Abstract
Acclimation to changing environmental conditions is mediated by proteins, the abundance of which is carefully tuned by an elaborate interplay of DNA-templated and post-transcriptional processes. To dissect the mechanisms that control and mediate cellular iron homeostasis, we conducted quantitative high-resolution iTRAQ proteomics and microarray-based transcriptomic profiling of iron-deficient Arabidopsis thaliana plants. A total of 13,706 and 12,124 proteins was identified with a quadrupole-Orbitrap hybrid mass spectrometer in roots and leaves, respectively. This deep proteomic coverage allowed accurate estimates of post-transcriptional regulation in response to iron deficiency. Similarly regulated transcripts were detected in only 13% (roots) and 11% (leaves) of the 886 proteins that differentially accumulated between iron-sufficient and iron-deficient plants, indicating that the majority of the iron-responsive proteins was post-transcriptionally regulated. Mutants harboring defects in the RING DOMAIN LIGASE1 (RGLG1)1 and RING DOMAIN LIGASE2 (RGLG2) showed a pleiotropic phenotype that resembled iron-deficient plants with reduced trichome density and the formation of branched root hairs. Proteomic and transcriptomic profiling of rglg1 rglg2 double mutants revealed that the functional RGLG protein is required for the regulation of a large set of iron-responsive proteins including the coordinated expression of ribosomal proteins. This integrative analysis provides a detailed catalog of post-transcriptionally regulated proteins and allows the concept of a chiefly transcriptionally regulated iron deficiency response to be revisited. Protein data are available via ProteomeXchange with identifier PXD002126.
With the exception of some bacteria that substitute manganese for iron or that rely on obligate parasitism (1, 2), iron is an essential element for all organisms, catalyzing numerous redox reactions by virtue of its unique electrochemical properties. The cellular iron concentration must be carefully balanced. An inadequate supply compromises essential cellular processes, whereas excess amounts trigger the formation of toxic reactive oxygen species through Fenton chemistry. Iron deficiency causes substantial yield losses and decreases the nutritional quality of crop plants. Low iron levels in edible plant parts are the major cause of iron deficiency-induced anemia, affecting approximately two billion people worldwide particularly in areas where plant-based diets are the major source of iron (3).
Iron shortage leads to pronounced changes in transcriptomic and proteomic profiles and causes reprogramming of metabolic and developmental pathways, which are aimed at improving iron acquisition and distribution (4–8). In Arabidopsis, massive up-regulation of the phenylpropanoid and scopoletin pathways leads to increased production of iron-binding coumarins that chelate poorly soluble Fe3+ oxyhyrates and thereby improve the acquisition of iron from pools of sparingly soluble iron (4, 9–11). Induction of the proton-translocating (H+)-ATPASE2 (AHA2) decreases the rhizospheric pH, thereby increasing the solubility of iron by a factor of 1000 for each pH unit (12). Fe3+ chelates are reduced by FERRIC REDUCTION OXIDASE2 (FRO2), and the released Fe2+ is taken up by IRON-REGULATED TRANSPORTER1 (IRT1) (13–15). In leaves, a rapid coordinated down-regulation of the protoporphyrin biosynthetic pathway and up-regulation of the expression of proteins involved in the detoxification of reactive oxygen species avoid the accumulation of potentially toxic tetrapyrrole intermediates and prevent photo-oxidative damage in anticipation of decreased chlorophyll production and diminished photosynthesis rates that occur during prolonged periods of iron deficiency.
Reciprocal grafting experiments with the pea mutant dgl, which displays constitutive expression of iron deficiency responses, indicate that the iron status of the leaves dictates the iron uptake rates of roots (16). In Arabidopsis, OLIGOPEPTIDE TRANSPORTER3 (OPT3) is critical in shoot-to-root signaling by mediating the loading of iron into the phloem (17–19). In roots, the expression of the key genes mediating the acquisition of iron from the rhizosphere, namely IRT1, FRO2, AHA2, and the feruloyl CoA ortho-hydroxylase 1 (F6′H1), which converts feruloyl CoA into 6′-hydroxyferuloyl CoA, a key step in the production of iron-binding compounds, are transcriptionally regulated by the bHLH transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT). FIT forms a heterodimer with the subgroup 1b bHLH proteins bHLH38, bHLH39, and probably also with bHLH100, and bHLH101 (20–26). Another bHLH transcription factor, POPEYE (PYE), is negatively regulating a separate set of genes by interacting with ILR3 and bHLH115 (27). The ubiquitin ligase BRUTUS (BTS) also interacts with IAA-LEUCINE RESISTANT3 (ILR3) and bHLH115 and has opposite effects on gene expression, thereby balancing PYE action (27, 28).
Limited information is available regarding post-transcriptional processes that control cellular iron homeostasis. A large set of genes is differentially alternatively spliced in response to iron deficiency, probably as a means to tune the amount of functional proteins to changing demand (29). The activity of two key players in iron uptake, FIT and IRT1, has been shown to be controlled both transcriptionally and by post-transcriptional processes. Proteolytic turnover and stabilization of FIT by interaction with the ethylene signaling transcription factors ETHYLENE-INSENSITIVE3 (EIN3) and ETHYLENE-INSENSITIVE3-LIKE (EIL1) is critical for FIT activity, which is rapidly degraded after binding to its targets (30, 31). Monoubiquitination and endocytic cycling are important for IRT1 function, calibrating the amount of IRT1 protein on the plasma membrane (32). Endocytic sorting of IRT1 is dependent on SORTING NEXIN1, a component in a retromer-like protein complex involved in endosome to lysosome protein transport (33). A RING-type ubiquitin ligase, IRT1 DEGRADATION FACTOR1, is involved in the proteolytic turnover of IRT1 (34).
The ubiquitin conjugase UBC13 is required for the formation of branched root hairs, a response of Arabidopsis to iron deficiency that has been interpreted as a strategy to increase the surface area of the roots (35, 36). The sequence of UBC13 is conserved among eukaryotes and its function has been related to the error tolerance branch of the DNA repair pathway in yeast, mammals, and plants. In Arabidopsis, UBC13 is encoded by two highly similar paralogs, UBC13A and UBC13B (37). ubc13a mutants fail to induce branched root hairs in response to iron deficiency and show deregulation of several iron-responsive genes (35). ubc13a ubc13b double mutants display severe defects in root hair formation also under iron-sufficient conditions (35, 38), indicating that functional UBC13 is critical in this process. UBC13 is the only known protein that can mediate the formation of ubiquitin chains linked to lysine 63 (K63) (39). The RING domain ligase RGLG1 and its close sequelog RGLG2 can interact with UBC13 and, together with UBC13, catalyze the formation of K63-linked polyubiquitin chains (40). RGLG1 and RGLG2 possess ubiquitin ligase activity and can also mediate the formation of canonical, K48-linked polyubiquitin chains that target proteins for degradation (41). Interestingly, rglg1 rglg2 double mutants show a constitutively branched root hair phenotype (35), which invited some speculation as to whether and how the double mutation can be linked to UBC13-mediated processes (42, 43). It has been suggested that under iron deficiency UBC13 recruits RGLG to the nucleus, leading to a decrease in RGLG protein in the cytoplasm and ultimately to branching of the root hairs (43).
To dissect post-transcriptional responses of Arabidopsis to iron deficiency and to determine the possible impact of RGLG on the regulation of these responses, we conducted genome-wide proteomic and transcriptomic surveys of leaves and roots from iron-sufficient and iron-deficient Col-0 wild-type plants and rglg1 rglg2 double mutants. This analysis showed that post-transcriptional regulation has a stronger influence on the proteomic readout than transcriptional control, affecting proteins that in turn control post-transcriptional processes. In particular, the expression of ribosomal proteins is strongly post-transcriptionally affected by iron deficiency, putatively leading to a bias in translation by prioritizing subsets of mRNAs that are critical to the acclimation to low iron availability. RGLG has a dramatic influence on the proteomic profile of iron-deficient plants, affecting both protein abundance and, most likely as a secondary affect, the transcriptional profile by targeting transcription factors and other proteins involved in transcriptional regulation. The combined analysis permits an integration of several regulatory layers involved in adapting plants to low iron availability and underscores the importance of protein turnover in this process.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis thaliana (L.) Heynh, Columbia (Col-0) ecotype was used as the wild-type control. rglg1 rglg2 mutants were kindly provided by A. Bachmair, University of Vienna. Plants were grown in a growth chamber on medium as described by Estelle and Somerville (44). Seeds were surface sterilized and germinated on a media containing, KNO3 (5 mm), MgSO4 (2 mm), Ca(NO3)2 (2 mm), KH2PO4 (2.5 mm), H3BO3 (70 μm), MnCl2 (14 μm), ZnSO4 (1 μm), CuSO4 (0.5 μm), CoCl2 (0.01 μm), Na2MoO4 (0.2 μm), and FeEDTA (40 μm), solidified with 0.4% Gelrite pure (Kelco), 1.5% sucrose and 1 g/L MES. The pH was adjusted to 5.5 with KOH. Seeds were sown on Petri plates and stratified for 1 day in 4 °C in the dark before being transferred to a growth chamber and grown at 21 °C under continuous illumination (50 μmol m−2 s−1). After 10 d of precultivation, plants were transferred to fresh agar medium either with 40 μm FeEDTA (+Fe plants) or without Fe and with 100 μm 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate (-Fe plants). For iron quantification and determination of trichome density, plants were grown for 14 d on media supplemented with either 40 μm or 0.5 μm FeEDTA.
Protein Extraction
Roots from control and iron-deficient plants (13-day-old) were ground in liquid nitrogen and suspended in 10× volume of precooled acetone (−20 °C) containing 10% (w/v) TCA and 0.07% (v/v) 2-mercaptoethanol. Proteins were then thoroughly mixed and precipitated for 2 h at −20 °C. Proteins were collected by centrifuging at 35,000 × g (JA-20 108 rotor; Beckman J2-HS) at 4 °C for 30 min. The supernatant was carefully removed, and the protein pellets were washed twice with cold acetone containing 0.07% (v/v) 2-mercaptoethanol and 1 mm phenylmethanesulfonyl fluoride and a third time with cold acetone without 2-mercaptoethanol. Protein pellets were dried by lyophilization and stored at −80 °C or immediately extracted using protein extraction buffer composed of 8 m urea, 50 mm Tris, pH 8.5, for 1 h at 6 °C under constant shaking. Protein extracts were centrifuged at 19,000 × g for 20 min at 10 °C. The supernatant was then collected, and the protein concentration was determined using a protein assay kit (Pierce).
In-Solution Trypsin Digestion and iTRAQ Labeling
Total protein (100 μg) was reduced by adding dithiothreitol to a final concentration of 10 mm and incubated for 1 h at room temperature. Subsequently, iodoacetamide was added to a final concentration of 50 mm, and the mixture was incubated for 30 min at room temperature in the dark. Then, dithiothreitol (30 mm) was added to the mixture to consume any free iodoacetamide by incubating the mixture for 1 h at room temperature in the dark. Proteins were then diluted by 50 mm Tris, pH 8.5, to reduce the urea concentration to 4 m and digested with 0.5 μg of Lys-C (Wako) for 4 h at room temperature. The Lys-C digested protein solution was further digested with 20 μg of modified trypsin (Promega) at 37 °C overnight after the solution was further diluted with 50 mm Tris, pH 8.0, to reduce the urea concentration to less than 1 m. The resulting peptide solution was acidified with 10% trifluoroacetic acid and desalted on a C18 solid-phase extraction cartridge.
Desalted peptides were then labeled with iTRAQ reagents (Applied Biosystems) according to the manufacturer's instructions. Control samples (proteins extracted from roots of control plants) were labeled with reagent 114; samples from iron-deficient roots were labeled with reagent 117. Three independent biological experiments with two technical repeats each were performed. The reaction was allowed to proceed for 1 h at room temperature. Subsequently, treated and control peptides were combined and further fractionated offline using high-resolution strong cation-exchange chromatography (PolySulfoethyl A, 5 μm, 200-Å beads). In total, 50 fractions were collected and combined into 16 final fractions. Each final fraction was lyophilized in a centrifugal speed vacuum concentrator. Samples were stored at −80 °C.
