Abstract
Macrophages are crucial in controlling infectious agents and tissue homeostasis. Macrophages require a wide range of functional capabilities in order to fulfill distinct roles in our body, one being rapid and robust immune responses. To gain insight into macrophage plasticity and the key regulatory protein networks governing their specific functions, we performed quantitative analyses of the proteome and phosphoproteome of murine primary GM-CSF and M-CSF grown bone marrow derived macrophages (GM-BMMs and M-BMMs, respectively) using the latest isobaric tag based tandem mass tag (TMT) labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS). Strikingly, metabolic processes emerged as a major difference between these macrophages. Specifically, GM-BMMs show significant enrichment of proteins involving glycolysis, the mevalonate pathway, and nitrogen compound biosynthesis. This evidence of enhanced glycolytic capability in GM-BMMs is particularly significant regarding their pro-inflammatory responses, because increased production of cytokines upon LPS stimulation in GM-BMMs depends on their acute glycolytic capacity. In contrast, M-BMMs up-regulate proteins involved in endocytosis, which correlates with a tendency toward homeostatic functions such as scavenging cellular debris. Together, our data describes a proteomic network that underlies the pro-inflammatory actions of GM-BMMs as well as the homeostatic functions of M-BMMs.
Macrophages are a heterogeneous population of immune cells that are essential for the initiation and resolution of pathogen- or tissue damage-induced inflammation (1). They show remarkable plasticity that allows them to respond efficiently to environmental signals and change their phenotype and physiology upon cytokine and microbial signaling (2). These changes can give rise to populations of cells with distinct functions that are phenotypically characterized by the production of pro-inflammatory and anti-inflammatory cytokines (3). Among the growth factors that affect macrophage activation states, two cytokines that appear to be important in controlling the functions of macrophage lineage populations in inflammatory conditions are granulocyte-macrophage colony stimulating factor (GM-CSF)1 and macrophage colony stimulating factor (M-CSF) (4). These CSFs are critical to the proper maintenance of steady-state macrophage development, although with different roles. GM-CSF has a role in inducing emergency hematopoiesis not in steady state, and influences the pathogenesis of various inflammatory as well as autoimmune diseases (5). In this line, in vitro generated GM-CSF grown macrophages are now considered as pro-inflammatory macrophages that display a robust immune responses upon LPS stimulation compared with M-CSF grown macrophages (6). In contrast, M-CSF contributes the maintenance of most resident macrophages including osteoclast in vivo and is known to affect homeostatic anti-inflammatory characteristics of macrophages. M-CSF grown macrophages are widely accepted as in vitro-generated macrophage sources because they showed relatively homogenous and stable macrophage phenotypes (7). A number of genomic studies have been performed to analyze macrophage activation in response to pro-inflammatory/anti-inflammatory stimuli. However, to date, there have been no clear reports on the global proteomic differences that govern the functional characteristics of differently differentiated or activated macrophages (8–11). To fully elucidate what enables pro-inflammatory macrophages to be poised for rapid and robust immune responses requires assessing their global intracellular proteomic network signatures.
There has long been an appreciation, especially in the cancer field, for how changes in cellular activation coincide with alterations in cellular metabolic states (12, 13). Importantly, over the last couple of years it is becoming increasingly clear that immune cell activation is also coupled to profound changes in cellular metabolism and that their fate and function are metabolically regulated (14). In line with this, in this study, we found that GM-CSF grown macrophages have a higher glycolytic capacity through up-regulated glycolytic enzymes, as well as high lipid/nitrogen compound biosynthetic enzymes compared with M-CSF grown macrophages. They produce robust inflammatory cytokines upon TLR ligand stimulation only when sufficient glucose is available.
Here we performed a quantitative analysis of the proteome/phosphoproteome of primary GM-CSF and M-CSF grown macrophages using the latest isobaric tag based TMT labeling and LC-MS/MS (15). This proteomic approach with high throughput technology is the first attempt to show the fundamental differences between primary GM-CSF and M-CSF grown macrophages and finally reveals that innate cellular anabolic metabolism paves the way for inducing robust immune responses. In this study, we describe individual differentially expressed proteins in the total network maps of GM-CSF and M-CSF grown macrophages and predict how they are specifically involved in initiating inflammation or resolution.
EXPERIMENTAL PROCEDURES
Macrophages Preparation
C57BL/6J mice were obtained from Jackson Laboratory (Bar Harbor, ME). Male mice of five to 10 weeks of age were used to isolate bone marrow cells. The mice were housed under specific pathogen-free (SPF) conditions and cared according to the Guide for the Care and Use of Laboratory Animals prepared by the Institution of Animal Care and Use Committee of Seoul National University. All of the experiments were approved by the Institute for Animal Care and Use Committee of Seoul National University (accession number SNU-130311-2-2). Bone marrow derived macrophages (BMMs) were isolated as described before (16). To enrich the macrophage population, we supplemented complete medium with 25 ng/ml murine GM-CSF (Miltenyi Biotech, Bergisch Gladbach, Germany) for GM-CSF grown bone marrow derived macrophages (GM-BMMs) or 20% L929 murine fibrosarcoma cell line culture supernatants for M-CSF grown bone marrow derived macrophage (M-BMMs). After 7 days of differentiation, GM-BMMs and M-BMMs were stimulated with lipopolysaccharide (E. coli LPS, Sigma) 100 ng/ml for 2 h. Four experimental groups (GM-BMM, GM-BMM/LPS, M-BMM, M-BMM/LPS) were processed for further analysis.
Flow Cytometry
Macrophages were scrapped and incubated for 20 min with appropriate antibodies diluted to optimal concentrations in FACS buffer (PBS, 5% FBS, 5 mm EDTA, and 1% NaN3). For cell sorting and proteomics analysis, antibodies included anti-CD45 (30-F11), F4/80 (BM8), CD11b (M1/70), and IA/IE (M5/114.15.2). For surface marker expression analysis, additional antibodies included CD11c (HL3), CD80 (L307.4), MerTK (clone 125518, R&D systems, Minneapolis, MN), and CD64 (X54–5/7.1, BD Biosciences, Franklin Lakes, NJ) and all were purchased from eBioscience unless indicated. Cells were sorted on a FACSAria or data were acquired on a LSRII flow cytometer (BD Biosciences). The surface marker expressions were analyzed using the FlowJo software.
Trypsin Digestion, TMT Labeling, and OFF-Gel Fractionation
Equal protein amounts were digested using Filter Aided Sample Preparation (FASP) method (17). Briefly, lysates were washed with 8 m urea, followed by alkylation with 50 mm iodoacetamide (20 min at RT). After alkylation, filters were washed with 8 m Urea two times and with 0.1 m TEAB two times for labeling with TMT reagents. Trypsin was added at a ratio of 1 μg trypsin : 50 μg protein and samples were incubated overnight at 37 °C. Tryptic digests of the four cell lysates were labeled with four different mass-tags among the TMT sixplex reagents respectively, per the protocol described by Thermo Fisher Scientific (Rockford, IL). Resulting TMT-labeled peptides were subject to peptide isoelectrofocusing (IEF) fractionation, the 3100 OFFGEL fractionator with a “Low Resolution Kit” pH 3–10 (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions.
