Abstract
Prostate cancer is the most common cancer among men in the U.S. and worldwide, and androgen-deprivation therapy remains the principal treatment for patients. Although a majority of patients initially respond to androgen-deprivation therapy, most will eventually develop castration resistance. An increased understanding of the mechanisms that underline the pathogenesis of castration resistance is therefore needed to develop novel therapeutics. LNCaP and PC3 prostate cancer cell lines are models for androgen-dependence and androgen-independence, respectively. Herein, we report the comparative analysis of these two prostate cancer cell lines using integrated global proteomics and glycoproteomics. Global proteome profiling of the cell lines using isobaric tags for relative and absolute quantitation (iTRAQ) labeling and two- dimensional (2D) liquid chromatography-tandem MS (LC-MS/MS) led to the quantification of 8063 proteins. To analyze the glycoproteins, glycosite-containing peptides were isolated from the same iTRAQ-labeled peptides from the cell lines using solid phase extraction followed by LC-MS/MS analysis. Among the 1810 unique N-linked glycosite-containing peptides from 653 identified N-glycoproteins, 176 glycoproteins were observed to be different between the two cell lines. A majority of the altered glycoproteins were also observed with changes in their global protein expression levels. However, alterations in 21 differentially expressed glycoproteins showed no change at the protein abundance level, indicating that the glycosylation site occupancy was different between the two cell lines. To determine the glycosylation heterogeneity at specific glycosylation sites, we further identified and quantified 1145 N-linked glycopeptides with attached glycans in the same iTRAQ-labeled samples. These intact glycopeptides contained 67 glycan compositions and showed increased fucosylation in PC3 cells in several of the examined glycosylation sites. The increase in fucosylation could be caused by the detected changes in enzymes belonging to the glycan biosynthesis pathways of protein fucosylation observed in our proteomic analysis. The altered protein fucosylation forms have great potential in aiding our understanding of castration resistance and may lead to the development of novel therapeutic approaches and specific detection strategies for prostate cancer.
Androgen is important for the development, function, and proliferation of both normal and cancerous prostate cells (1). At the earliest stage of prostate cancer, prostate cancer cells are dependent on the presence of androgen, and androgen-deprivation therapy (ADT)1 is used to treat prostate cancer (2). However, cells become androgen-independent as a result of androgen deprivation therapy, and they become more aggressive. This results in androgen-independent remission of prostate cancer (3). LNCaP and PC3 cell lines have been widely used as models of prostate cancer. LNCaP is an androgen-dependent cancer cell line, whereas PC3 is an androgen-independent cell line. The LNCaP cell line is less aggressive as compared with PC3 cells that have a high metastatic potential. LNCaP and PC3 cells have been previously studied by genomics and proteomics approaches to understand the mechanism(s) responsible for the aggressive and metastatic nature of prostate cancer (4–8).
Post-translational modifications (PTMs) such as phosphorylation are important in the function of the androgen-dependent pathway. Androgen receptors bind to androgen and are then phosphorylated before translocating into the nucleus (3). However, protein PTMs cannot be directly inferred from gene expression. Glycosylation is an abundant PTM and most cell surface or secreted proteins are expected to be glycosylated (9). Glycosylation is one of the more complex PTMs because of the fact that different glycosylation machineries are present in different cells, multiple glycosylation sites exist on many glycoproteins and each glycosylation site can be modified by several different glycans (10, 11). Such microheterogeneity of glycan structures at each glycosylation site with different site occupancy significantly increases the structural diversity of each glycoprotein that is specific to the microenvironment of the cells where each glycoprotein is produced. Although these characteristics of protein glycosylation pose considerable challenges to the structural and functional analyses of glycoproteins, we expect that cell and cell microenvironment-specific glycoproteins differ according to the physiological and pathological states of the cells. Aberrant glycosylation is the result of alterations in glycosylation genes that may lead to the development of cancer. A systematic approach to analyze proteins, glycoproteins, and glycosylation is expected to permit the identification of the glycoprotein alterations that are specific to each cell state and aid the understanding of the functions of glycosylation because alterations in glycosylation can affect glycoprotein abundance or function (12, 13). A detailed analysis of glycoproteins in cancer cells with different functions is needed to understand tumor biology and how glycoproteins can function as therapeutic targets or diagnostic biomarkers (14, 15).
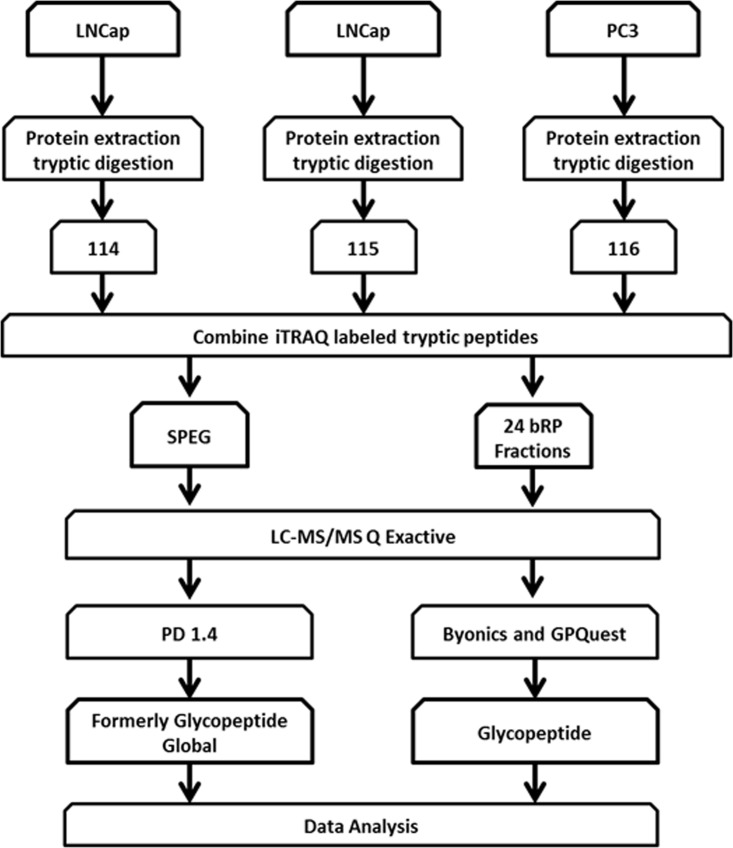
In this study, a comprehensive proteomic and glycoproteomic platform was designed to investigate the differences in proteins, glycoproteins, and site-specific glycosylation forms of glycoproteins between LNCaP and PC3 cells (Fig. 1). To our knowledge, this is the first report to characterize glycoproteins with respect to protein abundance, glycosylation occupancy, and glycosylation heterogeneity at specific glycosites. These altered glycosylation patterns among proteins between LNCaP and PC3 cell lines have a significant potential to aid our understanding of the altered glycoprotein expression in prostate cancer cells, thus leading to novel specific methods to detect aggressive prostate cancer.
Fig. 1.
