Abstract
Introduction: Human health is deemed to be maintained by the crosstalk among the body and probiotic bacteria. Disruptions in this composition are associated with the pathogenesis of numerous diseases. Through modernization, traditional foods consumption has been abandoned and native food starters have been substituted with industrial products. Hence, we aimed to isolate and evaluate probiotic bacteria from traditional fermented foods which can be used as probiotics as well as starter cultures in food industry and in medicine against antibiotic resistant pathogenes. To this end, an intact village was recognized in the Republic of Azerbaijan with traditional dairy products, yielding a variety of potential probiotics.
Methods: In this study, tvorog as a traditional dairy product from Dashkasan, Ismailli and khachmaz regions in Azerbaijan was characterized for the isolation of Lactobacilli with probiotic potentiality. The bacteria were tested for the resistance to the acid, bile, and the antagonistic effect of human pathogenic bacteria. The isolates were identified by 16s rDNA sequencing with a blast to the databank.
Results: Three species with higher homology to the L. planetarium, L. rhamnosus, and L. casei with high probiotic potentiality and antibacterial effects were isolated.
Conclusion: Homemade tvorog curd cheese in Azerbaijan harbor a variety of probiotics with industrial applications as well as potentiality to be preserved in a biobank for the future medicinal applications especially against antibiotic resistant pathogenes.
Keywords: Probiotic, Microbiome, Tvorog curd cheese, Enteropathogenic bacteria
Introduction
According to a definition by the World Health Organization (WHO), probiotics are distinguished as microorganisms with beneficial effects for human health.1 It is around millions years that the indigenous microflora has co-evolved with humans and they have preserved the inherited microbiomes via consumption of fermented foods and interactions with environmental microbes.2 Through modernization, traditional foods consumption has gradually been abandoned and native food starters have been substituted with industrial products. This gradual change has reduced human contact and exposure to microbial symbionts and led to the shrinkage of core microbiome.3 Besides, the openings of large numbers of antibiotics during the last three decades have caused anxiety about the threat of bacterial resistance. The widespread use of antibiotics by the population and in hospitals has inflamed this crisis. Mechanisms such as antibiotic control programs, better hygiene, and synthesis of agents with improved antimicrobial activity seem necessary to be adopted in order to restrict the bacterial resistance.4 In this regard, many research have been conducted on the isolation and evaluation of probiotic potential bacteria and in turn establishment of a biobank to save them from destruction, for future applications in medicine and food industry. Today, the probiotic human-friendly bacteria are isolated from foods, cheese,5 yogurt,6,7 fermented olives,8 and wine,9 as well as human himself, human milk,10,11 infant feces,12 women vagina,13 etc. According to WHO guidelines for evaluation of probiotics, putative strains should be screened for resistance to gastric acidity and bile salts, antimicrobial compound production and safety properties such as antibiotic resistance. Other recommendations are for the isolation with respect to the natives’ microbiome, indigenous acid- and bile-resistant bacteria14 and the traditional native food and environment. On the whole according to the literature, it has been emphasized that the isolation of probiotics should be done from native’s.15 Azerbaijan has many villages with traditional lifestyles that could be superb sources of super potential probiotics.16 In this research, we aimed to isolate lactobacilli from traditional dairy products named tvorog curd cheese, produced in the rural regions; Dashkasan, Ismailli, and khachmaz of Azerbaijan.
Materials and methods
Sampling and isolation of acid-bile resistant bacteria
We screened bacterial species isolated from six samples of tvorog collected from Dashkasan, Ismailli, and khachmaz regions of Azerbaijan for potential beneficial features. We focused in our search on lactobacilli and evaluated their ability for or against human enteropathogenic bacteria. Seventeen tvorog samples were collected from Dashkasan, Ismailli, and khachmaz regions and then 1.0 g of each sample was suspended in 10 ml PBS. Then, 500 µL of suspension was added to MRS broth (Fluka, Buchs, Switzerland) and incubated at 37°C for 48 h. For screening the tolerance of lactobacilli to harsh condition of stomach, 1.0 mLof each enriched culture was incubated in 10.0 mL PBS buffer (pH = 3) for 3 h. After centrifugation, pH resistant bacteria were resuscitated by addition of 10.0 mL MRS broth and incubation at 37°C for 4 h. Additionally, the second test was used for screening the bacteria against bile salt. Briefly, the acid resistant bacteria were inoculated in MRS broth containing 0.3% (w/v) oxgall (Sigma, Louis, USA) and incubated at 37°C for 4 h. The dilutions were prepared from acid and bile resistant enriched bacterial culture, then 100 µL of 10-5 dilution were spread onto MRS-agar plates and incubated at 37°C for 24-48 h. Different clonies were picked up and cultured in 10 mL MRS broth. First, the isolates were evaluated by gram staining and cell morphology. Then, he isolates were conserved in MRS broth with skim milk (10%) and glycerol (25%) at -70°C.