Phosphopeptide Enrichment Using TiO2
Phosphopeptide enrichment was carried out as described in the TitansphereTm Phos-TiO2 Kit manual. Briefly, phosphopeptides were eluted with 5% aqueous ammonium hydroxide and 5% aqueous pyrrolidine solutions from Phos-TiO2 (3 mg/200 μl, Titansphere, GL Sciences Inc, Tokyo, Japan) spin column.
Mass Spectrometric Analysis and Database Search
The extracted tryptic peptides were analyzed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled with an Easy-nLC system (Thermo Fisher Scientific, Odense, Denmark). Tryptic peptides were resuspended in 0.1% formic acid and separated on EASY-Spray column (C18, 2 μm particle size, 100 Å pore size, 75 μm id × 50 cm length, Thermo Fisher Scientific). Samples were resolved with a linear gradient of solvent B (100% ACN, 0.1% formic acid); 5–50% over 76 min, 50–90% over 12 min at a flow rate of 300 nL/min. The separated peptide ions eluted from the analytic column were entered into the mass spectrometer at an electrospray voltage of 2.1 kV. All MS/MS spectra were acquired in a data-dependent mode for fragmentation of the ten most abundant peaks from the full MS scan with 30% normalized collision energy. The dynamic exclusion duration was set at 20 s and the isolation mass width was 2.5 Da. MS spectra were acquired with a mass range of 400–1800m/z and 70,000 resolution at m/z 200. MS/MS resolution was acquired at a resolution of 17,500. The acquired MS/MS spectra were searched against the Universal Protein Resource mouse protein database (Uniprot release 2013_09, 50287 entries, http://www.uniprot.org/) with the Sequest algorithm in Proteome Discoverer 1.4 (Thermo Fisher Scientific, Bremen, Germany). Search parameters were as follows: tryptic specificity with up to two missed cleavage sites, mass tolerances for precursor ions and fragment ions were set to 10 ppm and 0.8 Da, respectively, fixed modification for carbamidomethyl-cysteine, TMT 4-plex of lysine and N terminus and variable modification for methionine oxidation. Following database searching, the output files were imported into Scaffold Q+ (version Scaffold_4.3.2, Proteome Software Inc. Portland, OR). Scaffold was used to organize all data, to quantitate protein and to validate peptide identifications using the Peptide Prophet algorithm (18). We selected peptides with cutoff of FDR = 0.01 and Peptide threshold 95%. We then identified the proteins that have Protein threshold 99.9%. Validated data were normalized between intrasample channels and TMT signals showed at least 1.5-fold change in abundance were used for quantitative analysis of protein. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (19) via the PRIDE partner repository with the dataset identifier PXD002582.
GO Analyses
Gene Ontology biological processes (GOBPs) represented by the sets of proteins were determined using the DAVID software (20). For each set of proteins, we identified the GOBPs represented by the genes with p value < 0.05 using STRING v9.1 (21). The degree of centrality (K) and shortest-path centrality (SP) were computed using CentiScaPe (22), and the nonseed proteins with SP = 0 or K = 1 were removed. The nodes with the same GOBPs were grouped into the same modules, each of which was named by the corresponding GOBP. The network was visualized using Cytoscape (v. 2.8.3) (23).
RNA Isolation and Gene Expression Profiling
Global gene expression analyses were performed using Affymetrix GeneChip® Mouse Gene 1.0 ST oligonucleotide arrays. The sample preparation was performed according to the instructions and recommendations provided by the manufacturer. Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Expression data were generated by Affymetrix Expression Console software version1.1. For normalization, RMA (Robust Multi-Average) algorithm was implemented in the Affymetrix Expression Console software.
Western blotting
Sorted GM-BMMs and M-BMMs were lysed and analyzed for protein expression as described earlier (24). Antilactate dehydrogenase B chain (Thermo), antiphosphofructokinse-1 (SantaCruz, TX), antiribose phosphate isomerase a (Abcam, Cambridge, UK), antitransferrin receptor (SantaCruz) and antibeta actin (SantaCruz) were used at 1/2000 dilutions.
Cytokine Production
Sorted GM-BMMs and M-BMMs were seeded in 96-well plates at 5 × 105/200 μl with complete media and rested for 6 h. Macrophages were stimulated with LPS (100 ng/ml) for 4 h and 24 h. Supernatants were collected and stored at −80 °C until TNFα, IL-6, and IL-10 quantification. For IL-1β quantitation, cells were lysed in RIPA buffer and stored at −80 °C. Cells in each well were analyzed for their protein concentration using the BCA assay kit (Thermo, Waltham, MA) and this was used as a normalization factor for cytokine quantitation. Cytokine concentrations were determined using the duoset ELISA kit (R&D systems, Minneapolis, MN).
Latex Bead Phagocytosis
Sorted GM-BMMs and M-BMMs were seeded in 12-well plates at 106/ml with complete media and rested for 6 h. Each well of cells were incubated with 107 Alexa 350-tagged latex beads (Molecular Probe, Eugene, OR) of 1 μm diameter for 2 h. The number of phagocytozed beads were analyzed on a LSRII flow cytometer (BD Biosciences) and analyzed using the FlowJo software.
ECAR and OCR Measurements
The Seahorse XF-24 metabolic extracellular flux analyzer (Seahorse Bioscience, Billerica, MA) was used to analyze the ECAR (in mpH/min) and the OCR (in pmol/min) (25). Briefly, GM-BMMs and M-BMMs were differentiated for 7 days in Seahorse cell plates (5 × 105 cells per well). Before plate reading, cells were washed three times with glucose free assay media (Seahorse Bioscience), and the OCR and ECAR were assessed in glucose-containing assay media. Perturbation profiling of GM-BMMs and M-BMMs metabolic pathways was achieved by the addition of glucose (10 mm), oligomycin (5 μm), 2-deoxyglucose (100 mm) or LPS. Experiments with the Seahorse system were done with the following assay conditions: 2 min mixture; 2 min wait; and 4–5 min measurement. Metabolic parameters were then calculated.
Statistical Analysis
All data unless otherwise indicated are shown as mean ± S.E. and were tested using two-tailed Student's t test or two-way ANOVA using GraphPad Prism 4.