Schematic representation of the workflow for the integrated analysis of glycosite-containing peptides, global protein expression, and intact glycopeptides. Proteins were obtained from LNCaP and PC3 cell lines followed by tryptic digestion and iTRAQ labeling. Labeled peptide samples were then combined and separated into two aliquots. One aliquot was enriched for glycosite-containing peptides using Solid Phase Extraction of Glycopeptides (SPEG) and the other aliquot was used for bRPLC separation followed by the analysis of global proteins and intact glycopeptides. Finally, peptides were analyzed using LC-MS/MS.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
Human prostate cancer cell lines were cultured as described in our previous publication (16). The cells were used for proteomic analysis upon reaching 80–90% confluence. The cells were washed six times with ice-cold phosphate buffered saline prior to cell lysis for protein extraction. Biological replicates were prepared using the same conditions.
Protein and Peptide Extraction from Cells for Proteomic Analysis
The cell pellet from two 10 cm dishes was first denatured in 1 ml of 8 m urea and 0.4 m NH4HCO3 and sonicated thoroughly. The protein concentration was measured using a BCA protein assay kit (Thermo Scientific, Rockford, IL). The proteins were then reduced by incubating in 120 mm Tris (2-carboxyethyl) phosphine for 30 min and alkylated by addition of 160 mm iodoacetamide at room temperature for 30 min in the dark. The sample was diluted with buffer (100 mm Tris-HCl, pH 7.5) containing 0.5 μg/μl sequence grade trypsin (Sigma-Aldrich, St Louis, MO) and incubated at 37 °C overnight. The completion of protein digestion was assessed using SDS-PAGE and silver staining. Peptides were purified with C18 desalting columns and dried using a SpeedVac concentrator.
iTRAQ Labeling of Global Tryptic Peptides from Cell Lines
Each iTRAQ (isobaric tags for relative and absolute quantitation) fourplex reagent was dissolved in 70 μl of methanol. One mg of each tryptic peptide sample was added into 250 μl of iTRAQ dissolution buffer, then mixed with iTRAQ fourplex reagent and incubated for one hour at room temperature. iTRAQ channels 114 and 115 were used to label two replicate LNCaP samples in order to determine the analytical reproducibility, iTRAQ channel 116 was used to label peptides from PC3 cells and iTRAQ channel 117 was used for labeling peptides from another cell line unrelated to this study. After iTRAQ labeling, the four sets of tagged peptides were combined and purified by SCX column. Then, 10% of the labeled peptides were dried and resuspended into 0.4% acetic acid solution prior to fractionation for mass spectrometry analysis. The remaining peptides were desalted for glycopeptide capture. In the biological replicate, channels 114 and 115 were used to label peptides from the PC3 cells and channel 116 was used to label peptides from LNCaP cells.
Glycopeptide Capture
Glycosite-containing peptides were extracted from tryptic peptides using solid-phase extraction of glycosite-containing peptides (SPEG) (17). Briefly, 90% of the iTRAQ-labeled tryptic peptides (3.6 mg) were dissolved in 5% ACN in 0.1% TFA followed by adding 1/10 of the final volume of 100 mm sodium periodate to the samples. The samples were incubated with hydrazide beads for one hour at room temperature in the dark with gentle shaking. Beads were then washed to remove any nonspecific binding. PNGaseF (New England Biolabs, Ipswich, MA) was used to detach glycosite-containing peptides from glycans conjugated on the beads.
Chromatography Fractionation
The global peptide mixture resulting from 100 μg of iTRAQ-labeled tryptic peptides was fractionated into 24 fractions by basic reverse phase liquid chromatography (bRPLC) using a 1200 Infinity LC system (Agilent Technology, Santa Clara, CA) equipped with a 4.6 × 100 mm BEH120 C-18 column (Waters, Milford, MA). Samples were adjusted to a basic pH using 1% ammonium hydroxide. Solvent A was 7 mm tri-ethyl ammonium bicarbonate (TEAB), whereas Solvent B was 7 mm TEAB in 90% acetonitrile. A total of 96 fractions were collected and concatenated into 24 fractions. The glycosite-containing peptides isolated by SPEG were not further fractionated but were analyzed directly by LC-MS/MS.
LC-MS/MS Analysis
The peptide samples were separated utilizing a Dionex Ultimate 3000 RSL C nano system (Thermo Scientific) with a 75 μm × 50 cm C18 PepMap RSLC column (Thermo Scientific) protected by a 5 mm guard column (Thermo Scientific). The flow rate was 0.350 μl/min with 0.1% formic acid and 2% acetonitrile in water (A) and 0.1% formic acid, 95% acetonitrile (B). The peptides were separated with a 5–40% B gradient in 104mins. MS analysis was performed using a Thermo Q Exactive mass spectrometer (Thermo Scientific). The AGC target for MS1 was set for 3 × 106 for MS1 in 60ms maximum injection time (IT). The AGC target for MS/MS was 5 × 104 (resolution of 17,500, High-energy collisional dissociation (HCD) and maximum IT of 100ms) of the 20 most abundant ions. Charge state screening was enabled to reject ions with z = 1, ≥8 and unassigned protonated ions. A dynamic exclusion time of 25 s was used to discriminate against previously selected ions. The mass spectrometry data is available via ProteomeXchange (18), and the assigned data set identifier is PXD002107.
Data Analysis
Data were searched using SEQUEST in Proteome Discoverer 1.4 (Thermo Scientific) against the Human RefSeq database downloaded on August 13, 2014 containing 55,926 proteins. Peptides were searched with two tryptic termini allowing two missed cleavages. The search parameters used were 10ppm precursor tolerance and 0.06Da fragment ion tolerance, fragmentation using HCD, static modification of fourplex iTRAQ at N terminus and lysine, carbamidomethylation of cysteine, and variable modification of oxidation at methionine. Deamidation at asparagine was applied as a variable modification to identify N-linked glycosylation sites of formerly glycosylated peptides. Filters used for global data analysis included peptide rank 1, two peptides per protein, and 2% FDR threshold. Filters used for glycopeptide analysis were peptide rank 1 and 1% FDR. The data were normalized by protein median using PD 1.4. Glycopeptides were required to contain NXS/T motifs.
For intact glycopeptide identification, the data were searched using Byonic 1.3 (19) and an in house developed glycopeptide analysis software, GPQuest 1.0 (20), with the same parameters as mentioned above for proteomic data using the human RefSeq database (Fig. 1). Additional parameters for database search were the mammalian N-glycans database and the offset was set for ≥ 1. Results were filtered using in-house software based on the following criteria: (1) presence of two oxonium ions, (2) intensity of the highest oxonium ion greater than the maximum reporter ion intensity, and (3) all the peptides with scores below the score of the highest reverse database match were removed. The data was normalized using the normalization factor obtained in global data.
The raw mass spectrometry data are publicly accessible in ProteomeXchange (ProteomeXchange.com) with the project accession# PXD002107.