LC-MS/MS Analysis
Liquid chromatography was performed on a Dionex UltiMate 3000 RSLCnano System coupled to a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Scientific) equipped with a Nanospray Flex Ion Source. Peptide mixtures were loaded onto a 75 μm × 250 mm Acclaim PepMap RSLC column (Thermo Scientific) and separated using a segmented gradient in 120 min from 5 to 40% solvent B (100% acetonitrile with 0.1% formic acid) at a flow rate of 300 nl/min. Solvent A was 0.1% formic acid in water. The samples were maintained at 8 °C in the autosampler. The LTQ Orbitrap was operated in the positive ion mode with the following acquisition cycle: a full scan (m/z 350∼1600) recorded in the Orbitrap analyzer at resolution R 70,000 was followed by MS/MS of the 10 most intense peptide ions with HCD acquisition of the same precursor ion. HDC was done with a collision energy of 30%. HCD-generated ions were detected in the Orbitrap at resolution 17,500.
Database Search
Two search algorithms, Mascot (version 2.4, Matrix Science) and SEQUEST, which is integrated in Proteome Discoverer software (version 1.4, Thermo Scientific), were used to simultaneously identify and quantify proteins. Searches were made against the Arabidopsis protein database (TAIR10 20110103, 27416 sequences; ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR10_blastsets/TAIR10 pep 20110103 representative gene model) concatenated with a decoy database containing the reversed sequences of the original database. The protein sequences in the database were searched with trypsin digestion at both ends and two missed cleavages allowed, fixed modifications of carbamidomethylation at Cys, iTRAQ 4plex at N terminus and Lys, variable modifications of oxidation at Met and iTRAQ 4plex at Tyr; peptide tolerance was set at 10 ppm, and MS/MS tolerance was set at 0.05 Da. iTRAQ 4plex was chosen for quantification during the search simultaneously. The search results were passed through additional filters, peptide confidence more than 95% (p < 0.05), before exporting the data. For protein quantitation, only unique peptides were used to quantify proteins. These filters resulted in a false discovery rate of less than 5% after decoy database searches were performed. For biological repeats, spectra from the two technical repeats were combined into one file and searched.
Statistical Analysis
Proteins identified and quantified in at least two biological repeats were considered to further analyze the abundance change in response to iron deficiency using a method described by Cox and Mann (45). In brief, the log2 ratios of the 9,110 (roots) and 8,303 (leaves) quantified proteins overlapping in at least two biological repeats were calculated and analyzed for normal distribution. For a given protein in one biological repeat, the ratio was calculated as the inverse log2 of the median of the log2 value of all peptide ratios and averaged across the biological replicates. Next, mean and S.D. were calculated and 95% confidence (Z score = 1.96) was used to select those proteins whose distribution was far from the main distribution. For the down-regulated proteins, the confidence interval was −0.475072001 (0.040469377, mean ratio of the 9,110 proteins in roots) −1.96 × 0.263031316, corresponding to a protein ratio of 0.83. Similarly, for the up-regulated proteins, the confidence interval was calculated (mean ratio +1.96× S.D.), corresponding to a protein ratio of 1.29. Protein ratios outside this range were defined as being significantly different at p = 0.05.
RNA Extraction
Total RNA was isolated from roots of 13-d-old plants with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. Nucleic acid quantity was analyzed with a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington).
Microarray Analysis
The Affymetrix gene chip Arabidopsis ATH1 Genome Array was used for microarray analysis. Total RNA samples were prepared as described above. All RNA samples were quality assessed by using the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). Complementary RNA synthesis was performed by use of the GeneChip One-Cycle Target Labeling Kit (Affymetrix, Santa Clara, CA). Hybridization, washing, staining, and scanning procedures were performed as described in the Affymetrix technical manual.
Gene expression data were imported directly into GeneSpring (version 11.5, Agilent). The software was used to normalize the data per chip to the 50th percentile and per gene to the control samples. Genes that were flagged as absent in two replicates were not considered in the analysis. p values for the Benjamini and Hochberg method (false discovery rates; FDRs) were calculated by GeneSpring. Transcripts were defined as differentially expressed that showed delta signal changes larger than the mean expression value of the whole data set or twofold changes upon iron deficiency with p < 0.05.
Microscopy
For cross-sections, root samples were fixed, dehydrated, and then embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim) resin in gelatin capsules. Transverse sections (30 μm) were cut using a RM 2255 Leica microtome (Leica, Nussloch, Germany). Sections were dried and stained with toluidine blue (0.05%) on glass slides and examined using bright-field on an Imager Z1 microscope (Zeiss, Jena, Germany). Trichomes were observed on rosette leaves that were attached to the stage and cooled in liquid nitrogen. Observations were made in low vacuum with a scanning electron microscope (FEI Quanta 200).
Iron Concentration
Plants were dried in an oven at 60 °C, mineralized with 225 μl 65% nitric acid (HNO) at 96 °C for 6 h, and oxidized with 150 μl 30% hydrogen peroxide (H2O2) at 56 °C for 2 h. Iron concentrations were determined against a standard curve made with FeCl3 that was treated in the same way as the plant materials. Samples were measured spectrophotometrically at A535 nm in an assay solution that contained 1 mm bathophenanthroline disulfonic, 0.6 m sodium acetate and 0.48 m hydroxylamine hydrochloride.
RESULTS
Protein and mRNA Expression are Largely Uncoupled in Iron-Deficient Plants
To dissect global transcriptional and post-transcriptional regulation of gene expression in iron-deficient plants, changes in proteomic and transcriptomic profiles were assessed by liquid chromatography combined with tandem mass spectroscopy on a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer and by Affymetrix ATH1 microarray analysis. To gain insights into the post-transcriptional regulation of iron-responsive genes, responses of Col-0 wild-type plants were compared with mutants defective in the RING domain ligases RGLG1 and RGLG2. We previously reported that homozygous rglg1 rglg2 double mutants showed a constitutive branched root hair phenotype, a trait that is inducible in the wild type by subjecting plants to iron deficiency (35). It can thus be assumed that RGLG is involved in the regulation of the activity of iron-responsive genes, presumably acting as a post-translational regulator. It was shown previously that neither rglg1 nor rglg2 mutants produce full length RGLG transcript (40).
Proteins were extracted, digested in solution, iTRAQ-labeled and identified using the Mascot and SEQUEST search algorithms. With this technology, we identified a total of 13,706 and 12,124 proteins in roots and leaves of Col-0 wild-type plants, respectively. Peptides from 9,110 proteins in roots and 8,303 proteins in leaves were determined in at least two of the three biological replicates and were defined as reliably detected (Fig. 1A, 1B, supplemental Data Set S1, supplemental Tables S1 and S2).
Fig. 1.
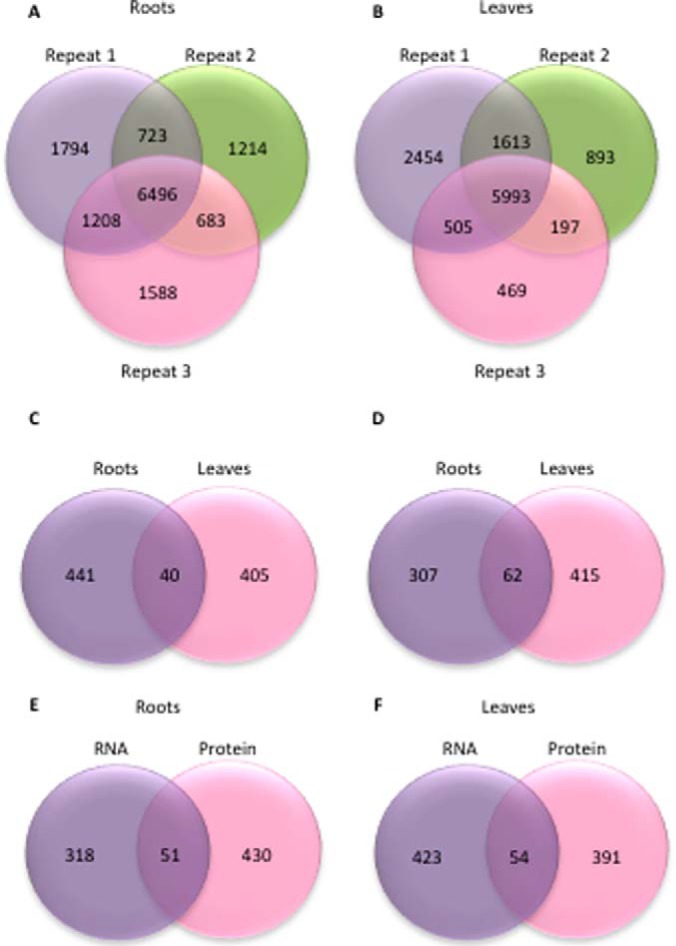
Identified and differentially expressed proteins and transcripts in roots and leaves of Col-0 plants. A, B, Number of proteins identified in roots, A, and leaves, B, of iron-sufficient and iron-deficient plants in three biological repeats. C, Differentially expressed proteins. D, Differentially expressed transcripts. E, F, Overlap of differentially expressed transcripts and proteins in leaves, E, and roots, F.
Based on a 95% confidence (Z score = 1.96), we defined subsets of 481 (roots) and 445 (leaves) reliably detected proteins as differentially expressed between iron-sufficient and iron-deficient plants (Fig. 1C). Only 40 proteins accumulated differentially in both roots and leaves, among them several proteins that have been previously associated with iron metabolism, including the Feruloyl CoA ortho-hydroxylase 1 (F6′H1), NICOTIANAMINE SYNTHASE4 (NAS4), IRON-RESPONSIVE PROTEIN3 (IRP3), a protein with unknown function that is highly expressed in leaves and roots of iron-deficient plants (5), and the METAL TOLERANCE PROTEIN A2 (MTPA2) (46). Notably, of the 40 differentially expressed proteins in common between roots and leaves, 10 are structural constituents of ribosomes.
Subsets of 369 and 477 transcripts showed either delta signal changes larger than the mean expression value of the whole data set or twofold changes upon iron deficiency with p < 0.05 and were defined as differentially expressed. Sixty-two mRNAs were iron-regulated in both roots and leaves (Fig. 1D; supplemental Table S2). A comparison with previously published RNA-seq data sets for roots and leaves of plants grown under similar conditions (4, 5) showed a comparable fold-change of the differentially expressed genes despite the use of different technologies, showing the robustness of the current transcriptional analysis (Fig. 2). A comparison of the differentially expressed proteins in leaves and roots revealed that only a relatively small subset of mRNAs had cognate proteins accumulated differentially between iron-deficient and iron-sufficient plants (51 and 54 mRNA/protein pairs in roots and leaves, respectively; Fig. 1E–1F).
Fig. 2.
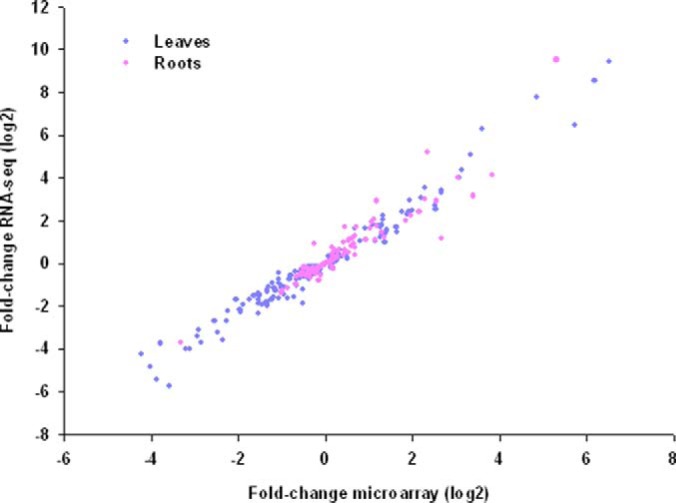
Comparison of gene expression changes between microarray data (this study) and RNA-seq data take from (4) (roots) and (5) (leaves).
rglg1 rglg2 Mutants Resemble Iron-Deficient Plants
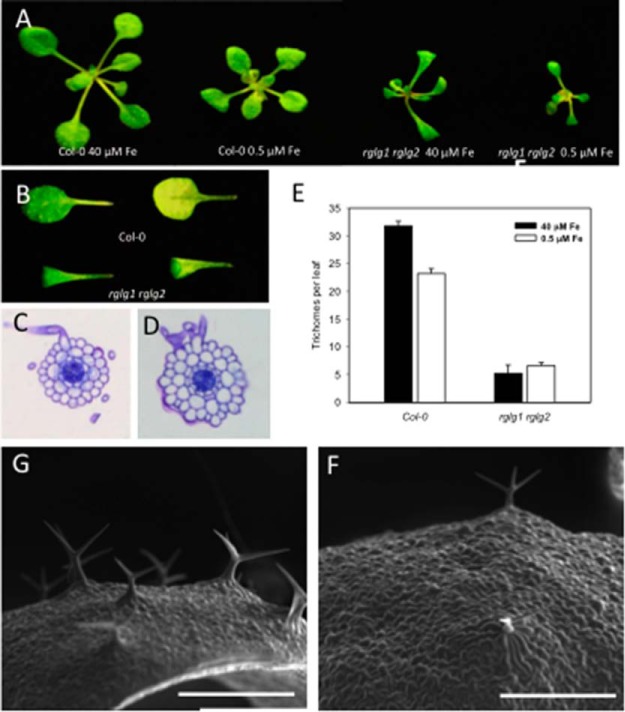
Mutants defective in the expression of the RING ligases RGLG1 and RGLG2 were considerably smaller than wild-type plants when grown on both iron-replete and low iron media (0.5 μm FeEDTA; Fig. 3A). The rglg1 rglg2 double mutants formed leaves that were curled and more pointed than those of the wild type (Fig. 3B), a trait also observed in mutants defective in the expression of genes encoding ribosomal proteins (47, 48). Constitutive root hair branching in rglg1 rglg2 plants was confirmed in the present study (Fig. 3C, 3D). In roots of rglg1 rglg2 plants, additional cell divisions were frequently observed (Fig. 3D) (35). In addition to the root hair phenotype, we observed a massive reduction in trichome density in rglg1 rglg2 plants (Fig. 3E–3G). Under control conditions, trichome density in rglg1 rglg2 plants was 84% lower when compared with wild-type plants. Iron deficiency significantly decreased the number of trichomes in the wild type (p < 0.05%, Fig. 3G), but did not further alter the trichome density in rglg1 rglg2 mutants (Fig. 3E).