RESULTS
Quantitative Analysis of Proteome and Phosphoproteome of GM-BMMs and M-BMMs
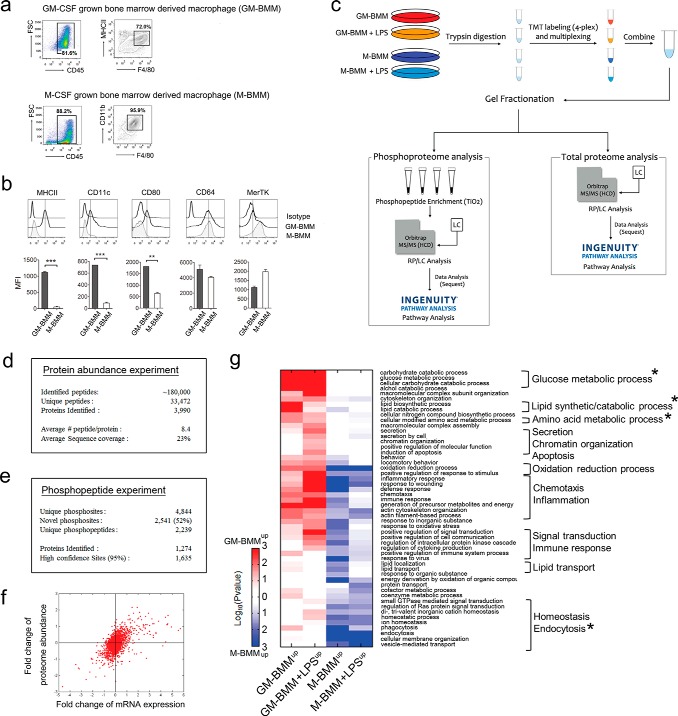
Macrophage sorting for proteomic analysis was performed according to the gating strategy shown in Fig. 1A. We first characterized in vitro GM-CSF grown bone marrow derived macrophages (GM-BMMs) and M-CSF grown bone marrow derived macrophages (M-BMMs). GM-GMMs were CD45+, F4/80+, and MHCII+. M-BMMs showed homogenous population and expressed CD45, F4/80, and CD11b. GM-BMMs expressed CD11c, CD80 as well as the macrophage marker CD64 (Fig. 1B) (26). In contrast, M-BMMs expressed MHCII, CD11c, and CD80 scarcely, indicating poor antigen presentation ability. Instead, they expressed relatively higher MerTK, which was further up-regulated by M-CSF (27). Further experiments used sorted GM-BMMs and M-BMMs according to the above gating strategy. In case of GM-CSF cultured cells, small population of CD45+F4/80lowMHCIIhigh dendritic cells were excluded throughout the experiments. To enrich early TLR-responsive proteins as well as core polarization specific proteins, we had four experimental groups of GM-BMMs and M-BMMs, with and without LPS stimulation for 2 h (Fig. 1C). Three biologic replicates were generated by combining the protein extracts from five mice per group. The TMT labeling (fourplex) quantitative proteomic method was used to profile changes in both protein abundance and phosphorylation stoichiometry in the whole cell extracts. Comparative MS analysis of these extracts on a Q-Exactive mass spectrometer led to the identification of 33,472 unique peptides from 3990 proteins with an average sequence coverage of 23% (Fig. 1D, supplemental table S12-S13). Although expression levels remained unaffected by TLR stimulation for most of the identified proteins of GM-BMMs and M-BMMs, a subset of 294 proteins showed more than a 1.5-fold change in abundance (supplemental Tables S1 and S2).
Fig. 1.
Quantitative proteomic/phosphoproteomic analysis of GM-BMMs/M-BMMs. A, GM-BMMs and M-BMMs were differentiated over 7 days from C57BL/6J male mice bone marrow cells with GM-CSF or M-CSF supplementation, respectively. Cells were sorted by CD45+F4/80+MHCII+ population for GM-BMMs, or CD45+F4/80+CD11b+ population for M-BMMs. B, Surface marker examination of GM-BMMs and M-BMMs. Attached cells were stained with the appropriate antibodies and analyzed for surface MHCII, CD11c, CD80, CD64, and MerTK expressions per gated populations in A. Three independent experiments were performed. **, p < 0.01, ***, p < 0.001 by Student t test. C, Experimental strategy. Macrophage proteins were pooled to generate three biological replicates for each of the GM-BMM/M-BMM/GM-BMM+LPS/M-BMM+LPS experimental groups. D, Results of protein expression experiments. E, Results of phosphoproteome experiments. F, Scatter plot of gene symbols detected both in proteome and microarray analyses. Data represents the fold change of log2(GM-BMM/M-BMM) of 2491 genes. Regression p value = 2.1987e-242, R-squre = 0.31352. G, GO analysis of differentially expressed proteins and phosphopeptides. Heat-map shows significant GO biological process terms (p < 0.05) for differentially expressed proteins/phosphoproteins between GM-BMM/M-BMM and GM-BMM+LPS/M-BMM+LPS. Red color indicates increased protein/phosphoprotein abundances in GM-BMM or GM-BMM/LPS, and blue color indicates increased abundances in M-BMM or M-BMM/LPS. Star markings show the most enriched GO category in a heatmap.
We next examined the differences in the status of protein phosphorylation between GM-BMMs and M-BMMs as well as GM-BMMs/LPS and M-BMMs/LPS (Fig. 1E). This analysis led to the identification of 2239 unique phosphopeptides from 1274 phosphoproteins (4844 phosphorylation sites, peptide probability>95% and protein probability>99%). A comparison of the identified phosphorylation sites with those from PHOSIDA (28), PhosphoSitePlus (29), and PhosphoELM (30) indicated that 2541 sites (∼52%) have not been previously reported. We also evaluated the confidence level of site localization using a probability score function and determined that 34% of the identified sites (1635 sites) corresponded to high confidence assignments (supplemental Table S11). Among them, 450 and 899 phosphopeptides, corresponding to 228 and 481 proteins, were differentially regulated between GM-BMMs and M-BMMs upon LPS stimulation, respectively (supplemental Table S8).
A scatter plot of protein abundance and mRNA expression (GEO accession number GSE63245) showed a general prediction ability of microarray data to project protein quantity (Fig. 1F), but is still limited in representing exact whole protein quantities of macrophages with a regression p value of 2.1987e−242 and R-square of 0.31352. This means that substantial macrophage proteins go through post-translational modification and our proteomics data could give valuable and precise insight into these global differences between opposite characterized macrophages.
GO Analysis Revealed Metabolism as a Key Regulator of Macrophage Function
To gain insight into the functional differences of each macrophage type, we conducted a comparison of Gene Ontology (GO) annotations of biological processes from differentially expressed proteins (DEPs) and differentially phosphorylated proteins (DPPs) in GM-BMMs/M-BMMs and GM-BMMs/LPS M-BMMs/LPS pairs (Fig. 1G). A total of 52 categories were enriched and are presented in a heatmap. Interestingly, the most obvious enriched categories in both GM-BMMs and GM-BMMs/LPS macrophages were metabolic processes (Table I). These include carbohydrate catabolic, glucose metabolic, alcohol catabolic, lipid biosynthetic, and amino acid metabolic processes (Table I). The GO term macromolecular complex subunit organization was enhanced in GM-BMMs/LPS compared with GM-BMMs. These energy consuming processes are potentially supported by glucose metabolic/nitrogen compound biosynthetic and lipid synthetic pathways in GM-BMMs, as with rapidly proliferating cancer cells (31). Secretion, chromatin organization, and apoptosis were specifically up-regulated in GM-BMMs/LPS.