RESULTS
Global Proteomic Analysis
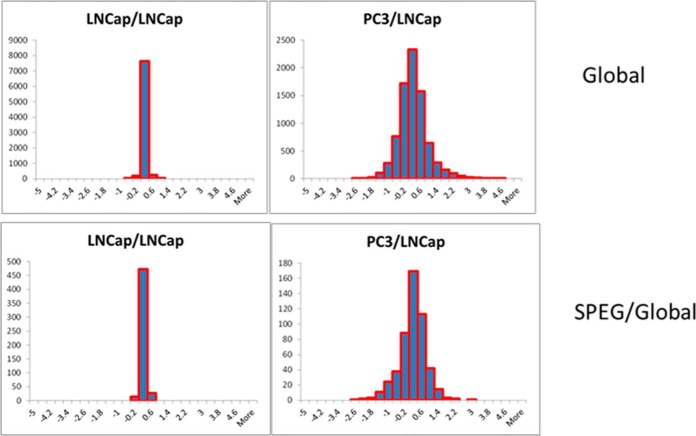
To determine the proteomic profiles of LNCaP and PC3 cells, global proteomics was performed using iTRAQ labeling (Fig. 1). Each bRPLC fraction was analyzed by LC-MS/MS to obtain both qualitative and quantitative information on the proteome of LNCaP and PC3 cells. At 2% spectral FDR, 8063 protein groups were identified with a minimum of two peptides per protein (supplemental Table S1). Twofold differences were considered to be significant changes. Six hundred and thirty seven protein groups were up-regulated in PC3 cells compared with LNCaP cells and 410 proteins were determined to be down-regulated in PC3 cells. In the replicate analyses of LNCaP cells, none of the proteins were up- or down-regulated, indicating a false discovery rate of less than 0.01%. The distribution of the fold-change expression ratios of the 8063 global proteins between LNCaP replicates and PC3/LNCaP cells is represented in the histogram in Fig. 2. For the replicate analysis of LNCaP cells, the protein ratios were distributed between −1 and +1 (on the log2 scale) (Fig. 2). For the PC3/LNCaP comparison, the presence of protein ratios < −1 and > +1 suggest that protein expression changed in PC3 cells compared with LNCaP cells.
Fig. 2.
Distribution of proteins and glycoproteins between replicate analyses of LNCaP (top panel) and the distribution of glycoproteins versus global proteins between LNCaP and PC3 cells indicating changes in glycosylation occupancy.
When comparing the identified global proteins with the genes that are known to play roles in glycan synthesis pathways, we identified 191 enzymes that were involved in glycan biosynthesis or degradation. Eleven proteins including hyaluronan synthase 3 (HAS3), bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase 2 (PAPSS2), phosphoglucomutase-1 (PGM1), and alpha-(1,6)-fucosyltransferase (FUT8) were found to be up-regulated with greater than a three fold increase in PC3 cells. Although 14 proteins in PC3 cells were identified with at least two-fold decreased expression when compared with the LNCaP proteome (supplemental Table S2 and Table I), a similar trend was observed in the biological replicate analysis (supplemental Data S1). The differentially expressed enzymes involved in glycosylation indicate that there may be significant differences in the glycan structures among proteins between the two cell lines.
Table I. Changes in glycosylation enzymes in PC3 cells.
| Accession # | Gene | Description | Protein Type | PC3/LNCaP |
|---|---|---|---|---|
| 20302153 | HAS3 | Hyaluronan synthase 3 | Glycan-transferase | 6.2704 |
| 34447231 | PAPSS2 | Bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | Nuc. Sugar | 3.4607 |
| 21361621 | PGM1 | Phosphoglucomutase-1 | Nuc. Sugar | 3.4496 |
| 30410726 | FUT8 | α-(1,6)-fucosyltransferase | Glycan-transferase | 3.4058 |
| 4502343 | B3galnt1 | UDP-GalNAc:β-1,3-N-acetylgalactosaminyltransferase 1 | Glycan-transferase | 2.7572 |
| 190014632 | GNE | Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase | Nuc. Sugar | 2.7074 |
| 11321585 | GNB1 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | Nuc. Sugar | 2.2892 |
| 194097330 | HK3 | hexokinase-3 | Nuc. Sugar | 2.2705 |
| 42516563 | UXS1 | UDP-glucuronic acid decarboxylase 1 | Nuc. Sugar | 2.1321 |
| 240255483 | SULF2 | Extracellular sulfatase Sulf-2 | Glycan Degradation | 2.1069 |
| 13027378 | GNPDA1 | Glucosamine-6-phosphate isomerase 1 | Nuc. Sugar | 2.0157 |
| 84798622 | MANBA | β-mannosidase | Glycan Degradation | 0.4966 |
| 189011548 | ASAH1 | Acid ceramidase isoform a preproprotein | Glycan Degradation | 0.4909 |
| 109148548 | NAAA | N-acylethanolamine-hydrolyzing acid amidase | Glycan Degradation | 0.4559 |
| 4505235 | MPI | Mannose-6-phosphate isomerase | Nuc. Sugar | 0.4540 |
| 189011550 | ASAH1 | acid ceramidase | Glycan Degradation | 0.4515 |
| 4504373 | HEXB | β-hexosaminidase subunit beta preproprotein | Glycan Degradation | 0.4304 |
| 119360348 | FUCA1 | Tissue α-l-fucosidase | Glycan Degradation | 0.4238 |
| 285002251 | AGA | N(4)-(β-N-acetylglucosaminyl)-l-asparaginase isoform 1 preproprotein | Glycan Degradation | 0.4218 |
| 38026892 | ALG6 | Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase | Glycan-transferase | 0.3808 |
| 4507303 | SULT1A2 | Sulfotransferase 1A2 | xSulfotransferase | 0.3672 |
| 4758092 | CTBS | Di-N-acetylchitobiase | Glycan Degradation | 0.3590 |
| 189181666 | HEXA | β-hexosaminidase subunit alpha preproprotein | Glycan Degradation | 0.3405 |
| 66346698 | NAGLU | α-N-acetylglucosaminidase | Glycan Degradation | 0.3185 |
| 29550921 | SULT1A3 | Sulfotransferase 1A3/1A4 | xSulfotransferase | 0.2449 |
Glycoproteomic Analysis
To determine the changes in glycoproteins, N-glycosite-containing peptides from the same iTRAQ-labeled tryptic peptides that were used for global proteomic analysis were isolated using SPEG. This eliminated variation because of sample preparation between the global proteomic and glycoproteomic analyses. In this study, we identified 1810 N-linked glycosite-containing peptides (supplemental Table S3) from 653 glycoproteins (supplemental Table S4). The spontaneous deamidation of asparagine residues has been previously reported (21). However, the N-linked glycosite-containing peptides identified in this study were from the chemical immobilization of glycopeptides by conjugating oxidized glycans to a solid support and the release of formerly N-linked glycopeptides by PNGase F. Therefore, the N-linked deamidated asparagine residues identified from the N-linked glycosite-containing peptides were most from formerly N-linked glycopeptides. In order to increase the confidence in identification of glycosites in current study, subcellular location of glycoprotein was determined (supplemental Data S4).