Fig. 3.
Phenotype of the rglg1 rglg2 mutant. A, Two-week-old seedlings grown on iron-replete (40 μm FeEDTA) or low Fe (0.5 μm FeEDTA) media. B, Leaves of 2-week-old plants grown on iron-replete media. Adaxial surface (left panel) and abaxial surface (right panel). C, D, Cross-sections in the root hair zone of Col-0 plants, C, and rglg1 rglg2 mutants on Fe-replete media, D. E, Quantification of trichome density on leaves of Col-0 and rglg1 rglg2 plants. F, G, Scanning electron micrographs of the leaf surface of Col-0 plants, G, and rglg1 rglg2 mutants, F. Scale bars = 500 μm.
The Lack of Functional RGLG Proteins has Far-Reaching Consequences on Global Gene Expression
A total of 443 and 1,351 transcripts were differentially expressed between the wild type and the mutant in roots and leaves, respectively (Fig. 4). Subsets of 285 and 445 proteins accumulated differentially between the two genotypes in roots and leaves. Notably, several transcripts that were iron-responsive in the wild type were differentially expressed in rglg1 rglg2 plants independent of the iron supply. In roots, this set comprises 124 genes and includes genes with well-defined roles in iron metabolism such as AHA2, PAL1, SAM2, and the superoxide dismutases FSD1 and CSD1. Also, the metallothionein MT2B was constitutively (up)-regulated in rglg1 rglg2 plants. In leaves, 289 iron-responsive genes were differentially expressed between Col-0 and rglg1 rglg2 plants under control conditions. Among them are several key genes involved in photosynthesis such as PSI/PSII light harvesting complexes, PSI/PSII subunits, as well as genes encoding proteins involved in plastid-to-nucleus signal transduction (GUN5) and protoporphyrin synthesis (PORB).
Fig. 4.
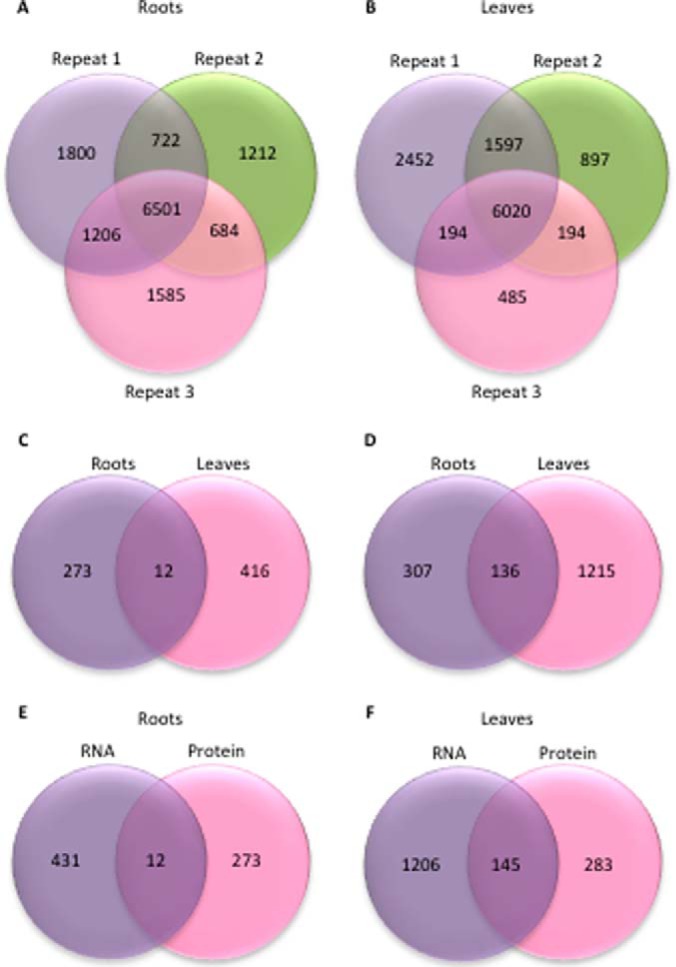
Identified and differentially expressed proteins and transcripts in roots and leaves of rglg1 rglg2 double mutants. A, B, Number of proteins identified in roots, A, and leaves, B, of iron-sufficient and iron-deficient plants in three biological repeats. C, Differentially expressed proteins. D, Differentially expressed transcripts. E, F, Overlap of differentially expressed transcripts and proteins in leaves, E, and roots, F.
Similar to what has been observed at the transcript level, relatively large subsets of proteins that were iron-responsive in the wild type had a constitutive iron response in rglg1 rglg2 mutants grown under iron-sufficient conditions (Fig. 4; supplemental Data Set S1, supplemental Tables S1 and S2). Although some constitutively regulated proteins showed similarly increased or decreased abundance in iron-deficient wild type and iron-sufficient rglg1 rglg2 plants, several exceptions to this pattern were observed. In particular, in roots, several proteins accumulated massively in rglg1 rglg2 plants under control conditions whereas, similar to iron-deficient wild-type plants, they were down-regulated when mutant plants were subjected to iron deficiency. All three ferritin genes showed reduced expression in roots and leaves of rglg1 rglg2 plants at both the RNA and protein level, indicative of a higher iron status of rglg1 rglg2 relative to Col-0 plants in both organs and growth types.
Severe repression of two transcription factors in the leaves of rglg1 rglg2 plants, the class IV homeodomain leucine zipper GLABRA2 (GL2) and the R2R3 MYB-family protein MYB23, may account for the reduced trichome phenotype of the mutant. Both proteins are critical for proper trichome development (49, 50) and act together in a positive feedback loop in which GL2 is required for transcriptional activation of MYB23 (51). In rglg1 rglg2 plants, expression of both genes was decreased by more than fourfold. In roots, the expression of three genes encoding proteins involved in cell wall loosening, the β-galactosidase glycosyl hydrolases BGAL1, BGAL4, and BGAL8 were strongly decreased in rglg1 rglg2 plants when compared with the wild type. BGAL4 and BGAL8 transcripts are enriched in root hairs (29) and showed decreased abundance in iron-deficient wild-type plants that also develop branched root hairs, albeit with a lower frequency (35, 36). Another cell wall modifying enzyme, PECTIN METHYLESTERASE17 (PME17), was down-regulated in iron-sufficient rglg1 rglg2 and iron-deficient wild-type plants. Similar to BGAL4 and BGAL8, PME17 is predominantly expressed in root hairs (29). In addition, PME17 is part of a core set of root hair genes that have been compiled from genetic, genomic, and computational analyses (52) and are thus putatively involved in root hair morphogenesis. Decreased expression of PME17, possibly in combination with a lack of functional BGAL protein, may contribute to the branched root hair phenotype.
The Majority of Iron-Responsive Proteins is Post-Transcriptionally Regulated
Proteins that are differentially expressed under iron-deficient conditions can be divided into populations of chiefly transcriptionally regulated and mainly or entirely post-transcriptionally regulated proteins. This distinction is based on the detection of a cognate, differentially expressed transcript that is similarly regulated in the protein. Transcription data are taken from either the current microarray analysis or from previous RNA-seq-based transcriptomic surveys conducted on plants grown under similar conditions (4, 5) that were surveyed to avoid false negatives because of ambiguous or absent probe sets in the ATH1 gene chip. It should be noted that the term “transcriptionally regulated” as it is used here does not exclude the possibility of additional control of protein activity at the post-transcriptional or post-translational levels.
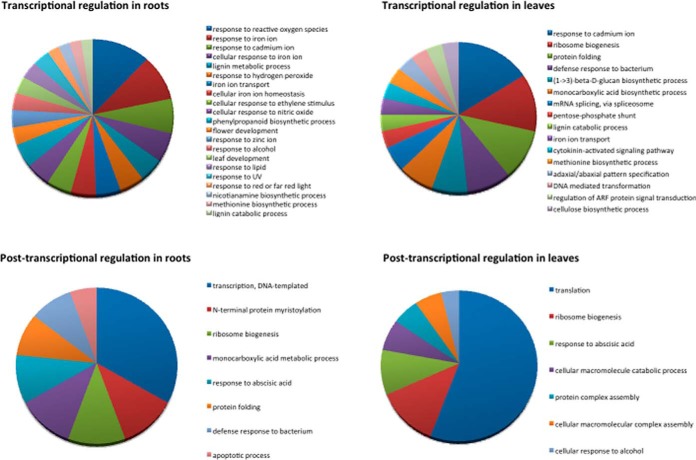
Categorizing the responses in roots and leaves of wild-type plants at the protein level revealed that in both organs transcriptional regulation appears to be more diverse, i.e. more gene ontology (GO) categories are represented when compared with post-transcriptionally regulated proteins (Fig. 5). In roots, GOs related to cellular iron homeostasis such as “response to reactive oxygen species,” “response to iron homeostasis,” “iron transport,” “response to nitric oxide,” “ethylene stimulus,” and “phenylpropanoid biosynthetic pathway” are represented by several proteins. Post-transcriptionally regulated proteins in roots comprise the GOs “DNA-templated transcription,” “N-terminal myristoylation,” and “ribosome biogenesis.” In leaves, “response to cadmium ion,” “ribosome biogenesis,” and “protein folding” represent the GOs comprising most transcriptionally regulated proteins. Among the post-transcriptionally regulated proteins, “translation” and “ribosome biogenesis” are most dominant.
Fig. 5.
Gene Ontology (GO) categorization of differentially expressed proteins.
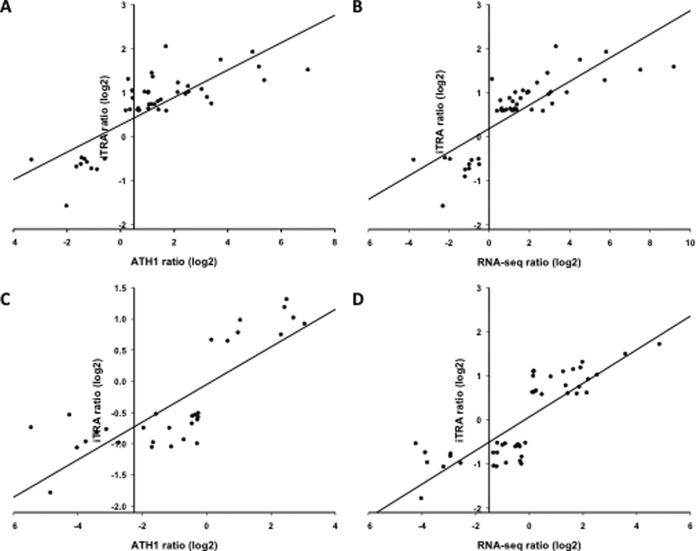
Fifty-seven and 51 similarly regulated transcripts and proteins were detected in roots and leaves of wild-type plants, respectively. These subsets account for 13 and 11% of the differentially expressed proteins, indicative of a pronounced post-transcriptional component in the “proteoferrome.” To estimate the involvement of transcriptional regulation, we analyzed the concordance of mRNA and protein expression for genes in which both the mRNA and the protein accumulated differentially between iron-sufficient and iron-deficient plants. Using the microarray-derived transcript data, in roots a Pearson correlation coefficient of r = 0.79 was calculated (Fig. 6). A similar correlation was determined using the RNA-seq data set (r = 0.81). In leaves, almost identical correlation coefficient values were observed with data derived from microarray (r = 0.83) and RNA-seq analysis (r = 0.81), indicating that changes in mRNA abundance account for ∼80% of the changes in protein abundance in the wild type for transcriptionally regulated proteins. When all protein data were taken into account for which a differentially expressed transcript was detected regardless of the direction of regulation, Pearson correlation values for roots were r = 0.62 and r = 0.65 for ATH1 and RNA-seq data, respectively, those for leaves were r = 0.74 and r = 0.68 (Fig. 6).