Table I. List of proteins enriched in Gene Ontology Biological Process category from differentially expressed protein abundances between GM-BMMs versus M-BMMs and GM-BMMs+LPS vs M-BMMs+LPS: upregulated in GM-BMMs.
| Gene symbol | Protein Name | Ratio GM-BMM/M-BMM | p value | Fold enrichment |
|---|---|---|---|---|
| Glucose metabolic process | 0.00006 | 7.84 | ||
| ALDOC | Fructose-bisphosphate aldolase C | 2.22 | ||
| ENO1 | Alpha-enolase | 1.57 | ||
| FABP5 | Fatty acid-binding protein, epidermal | 2.64 | ||
| GYS1 | Glycogen [starch] synthase, muscle | 1.68 | ||
| LDHB | l-lactate dehydrogenase B chain | 2.00 | ||
| PFKP | 6-phosphofructokinase | 1.80 | ||
| PYGL | Glycogen phosphorylase, liver form | 1.62 | ||
| RPIA | Ribose-5-phosphate isomerase | 1.62 | ||
| Nitrogen compound biosynthetic process | 0.02197 | 3.18 | ||
| ASS1 | Argininosuccinate synthetase 1 | 3.03 | ||
| ATP6V0D2 | V-type proton ATPase subunit d2 | 2.93 | ||
| KYNU | Kynureninase | 1.62 | ||
| PADI4 | Protein-arginine deiminase type-4 | 1.74 | ||
| PHGDH | d-3-phosphoglycerate dehydrogenase | 2.00 | ||
| RRM1 | Ribonucleoside-diphosphate reductase large subunit | 1.93 | ||
| SLC7A2 | Low affinity cationic amino acid transporter 2 | 1.68 | ||
| Lipid biosynthetic process | 0.00021 | 4.80 | ||
| ACLY | ATP-citrate synthase | 1.62 | ||
| ALOX5 | Arachidonate 5-lipoxygenase | 1.68 | ||
| CD74 | H-2 class II histocompatibility antigen gamma chain | 2.46 | ||
| FABP5 | Fatty acid-binding protein, epidermal | 2.64 | ||
| FASN | Fatty acid synthase | 1.62 | ||
| FDPS | Farnesyl pyrophosphate synthase | 2.00 | ||
| HMGCS1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 1.68 | ||
| IDI1 | Isopentenyl-diphosphate Delta-isomerase 1 | 1.93 | ||
| LTA4H | Leukotriene A4 hydrolase | 2.00 | ||
| MVD | Diphosphomevalonate decarboxylase | 2.00 | ||
In contrast, M-BMMs and M-BMMs/LPS had different enriched functions, in that they showed higher endocytosis and homeostasis processes. Of note, “energy derivation by oxidation of organic compounds” was highly up-regulated in M-BMMs (Fig. 1G), showing a metabolic state that is the converse of GM-BMMs (Table II). This decreased upon LPS stimulation, implying a metabolic shift from oxidation to other processes. Subcellular distribution of DEPs also showed that mitochondrial components were highly enriched in M-BMMs compared to GM-BMMs, suggesting a bias for oxidation processes as their cellular energy source (supplemental Table S9). Several signaling pathways related to endocytosis/phagocytosis, including small GTPase and Ras, were enriched in both M-BMMs and M-BMMs/LPS.
Table II. List of proteins enriched in Gene Ontology Biological Process category from differentially expressed protein abundances between GM-BMMs versus M-BMMs and GM-BMMs+LPS vs M-BMMs+LPS: upregulated in M-BMMs.
| Gene symbol | Protein Name | Ratio M-BMM/GM-BMM | p value | Fold enrichment |
|---|---|---|---|---|
| Energy derivation by oxidation of organic compounds | 0.00180 | 6.82 | ||
| CAT | Catalase | 1.80 | ||
| GAA | Lysosomal alpha-glucosidase | 1.57 | ||
| SDHC | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | 1.74 | ||
| SLC37A2 | Sugar phosphate exchanger 2 | 1.74 | ||
| SOD2 | Superoxide dismutase [Mn], mitochondrial | 2.46 | ||
| SUCLG2 | Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial | 1.57 | ||
| Endocytosis | 0.00004 | 5.9 | ||
| ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 1.57 | ||
| ARHGAP27 | Rho GTPase activating protein 27 | 1.57 | ||
| BET1 | Bet1 golgi vesicular membrane trafficking protein | 1.52 | ||
| CD36 | CD36 molecule (thrombospondin receptor) | 1.80 | ||
| EHD1 | EH-domain containing 1 | 2.22 | ||
| ELMO1 | engulfment and cell motility 1 | 1.52 | ||
| FNBP1L | formin binding protein 1-like | 2.00 | ||
| HCK | hemopoietic cell kinase | 1.74 | ||
| ITSN1 | intersectin 1 (SH3 domain protein) | 2.14 | ||
| TFRC | transferrin receptor | 2.46 | ||
| VAV1 | vav 1 guanine nucleotide exchange factor | 1.57 | ||
Similar results were also obtained in our integrated pathway analysis (IPA) of DPPs between GM-BMMs and GM-BMMs/LPS or M-BMMs and M-BMMs/LPS. GM-BMMs/LPS showed enhanced protein phosphorylations for cytokine signaling, MAPK signaling, TLR4 cascade as well as mTOR signaling events compared with basal GM-BMMs (supplemental Table S4). In line with this, 58 proteins were up-regulated in GM-BMMs/LPS compared with GM-BMMs, mostly related to immune responses including IL-1β, lactotransferrin, TNF, copper transporter protein ATOX-1, and IL-1α (supplemental Table S3). M-BMMs/LPS showed enhanced Rho GTPase and RAC1 activity in IPA of DPPs, which correlate with endocytic activity. M-BMMs/LPS showed only seven up-regulated proteins relative to basal M-BMMs (supplemental Table S7), suggesting remarkable down-regulated immune responses compared with GM-BMMs. Taken together, GM-BMMs had enhanced anabolic metabolism and responded to LPS with inflammatory activities. On the contrary, M-BMMs had enhanced endocytic processes and did not show marked inflammatory activities upon LPS stimulation.