Based on the replicate analysis of LNCaP cells, a two-fold threshold was set to determine the number of glycoproteins that were differentially expressed between the PC3 and LNCaP prostate cancer cells. Among the 653 identified glycoproteins, 97 glycoproteins containing glycosites were quantified with significantly higher expression levels in PC3 cells, whereas 79 glycoproteins were quantified with down-regulated expression when compared with the LNCaP, with an FDR of ∼0.1% based on the changes in the replicate analyses between LNCaP glycoproteins (Supplemental Table S5). A slightly higher number of up-regulated glycoproteins in PC3 cells indicated a moderate increase in glycoprotein expression and/or glycosylation occupancy in PC3 cells. Differentially expressed glycoproteins were compared with glycoproteins without changes using the DAVID annotation tool to facilitate biological interpretation in a network context (22). Down-regulated glycoproteins in PC3 cells are highly represented in the lysosome pathway. These down-regulated glycoproteins include the lysosomal acid hydrolases CTSL2 and LGMN, the glucosidases IDUA, NAGA, FUCA1, HEXA, and MANB, and AGA, PSAP, GM2A, NRAMP, and CLN5. Other glycoproteins that belong to the N-glycan degradation pathway are also enriched among the down-regulated glycoproteins in PC3 cells and these glycoproteins include AGA, FUCA1, FUCA2, HEXA, and MANBA. Up-regulated glycoproteins in PC3 compared with LNCaP cells are enriched in the extracellular matrix (ECM) pathway and these glycoproteins include LAMB1, CD44, CD47, ANPEP, ITGB4, ITGA1, ITGA2, ITGA3, RELN, and HSPQ2. Hematopoietic cell lineage, cell adhesion molecule and focal adhesion pathway proteins were also enriched among the up-regulated glycoproteins with p values < 0.005.
Although 176 glycoproteins underwent ≥two-fold changes between PC3 and LNCaP cells, it was not clear whether the changes were because of changes in protein abundance or in glycosylation site occupancy. To resolve the above dilemma, we compared the glycoprotein changes identified from our glycoproteomic analysis to those identified from our global proteomic analysis. Comparing the glycoprotein versus global protein ratio between the two cell lines indicates the differential glycosylation occupancy on the glycoproteins. Plotting the histogram of glycoprotein/protein ratios between the PC3 and LNCaP cells, the distribution revealed that the vast majority of the proteins (155 glycoprotein changes; 88.1%) were located within a range of ±1 (log2 scale) of the glycoprotein/protein ratio, indicating that these proteins were regulated at the protein abundance level (Fig. 2). However the ratios of the remaining 21 glycoproteins (11.9%) were outside of the −1 and +1 interval suggesting significant changes in the glycosylation occupancy of these proteins (Fig. 2). The data show that the majority of the glycoprotein changes were caused by differential protein expression and there was a subset of glycoproteins where the changes resulted from differential glycosylation occupancy. The CD63 protein, which is mainly associated with the membranes of intracellular vesicles, was observed to have a protein abundance ratio of 1.17 between the LNCaP and PC3 cells, indicating a minor change at the protein level. However, the observed CD63 glycosite ratio between the two cell lines was determined to be 3.45 with a total of 90 MS/MS spectra showing a significant change at the glycosylation site occupancy level.
To determine whether the differences in glycoprotein ratio compared with global protein levels were attributed to partial glycosylation of the glycosylation site, the global data was analyzed to identify the nonglycosylated peptides that contained nonglycosylated sequences of the glycosite-containing peptides that were identified from our glycoproteomic analysis using the SPEG method. We identified 73 unique peptides containing the same N-linked glycosylation sites but without an attached glycan or Asn-to-Asp conversion because they were identified in the global proteomic data (supplemental Table S6). The identification of nonglycosylated peptides existing in their unmodified and glycosylated forms indicated that these peptides were partially glycosylated and their glycosylation sites were not fully occupied. This could be part of the reason why different ratios were observed for glycoproteins compared with the ratios that were obtained in the global proteomics analysis. The extent of glycosylation of a particular site could vary in different cell lines.
Glycopeptide Identification
To determine the glycosylation heterogeneity at specific glycosylation sites, we further analyzed the global proteomics data to identify the intact glycopeptides. MS/MS spectra from intact glycopeptides have unique fragmentation signatures following HCD fragmentation: oxonium ions (m/z 138, 163, 204, 274, 292, and 366) are generated along with peptide and peptide + HexNAc fragment ions, and these ions were used to identify glycopeptides. In the global proteomics data analysis, the glycopeptide spectra had unique signatures. Interestingly, in the MS/MS spectra of nonglycosylated peptides, the iTRAQ reporter ions at m/z 114,115,116, and 117 were generally the most intense ions because of HCD fragmentation. For glycopeptides with attached glycans, oxonium ion intensities were observed to be higher than the intensities of the iTRAQ reporter ions, and the diagnostic oxonium ions were the base peak in MS/MS. In previous studies, a similar phenomenon was observed when tandem mass tags (TMT) were used for the analysis of bovine fetuin/glycosidic bonds were preferentially cleaved versus amides in HCD fragmentation (23, 24). To extract the MS/MS spectra of glycopeptides from the global proteomic data, we required the oxonium ion intensities to be greater than the highest iTRAQ reporter ion intensity. Another filter was applied to the MS/MS data wherein at least two of the top five most abundant peaks were required to be oxonium ions. This reduced and almost eliminated the false positive glycopeptide identifications.
With these stringent filters, we identified 3670 MS/MS spectra meeting our glycopeptide identification threshold, and they were assigned to glycopeptides using Byonic and GPQuest for searches against a forward human protein database, and 469 MS/MS spectra were matched to peptides from the reversed database. For glycopeptide identification using the highest score of a spectrum matched to the reversed database as a cutoff score, we considered glycopeptides with a score greater than the score for a reversed database match as identified glycopeptides (with < 0.05% FDR at the spectral level, considering one reverse database match). We assigned 2021 MS/MS spectra to 1145 unique N-glycopeptides from 227 protein groups (supplemental Table S7). These glycopeptides were comprised of 67 glycan compositions (supplemental Table S7).