Fig. 6.
Concordance between changes in the abundance of mRNAs and the encoded proteins. A–D, Correlation between protein and transcript fold-changes upon iron deficiency for significantly changed transcript/protein pairs in roots, A, B, and leaves, C, D, with mRNA data taken from mircoarray analysis, A, C, or an RNA-seq survey B, D (5).
The Expression of Key Proteins in Iron Homeostasis is Transcriptionally Regulated
In roots of the wild type, the group of transcriptionally regulated proteins is composed of several enzymes that mediate the uptake of iron (IRT1 and FRO2), proteins involved in the production of iron-binding phenylpropanoids (PAL1, PAL2, F6′H1, 4CL1, 4CL2, BGLU42), enzymes mediating the biosynthesis of the iron chelator nicotianamine (NAS1, NAS4), iron storage proteins (FER1, FER3), and key players in the metabolism of iron-deficient plants (SAM1, MS2, G6PD6, PEPC1). Several of the genes encoding these proteins in roots are regulated by the transcription factor FIT (21). The transcriptionally regulated proteins contain 20 members of the “ferrome,” a core set of iron-responsive genes inferred from public microarray data (53). The function of transcriptionally regulated iron-responsive proteins and the mechanisms controlling the expression of the corresponding genes in roots have been reviewed extensively and will not be further discussed here (26, 54–65).
In leaves, the iron transporter OPT3, all three expressed ferritins (FER1, FER3, FER4), proteins involved in protoporphyrin biosynthesis (HEMA1, PORB), as well as proteins with putative roles in antioxidative stress responses (ENH1, CGLD27) accumulated differentially with similarly directed changes at the mRNA level. Also, NONINTRINSIC ABC PROTEIN1 (NAP1), a protein with homology to prokaryotic SufB proteins (66), is a member of this group. Fold-changes of the differentially expressed iron-responsive proteins discussed in the text and changes in the abundance of their cognate mRNAs are shown in Table I.
Table I. Fold-changes (log2) and regulation mode of selected iron-responsive proteins. R/L = roots/leaves; n.d. = not defined; n.c. = not significantly changed.
| Locus | Name | Biol. process | iTRAQ −Fe/+Fe | mRNA −Fe/+Fe | R/L |
|---|---|---|---|---|---|
| Transcriptionally regulated proteins | |||||
| At4g19690 | IRT1, IRON-REGULATED TRANSPORTER1 | Metal ion transport | 1.28 | 5.37 | R |
| At1g01580 | FRO2, FERRIC REDUCTION OXIDASE2 | Iron chelate reduction | 1.16 | 3.69a | R |
| At2g37040 | PAL1, PHE AMMONIA LYASE1 | Coumarin biosynthesis | 0.61 | 0.36 | R |
| At3g53260 | PAL2, PHE AMMONIA LYASE2 | Coumarin biosynthesis | 1.01 | 0.90 | R |
| At3g13610 | F6'H1, feruloyl CoA ortho-hydroxylase1 | Coumarin biosynthesis | 2.04 | 1.70 | R |
| At5g36890 | BGLU42, BETA GLUCOSIDASE42 | Coumarin biosynthesis | 1.14 | 2.53 | R |
| At5g04950 | NAS1, NICOTIANAMINE SYNTHASE1 | Nicotianamine biosynthesis | 0.61 | 1.42 | R |
| At1g56430 | NAS4, NICOTIANAMINE SYNTHASE4 | Nicotianamine biosynthesis | 1.22 | 2.15 | R |
| At5g01600 | FER1, FERRETIN 1 | Iron homeostasis | −1.58 | −2.01 | R |
| At3g56090 | FER3, FERRETIN 3 | Iron homeostasis | −0.63 | −0.98a | R |
| At1g02500 | SAM1, S-ADENOSYLMETHIONINE SYNTHETASE1 | S-adenosylmethionine biosynthesis | 1.01 | 1.05 | R |
| At3g03780 | MS2, METHIONINE SYNTHASE 2 | Methionine biosynthesis | 0.82 | 0.58a | R |
| At5g40760 | G6PD6, GLUCOSE-6-PHOSPHATE DEHYDROGENASE 6 | Glucose metabolism | 0.59 | 0.75a | R |
| At1g53310 | PEPC1, PHOSPHOENOLPYRUVATE CARBOXYLASE1 | Tricarboxylic acid cycle | 0.60 | 0.64 | R |
| At4g16370 | OPT3, OLIGOPEPTIDE TRANSPORTER | Iron homeostasis | 1.31 | 1.99 | L |
| At2g40300 | FER4, FERRITIN4 | Iron homeostasis | −1.07 | −3.20 | L |
| At1g58290 | HEMA1, glutamyl-tRNA reductase | Chlorophyll biosynthesis | −0.77 | −2.93 | L |
| At4g27440 | PORB, PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B | Chlorophyll biosynthesis | −0.94 | −0.34 | L |
| At5g17170 | ENH1, ENHANCER OF SOS3–1 | Plastid organization | −0.97 | −3.80 | L |
| At5g67370 | CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS27 | n.d. | 0.92 | 2.20 | L |
| At4g04770 | NAP1, NUCLEOSOME ASSEMBLY PROTEIN1 | Iron-sulfur cluster assembly | −0.98 | −2.55 | L |
| At1g53310 | PEPC1, PHOSPHOENOLPYRUVAT CARBOXYLASE1 | Tricarboxylic acid cycle | 0.64 | 0.60 | R |
| Post-transcriptionally regulated proteins (RGLG-independent) | |||||
| At1g01410 | PUM22, PUMILIO22 | Regulation of mRNA stability and translation | 0.99 | n.c. | R |
| At1g28060 | RDM16, RNA-DIRECTED DNA METHYLATION16 | DNA methylation | 1.34 | n.c. | R |
| At5g20320 | DCL4, DICER-LIKE4 | Small RNA biogenesis | −0.77 | n.c. | R |
| At5g22880 | HTB2, HISTONE B2 | Nucleosome assembly | 1.09 | n.c. | L |
| At5g01770 | RAPTOR2 | Cell growth | 0.83 | n.c. | L |
| At1g32500 | NAP6, NON-INTRINSIC ABC PROTEIN6 | Iron-sulfur cluster assembly | −0.60 | n.c. | L |
| At3g10670 | NAP7, NON-INTRINSIC ABC PROTEIN7 | Iron-sulfur cluster assembly | −0.78 | n.c. | L |
| At5g57030 | LUT2, LUTEIN DEFICIENT2 | Carotene biosynthesis | −0.73 | n.c. | L |
| At4g31560 | HCF153, HIGH CHLOROPHYLL FLUORESCENCE153 | Cytochrome b6f complex assembly | −0.57 | n.c. | L |
| At5g51720 | AT-NEET, NEET GROUP PROTEIN | Iron homeostasis | −0.52 | n.c. | L |
| At1g55140 | Ribonuclease III family protein | RNA processing | −0.60 | n.c. | L |
| At1g44446 | CH1, CHLORINA1 | Chlorophyll biosynthesis | −1.21 | n.c. | L |
| At4g14890 | FERREDOXIN C1 | Electron transport chain | −1.27 | n.c. | L |
| At1g59760 | ATMTR4 | rRNA processing | 0.58 | n.c. | R |
| At3g55460 | SCL30, SC35-LIKE SPLICING FACTOR30 | mRNA splicing | 0.58 | n.c. | R |
| At5g09230 | SRT2, SIRTUIN2 | Chromatin silencing | 0.83 | n.c. | R |
| At1g31280 | AGO2, ARGONAUTE2 | Small RNA biogenesis | 0.77 | n.c. | R |
| At1g31970 | STRS1, STRESS RESPONSE SUPPRESSOR1 | DNA methylation | 0.58 | n.c. | R |
| At1g04950 | TAF6, TBP-ASSOCIATED FACTOR6 | Transcription | 0.66 | n.c. | R |
| At3g18380 | DTF2, DNA-BINDING TRANSCRIPTION FACTOR2 | Transcription | 0.94 | n.c. | R |
| At1g17600 | Disease resistance protein | Defense response | −0.53 | n.c. | R |
| At4g23190 | CRK11, CYSTEINE-RICH RLK | Defense response | −0.71 | n.c. | R |
| At3g47960 | GTR1, GLUCOSINOLATE TRANSPORTER-1 | Defense response | 2.29 | n.c. | R |
| At3g50930 | BCS1, CYTOCHROME BC1 SYNTHESIS | Defense respose | 0.89 | n.c. | R |
| At5g40650 | SDH2–2, SUCCINATE DEHYDROGENASE2–2 | Mitochondrial electron transport | 0.83 | n.c. | R |
| At5g25450 | Cytochrome bd ubiquinol oxidase | Mitochondrial electron transport | 0.56 | n.c. | R |
| At4g36390 | Methylthiotransferase | Iron-sulfur cluster assembly | −0.72 | n.c. | L |
| RGLG-dependent regulation | |||||
| At3g10360 | PUM4, PUMILLO4 | n.d. | 0.68 | n.c. | R |
| At5g08450 | HDC1, HISTONE DEACETYLATION COMPLEX1 | Histone deacetylation | 0.64 | n.c. | L |
| At4g39100 | SHL1, SHORT LIFE | Chromatin assembly/disassembly | 0.65 | n.c. | L |
| At3g42790 | AL3, ALFIN-LIKE3 | Regulation of transcription | 0.59 | n.c. | L |
| At5g59870 | HTA6, HISTONE H2A 6 | Nucleosome assembly | 0.90 | n.c. | L |
| At1g10500 | CPISCA, CHLOROPLAST-LOCALIZED ISCA-LIKE PROTEIN | Iron-sulfur cluster assembly | −0.63 | n.c. | L |
| At4g03280 | PETC, PHOTOSYNTHETIC ELECTRON TRANSFER C | Photosystem II assembly | −1.0 | −0.29 | L |
| At5g52470 | FIB1, FIBRILLARIN1 | rRNA processing | 0.86 | n.c. | R |
| At5g52490 | Fibrillarin family protein | rRNA processing | −0.73 | n.c. | R |
| At1g48920 | NUC-L1NUCLEOLIN1 | rRNA processing | 0.70 | n.c. | R |
| At1g17070 | STIPL1, SPLICEOSOMAL TIMEKEEPER LOCUS1 | mRNA splicing | 0.63 | n.c. | R |
| At3g47120 | RNA recognition motif (RRM)-containing protein | n.d. | 0.76 | n.c. | R |
| At4g21660 | Proline-rich spliceosome-associated (PSP) family protein | mRNA processing | 0.71 | n.c. | R |
| At4g31580 | SRZ22, SERINE/ARGININE-RICH22 | mRNA splicing | 0.76 | n.c. | R |
| At3g61860 | RSP31, ARGININE/SERINE-RICH SPLICING FACTOR31 | mRNA splicing | 0.87 | n.c. | R |
| At1g55310 | SCL33, SC35-LIKE SPLICING FACTOR33 | mRNA splicing | 0.57 | n.c. | R |
| At3g44750 | HDA3, HISTONE DEACETYLASE3 | Histone deacetylation | 0.63 | n.c. | R |
| At5g50320 | HISTONE ACETYLTRANSFERASE8 | Histone acetylation | 0.73 | n.c. | R |
| At1g01040 | DCL1, DICER-LIKE1 | Small RNA biogenesis | 0.73 | n.c. | R |
| At4g13830 | J20, DNA J PROTEIN C26 | Protein folding | 0.59 | n.c. | R |
| At5g22060 | J2, DNAJ HOMOLOGUE2 | Protein folding | 0.61 | n.c. | R |
| At3g44110 | J3, DNAJ HOMOLOGUE3 | Protein folding | 0.64 | n.c. | R |
| At5g40820 | ATR, ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED | DNA repair | 0.62 | n.c. | R |
| At2g45280 | RAD51C, RAS ASSOCIATED WITH DIABETES PROTEIN 51C | DNA repair | −0.52 | n.c. | R |
| At3g24340 | CHR40, CHROMATIN REMODELING40 | Histone lysine methylation | 1.09 | n.c. | R |
| At1g16700 | Alpha-helical ferredoxin | Mitochondrial electron transport | −0.48 | n.c. | R |
| At3g18970 | MEF20, MITOCHONDRIAL EDITING FACTOR20 | Mitochondrial mRNA modification | −0.51 | n.c. | R |
| At1g52230 | PSAH2, PHOTOSYSTEM I SUBUNIT H2 | Photosynthesis | −0.66 | n.c. | L |
| At5g42070 | Unknown protein | n.d. | −0.62 | n.c. | L |
| At5g49940 | NFU2, CHLOROPLAST-LOCALIZED NIFU-LIKE PROTEIN2 | Iron-sulfur cluster assembly | −0.52 | n.c. | L |
| At1g67810 | SUFE2, SULFUR E2 | Iron-sulfur cluster assembly | 0.73 | n.c. | L |
| At3g24430 | HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE101 | Iron-sulfur cluster assembly | −0.51 | n.c. | L |
| At1g12110 | NRT1, NITRATE TRANSPORTER1 | Nitrate transport | −0.58 | n.c. | R |
| At2g38940 | PT2, PHOSPHATE TRANSPORTER2 | Phosphate transport | −0.48 | n.c. | R |
| At3g58810 | MTPA2, METAL TOLERANCE PROTEIN A2 | Zinc transport | −0.57 | 3.93 | R |
| At5g03570 | IREG2, IRON REGULATED2 | Nickel transport | −0.64 | 3.08 | R |
| At3g06450 | HCO3- transporter family | Anion transport | 0.99 | n.c. | R |
| At1g14400 | UBC1, UBIQUITIN CARRIER PROTEIN1 | Ubiquitin-dependent protein catabolic process | −0.59 | n.c. | L |
| At3g17000 | UBC32, UBIQUITIN-CONJUGATING ENZYME32 | Ubiquitin-dependent protein catabolic process | 0.76 | n.c. | L |
| At1g49780 | PUB26, PLANT U-BOX26 | Protein ubiquitination | 0.78 | n.c. | L |
| At2g42160 | BRIZ1, BRAP2 RING ZNF UBP DOMAIN-CONTAINING PROTEIN1 | Protein ubiquitination | −0.66 | n.c. | L |
| At2g26000 | BRIZ2, BRAP2 RING ZNF UBP DOMAIN-CONTAINING PROTEIN1 | Protein ubiquitination | −0.52 | n.c. | L |
| At2g19410 | U-box domain-containing protein kinase family protein | Protein ubiquitination | −0.58 | n.c. | L |
| At1g32850 | UBP11, UBIQUITIN-SPECIFIC PROTEASE11 | Ubiquitin-dependent protein catabolic process | −0.55 | n.c. | L |
| At4g22285 | Ubiquitin C-terminal hydrolases superfamily protein | Ubiquitin-dependent protein catabolic process | 0.60 | n.c. | L |
a RNAseq data, taken from Rodríguez-Celma et al. (4).