Glycolysis Signatures GM-CSF Grown Macrophages
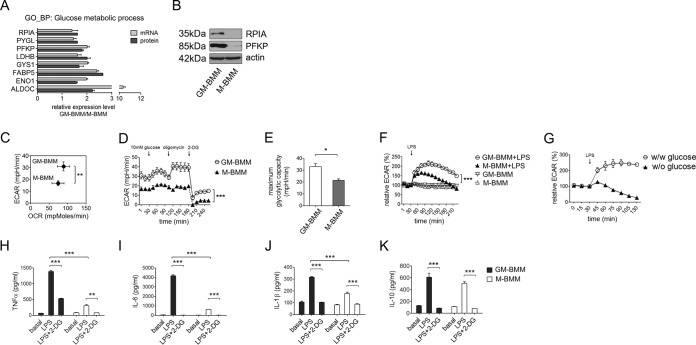
Among the eight proteins involved in glucose metabolism up-regulated in GM-BMMs, five were direct glycolytic enzymes (RPIA, PFKP, LDHB, ENO1, ALDOC) and one, PYGL, that is indirectly related to glycolysis (Fig. 2A). Among these, we confirmed the abundant protein expressions of ribose 5 phosphate isomerase (RPIA) and 6-phosphofructokinase (PFKP) in GM-BMMs compared with M-BMMs by Western blot, indicating the reliability of our proteomic methodology (Fig. 2B).
Fig. 2.
GM-BMMs have higher glycolytic capacities than M-BMMs. A, Expression profiles of glycolytic genes and proteins in GM-BMMs compared with M-BMMs. ***, p < 0.001 by two-way ANOVA. B, Western blot results for RPIA, PFKP, and actin. C, Basal OCR and ECAR of GM-BMMs and M-BMMs. Data obtained using XF-24 extracellular flux analyzer. Three independent experiments were performed. **, p < 0.01 by Student t test. D, Maximum glycolytic capacities of GM-BMMs and M-BMMs. Perturbation profiling of lactate production (ECAR) was achieved by the addition of glucose (10 mm), oligomycin (5 μm), and 2-deoxyglucose (2-DG, 100 mm). Three independent experiments were performed. ***, p < 0.001 by two-way ANOVA. E, Quantitative graph representing mean maximum glycolytic capacities of GM-BMMs and M-BMMs. Three independent experiments were performed. *, p < 0.05 by Student t test. F, LPS induced acute glycolysis. GM-BMMs and M-BMMs were stimulated with 100 ng/ml E. coli LPS and recorded for the ECAR changes. Data showing relative ECAR (%) changes from baseline. Three independent experiments were performed. ***, p < 0.001 by two-way ANOVA. G, Glucose effect on acute glycolysis upon LPS stimulation. GM-BMMs were stimulated with LPS with or without media glucose and ECAR changes were recorded. H–K, GM-BMMs produced more inflammatory cytokines than M-BMMs in a glycolysis-dependent manner. GM-BMMs and M-BMMs were sorted and replated in 96-well plates at 5 × 105 density. Cells were treated with LPS (100 ng/ml) with or without 2-DG (100 mm) and cytokines were measured at 4 h (TNFα) and 24 h (IL-6, IL-1β, IL-10). ELISA was performed to quantify TNFα (H), IL-6 (I), IL-10 (K) production in culture supernatants, or IL-1β synthesis (J) in cell lysates. Three independent experiments were performed. **, p < 0.01, ***, p < 0.001 by two-way ANOVA.
Next we hypothesized that GM-BMMs have a much higher maximum glycolytic capacity than M-BMMs. To assess metabolic differences, we recorded extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of GM-BMMs and M-BMMs. GM-BMMs had significantly higher basal ECAR than M-BMMs, although OCR was similar (Fig. 2C). Next we treated the macrophages with glucose and oligomycin to maximize glycolysis as shown in Fig. 2D. This confirmed that GM-BMMs have a higher glycolytic capacity than M-BMMs (Fig. 2E). This difference contributes to the varied extent of acute glycolysis upon LPS stimulation, with GM-BMMs showing a more prominent and prolonged ECAR curve than M-BMMs after LPS treatment (Fig. 2F). Acute glycolysis in macrophages following LPS was totally dependent on glucose uptake as ECAR does not increase without glucose in the media (Fig. 2G). Taken together, these results indicate that GM-CSF grown macrophages have inherently higher glycolytic capacities than M-CSF grown macrophages to enable heightened glucose conversion to lactate in response to LPS.
Metabolic Differences are Linked to Cytokine Responses in Macrophages
Finally, to test the impact of metabolic construct of GM-BMMs and M-BMMs on cytokine production, we employed 2-deoxyglucose (2-DG) to interfere with glucose metabolism and measured cytokine production as a functional readout. Consistent with previous reports (6), GM-BMMs produced more TNFα, IL-6, and IL-1β than M-BMMs (Fig. 2H, 2I, and 2J). Importantly, blockade of glycolysis significantly reduced LPS-induced TNFα, IL-6, and IL-1β production to that of basal levels both in GM-BMMs and M-BMMs. Both macrophages produced similar levels of IL-10 upon LPS stimulation, but this was also inhibited by 2-DG (Fig. 2K). These results confirm the vital role of glycolysis in LPS induced pro-inflammatory cytokine responses. Given that LPS induced cytokine production is largely dependent on glycolysis, it goes to reason that the more glycolytically active GM-BMMs can synthesize considerably more inflammatory cytokines than homeostatic M-BMMs.
Endocytosis Predominates in M-CSF Grown Macrophages
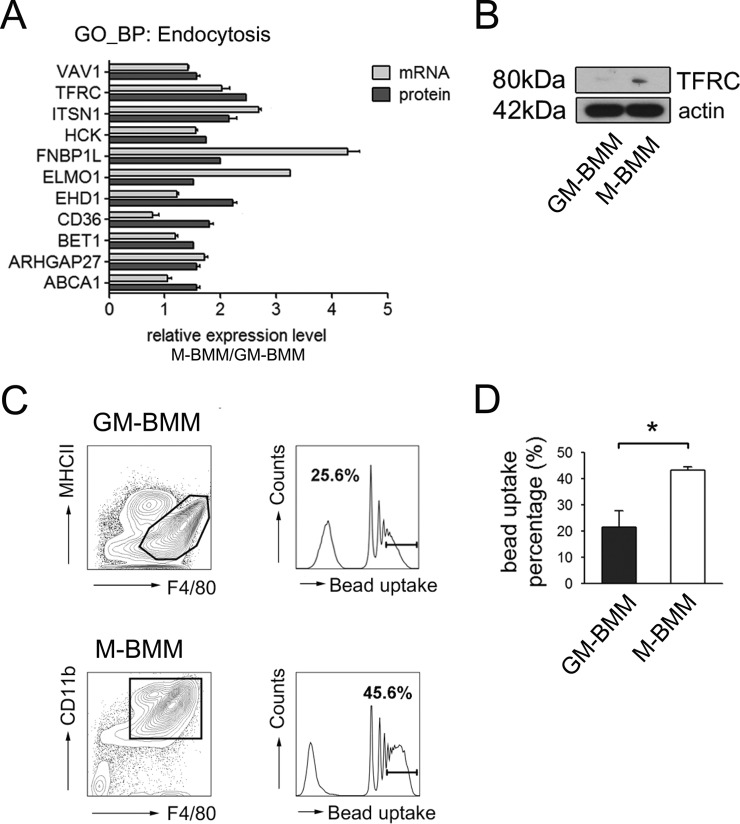
Eleven proteins related to endocytosis were up-regulated in M-BMMs compared to GM-BMMs (Fig. 3A). LPS further upregulated endocytosis-relating proteins in M-BMMs (supplemental Table S5). Among the upregulated 11 proteins in M-BMMs compared to GM-BMMs, we confirmed that transferrin receptor, CD71 (TFRC), which transports transferrin inside the cell and is up-regulated by CSF1 (32), expression was higher in M-BMMs than GM-BMMs (Fig. 3B). M-BMMs showed enhanced endocytic processes both in protein expression as well as phosphoprotein enrichment. Thus, we performed latex bead uptake experiments (Fig. 3C). After GM-BMMs and M-BMMs sorting according to Fig. 1A, replated macrophages were incubated with Ax350-tagged latex beads of 1 μm diameter for 2 h. As expected 40% of M-BMMs had more than four beads per cell, but only 20% of GM-BMMs did (Fig. 3D). These results confirmed the relationship between our proteome/phosphoproteome analysis and macrophage functional characteristics described above.