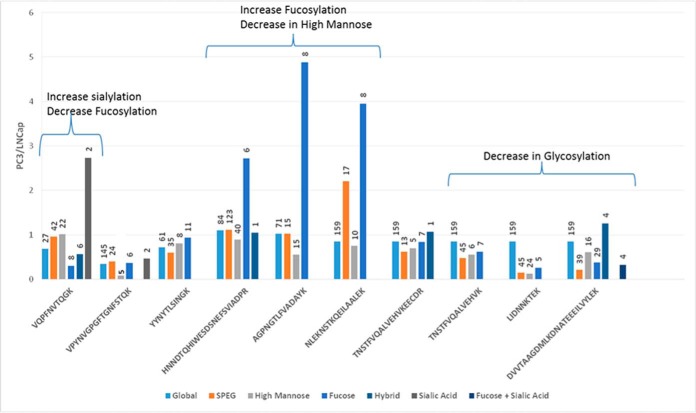
We identified intact N-glycopeptides at glycosites containing different glycan structures. To determine the glycosite occupied by different type of glycans, the MS/MS spectra with high mannose structures on a specific glycosite were combined and the median iTRAQ ratio was used to represent the high mannose glycan structures (HM). The MS/MS spectra of other glycan structures were classified in a similar manner into the following groups: fucosylated glycans (F), hybrid glycans (H), sialylated glycans (S), and sialylated + fucosylated glycans (SF). Most of the MS/MS spectra were assigned to glycosites containing either fucosylated or high mannose type N-glycans. To increase the accuracy of the iTRAQ-based quantitation of the glycopeptides, only intact glycopeptides with least five MS/MS spectral assignments were used for quantification purpose to determine the ratios for the glycans belonging to any of the five categories described above (HM, F, H, S, and SF). The glycopeptide quantification was also normalized based on the same factor determined in global proteomic analysis because the glycopeptide data were extracted from the global proteomic analysis. Glycosites exhibiting both high mannose and fucosylated glycan structures are represented in Fig. 3. The data indicate that the relative ratios of high mannose and fucosylated glycans were not always correlated to the ratios of protein abundance or total glycosylation level of each glycosite, indicating the glycosylation heterogeneity at each glycosylation site.
Fig. 3.
Quantitative analysis of intact glycopeptides. The intact glycopeptides with a minimum of five MS/MS spectra matching with both high mannose and complex fucosylated glycans assigned to the same N-glycosites are presented here. VQPFN*VTQGK (lysosome associated membrane protein 2), HNN*DTQHIWESDSNEFSVIADPR (heat shock protein 90 B1), VPYNVGPGFTGN*FSTQK (glutamate carboxypeptidase), YYN*YTLSINGK (glucosamine 6 sulfatase), AGPN*GTLFVADAYK (adipocyte plasma membrane protein) and NLEKN*STKQEILAALEK, TN*STFVQALVEHVKEECDR, DVVTAAGDMLKDN*ATEEEILVYLEK, TN*STFVQALVEHVK, LIDNN*KTEK (prosaposin). The y axis represents the ratio observed between PC3/LNCaP cells using iTRAQ-based quantification. The number above each bar indicates the number of MS/MS spectra used to determine the glycopeptide ratio between PC3 and LNCaP cells.
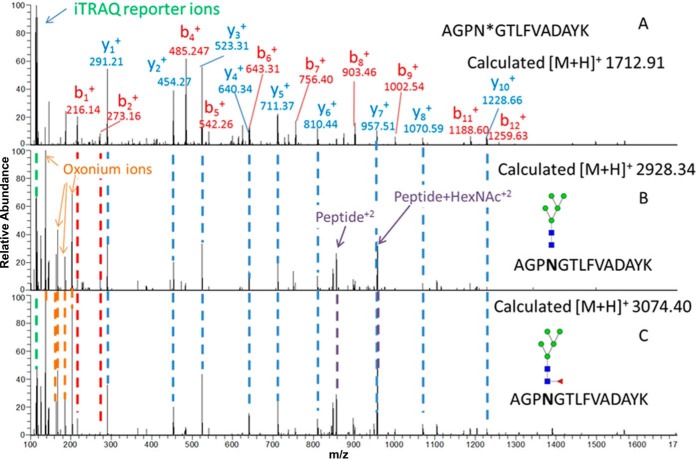
A change in fucosylation site occupancy was observed for several glycosites including adipocyte plasma membrane associated protein (APMAP). The relative global protein and glycosylation site levels of the AGPNGTLFVADAYK peptide of APMAP were unchanged between the LNCaP and PC3 cells based on identification and quantification by global proteomics and glycosite analysis using SPEG and iTRAQ (Fig. 4A). Using the b and y ions of the fragmented glycosite-containing peptide, additional glycopeptides having the same peptide fragmentation pattern along with high mannose glycans and fucosylated glycans were identified. Glycopeptides with Man5GlcNAc2 were identified, but their iTRAQ reporter ion intensity suggested a decrease in Man5GlcNAc2 occupancy at the glycosite in PC3 cells (Fig. 4B). Fig. 4C is an MS/MS spectrum of the same glycosite containing Fuc1Man5GlcNAc2 glycans; however, in this case the intensity of the iTRAQ reporter ions suggested an increase in Fuc1Man5GlcNAc2 glycan occupancy at the same glycosite. Fuc1Man5GlcNAc2 identified in the current study has an atypical N-glycan structure which might be a result of biodegradation or synthetic process as discussed in previous studies (25–27). We identified eight additional glycopeptides on the same N-linked glycosite from APMAP, and we observed various high mannose glycoforms with decreased relative abundance in PC3 compared with LNCaP cells. However, the fucosylated glycopeptide ratio was increased in PC3 cells (Table II) as observed in the biological replicate analysis (supplemental Data S1).
Fig. 4.
MS/MS spectra of glycosite-containing peptides (glycans were removed) and glycopeptides containing different glycans at the same glycosite. A, An MS/MS spectrum of a glycosite-containing peptide, AGPN*GTLFVADAYK, identified by SPEG method after PNGaseF treatment. B, An MS/MS spectrum of a glycopeptide, AGPN*GTLFVADAYK, containing HexNAc2Hex5. C, An MS/MS spectrum of an identified glycopeptide, AGPN*GTLFVADAYK, containing fucosylated glycan HexNAc 2 Hex5 Fuc1. The glycan cartoon structures are the most plausible structures based based on accurate masses and biosynthetic knowledge of N-linked glycans. The peptide with same backbone was observed in all three spectra. The oxonium ions represent the presence of the glycan structures. The reporter ion intensity represents a decrease in high mannose structures and an increase in fucosylation on the peptide AGPN*GTLFVADAYK in PC3 cells (116) compared with LNCaP cells (114, 115). * indicates deamidation of N because of PNGase F treatment.