Post-Transcriptionally Regulated Proteins are Involved in Disparate Cellular Processes
A large subset of post-transcriptionally regulated proteins can be assigned to processes that themselves regulate post-transcriptional processes such as premRNA processing or translation. This positive feedback loop amplifies the extent of nontranscriptional regulation of gene expression and largely disconnects transcription from the proteomic readout. Several of the strongly up-regulated proteins harbor RNA-binding domains and may play putative regulatory roles. PUMILO22 (PUM22), a Puf-family protein that controls gene expression post-transcriptionally by promoting RNA decay (67, 68), showed increased abundance in iron-deficient plants. Another member of this family, PUM4, was also up-regulated by iron-deficiency. Indicative of a regulatory role in iron homeostasis, the RRM/RBD/RNP motif containing protein At4g35785 was differentially phosphorylated upon iron deficiency in a phosphoproteomic survey (7), and was post-transcriptionally up-regulated in the present study. Increased abundance upon iron deficiency was also observed for the premRNA splicing factor RNA-DIRECTED DNA METHYLATION16 (RDM16). DICER-LIKE4 (DCL4), acting in processing tasiRNAs in a small RNA generating pathway, showed markedly decreased abundance in roots of iron-deficient plants.
Similar to what has been observed in roots, a large proportion of post-transcriptionally regulated, iron-responsive proteins in leaves is involved in post-transcriptional processes. The most obvious change in post-transcriptionally regulated proteins in leaves is the large number of ribosomal proteins (r-proteins) that showed increased abundance in response to iron deficiency. Furthermore, up-regulation of HISTONE DEACETYLATION COMPLEX1, two so-called “histone readers” that bind to histone H3 tails (the PHD finger containing proteins SHORT LIFE and ALFIN-LIKE3), and strong induction of the histone variants HTA6 and HTB2 indicates epigenetic changes induced by iron deficiency. Interestingly, the pool of up-regulated proteins also contains RAPTOR2, a key node in stress signal integration and cell growth (69, 70).
The majority of the post-transcriptionally down-regulated proteins is located in the chloroplasts. Two partners of NAP1, the plastidic SufD homolog NAP6 and the SufC homolog NAP7 that form a complex in chloroplasts involved in the transfer of Fe-S clusters to apoproteins (66, 71), were among the post-transcriptionally regulated proteins. Interestingly, NAP1 was transcriptionally regulated, whereas NAP6 and NAP7 were post-transcriptionally regulated, indicating that the assembly of functional modules could be regulated at different levels. The abundance of another protein involved in chloroplast Fe-S cluster assembly, the scaffold protein CHLOROPLAST-LOCALIZED ISCA-LIKE PROTEIN (CPISCA) (72), was also decreased upon iron deficiency.
Similar to the transcriptionally regulated CGLD27 (73, 74), LUTEIN DEFICIENT2 (LUT2) was predicted to function in carotenoid-xanthophyll metabolism related to the detoxification of reactive oxygen species. The down-regulated proteins also comprise HIGH CHLOROPHYLL FLUORESCENCE153 (HCF153), which is involved in the post-translational steps of cytochrome b6f complex biogenesis (75) and the NEET GROUP PROTEIN (AT-NEET) for which a role in iron metabolism and reactive oxygen species metabolism was proposed (76). Also down-regulated were the ribonuclease III family protein At1g55140, which was associated with RNA metabolism in the chloroplast (77), and the chlorophyllide a oxygenase CHLORINA1 (CH1), which is required for chlorophyll b synthesis. Consistent with the high iron content of PS1, many of the down-regulated proteins are associated with this photosystem. Also, PHOTOSYNTHETIC ELECTRON TRANSFER C, encoding the Rieske Fe-S center of cytochrome b6f complex (78), showed decreased abundance in leaves of iron-deficient plants. The strongest post-transcriptional down-regulation was observed for FERREDOXIN C1.
RNA-Binding Proteins Occupy Central Hubs in PPI Networks of Post-Transcriptionally Regulated Proteins
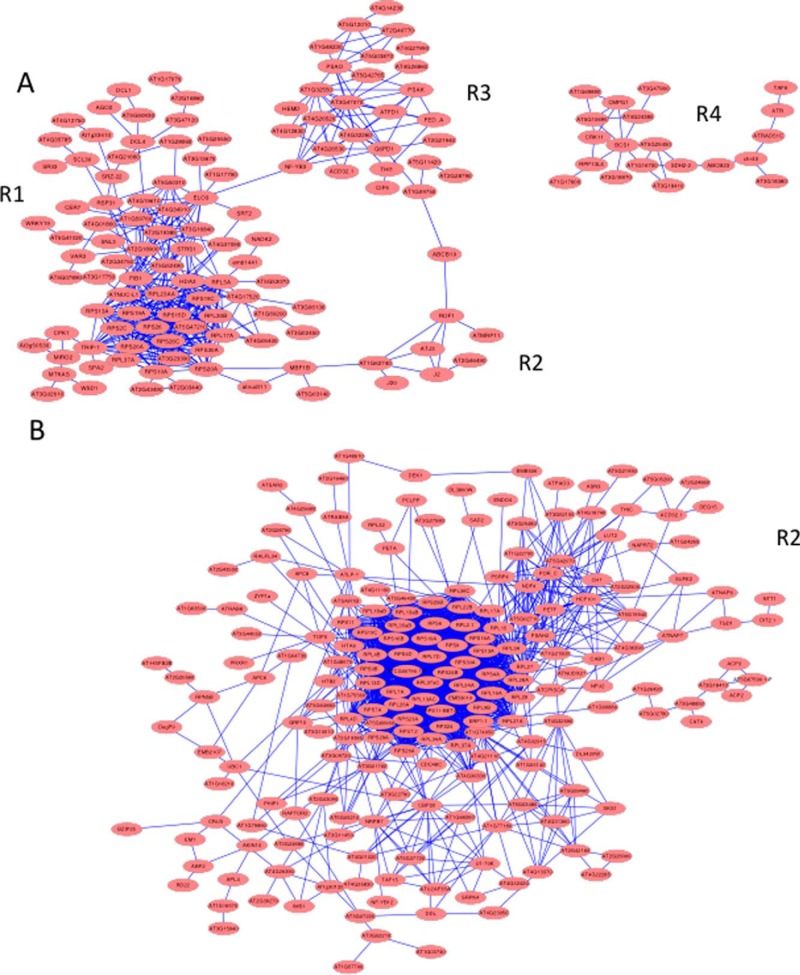
Constructing protein-protein interaction (PPI) networks of post-transcriptionally regulated proteins resulted in large clusters dominated by r-proteins in both roots and shoots (Fig. 7). Although a high connectivity of r-proteins can be anticipated from their cooperativity in ribosomal function, smaller neighboring clusters and differentially expressed proteins associated with these central clusters are of particular interest.
Fig. 7.
Protein–protein interaction (PPI) network of post-transcriptionally regulated proteins. A, PPIs of differentially expressed proteins in roots. B, PPIs of differentially expressed proteins in leaves. Networks were generated using the STRING (http://string-db.org) algorithm.
In roots, several genes involved in rRNA processing, including FIBRILLARIN1 (FIB1), the fibrillarin family protein At5g52490, NUCLEOLIN1 (NUC-L1) and a homolog of yeast MTR4, ATMTR4, are associated with this cluster (R1; Fig. 7A). Also, several RNA-binding proteins are part of this subcluster, many with annotated or validated function in premRNA splicing (STIPL1, At3g47120, RDM16, At4g21660, SRZ22, RSP31, SCL30, SCL33). All of these proteins showed increased abundance under iron deficiency. The histone deacetylases SRT2 and HDA3, as well as the HISTONE ACETYLTRANSFERASE8 are within this subcluster, indicative of post-translational modifications of histone tails by iron deficiency (Fig. 7A). This subcluster also comprises several genes involved in microRNA processing (DCL1, DCL4, and AGO2). Two genes, RDM16 and STRESS RESPONSE SUPPRESSOR1 (STRS1) have been associated with epigenetic silencing via the RNA-directed DNA methylation pathway (79, 80).
Subcluster R2 contains, among other stress-responsive proteins, three DNAJ-like proteins, J2, J3, and J20, homologs of the cochaperone DNAJ protein from E. coli that were all up-regulated upon iron deficiency. J3 acts as an activator of the plasma membrane H+-ATPase by interacting with and inhibiting the protein kinase PKS5 (81). We showed previously that pks5 mutants have increased proton pumping activity (12), supporting a scenario in which J3 activates AHA2 activity under iron-deficient conditions.
Subcluster R3 contains several DNA-binding proteins (TAF6, ATR, RAD51C, CHR40, DTF2), which are involved in the regulation of transcription, chromatin remodeling and DNA repair, and proteins related to pathogen defense (At1g17600, CRK11, GTR1, BCS1). Several mitochondrial proteins associated with respiration (SDH2–2, At5g25450, At1g16700) and MITOCHONDRIAL EDITING FACTOR20 are part of this cluster. ABC TRANSPORTER OF THE MITOCHONDRION1 (ATM1), a mitochondrial ATP-binding cassette transporter homologous to the yeast mitochondrial membrane protein ScATM1 involved in the mitochondrial export of Fe-S clusters or their precursors (82), was down-regulated upon Fe-deficiency but SDH2–2 and At5g25450 were up-regulated, probably compensating for the decrease in mitochondrial respiration under iron-deficient conditions as a consequence of impaired heme and Fe-S cluster synthesis (83, 84). The Fe-S cluster-binding protein At1g16700, a component of complex I, showed decreased abundance upon iron deficiency. A smaller cluster, R4, contains mainly plastid genes.