Fig. 3.
M-BMMs phagocytosed more beads than GM-BMMs. A, Relative mRNA and protein expression levels of endocytosis participants in M-BMMs compared with GM-BMMs. **, p < 0.01, ***, p < 0.001 by two-way ANOVA. B, Western blot results for TFRC and actin. C, Histograms showing the number of beads per cell. Sorted macrophages were replated and incubated with Alexa350 tagged latex beads for 2 h. Bead phagocytosis was analyzed per macrophage population (GM-BMMs: F4/80+MHCII+, M-BMMs: F4/80+CD11b+) using FACS. Cell percentages containing more than four beads are indicated in both histograms. D, Quantitative graph showing cell percentages with more than four beads per cells. Three independent experiments were performed. *, p < 0.05 by Student t test.
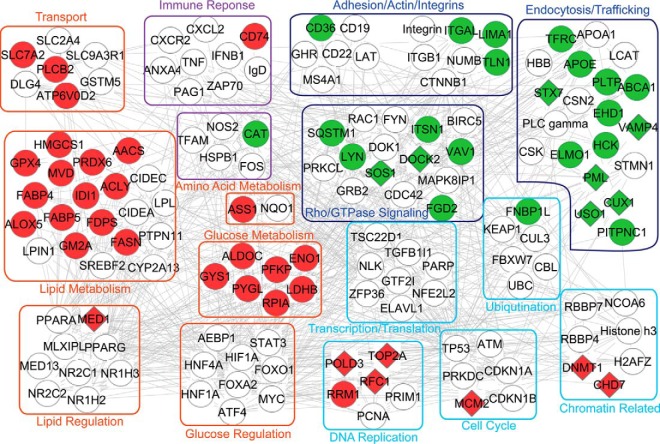
Protein Interaction Network Analysis of Macrophages
To gain insight into the proteome and phosphoproteome of GM-BMMs and M-BMMs, we conducted an Ingenuity Pathway Analysis (IPA) using DEPs and DPPs involved in differentially regulated biological processes of six major groups between GM-BMMs and M-BMMs: (1) Glucose metabolism, (2) Lipid metabolism, (3) Amino acid metabolism, (4) DNA replication, (5) Endocytosis, and (6) Rho/GTPase signaling. The total network map is presented in Fig. 4, which depicts 145 DEPs or DPPs in colored figures as well as predicted regulating transcription factors, transporters, enzymes, and signaling molecules denoted with white circles. Of note, transcription factors that regulate glycolysis such as HIF1A, MYC, FOXO1, FOXA2, ATF4, HNF1A, and HNF1B, underpin various other processes in GM-CSF grown macrophages including glucose/lipid/amino acid metabolism, immune response as well as DNA replication. In M-CSF grown macrophages, Rho/GTPase signaling is well interconnected with endocytosis and adhesion.
Fig. 4.
Regulated protein networks in GM-BMMs versus M-BMMs. Protein abundance and phosphorylation data were analyzed with the Ingenuity Pathway Analysis (IPA) using 145 differentially expressed proteins and phosphoproteins involved in differentially regulated biological process between GM-BMMs and M-BMMs. Glucose metabolism, lipid metabolism, amino acid metabolism, DNA replication, endocytosis, and Rho/GTPase signaling were the subnetworks most significantly affected. Proteins/nodes are grouped according to their function. Red color indicates GM-BMMs up-regulation and green is M-BMMs up-regulation. Circle means DEP, diamond means DPP. White circles are predicted interacting signaling molecules as well as transcription factors. All indicated by gene symbol.
DISCUSSION
In this study, we used the latest isobaric tag based multiplex proteomic/phosphoproteomic quantitative analysis method to identify key molecular differences between GM-CSF and M-CSF grown macrophages. We found that GM-BMMs have potentiative glycolytic/lipid biosynthetic pathways as well as nitrogen compound biosynthesis processes. These enhanced anabolic pathways directly link to their pro-inflammatory cytokine production capacity. We also presented valuable targets for disrupting macrophage functions, as well as novel phosphosites of macrophage proteins (supplemental Table S11). To our knowledge, this is the first large scale proteome/phosphoproteome analysis of GM-CSF and M-CSF grown macrophages. Our reliable data gives insight into the importance of cellular metabolic polarization and their functional outcomes in macrophages.
Aberrant carbohydrate metabolism is classically a distinctive feature of tumor cells (33). In general, normal cells produce most of their ATP from glucose through oxidative phosphorylation. However, many cancer cells produce ATP by conversion of glucose to lactate and therefore exhibit lower oxidative phosphorylation activity. This “glycolytic phenotype” ensures sufficient macromolecule biosynthesis necessary for rapid cell growth and division (34). Compellingly, we found that GM-BMMs showed a metabolic state similar to cancer cells. We confirmed that the up-regulated glycolytic enzymes are collectively involved in LPS-induced glycolysis and consequently connected with inflammatory cytokine productions. These data give us profound implications in that GM-CSF affected macrophages are usually generated when our body has been influenced by microbial or aseptic inflammatory stimuli (35). Given that GM-CSF therapy has been considered to alleviate inflammatory conditions such as arthritis (36), enhanced glycolytic pathway of GM-CSF grown macrophages might be an alternative specific valuable target for macrophage mediated inflammatory diseases.