Screening for antagonistic activity
Antagonistic activity of isolates was evaluated as described in literature.17 Overnight cultures of the isolates were centrifuged. Then the supernatant was filtered by 0.25 µm microfilter. Two-hundred microliters of the supernatant were spotted onto the blank disk (Pad Tan Teb, Tehran, Iran) that was created on Muler Hinton Agar (MHA) plate with enteropathogenic bacteria spread and incubated at 37°C for 24 h. The plates were checked for the inhibition zones. Inhibition was scored positive if the width of the clear zone around the clonies of the producer strain was 7 mm or larger. The following enteropathogenic bacteria were used as indicator strains: Escherichia coli PTCC 1399, Yersinia enterocolitica PTCC 1151, Shigella flexneri PTCC 1234, Klebsiella pneumoniae PTCC 1290.
DNA extraction and molecular identification
The bacterial genomic DNA was extracted according to a previously published method.18 Amplification of the 16s rDNA was carried out using the primer pair reported previously as: 16 16lacF5ˊ-AGAGTTTGATCMTGGCTCAG-3ˊ16lacR5ˊ-TACCTTGTTAGGACTTCACC-3ˊ.19 Reactions were performed in an automatic thermal cycler (Bio-Rad, Hercules, CA, USA) under the following conditions: initial denaturation at 94°C for 4 min; 32 cycles of 94°C for 50 s, 59°C for 50 s and 72°C for 90 s and final extension at 72°C for 10 min and holding at 4°C. PCR products were ligated to the pGEM T/A cloning vector (Promega, Madison, WI, USA) according to the manufacturing instruction. Then, they were transformed to the E. coli DH5α according to the literature.20
The plasmids were then sent to a commercial sequencing facility (Macrogene, Seoul, Korea). The sequences were compared to those reported in GenBank, using Basic Local Alignment Search Tool (BLAST) algorithm. The isolates were identified by similarity with standard strains in GenBank.
Result
Screening of acid and bile resistant isolates
The isolates were screened against simulated harsh condition of human gastrointestinal system (i.e., pH=3 for 3 h and 0.3% bile salts for 4 h) as shown in Fig. 1.
Fig. 1 .
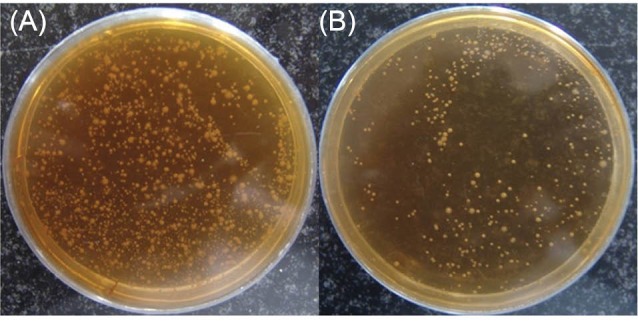
Clony of bacteria before and after enrichment in 3% bile and low pH. A: the clonies are mix of resistant and unresistant bacteria that many of them should be removed in screening process.
Analysis of antimicrobial activity
The antagonistic activity against human enteropathogenic bacteria was tested. The results are shown in Fig. 2. These 17 isolates were able to inhibit the indicator bacteria, but the strains TA5 (L. casei) and TB2 (L. rhamnosus) showed antibacterial effects for all of indicators with high efficiency (Fig. 3). The probiotic potential and physiological characterization of selected Lactobacillus is shown in Table 1. Overnight culturing of selected isolates in oxgal (0.3%) is shown in Fig. 4.
Fig. 2 .
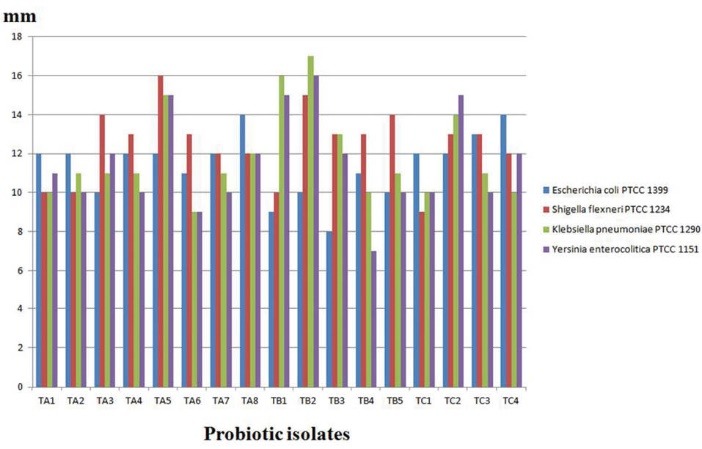
Antagonistic effect of selected isolates for human enteropathogenic bacteria.