Table II. Summary of the glycopeptide, AGPN#GTLFVADAYK, with different glycoforms.
| Accession | Spectra | HexNAc | Hexose | Fucose | Sialic Acid | Glycopeptide |
Global |
SPEG |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LNCaP/LNCaP | PC3/LNCaP | LNCaP/LNCaP | PC3/LNCaP | LNCaP/LNCaP | PC3/LNCaP | ||||||
| 24308201 | 1 | 3 | 3 | 1 | 0 | 1.46 | 5.44 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 2 | 2 | 3 | 1 | 0 | 1.04 | 2.37 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 2 | 4 | 3 | 1 | 0 | 1.42 | 7.37 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 1 | 2 | 5 | 1 | 0 | 1.14 | 4.33 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 2 | 5 | 3 | 1 | 0 | 1.09 | 8.47 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 5 | 2 | 5 | 0 | 0 | 1.17 | 0.74 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 5 | 2 | 6 | 0 | 0 | 1.09 | 0.39 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 2 | 2 | 8 | 0 | 0 | 0.94 | 0.51 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 2 | 2 | 9 | 0 | 0 | 1.48 | 0.48 | 0.99 | 1.02 | 0.98 | 1.07 |
| 24308201 | 1 | 2 | 7 | 0 | 0 | 1.12 | 0.94 | 0.99 | 1.02 | 0.98 | 1.07 |
Another protein, prosaposin (PSAP), was identified in the SPEG analysis with six different glycopeptides from four unique glycosylation sites. Among these glycopeptides, five were down-regulated in PC3 cells. The overall protein ratio of PSAP between PC3 and LNCaP cells was 0.85 suggesting no significant change at the protein expression level. Upon glycopeptide analysis, the glycosite NLEKNSTKQEILAALEK had an increased level of fucosylation and the ratio of PC3 versus LNCaP was 3.95 with eight matched MS/MS spectra indicating an alteration of glycosylation in a site-specific manner.
The change in glycoforms could be a result of changes in the expression of glycosylated proteins or changes in the protein expression of enzymes that are involved in glycosylation (supplemental Table S2). Quantitative analysis identified 191 proteins that were involved in the substrate synthesis, glycan branching, elongation, or degradation of N-glycans (supplemental Table S2). One of the glycosylation enzymes identified, alpha-(1,6)-fucosyltransferase (FUT8), was up-regulated 3.4-fold at the protein level in PC3 cells compared with LNCaP cells. Alpha-l-fucosidase (FUCA1), which is involved in glycan degradation, was down-regulated in PC3 cells by more than twofold (Table I) and this observation was validated in the replicate analysis (supplemental Fig. S1). This indicates that the change in fucosylation observed in PC3 cells might be caused by changes in the expression of fucosyltransferase and alpha-l-fucosidase.
DISCUSSION
In this report, we have provided the most comprehensive proteomic and glyoproteomic characterization of LNCaP and PC3 cells to-date. These two prostate cancer cell lines are widely used models to study androgen-dependent and androgen-resistant disease. Previous studies have elucidated the differences among the two cell types using differential mRNA arrays and proteomic approaches. However, the elucidation of these complex biological systems remains limited by the available analytical technologies. Besides morphological differences, the two cell lines have been shown to have marked differences in Androgen receptor, Prostate specific antigen, Glutamate carboxypeptidase, Kallikrein 2, Prostatic acid phosphate, and Spondin 2 levels. The levels of these proteins have been previously reported to be elevated in LNCaP cells (4, 28). However, these studies either evaluated the transcriptional activity or the protein expression of the cells with little to no information about the post-translational mechanisms.
In the current study, a total of 8063 proteins were identified and quantified from LNCaP and PC3 prostate cancer cell lines. As expected, most of the changes in glycoproteins observed at specific glycosites were accompanied by changes in protein expression. However, a few glycoproteins underwent changes in glycosylation without associated changes in the protein level between the two cell lines. The N-glycosite-containing peptides identified from the CD63 protein were detected at relatively high levels in the PC3 cells compared with the LNCaP cells, where no significant changes were observed in the global expression of the same protein, suggesting an overall increase in the occupancy of CD63 glycosylation in PC3 cells. CD63 N-linked glycosylation has been shown to be important for its interaction with CXCR4 and the regulation of CXCR4 trafficking. (29) CXCR4 is a chemokine receptor type 4, whose expression is related to the metastasis of cancer cells (30). Altered CD63 glycosylation could play a significant role in prostate cancer metastasis, but this hypothesis needs to be validated by further studies.
Analysis using the DAVID annotation tool indicated that among the expressed glycoproteins, the “Other N-glycan degradation” pathway was enriched by up-regulation in LNCaP cells. This enriched pathway contained five enzymes that were differentially expressed. These five enzymes are responsible for the removal of glycans, which impacts glycosylation occupancy or changes the glycan structures of glycoproteins in the cells. Among the five enzymes, two are fucosidases that are involved in removing fucose from glycoproteins and glycolipids. These fucosidases were down-regulated in PC3 cells, along with the up-regulation of Fut8 and Fut11 as observed in the global analysis, which explains the change in fucosylation observed in PC3 cells. In prostate tissues, three fucosyltransferases (FUT3, FUT6, and FUT7) have been shown to have elevated gene expression, and the overexpression of these fucosyltransferases in PC3 cells showed that they may serve as master regulators of prostate cancer cell trafficking and explain the aggressive nature of PC3 cells (31). In our global proteomic study, only two fucosyltransferases (FUT8 and FUT11) were identified as overexpressed in PC3 cells. In the case of LNCaP and PC3 cells, this appears to be caused by a change in the expression of enzymes of the fucosylation machinery based on our RNA microarray analysis (data not shown). In our previous study, we showed that FUT8 expression is associated with aggressive prostate cancer tissue (16).
Altered enzymes involved in fucosylation resulted in changes in protein fucosylation in a site-specific manner. In the case of the PSAP glycosite NLEKNSTKQEILAALEK, both fucosylation and N-glycosylation occupancy increased. However, we did not observe an increase in fucosylation or glycosylation occupancy for any of the other PSAP glycosites: TNSTFVQALVEHVKEECDR, DVVTAAGDMLKDNATEEEILVYLEK, TNSTFVQALVEHVK, and LIDNNKTEK. PSAP has been shown to be elevated in the prostate tissue of patients with advanced prostate cancer (32, 33). Altered PSAP expression along with altered specific glycosylation occupancy could be a good indicator of prostate cancer. PSAP has a signal peptide, which indicates that it can be secreted. Altered fucosylation on a secreted protein can be used as a biomarker similar to alpha-Fetoprotein (AFP). Altered AFP and fucosylated AFP have been shown to differentiate various pathologies of liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). In 2006, the FDA approved AFP-L3, a fucosylated form of AFP, for the early detection of primary HCC (34). Changes in specific glycosylation occupancy could be an indicator of an aggressive prostate disease, but this hypothesis requires further investigation to explore whether secretory serum proteins such as PAP and PSA have higher site-specific fucosylation levels in cancer patients compared with healthy individuals.
Similar to the case of PC3 and LNCaP cells, the expression of FUT8 is low in normal liver but increased in HCC (35, 36). Secreted fucosylated proteins from prostate cancer tissues need to be studied, which could lead to the identification of specific glycosylated forms of glycoproteins as novel biomarkers. Recently, we have shown that fucosylation of PSA can be used to differentiate aggressive from nonaggressive prostate cancer (37). Altered fucosylated glycans have been reported in the serum glycome from 10 normal patients and 24 prostate cancer patients (38). However, the cause of the glycan changes was not clear in the previous reports. Our results from this study and our previous report show that the altered expression of fucosylation enzymes in prostate cancer cells or tissues could contribute to the increased fucosylation of prostate cancer glycoproteins detected in serum (16).