Of note, the role of glycolysis in GM-BMMs differs from the acute glycolytic switch following LPS stimulation in dendritic cells, which results from IKKε/TBK1 mediated Akt activation (37), because basal enhanced glycolytic state exists in GM-CSF primed macrophages in the absence of TLR signaling. It also appears that this bias toward glycolysis does not stem from TLR mediated inducible nitric oxide synthase (iNOS) expression, which produces nitric oxide (NO) and inhibit oxidative phosphorylation (38). Instead, our network analysis predicts that GM-BMMs have up-regulated upstream glucose regulatory factors such as HIF1A, MYC, HNF1A (39), or FOXA2 (40). Given that GM-CSF is known to increase l-Myc expression in dendritic cells after 24 h (41), glycolysis in GM-CSF grown macrophages may be possibly further up-regulated by Myc because many glycolytic enzymes have myc-binding sites in their promoters (42). In 1995, Brissette et al. reported that GM-CSF primes mice for enhanced cytokine production in response to LPS, and our results suggest a possible underlying mechanism (43).
Another interesting result was that GM-BMMs have up-regulated enzymes in the mevalonate pathway. They are included in the GO term lipid biosynthetic process: diphosphomevalonate decarboxylase (MVD), farnesyl pyrophosphate synthase (FDPS), hydroxymethylglutaryl-CoA synthase (HMGCS1), and isopentenyl-diphosphate delta-isomerase 1 (IDI1) (Table I). Similar to glycolysis, the mevalonate pathway is dysregulated in tumor cells (44) and several researchers have tried to target this pathway to inhibit cancer cell malignancy (45). The mevalonate pathway is a complex biochemical pathway that generates several fundamental end-products including cholesterol, isoprenoids, dolichol, ubiquinone, and isopentenyladenine (45). In fact, glycolysis cascades into the mevalonate pathway when macrophages are activated because glycolysis induced by TLR activation produces abundant pyruvate and acetyl-CoA followed by lipid accumulation inside cells (46). Thus, it would be interesting to investigate whether the mevalonate pathway contributes to the macrophage pro-inflammatory phenotype.
The immune system is comprised of a heterogeneous population of cells that, for the most part, are relatively quiescent in the steady state but share the ability to rapidly respond to infection, inflammation, and other perturbations. There is a growing appreciation of the fact that transitions between quiescent and activated states require the apportioning of nutrients into different pathways, and, therefore, there is a strong interest in how metabolic pathways are regulated to support or direct functional changes. Several reports have been already noted the importance of macrophage metabolic states on polarization and attempted to target various metabolic pathways, such as glucose-6-phosphate dehydrogenase (47), CARKL (48), PGC-1β (49), TRAP1 (50), or glucose transporter 1 (51), to modulate macrophage phenotype. Our data clearly reveals a systemic link between macrophage activation and metabolic state. In addition, this study identifies novel opportunities for targeting macrophage mediated immune responses.
Supplementary Material
Footnotes
Authorship: Contribution: S.H.S., K.P.K. and Y.R.N. designed the experiments. J.H.H. and J.H.J. performed the proteomic experiments. Y.R.N., D.J. and D.S. performed macrophage functional experiments. M.Y.L. and Y.R.N. analyzed the proteomic data. M.C. constructed figures and Y.W.K. described the schematic image. Y.R.N. and M.Y.L. wrote the manuscript.
* This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2014R1A1A1008012, NRF-2012R1A1A3013393) and supported by Proteogenomic Research Program, and the Bio- and Medical Technology Development Program (Project No. 2012M3A9B6055305) through the National Research Foundation of Korea funded by the Korean Ministry of Education, Science and Technology (to K.P.K.).
 This article contains supplemental Tables S1 to S13.
This article contains supplemental Tables S1 to S13.
1 The abbreviations used are:
- GM-CSF
- granulocyte-macrophage colony stimulating factor
- M-CSF
- macrophage colony stimulating factor
- DEP
- differentially expressed proteins
- DPP
- differentially phosphorylated proteins
- GOBP
- Gene Ontology biological process.
REFERENCES
- 1. Gordon S., Taylor P. R. (2005) Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 [DOI] [PubMed] [Google Scholar]
- 2. Mosser D. M., Edwards J. P. (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon S. (2003) Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 [DOI] [PubMed] [Google Scholar]
- 4. Fleetwood A. J., Cook A. D., Hamilton J. A. (2005) Functions of granulocyte-macrophage colony-stimulating factor. Crit. Rev. Immunol. 25, 405–428 [DOI] [PubMed] [Google Scholar]
- 5. Hamilton J. A. (2008) Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 8, 533–544 [DOI] [PubMed] [Google Scholar]
- 6. Fleetwood A. J., Lawrence T., Hamilton J. A., Cook A. D. (2007) Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178, 5245–5252 [DOI] [PubMed] [Google Scholar]
- 7. Murray P. J., Allen J. E., Biswas S. K., Fisher E. A., Gilroy D. W., Goerdt S., Gordon S., Hamilton J. A., Ivashkiv L. B., Lawrence T., Locati M., Mantovani A., Martinez F. O., Mege J. L., Mosser D. M., Natoli G., Saeij J. P., Schultze J. L., Shirey K. A., Sica A., Suttles J., Udalova I., van Ginderachter J. A., Vogel S. N., Wynn T. A. (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez F. O., Gordon S., Locati M., Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]
- 9. McDermott J. E., Archuleta M., Thrall B. D., Adkins J. N., Waters K. M. (2011) Controlling the response: predictive modeling of a highly central, pathogen-targeted core response module in macrophage activation. PloS One 6, e14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nau G. J., Richmond J. F., Schlesinger A., Jennings E. G., Lander E. S., Young R. A. (2002) Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 99, 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramsey S. A., Klemm S. L., Zak D. E., Kennedy K. A., Thorsson V., Li B., Gilchrist M., Gold E. S., Johnson C. D., Litvak V., Navarro G., Roach J. C., Rosenberger C. M., Rust A. G., Yudkovsky N., Aderem A., Shmulevich I. (2008) Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput. Biol. 4, e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raffaello A., Rizzuto R. (2011) Mitochondrial longevity pathways. Biochim Biophys Acta. 1813, 260–268 [DOI] [PubMed] [Google Scholar]
- 14. Pearce E. L., Pearce E. J. (2013) Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dayon L., Sanchez J. C. (2012) Relative protein quantification by MS/MS using the tandem mass tag technology. Methods Mol. Biol. 893, 115–127 [DOI] [PubMed] [Google Scholar]
- 16. Na Y. R., Yoon Y. N., Son D. I., Seok S. H. (2013) Cyclooxygenase-2 inhibition blocks M2 macrophage differentiation and suppresses metastasis in murine breast cancer model. PloS One 8, e63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. AM V. O. s., Dominin S. G., Kutepov E. N., Leonov A. V., Lobov A. V., Maimulov V. G., Nesvizhskii Iu V., Semenova V. V., Fokin M. V., Tselykovskaia N. (2003) [Training physicians in medical prevention specialty: problems and prospects]. Gigiena i Sanitariia, 1, 13–15 [PubMed] [Google Scholar]