In leaves, the network is more complex. A larger cluster (L1) is dominated by r-proteins and also contains proteins related to photosynthesis, including, among others, a PSI subunit (PSAH2) and the chlorophyllide a oxygenase CH1 (Fig. 7B). Interestingly, a putative positive regulator of transcription (At5g42070) has a central position in this cluster. LUT2, several proteins involved in Fe-S cluster assembly (NAP6, NAP7, NFU2, SUFE2, HCF101, and CPISCA) and the Fe-S binding protein At4g36390, which was strongly down-regulated and highly interconnected with the Fe-S cluster assembly machinery, are members of this cluster.
Post-Transcriptional Regulation of Iron-Responsive Proteins is Partly Dependent on RGLG
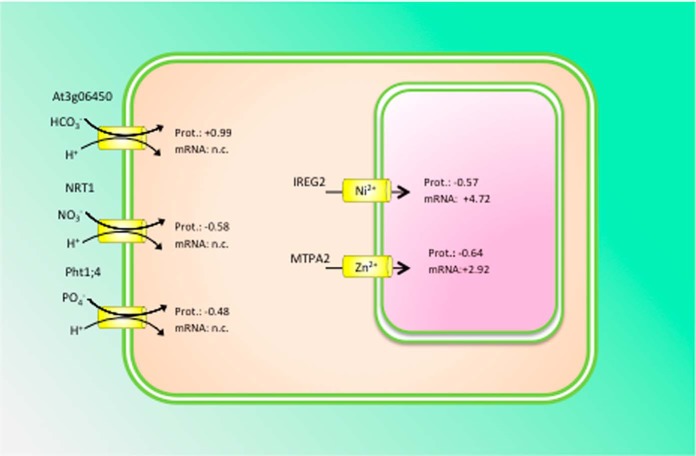
Relatively large sets of proteins were iron-regulated solely in wild-type plants but not in rglg1 rglg2 mutants (275 proteins in roots and 233 in leaves) and were thus classified as RGLG-dependent. In addition, iron-responsive proteins that were deregulated in rglg1 rglg2 plants under iron-deficient conditions (i.e. regulated in the opposite direction) were included in this group. In roots of wild-type plants, 164 RGLG-dependent proteins were up-regulated by iron deficiency, among them are many proteins with RNA-binding properties including r-proteins and proteins involved in RNA processing or metabolism. Several transporters for mineral nutrients localized on the plasma or vacuolar membrane were among the 114 down-regulated proteins, including NITRATE TRANSPORTER1 (NRT1), PHOSPHATE TRANSPORTER2 (PT2), MTPA2, and IRON-REGULATED PROTEIN2 (IREG2) (Fig. 8). The major plasma membrane transporters mediating the uptake of nitrate and phosphate in roots, NRT1 and PT2, were also down-regulated in iron-deficient wild-type plants. Uptake of anions that are acquired by proton-anion cotransport (nH+/A−) such as nitrate and phosphate (85, 86) decreases the electrochemical potential of the plasma membrane and counteracts iron solubilization in the rhizosphere via ATPase-mediated proton extrusion (87). A more acidic uptake pattern (i.e. an increased cation/anion ratio), achieved by decreased anion uptake is thus advantageous for the plant under iron-deficient conditions. Remarkably, the regulation of these transporters at the protein level was not observed in rglg1 rglg2 mutants.
Fig. 8.
RGLG-dependent regulation of transport proteins. Proton-cotransport of bicarbonate, nitrate and phosphate at the plasma membrane is post-transcriptionally down-regulated to maintain an acidic uptake pattern that facilitates the mobilization of Fe(III) oxides in the rhizosphere by ATPase-mediated proton extrusion. The transcript levels of these transporters remained unchanged. At the tonoplast, IREG2 and MTPA2 protein was decreased under iron-deficient conditions, whereas the genes were highly up-regulated at the transcriptional level. Numbers denote log2 fold changes of the respective proteins. n.c. = not changed.
An uncharacterized bicarbonate (HCO3−) transporter, At3g06450, was highly up-regulated in iron-deficient plants at the protein level, but the transcript abundance remained unchanged (Fig. 8). No regulation of the protein abundance was observed in the mutant. Dark fixation of CO2 via phosphoenolpyruvate carboxylase (PEPC), catalyzing the assimilation of bicarbonate to phosphoenolpyruvate to form oxaloacetate and inorganic phosphate, has been described as an adaptive response of roots to iron deficiency as a consequence of increased ATPase-mediated proton extrusion and a higher demand for organic acids (88, 89). Similar to what was reported previously for Arabidopsis and other strategy I plants, PEPC1 was highly induced upon iron starvation both at the transcript and protein level. An iron-responsive bicarbonate transporter has not yet been described. It is thus tempting to speculate that At3g06450 fulfills such a function and has escaped detection in transcriptional surveys because of a lack of transcriptional regulation of the gene.
In contrast to their cognate proteins that showed decreased abundance upon iron deficiency, IREG2 and MTPA2 transcripts were strongly enriched in roots of iron-deficient plants, indicating post-transcriptional regulation of these transporters. Reduced abundance of MTPA2 protein was not observed in rglg1 rglg2 mutants, indicating that post-transcriptional regulation of MTPA2 is dependent on functional RGLG protein. MTPA2, a vacuolar zinc transporter, is critical for the detoxification of surplus zinc in root cells of iron-deficient plants that accumulates intracellularly as a result of the low substrate specificity of the iron transporter IRT1 (46). We previously showed that the transcriptional regulation of MTPA2 is independent of the zinc supply and solely regulated by the iron status of the plants (81). Together, the present results that iron-dependent regulation of MTPA2 occurs at the transcriptional level, whereas protein abundance is post-transcriptionally controlled, presumably by the zinc status of the cell (Fig. 8).
The tonoplast transport protein IREG2, an ortholog of the mammalian iron efflux transporter ferroportin, was regulated in a similar manner, i.e. massive up-regulation at the transcriptional level and decreased protein abundance in response to iron deficiency. IREG2 sequesters nickel into the vacuole, but increased concentrations of nickel did not alter the expression of IREG2 (90), indicating that transcriptional regulation is independent of the actual concentration of potentially toxic metal ions. Co-expression analysis revealed closely coregulated, iron-dependent expression of IRT1, MTPA2, and IREG2 (81), suggesting that an “iron regulon” dictates transcriptional expression according to the iron status. All three genes are regulated by FIT (21), supporting this supposition. The pronounced up-regulation of MTPA2 and IREG2 mRNA may be interpreted in terms of “priming” the plant to a rapid production of the cognate proteins when the metal concentrations reach a critical level. Tuning of protein abundance and thus transporter activity to the prevailing metal concentration occurs post-transcriptionally and is dependent on RGLG (Fig. 8).
In leaves, 141 RGLG-dependent proteins were up-regulated and 99 proteins showed decreased abundance under iron deficiency. Notably, the ubiquitin carrier protein UBC1, the ubiquitin-conjugase UBC32, ubiquitin ligases (PUB26, BRIZ1, BRIZ2, and At2g19410), an ubiquitin-specific protease (UBP11), and an ubiquitin C-terminal hydrolases superfamily protein (At4g22285) were regulated by RGLG, indicating regulatory intervention of protein turnover by RGLG.
Does Iron Deficiency Induce Ribosome Specialization?
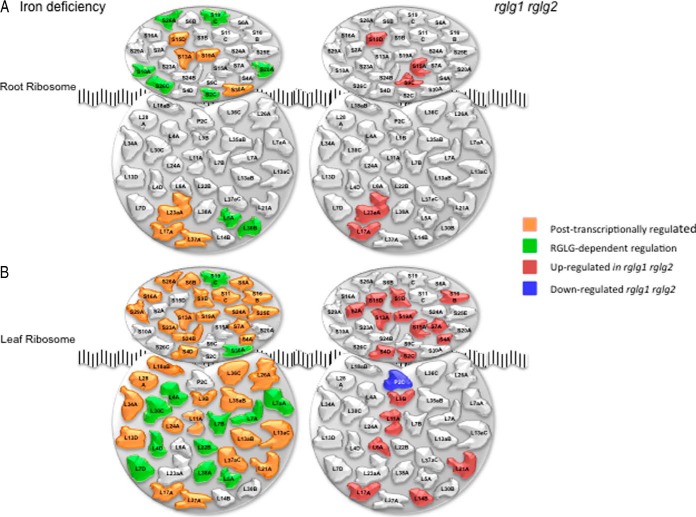
The present data show that iron deficiency post-transcriptionally induces changes in r-protein composition, leading to the formation of iron deficiency-specific ribosomes, a response that could not be deduced from a transcriptional analysis. Changes in r-protein expression were mainly post-transcriptionally controlled and thus masked in transcriptional surveys. Under iron-sufficient conditions, a large set of r-proteins showed significantly higher abundance in rglg1 rglg2 mutant plants compared with the wild type (five in roots and 16 in leaves, one r-protein down-regulated; Fig. 9). With one exception, the higher abundance of r-proteins relative to the wild type was not accompanied by increased transcript abundance, indicating that RGLG is a negative, post-transcriptional regulator of r-protein expression.
Fig. 9.
Differential expression of r-proteins. A, Changes induced by iron deficiency, B, Altered expression of r-proteins in rglg1 rglg2 double mutants.
In roots of wild-type plants, 15 r-proteins showed increased abundance upon iron deficiency, all of which were not accompanied by changes in mRNA level. In leaves, a subset of 54 r-proteins showed increased expression in iron-deficient relative to iron-sufficient plants. Similar to roots, all r-proteins were post-transcriptionally regulated. Several of the r-proteins that were differentially expressed in the wild type were either not iron-responsive in rglg1 rglg2 mutants (i.e. positively regulated by RGLG), or constitutively up-regulated, a group that we defined as negatively RGLG-regulated. With these criteria, we defined 50 r-proteins (eight in roots and 42 in leaves) as positively RGLG-regulated and nine r-proteins (two in roots and seven in leaves) as negatively regulated by RGLG (Fig. 9).
DISCUSSION
Transcriptional Regulation of Iron Homeostasis: The Tip of the Iceberg?
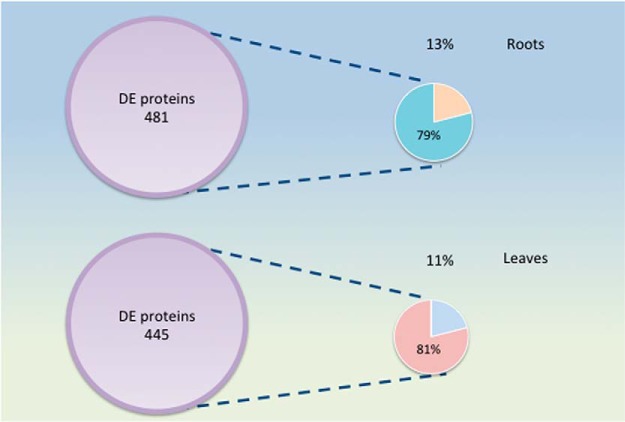
In summary, here we provide a high-resolution, nearly complete inventory of the Arabidopsis “proteoferrome”: proteins that accumulate differentially in roots and leaves between iron-sufficient and iron-deficient plants. A graphical overview of the mode of regulations of the proteins discussed here is given in Fig. 10. By uncovering an extensive and largely unexplored suite of iron-responsive proteins that are not or only partially transcriptionally regulated, we emphasize that a broad understanding of plant behavior in response to changing iron supply can only be achieved when post-transcriptional processes are taken into consideration. We show further that estimates made from changes in transcript levels may lead to incorrect predictions regarding the abundance of the cognate proteins. Concordance between mRNA and protein expression has often been used to estimate the contribution of transcriptional regulation on changes in protein abundance, and values around 40% have been inferred for several organisms from such calculations reviewed by Vogel and Marcotte (91). However, without a near-complete coverage of changes in the proteomic readout, such estimates rely on a relatively small proportion of transcripts and proteins for which the corresponding partners have been detected. In the present study, this fraction was estimated to account for 13 and 11% of the differentially expressed proteins in roots and leaves, respectively, resulting in a total contribution of transcription of 11 and 9% when the whole subset of differentially accumulated proteins and the concordance of mRNA and protein expression of this subset are taken into consideration. Thus, ∼90% of the differentially expressed genes are not surveyed in transcriptional profiling studies, that several critical regulatory mechanisms and key players are yet to be uncovered (Fig. 11).