HSP90B1 is another glycoprotein that exhibited an increase in fucosylation. HSP90B1 purified from tumors initiates a tumor-specific cytotoxic T-cell lymphocyte response (39, 40). The amino acid sequences of HSP90B1 from tumor and normal tissues are identical; (41) however, fucosylation differences between cancerous and normal tissue have been observed (42), suggesting a role of glycosylation in the cancer-specific immune system response and also emphasizing the importance of studying glycosylation.
Rapid improvements in LC-MS/MS instrumentation have enabled high-throughput proteomics to identify thousands of proteins. In the current study, the increased sensitivity and speed of LC-MS/MS system have enabled the identification and quantitation of intact glycopeptides from global proteomic analysis. Identifying all the possible glycoforms attached to each glycopeptide is still a challenge. The iTRAQ reporter ion intensities from glycopeptides were lower than the intensities of the unmodified peptides, resulting in decreased confidence of quantitation. Yet, multiple spectra can convey more reliable information to quantify glycopeptides. In the current study, the fragment ions from glycosite-containing peptides were used to identify the intact glycopeptides from the same glycosite, and oxonium ions were used to select the MS/MS spectra with intact glycopeptides. In MS/MS spectra, limited information was available for glycosite localization and glycoform identification. Based on accurate precursor mass and the glycosite-containing peptide identified using SPEG, the glycopeptides with different glycoforms can be extrapolated. However, there were a limited number of glycan structures and there is no information about the glycan structure or the linkages between the monosaccharides forming the glycan.
A thorough investigation of glycan structures on the glycoproteins is required to understand the progression of cancer in relation to changes in glycosylation. Further technological advancements in high throughput LC-MS/MS systems and data analysis are needed; however, progress is continuously being made. Quantitative assays need to be developed to validate the identified glycosylation changes. Site-specific glycosylation changes need to be researched to gain a better understanding of the roles of different glycoforms and how they affect biological functions. Multiple integrated proteomic strategies are ideal approaches for future studies to provide a holistic view to understand the biological changes within a system, and these strategies could readily be translated to tumor tissues and other cancer systems.
CONCLUSION
In this study, we report an integrated proteomic and glycoproteomic comparison of LNCaP and PC3 cell lines, which are the two most widely used cell culture models for androgen-dependent and androgen-independent prostate cancer with different metastatic potential. The two cell lines were vastly different, with several thousand glycoproteins and proteins being expressed. We not only identified differential glycoprotein expression, but we also identified changes in glycosylation occupancy and glycosite-specific heterogeneity in glycoproteins. We showed that an integrated omics approach for the quantification of global peptides and glycopeptides in combination with iTRAQ labeling could be useful in the determination of altered glycosylation. The results showed that altered fucosylation between the two cell lines was associated with altered fucosyltransferase and fucosidase expression. These glycoproteins can be pursued as targets for specific biomarkers for the early detection of prostate cancer and cancer progression. This glycoprotein analysis provides a detailed perspective of the post-translational modification of N-glycosylation in LNCaP and PC3 cells, and the results could benefit future cancer research.
Supplementary Material
Acknowledgments
We thank Dr Carl Bergmann of Complex Carbohydrate Research Center (CCRC) for reviewing the manuscript.
Footnotes
Author contributions: H.Z. designed research; P.S., X.W., L.C., and S.Y. performed research; P.S., W.Y., S.T.E., S.S., J.P., A.R., and H.Z. analyzed data; P.S., N.H., and H.Z. wrote the paper.
* This work was supported by National Institute of Health, National Cancer Institute, the Early Detection Research Network (EDRN, U01CA152813), the Clinical Proteomics Tumor Analysis Consortium (CPTAC, U24CA160036), National Heart Lung and Blood Institute, Program of Excellence in Glycosciences (PEG, P01HL107153).
 This article contains supplemental Data S1 and Tables S1 to S11.
This article contains supplemental Data S1 and Tables S1 to S11.
1 The abbreviations used are:
- ADT
- androgen-deprivation therapy
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LC-MS/MS
- liquid chromatography-tandem MS
- PTM
- post-translational modification
- ECM
- extracellular matrix
- TMT
- tandem mass tags
- APMAP
- adipocyte plasma membrane associated protein.
REFERENCES
- 1. Wang D., Tindall D. J.. Androgen action during prostate carcinogenesis. In Androgen Action, Springer: 2011, pp 25–44 [DOI] [PubMed] [Google Scholar]
- 2. Karantanos T., Corn P., Thompson T. (2013) Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 32, 5501–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris W. P., Mostaghel E. A., Nelson P. S., Montgomery B. (2009) Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat. Clin. Pract. Urol. 6, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dozmorov M. G., Hurst R. E., Culkin D. J., Kropp B. P., Frank M. B., Osban J., Penning T. M., Lin H. K. (2009) Unique patterns of molecular profiling between human prostate cancer LNCaP and PC 3 cells. Prostate 69, 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hasegawa N., Mizutani K., Suzuki T., Deguchi T., Nozawa Y. (2006) A comparative study of protein profiling by proteomic analysis in camptothecin-resistant PC3 and camptothecin-sensitive LNCaP human prostate cancer cells. Urol. Int. 77, 347–354 [DOI] [PubMed] [Google Scholar]
- 6. Glen A., Gan C. S., Hamdy F. C., Eaton C. L., Cross S. S., Catto J. W., Wright P. C., Rehman I. (2008) iTRAQ-facilitated proteomic analysis of human prostate cancer cells identifies proteins associated with progression. J. Proteome Res. 7, 897–907 [DOI] [PubMed] [Google Scholar]
- 7. Sardana G., Jung K., Stephan C., Diamandis E. P. (2008) Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J. Proteome Res. 7, 3329–3338 [DOI] [PubMed] [Google Scholar]
- 8. Whitaker H. C., Stanbury D. P., Brinham C., Girling J., Hanrahan S., Totty N., Neal D. E. (2007) Labeling and identification of LNCaP cell surface proteins: a pilot study. Prostate 67, 943–954 [DOI] [PubMed] [Google Scholar]
- 9. Apweiler R., Hermjakob H., Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 10. An H. J., Froehlich J. W., Lebrilla C. B. (2009) Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 13, 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kolarich D., Jensen P. H., Altmann F., Packer N. H. (2012) Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 7, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 12. Daniels M. A., Hogquist K. A., Jameson S. C. (2002) Sweet‘n’sour: the impact of differential glycosylation on T cell responses. Nat. Immunol. 3, 903–910 [DOI] [PubMed] [Google Scholar]