- 18. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 19. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 21. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L. J. (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scardoni G., Petterlini M., Laudanna C. (2009) Analyzing biological network parameters with CentiScaPe. Bioinformatics 25, 2857–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Son D., Na Y. R., Hwang E. S., Seok S. H. (2014) Platelet-derived growth factor-C (PDGF-C) induces anti-apoptotic effects on macrophages through Akt and Bad phosphorylation. J. Biol. Chem. 289, 6225–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spolarics Z., Wu J. X. (1997) Role of glutathione and catalase in H2O2 detoxification in LPS-activated hepatic endothelial and Kupffer cells. Am. J. Physiol. 273, G1304–1311 [DOI] [PubMed] [Google Scholar]
- 26. Gautier E. L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K. G., Gordonov S., Mazloom A. R., Ma'ayan A., Chua W. J., Hansen T. H., Turley S. J., Merad M., Randolph G. J., and Immunological Genome, C. (2012) Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zizzo G., Hilliard B. A., Monestier M., Cohen P. L. (2012) Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J. Immunol. 189, 3508–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gnad F., Gunawardena J., Mann M. (2011) PHOSIDA 2011: the post-translational modification database. Nucleic Acids Res. 39, D253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dinkel H., Chica C., Via A., Gould C. M., Jensen L. J., Gibson T. J., Diella F. (2011) Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 39, D261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Imamura K., Ogura T., Kishimoto A., Kaminishi M., Esumi H. (2001) Cell cycle regulation via p53 phosphorylation by a 5′-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem. Biophys. Res. Commun. 287, 562–567 [DOI] [PubMed] [Google Scholar]
- 32. Lokeshwar B. L., Lin H. S. (1990) Growth factor-dependent regulation of transferrin receptor in proliferating and quiescent macrophages. Cell. Immunol. 130, 401–415 [DOI] [PubMed] [Google Scholar]
- 33. Rolfe D. F., Brown G. C. (1997) Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 77, 731–758 [DOI] [PubMed] [Google Scholar]
- 34. Mazurek S., Eigenbrodt E. (2003) The tumor metabolome. Anticancer Res. 23, 1149–1154 [PubMed] [Google Scholar]
- 35. Becker L., Liu N. C., Averill M. M., Yuan W., Pamir N., Peng Y., Irwin A. D., Fu X., Bornfeldt K. E., Heinecke J. W. (2012) Unique proteomic signatures distinguish macrophages and dendritic cells. PloS One 7, e33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cook A. D., Pobjoy J., Steidl S., Dürr M., Braine E. L., Turner A. L., Lacey D. C., Hamilton J. A. (2012) Granulocyte-macrophage colony-stimulating factor is a key mediator in experimental osteoarthritis pain and disease development. Arthritis Res. Ther. 14, R199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Neill L. A. (2014) Glycolytic reprogramming by TLRs in dendritic cells. Nat. Immunol. 15, 314–315 [DOI] [PubMed] [Google Scholar]
- 38. Everts B., Pearce E. J. (2014) Metabolic control of dendritic cell activation and function: recent advances and clinical implications. Front. Immunol. 5, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Antinozzi P. A., Hagenfeldt K. A., Maechler P., Wollheim C. B. (2000) Molecular targets of a human HNF1 alpha mutation responsible for pancreatic beta-cell dysfunction. EMBO J. 19, 4257–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wolfrum C., Shih D. Q., Kuwajima S., Norris A. W., Kahn C. R., Stoffel M. (2003) Role of Foxa-2 in adipocyte metabolism and differentiation. J. Clin. Invest. 112, 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kc W., Satpathy A. T., Rapaport A. S., Briseño C. G., Wu X., Albring J. C., Russler-Germain E. V., Kretzer N. M., Durai V., Persaud S. P., Edelson B. T., Loschko J., Cella M., Allen P. M., Nussenzweig M. C., Colonna M., Sleckman B. P., Murphy T. L., Murphy K. M. (2014) L-Myc expression by dendritic cells is required for optimal T-cell priming. Nature 507, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gordan J. D., Thompson C. B., Simon M. C. (2007) HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 12, 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brissette W. H., Baker D. A., Stam E. J., Umland J. P., Griffiths R. J. (1995) GM-CSF rapidly primes mice for enhanced cytokine production in response to LPS and TNF. Cytokine 7, 291–295 [DOI] [PubMed] [Google Scholar]
- 44. Clendening J. W., Pandyra A., Boutros P. C., El Ghamrasni S., Khosravi F., Trentin G. A., Martirosyan A., Hakem A., Hakem R., Jurisica I., Penn L. Z. (2010) Dysregulation of the mevalonate pathway promotes transformation. Proc. Natl. Acad. Sci. U.S.A. 107, 15051–15056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Swanson K. M., Hohl R. J. (2006) Anti-cancer therapy: targeting the mevalonate pathway. Curr. Cancer Drug Targets 6, 15–37 [DOI] [PubMed] [Google Scholar]
- 46. Feingold K. R., Shigenaga J. K., Kazemi M. R., McDonald C. M., Patzek S. M., Cross A. S., Moser A., Grunfeld C. (2012) Mechanisms of triglyceride accumulation in activated macrophages. J. Leukocyte Biol. 92, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ham M., Lee J. W., Choi A. H., Jang H., Choi G., Park J., Kozuka C., Sears D. D., Masuzaki H., Kim J. B. (2013) Macrophage glucose-6-phosphate dehydrogenase stimulates proinflammatory responses with oxidative stress. Mol. Cell. Biol. 33, 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haschemi A., Kosma P., Gille L., Evans C. R., Burant C. F., Starkl P., Knapp B., Haas R., Schmid J. A., Jandl C., Amir S., Lubec G., Park J., Esterbauer H., Bilban M., Brizuela L., Pospisilik J. A., Otterbein L. E., Wagner O. (2012) The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15, 813–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vats D., Mukundan L., Odegaard J. I., Zhang L., Smith K. L., Morel C. R., Wagner R. A., Greaves D. R., Murray P. J., Chawla A. (2006) Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshida S., Tsutsumi S., Muhlebach G., Sourbier C., Lee M. J., Lee S., Vartholomaiou E., Tatokoro M., Beebe K., Miyajima N., Mohney R. P., Chen Y., Hasumi H., Xu W., Fukushima H., Nakamura K., Koga F., Kihara K., Trepel J., Picard D., Neckers L. (2013) Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc. Natl. Acad. Sci. U.S.A. 110, E1604–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Freemerman A. J., Johnson A. R., Sacks G. N., Milner J. J., Kirk E. L., Troester M. A., Macintyre A. N., Goraksha-Hicks P., Rathmell J. C., Makowski L. (2014) Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289, 7884–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.