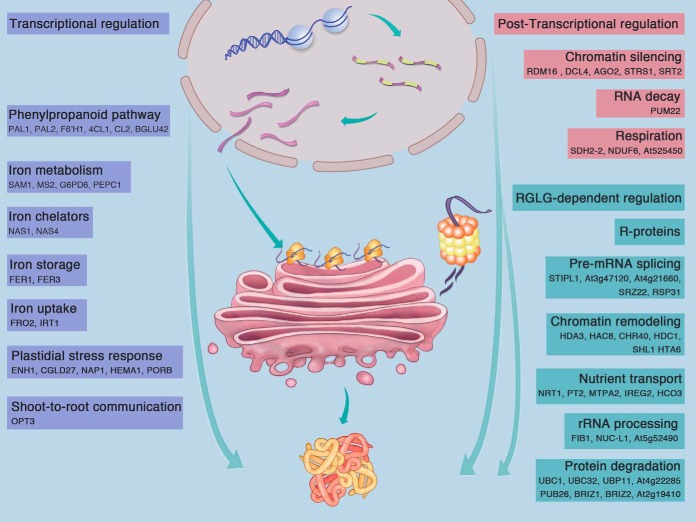
Fig. 10.
Regulatory control of iron-responsive proteins. Processes directly related to iron homeostasis such as the acquisition, uptake, chelation, and storage of iron, as well as adaptive responses in the plastid and shoot-to-root communication are chiefly transcriptional regulated. Proteins involved in RNA-directed DNA methylation, RNA decay and changes in oxidative phosphorylation are primarily post-transcriptionally regulated. RGLG affects predominantly processes involved in protein translation and degradation, but also post-transcriptional modifications of histones. RGLG is also critical for tuning the abundance of transport proteins. Proteins listed in the figure are discussed in the text.
Fig. 11.
Estimate of the concordance between mRNA and protein expression in iron-deficient plants. Only 13% (roots) and 11% (leaves) of the proteins that are differentially expressed between iron-deficient and iron-sufficient plants (DE proteins) are associated with similarly regulated transcripts. For proteins in this subset, changes in transcript level account for circa 80% of the observed changes in protein abundance.
Ribosome Composition - A novel Regulator of Translation?
In Arabidopsis, r-proteins are encoded by gene families comprising two to seven members, producing orthologs that for the most part are sufficiently unique to be distinguished by mass spectrometric analysis and that may have different properties. This stands in juxtaposition to mammals where r-proteins are encoded by a single gene and yeast where the diversity of r-proteins is less pronounced compared with plants. The heterogeneity of r-protein orthologs gives rise to speculations that specific, “made-to-order” ribosomes are generated that can capture mRNAs differentially and adapt the plant to the prevailing conditions by prioritizing a subset of mRNAs for translation (92–96). This hypothesis is particularly appealing from the perspective that translation in general and ribosome synthesis in particular are highly energy-consuming processes that need to be calibrated under stress conditions to secure plant vigor and productivity. Changes in r-gene expression have been described as being stress-specific (5, 97), but the current analysis shows that alterations in r-protein expression are post-transcriptionally regulated, making a proteomic analysis mandatory to identify such changes. At the protein level, it was shown that growth condition can alter the relative incorporation of r-protein paralogs into ribosomes (98). Regulation of protein abundance at the level of translation was also supported by translatome profiling studies (99, 100). These studies also support the assumption of a strong effect of “translational fitness” of specific mRNAs, a trait that is not necessarily associated with high steady-state mRNA levels (101, 102). Several mechanisms at the translation level such as internal ribosome entry site (IRES)-mediated translation, ribosome pausing, and modification of translation initiation factors (103–105) may contribute to post-transcriptional control of gene activity, although their individual contribution in plants remains to be shown. The present data strongly support the idea of a regulatory intervention at the level of translation, aiding in translating a subset of mRNAs that are critical in adapting proteomic profiles to the prevailing conditions. Changes in ribosome composition, resulting in altered mRNA efficiency, may represent a key component in a sophisticatedly coordinated response to environmental cues.
RGLG Plays Critical Roles in The Orchestration of The Iron Deficiency Responses
Our results further show that the RING ligases RGLG1 and RGLG2 are required for the orchestration of the post-transcriptional responses, underlining the complexity of acclimation to abiotic stresses. This includes the whole repertoire of post-transcriptional processes comprising mRNA maturation, generation of small RNAs, mRNA export from the nucleus as well as changes at the epigenetic level such as DNA and histone modifications that are mediated by post-transcriptionally regulated proteins. RGLG can affect protein abundance directly by targeting proteins for proteasome-mediated degradation, indirectly via nonproteolytic reversible modifications such as monoubiquitilation and, in conjunction with UBC13, by promoting the formation of K63-linked polyubiquitin chains, which can affect diverse cellular processes such as transcription, histone function, membrane trafficking, DNA repair, and DNA replication (106–109).
Interestingly, the percentage of proteins that were regulated without changes in corresponding mRNA abundance was lower for RGLG-regulated proteins than for the proteins that were regulated in common in both genotypes. In leaves, cognate transcripts for only 13 of the 240 RGLG-dependent proteins were identified. The concordance between mRNA and proteins was also markedly lower than that observed for the common set of genes (r = 0.03). In roots, a similar picture was observed. Only 6% of the proteins (18 out of 278) were associated with a differentially expressed transcript, with a weak correlation between mRNAs and proteins (r = 0.07). Together, these data indicate that RGLG is more engaged in post-transcriptional regulation of iron-responsive proteins and plays a minor and rather indirect role in the control of transcription.
Although the iceberg analogy would discredit the big steps forward already made by approaches targeting transcriptional regulation, the current investigation invites the exploration of a large portion of genes with altered expression upon iron starvation that may aid in generating iron-efficient germplasm and to combat iron deficiency-induced anemia.
Supplementary Material
Acknowledgments
We thank Drs. Louis Grillet and Isabel Cristina Vélez-Bermúdez for critical reading of the manuscript.
Footnotes
Author contributions: I.P. and W.S. designed research; I.P., H.T., Y.C., and W.S. performed research; I.P., H.T., Y.C., T.W., T.J.B., and W.S. analyzed data; W.S. and I.P. wrote the paper.
* Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/) supported by Academia Sinica. Work in the Schmidt laboratory is supported by grants from the Ministry of Science and Technology (MoST) and Academia Sinica. rglg1 rglg2 mutants were kindly provided by Andreas Bachmair (University of Vienna). Proteomic mass spectrometry analyses and in-gel digestion was performed by the Proteomics Core Laboratory sponsored by the Institute of Plant and Microbial Biology and the Agricultural Biotechnology Research Center, Academia Sinica. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [110] via the PRIDE partner repository with the dataset identifier PXD002126.
 This article contains supplemental Data Set S1 and Tables S1 and S2.
This article contains supplemental Data Set S1 and Tables S1 and S2.
1 The abbreviations used are:
- RGLG
- ring domain ligase
- FRO
- ferric reduction oxidase
- IRT
- iron-regulated transporter.
REFERENCES
- 1. Archibald F. S., Fridovich I. (1981) Manganese and defenses against oxygen-toxicity in Lactobacillus plantarum. J. Bacteriol. 145, 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Posey J. E., Gherardini F. C. (2000) Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653 [DOI] [PubMed] [Google Scholar]
- 3. Briat J. F., Dubos C., Gaymard F. (2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20, 33–40 [DOI] [PubMed] [Google Scholar]
- 4. Rodríguez-Celma J., Lin W. D., Fu G. M., Abadía J., López-Millán A. F., Schmidt W. (2013a) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol. 162, 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodríguez-Celma J., Pan I. C., Li W., Lan P., Buckhout T. J., Schmidt W. (2013) The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant Sci. 4, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng L. Q., Huang F. L., Narsai R., Wu J. J., Giraud E., He F., Cheng L. J., Wang F., Wu P., Whelan J., Shou H. X. (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lan P., Li W. F., Wen T. N., Schmidt W. (2012) Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiol. 159, 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen L., Ding C., Zhao X., Xu J., Mohammad A. A., Wang S., Ding Y. (2015) Differential regulation of proteins in rice (Oryza sativa L.) under iron deficiency. Plant Cell Rep. 34, 83–96 [DOI] [PubMed] [Google Scholar]
- 9. Fourcroy P., Siso-Terraza P., Sudre D., Saviron M., Reyt G., Gaymard F., Abadía A., Abadía J., Alvarez-Fernandez A., Briat J. F. (2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167 [DOI] [PubMed] [Google Scholar]
- 10. Schmid N. B., Giehl R. F., Doll S., Mock H. P., Strehmel N., Scheel D., Kong X., Hider R. C., von Wirén N. (2014) Feruloyl-CoA 6′-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 164, 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmidt H., Gunther C., Weber M., Sporlein C., Loscher S., Bottcher C., Schobert R., Clemens S. (2014) Metabolome analysis of Arabidopsis thaliana roots identifies a key metabolic pathway for iron acquisition. Plos One 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santi S., Schmidt W. (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183, 1072–1084 [DOI] [PubMed] [Google Scholar]
- 13. Robinson N. J., Procter C. M., Connolly E. L., Guerinot M. L. (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697 [DOI] [PubMed] [Google Scholar]
- 14. Eide D., Broderius M., Fett J., Guerinot M. L. (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vert G., Grotz N., Dedaldechamp F., Gaymard F., Guerinot M. L., Briat J. F., Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grusak M. A., Pezeshgi S. (1996) Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 110, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stacey M. G., Patel A., McClain W. E., Mathieu M., Remley M., Rogers E. E., Gassmann W., Blevins D. G., Stacey G. (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhai Z. Y., Gayomba S. R., Jung H. I., Vimalakumari N. K., Pineros M., Craft E., Rutzke M. A., Danku J., Lahner B., Punshon T., Guerinot M. L., Salt D. E., Kochian L. V., Vatamaniuk O. K. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26, 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendoza-Cozatl D. G., Xie Q. Q., Akmakjian G. Z., Jobe T. O., Patel A., Stacey M. G., Song L. H., Demoin D. W., Jurisson S. S., Stacey G., Schroeder J. I. (2014) OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol. Plant 7, 1455–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bauer P., Bereczky Z., Brumbarova T., Klatte M., Wang H. Y. (2004) Molecular regulation of iron uptake in the dicot species Lycopersicon esculentum and Arabidopsis thaliana. Soil Sci. Plant Nutr. 50, 997–1001 [Google Scholar]
- 21. Colangelo E. P., Guerinot M. L. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jakoby M., Wang H. Y., Reidt W., Weisshaar B., Bauer P. (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534 [DOI] [PubMed] [Google Scholar]
- 23. Wang H. Y., Klatte M., Jakoby M., Baumlein H., Weisshaar B., Bauer P. (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908 [DOI] [PubMed] [Google Scholar]
- 24. Yuan Y. X., Zhang J., Wang D. W., Ling H. Q. (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15, 613–621 [DOI] [PubMed] [Google Scholar]
- 25. Yuan Y. X., Wu H. L., Wang N., Li J., Zhao W. N., Du J., Wang D. W., Ling H. Q. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397 [DOI] [PubMed] [Google Scholar]
- 26. Ivanov R., Brumbarova T., Bauer P. (2012) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5, 27–42 [DOI] [PubMed] [Google Scholar]
- 27. Long T. A., Tsukagoshi H., Busch W., Lahner B., Salt D. E., Benfey P. N. (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selote D., Samira R., Matthiadis A., Gillikin J. W., Long T. A. (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 167, 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lan P., Li W. F., Lin W. D., Santi S., Schmidt W. (2013) Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sivitz A., Grinvalds C., Barberon M., Curie C., Vert G. (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J. 66, 1044–1052 [DOI] [PubMed] [Google Scholar]
- 31. Lingam S., Mohrbacher J., Brumbarova T., Potuschak T., Fink-Straube C., Blondet E., Genschik P., Bauer P. (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23, 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barberon M., Zelazny E., Robert S., Conejero G., Curie C., Friml J., Vert G. (2011) Monoubiquitin-dependent endocytosis of the IRON-REGULATED TRANSPORTER 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108, E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ivanov R., Brumbarova T., Blum A., Jantke A. M., Fink-Straube C., Bauer P. (2014) SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26, 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin L. J., Lo J. C., Chen G. H., Callis J., Fu H. Y., Yeh K. C. (2013) IRT1 DEGRADATION FACTOR1, a RING E3 ubiquitin ligase, regulates the degradation of IRON-REGULATED TRANSPORTER1 in Arabidopsis. Plant Cell 25, 3039–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li W. F., Schmidt W. (2010) Nonproteolytic protein ubiquitination is crucial for iron deficiency signaling. Plant Signal Behav. 5, 561–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Müller M., Schmidt W. (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 134, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wen R., Newton L., Li G. Y., Wang H., Xiao W. (2006) Arabidopsis thaliana UBC13: implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Biol. 61, 241–253 [DOI] [PubMed] [Google Scholar]
