. Mol. Neurodegener. 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Van Damme P., Van Hoecke A., Lambrechts D., Vanacker P., Bogaert E., van Swieten J., Carmeliet P., Van Den Bosch L., Robberecht W. (2008) Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J. Cell Biol. 181, 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ahrne E., Molzahn L., Glatter T., Schmidt A. (2013) Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 13, 2567–2578 [DOI] [PubMed] [Google Scholar]
- 71. Bonner J., O'Connor T. P. (2001) The permissive cue laminin is essential for growth cone turning in vivo. J. Neurosci. 21, 9782–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Falo M. C., Reeves T. M., Phillips L. L. (2008) Agrin expression during synaptogenesis induced by traumatic brain injury. J. Neurotrauma 25, 769–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jones L. L., Sajed D., Tuszynski M. H. (2003) Axonal regeneration through regions of chondroitin sulfate proteoglycan deposition after spinal cord injury: a balance of permissiveness and inhibition. J. Neurosci. 23, 9276–9288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lu P., Jones L. L., Tuszynski M. H. (2007) Axon regeneration through scars and into sites of chronic spinal cord injury. Exp. Neurol. 203, 8–21 [DOI] [PubMed] [Google Scholar]
- 75. Whetstone W. D., Hsu J. Y., Eisenberg M., Werb Z., Noble-Haeusslein L. J. (2003) Blood-spinal cord barrier after spinal cord injury: relation to revascularization and wound healing. J. Neurosci. Res. 74, 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meletis K., Barnabe-Heider F., Carlen M., Evergren E., Tomilin N., Shupliakov O., Frisen J. (2008) Spinal cord injury reveals multilineage differentiation of ependymal cells. PLos Biol. 6, e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gottle P., Kremer D., Jander S., Odemis V., Engele J., Hartung H. P., Kury P. (2010) Activation of CXCR7 receptor promotes oligodendroglial cell maturation. Ann. Neurol. 68, 915–924 [DOI] [PubMed] [Google Scholar]
- 78. Jaerve A., Bosse F., Muller H. W. (2012) SDF-1/CXCL12: its role in spinal cord injury. Int. J. Biochem. Cell Biol. 44, 452–456 [DOI] [PubMed] [Google Scholar]
- 79. Knerlich-Lukoschus F., von der Ropp-Brenner B., Lucius R., Mehdorn H. M., Held-Feindt J. (2010) Chemokine expression in the white matter spinal cord precursor niche after force-defined spinal cord contusion injuries in adult rats. Glia 58, 916–931 [DOI] [PubMed] [Google Scholar]
- 80. Furnish E. J., Zhou W., Cunningham C. C., Kas J. A., Schmidt C. E. (2001) Gelsolin overexpression enhances neurite outgrowth in PC12 cells. FEBS Lett. 508, 282–286 [DOI] [PubMed] [Google Scholar]
- 81. Bampton E. T., Ma C. H., Tolkovsky A. M., Taylor J. S. (2005) Osteonectin is a Schwann cell-secreted factor that promotes retinal ganglion cell survival and process outgrowth. Eur. J. Neurosci. 21, 2611–2623 [DOI] [PubMed] [Google Scholar]
- 82. Ma C. H., Bampton E. T., Evans M. J., Taylor J. S. (2010) Synergistic effects of osteonectin and brain-derived neurotrophic factor on axotomized retinal ganglion cells neurite outgrowth via the mitogen-activated protein kinase-extracellular signal-regulated kinase 1/2 pathways. Neuroscience 165, 463–474 [DOI] [PubMed] [Google Scholar]
- 83. Ma C. H., Palmer A., Taylor J. S. (2009) Synergistic effects of osteonectin and NGF in promoting survival and neurite outgrowth of superior cervical ganglion neurons. Brain Res. 1289, 1–13 [DOI] [PubMed] [Google Scholar]
- 84. Becerra S. P., Palmer I., Kumar A., Steele F., Shiloach J., Notario V., Chader G. J. (1993) Overexpression of fetal human pigment epithelium-derived factor in Escherichia coli. A functionally active neurotrophic factor. J. Biol. Chem. 268, 23148–23156 [PubMed] [Google Scholar]
- 85. Taniwaki T., Becerra S. P., Chader G. J., Schwartz J. P. (1995) Pigment epithelium-derived factor is a survival factor for cerebellar granule cells in culture. J. Neurochem. 64, 2509–2517 [DOI] [PubMed] [Google Scholar]
- 86. Houenou L. J., D'Costa A. P., Li L., Turgeon V. L., Enyadike C., Alberdi E., Becerra S. P. (1999) Pigment epithelium-derived factor promotes the survival and differentiation of developing spinal motor neurons. J. Comp. Neurol. 412, 506–514 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.