- 13. Takahashi M., Tsuda T., Ikeda Y., Honke K., Taniguchi N. (2003) Role of N-glycans in growth factor signaling. Glycoconj. J. 20, 207–212 [DOI] [PubMed] [Google Scholar]
- 14. Drake P. M., Cho W., Li B., Prakobphol A., Johansen E., Anderson N. L., Regnier F. E., Gibson B. W., Fisher S. J. (2010) Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin. Chem. 56, 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kobata A., Amano J. (2005) Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumors. Immunol. Cell Biol. 83, 429–439 [DOI] [PubMed] [Google Scholar]
- 16. Wang X., Chen J., Li Q. K., Peskoe S. B., Zhang B., Choi C., Platz E. A., Zhang H. (2014) Overexpression of alpha (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology. 24, 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang H., Li X.-j., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 18. VizcaÃno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bern M., Cai Y., Goldberg D. (2007) Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 79, 1393–1400 [DOI] [PubMed] [Google Scholar]
- 20. Yang W., Shah P., Toghi Eshghi S., Yang S., Sun S., Ao M., Rubin A., Jackson J. B., Zhang H. (2014) Glycoform analysis of recombinant and human immunodeficiency virus envelope protein GP120 via higher energy collisional dissociation and spectral-aligning strategy. Anal. Chem. 86, 6959–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palmisano G., Melo-Braga M. N., Engholm-Keller K., Parker B. L., Larsen M. R. (2012) Chemical deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 22. Huang D. W., Sherman B. T., Lempicki R. A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 23. Ye H., Boyne M. T., Buhse L. F., Hill J. (2013) Direct approach for qualitative and quantitative characterization of glycoproteins using tandem mass tags and an LTQ Orbitrap XL electron transfer dissociation hybrid mass spectrometer. Anal. Chem. 85, 1531–1539 [DOI] [PubMed] [Google Scholar]
- 24. Lee H.-J., Cha H.-J., Lim J.-S., Lee S. H., Song S. Y., Kim H., Hancock W. S., Yoo J. S., Paik Y.-K. (2014) Abundance Ratio-Based Semiquantitative Analysis of Site-Specific N-linked Glycopeptides Present in the Plasma of Hepatocellular Carcinoma Patients. J. Proteome Res. 13, 2328–2338 [DOI] [PubMed] [Google Scholar]
- 25. Balog C. I., Stavenhagen K., Fung W. L., Koeleman C. A., McDonnell L. A., Verhoeven A., Mesker W. E., Tollenaar R. A., Deelder A. M., Wuhrer M. (2012) N-glycosylation of colorectal cancer tissues a liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics 11, 571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin A. I., Philipsberg G. A., Haltiwanger R. S. (1994) Core fucosylation of high-mannose-type oligosaccharides in GlcNAc transferase I-deficient (Lec1) CHO cells. Glycobiology 4, 895–901 [DOI] [PubMed] [Google Scholar]
- 27. Hoja ukowicz D., Ciolczyk D., Bergquist J., Litynska A., Laidler P. (2000) High-mannose-type oligosaccharides from human placental arylsulfatase A are core fucosylated as confirmed by MALDI MS. Glycobiology 10, 551–557 [DOI] [PubMed] [Google Scholar]
- 28. Uchida A., OKeefe D. S., Bacich D. J., Molloy P. L., Heston W. D. (2001) In vivo suicide gene therapy model using a newly discovered prostate-specific membrane antigen promoter/enhancer: a potential alternative approach to androgen deprivation therapy. Urology 58, 132–139 [DOI] [PubMed] [Google Scholar]
- 29. Yoshida T., Ebina H., Koyanagi Y. (2009) N-linked glycan dependent interaction of CD63 with CXCR4 at the Golgi apparatus induces downregulation of CXCR4. Microbiol. Immunol. 53, 629–635 [DOI] [PubMed] [Google Scholar]
- 30. Sun X., Cheng G., Hao M., Zheng J., Zhou X., Zhang J., Taichman R. S., Pienta K. J., Wang J. (2010) CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 29, 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barthel S. R., Wiese G. K., Cho J., Opperman M. J., Hays D. L., Siddiqui J., Pienta K. J., Furie B., Dimitroff C. J. (2009) Alpha 1, 3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc. Natl. Acad. Sci. 106, 19491–19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koochekpour S., Hu S., Vellasco Gonzalez C., Bernardo R., Azabdaftari G., Zhu G., Zhau H. E., Chung L. W., Vessella R. L. (2012) Serum prosaposin levels are increased in patients with advanced prostate cancer. Prostate 72, 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koochekpour S., Zhuang Y. J., Beroukhim R., Hsieh C. L., Hofer M. D., Zhau H. E., Hiraiwa M., Pattan D. Y., Ware J. L., Luftig R. B. (2005) Amplification and overexpression of prosaposin in prostate cancer. Genes Chromosomes Cancer 44, 351–364 [DOI] [PubMed] [Google Scholar]
- 34. Adamczyk B., Tharmalingam T., Rudd P. M. (2012) Glycans as cancer biomarkers. Biochim. Biophys. Acta 1820, 1347–1353 [DOI] [PubMed] [Google Scholar]
- 35. Miyoshi E., Uozumi N., Noda K., Hayashi N., Hori M., Taniguchi N. (1997) Expression of 1–6 fucosyltransferase in rat tissues and human cancer cell lines. Int. J. Cancer 72, 1117–1121 [DOI] [PubMed] [Google Scholar]
- 36. Noda K., Miyoshi E., Uozumi N., Gao C. X., Suzuki K., Hayashi N., Hori M., Taniguchi N. (1998) High expression of 1,6 fucosyltransferase during rat hepatocarcinogenesis. Int. J. Cancer 75, 444–450 [DOI] [PubMed] [Google Scholar]
- 37. Li Q. K., Chen L., Ao M.-H., Chiu J. H., Zhang Z., Zhang H., Chan D. W. (2015) Serum Fucosylated prostate-specific antigen (PSA) improves the differentiation of aggressive from nonaggressive prostate cancers. Theranostics 5, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kyselova Z., Mechref Y., Al Bataineh M. M., Dobrolecki L. E., Hickey R. J., Vinson J., Sweeney C. J., Novotny M. V. (2007) Alterations in the serum glycome due to metastatic prostate cancer. J. Proteome Res. 6, 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suto R., Srivastava P. K. (1995) A mechanism for the specific immunogenicity of heat shock protein-chaperoned peptides. Science 269, 1585–1588 [DOI] [PubMed] [Google Scholar]
- 40. Janetzki S., Palla D., Rosenhauer V., Lochs H., Lewis J. J., Srivastava P. K. (2000) Immunization of cancer patients with autologous cancer derived heat shock protein gp96 preparations: a pilot study. Int. J. Cancer 88, 232–238 [DOI] [PubMed] [Google Scholar]
- 41. Srivastava P. K. (1993) Peptide-binding heat shock proteins in the endoplasmic reticulum: role in immune response to cancer and in antigen presentation. Adv. Cancer Res. 62, 154–179 [DOI] [PubMed] [Google Scholar]
- 42. Suriano R., Ghosh S. K., Ashok B. T., Mittelman A., Chen Y., Banerjee A., Tiwari R. K. (2005) Differences in glycosylation patterns of heat shock protein, gp96: implications for prostate cancer prevention. Cancer Res. 65, 6466–6475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.