- 38. Wen R., Wang S., Xiang D. Q., Venglat P., Shi X. Z., Zang Y. P., Datla R., Xiao W., Wang H. (2014) UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 80, 424–436 [DOI] [PubMed] [Google Scholar]
- 39. Hofmann R. M., Pickart C. M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 40. Yin X. J., Volk S., Ljung K., Mehlmer N., Dolezal K., Ditengou F., Hanano S., Davis S. J., Schmelzer E., Sandberg G., Teige M., Palme K., Pickart C., Bachmair A. (2007) Ubiquitin lysine 63 chain-forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19, 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng M. C., Hsieh E. J., Chen J. H., Chen H. Y., Lin T. P. (2012) Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li W. F., Schmidt W. (2010) A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 62, 330–343 [DOI] [PubMed] [Google Scholar]
- 43. Pan I. C., Schmidt W. (2014) Functional implications of K63-linked ubiquitination in the iron deficiency response of Arabidopsis roots. Front. Plant Sci. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Estelle M. A., Somerville C. (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206 [Google Scholar]
- 45. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 46. Arrivault S., Senger T., Kramer U. (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46, 861–879 [DOI] [PubMed] [Google Scholar]
- 47. Horiguchi G., Molla-Morales A., Perez-Perez J. M., Kojima K., Robles P., Ponce M. R., Micol J. L., Tsukaya H. (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736 [DOI] [PubMed] [Google Scholar]
- 48. Byrne M. E. (2009) A role for the ribosome in development. Trends Plant Sci. 14, 512–519 [DOI] [PubMed] [Google Scholar]
- 49. Kirik V., Lee M. M., Wester K., Herrmann U., Zheng Z. G., Oppenheimer D., Schiefelbein J., Hülskamp M. (2005) Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1749–1749 [DOI] [PubMed] [Google Scholar]
- 50. Rerie W. G., Feldmann K. A., Marks M. D. (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399 [DOI] [PubMed] [Google Scholar]
- 51. Khosla A., Paper J. M., Boehler A. P., Bradley A. M., Neumann T. R., Schrick K. (2014) HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26, 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bruex A., Kainkaryam R. M., Wieckowski Y., Kang Y. H., Bernhardt C., Xia Y., Zheng X. H., Wang J. Y., Lee M. M., Benfey P., Woolf P. J., Schiefelbein J. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. Plos Genet. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmidt W., Buckhout T. J. (2011) A hitchhiker's guide to the Arabidopsis ferrome. Plant Physiol. Biochem. 49, 462–470 [DOI] [PubMed] [Google Scholar]
- 54. Curie C., Briat J. F. (2003) Iron transport and signaling in plants. Annu. Rev. Plant Biol. 54, 183–206 [DOI] [PubMed] [Google Scholar]
- 55. Schmidt W. (2003) Iron solutions: acquisition strategies and signaling pathways in plants. Trends Plant Sci. 8, 188–193 [DOI] [PubMed] [Google Scholar]
- 56. Schmidt W. (2003) Iron homeostasis in plants: Sensing and signaling pathways. J. Plant Nutr. 26, 2211–2230 [Google Scholar]
- 57. Schmidt W. (2006) Iron stress responses in roots of Strategy I Plants. Iron Nutrition in Plants and Rhizospheric Microorganisms, pp. 229–250, eds Barton L.L., Abadía J., Springer, Dordrecht, The Netherlands [Google Scholar]
- 58. Kim S. A., Guerinot M. L. (2007) Mining iron: iron uptake and transport in plants. FEBS Lett. 581, 2273–2280 [DOI] [PubMed] [Google Scholar]
- 59. Walker E. L., Connolly E. L. (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11, 530–535 [DOI] [PubMed] [Google Scholar]
- 60. Jeong J., Guerinot M. L. (2009) Homing in on iron homeostasis in plants. Trends Plant Sci. 14, 280–285 [DOI] [PubMed] [Google Scholar]
- 61. Morrissey J., Guerinot M. L. (2009) Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 109, 4553–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Conte S. S., Walker E. L. (2011) Transporters contributing to iron trafficking in plants. Mol. Plant 4, 464–476 [DOI] [PubMed] [Google Scholar]
- 63. Hindt M. N., Guerinot M. L. (2012) Getting a sense for signals: regulation of the plant iron deficiency response. BBA-Mol. Cell Res. 1823, 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kobayashi T., Nishizawa N. K. (2012) Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152 [DOI] [PubMed] [Google Scholar]
- 65. Buckhout T. J., Schmidt W. (2013) Iron in plants. Wiley Online Library [Google Scholar]
- 66. Xu X. M., Adams S., Chua N. H., Moller S. G. (2005) AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J. Biol. Chem. 280, 6648–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Francischini C. W., Quaggio R. B. (2009) Molecular characterization of Arabidopsis thaliana PUF proteins - binding specificity and target candidates. FEBS J. 276, 5456–5470 [DOI] [PubMed] [Google Scholar]
- 68. Tam P. P. C., Barrette-Ng I. H., Simon D. M., Tam M. W. C., Ang A. L., Muench D. G. (2010) The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity, and subcellular localization. BMC Plant Biol. 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anderson G. H., Veit B., Hanson M. R. (2005) The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mahfouz M. M., Kim S., Delauney A. J., Verma D. P. S. (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu X. M., Moller S. G. (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 9143–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Abdel-Ghany S. E., Ye H., Garifullina G. F., Zhang L. H., Pilon- Smits E. A. H., Pilon M. (2005) Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 138, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Urzica E. I., Casero D., Yamasaki H., Hsieh S. I., Adler L. N., Karpowicz S. J., Blaby-Haas C. E., Clarke S. G., Loo J. A., Pellegrini M., Merchant S. S. (2012) Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24, 3921–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kourmpetis Y. A. I., van Dijk A. D. J., van Ham R. C. H. J., ter Braak C. J. F. (2011) Genome-wide computational function prediction of Arabidopsis proteins by integration of multiple data sources. Plant Physiol 155, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lennartz K., Bossmann S., Westhoff P., Bechtold N., Meierhoff K. (2006) HCF153, a novel nuclear-encoded factor necessary during a post-translational step in biogenesis of the cytochrome b6f complex. Plant J. 45, 101–112 [DOI] [PubMed] [Google Scholar]
- 76. Nechushtai R., Conlan A. R., Harir Y., Song L., Yogev O., Eisenberg-Domovich Y., Livnah O., Michaeli D., Rosen R., Ma V., Luo Y., Zuris J. A., Paddock M. L., Cabantchik Z. I., Jennings P. A., Mittler R. (2012) Characterization of Arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell 24, 2139–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stoppel R., Manavski N., Schein A., Schuster G., Teubner M., Schmitz-Linneweber C., Meurer J. (2012) RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res. 40, 8593–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Okegawa Y., Tsuyama M., Kobayashi Y., Shikanai T. (2005) The pgr1 mutation in the Rieske subunit of the cytochrome b6f complex does not affect PGR5-dependent cyclic electron transport around photosystem I. J. Biol. Chem. 5, 28332–28336 [DOI] [PubMed] [Google Scholar]
- 79. Huang C. F., Miki D., Tang K., Zhou H. R., Zheng Z. M., Chen W., Ma Z. Y., Yang L., Zhang H., Liu R. Y., He X. J., Zhu J. K. (2013) A premRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. Plos Genet. 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Khan A., Garbelli A., Grossi S., Florentin A., Batelli G., Acuna T., Zolla G., Kaye Y., Paul L., Zhu J., Maga G., Grafi G., Barak S. (2014) The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J. 79, 28–43 [DOI] [PubMed] [Google Scholar]
- 81. Yang T. J. W., Lin W. D., Schmidt W. (2010) Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol. 152, 2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen S., Sanchez-Fernandez R., Lyver E. R., Dancis A., Rea P. A. (2007) Functional characterization of AtATM1, AtATM2, and AtATM3, a subfamily of Arabidopsis half-molecule ATP-binding cassette transporters implicated in iron homeostasis. J. Biol. Chem. 282, 21561–21571 [DOI] [PubMed] [Google Scholar]
- 83. Pascal N., Douce R. (1993) Effect of iron-deficiency on the respiration of sycamore (Acer pseudoplatanus L) Cells. Plant Physiol. 103, 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Vigani G., Maffi D., Zocchi G. (2009) Iron availability affects the function of mitochondria in cucumber roots. New Phytol. 182, 127–136 [DOI] [PubMed] [Google Scholar]
- 85. Krouk G., Crawford N. M., Coruzzi G. M., Tsay Y. F. (2010) Nitrate signaling: adaptation to fluctuating environments. Curr. Opin. Plant Biol. 13, 266–273 [DOI] [PubMed] [Google Scholar]
- 86. Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., Nakanishi T. M., Thibaud M. C. (2011) Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schmidt W. (1999) Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol. 141, 1–26 [Google Scholar]
- 88. De Nisi P., Zocchi G. (2000) Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: activity and kinetic characterization. J. Exp. Bot. 51, 1903–1909 [DOI] [PubMed] [Google Scholar]
- 89. Abadía J., López-Millán A. F., Rombola A., Abadía A. (2002) Organic acids and Fe deficiency: a review. Plant Soil 241, 75–86 [Google Scholar]
- 90. Schaaf G., Honsbein A., Meda A. R., Kirchner S., Wipf D., von Wirén N. (2006) AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem. 281, 25532–25540 [DOI] [PubMed] [Google Scholar]
- 91. Vogel C., Marcotte E. M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mauro V. P., Edelman G. M. (2002) The ribosome filter hypothesis. Proc. Natl. Acad. Sci. U.S.A. 99, 12031–12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Komili S., Farny N. G., Roth F. P., Silver P. A. (2007) Functional specificity among ribosomal proteins regulates gene expression. Cell 131, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xue S. F., Barna M. (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Bio. 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Filipovska A., Rackham O. (2013) Pentatricopeptide repeats modular blocks for building RNA-binding proteins. RNA Biol. 10, 1426–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Carroll A. J. (2013) The Arabidopsis cytosolic ribosomal proteome: from form to function. Front Plant Sci. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang J. Y., Lan P., Gao H. M., Zheng L., Li W. F., Schmidt W. (2013) Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hummel M., Cordewener J. H. G., de Groot J. C. M., Smeekens S., America A. H. P., Hanson J. (2012) Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MSE proteomics. Proteomics 12, 1024–1038 [DOI] [PubMed] [Google Scholar]
- 99. Mustroph A., Zanetti M. E., Jang C. J. H., Holtan H. E., Repetti P. P., Galbraith D. W., Girke T., Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014) Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, E203–E212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lee M. V., Topper S. E., Hubler S. L., Hose J., Wenger C. D., Coon J. J., Gasch A. P. (2011) A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yanguez E., Castro-Sanz A. B., Fernandez-Bautista N., Oliveros J. C., Castellano M. M. (2013) Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. Plos One 8, e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chan C. P., Kok K. H., Tang H. M. V., Wong C. M., Jin D. Y. (2013) Internal ribosome entry site-mediated translational regulation of ATF4 splice variant in mammalian unfolded protein response. BBA-Mol. Cell Res. 1833, 2165–2175 [DOI] [PubMed] [Google Scholar]
- 104. Muñoz A., Castellano M. M. (2012) Regulation of translation initiation under abiotic stress conditions in plants: is it a conserved or not so conserved process among eukaryotes? Comp. Funct. Genom. 2012, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu B. T., Qian S. B. (2014) Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA 5, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sigismund S., Polo S., Di Fiore P. P. (2004) Signaling through monoubiquitination. Curr. Top Microbiol. 286, 149–185 [DOI] [PubMed] [Google Scholar]
- 107. Sadowski M., Suryadinata R., Lai X. N., Heierhorst J., Sarcevic B. (2010) Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol. Cell. Biol. 30, 2316–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ramanathan H. N., Ye Y. H. (2012) Cellular strategies for making monoubiquitin signals. Crit. Rev. Biochem. Mol. 47, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mattiroli F., Sixma T. K. (2014) Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat. Struct. Mol. Biol. 21, 308–316 [DOI] [PubMed] [Google Scholar]
- 110. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nature Biotechnol. 30, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.