Fig. 3 .

The inhibition zone surrounding the Lactobacillus isolated against human enteropathogenic bacteria.
Table 1 . The probiotic potential and physiological characterization of selected Lactobacillus .
| Isolates | Growth | pH resistancy | Bile resistancy | Antibiotic sensitivity | Antagonistic activity |
| TA5 | Fast | %80< | + | + | + |
| TB2 | Slow | %60< | + | + | + |
| TC2 | Medium | %60< | + | + | + |
Fig. 4 .
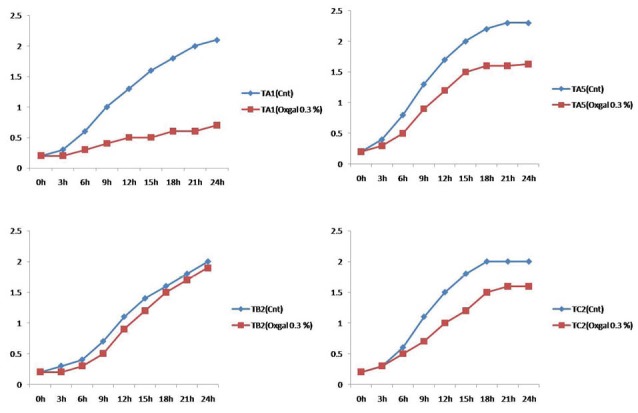
The growth inhibitory of oxgal (0.3%) for candidate probiotics at overnight incubation. TA1 isolate was very sensitive to bile compared with TB2, TC2, and TA5.
Identification of Lactoacilli by 16s rDNA pattern
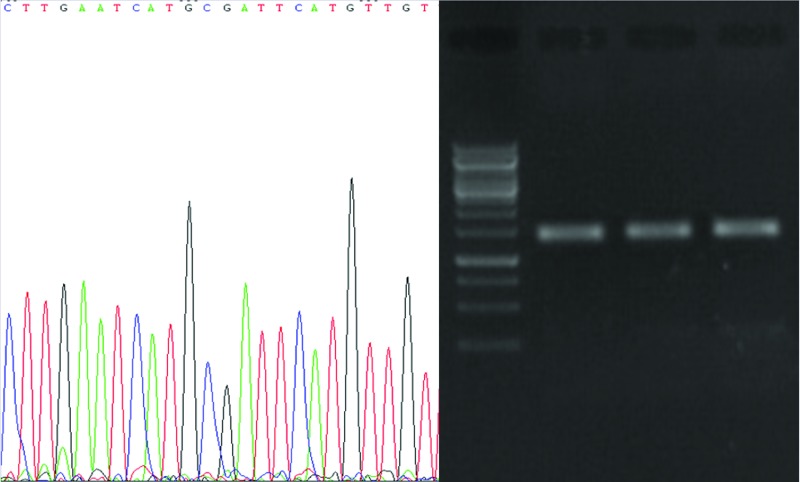
The 16s rDNA PCR product of the isolates with high antibacterial activity, namely, TA5, TB2, and TC2 were cloned in plasmids and sequenced (Fig. 5). Then the sequencing results were aligned using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared with the sequences deposited in NCBI GenBank for different lactobacillus species. The isolates had 100% similarity with L. Casei, L. plantarum, and L. rhominos. All these species represented antibacterial effects on the indicator bacteria.
Fig. 5 .
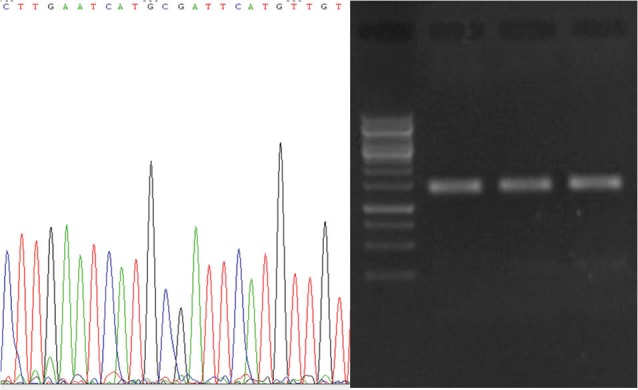
PCR electrophoresis of 16s rDNA. After amplification the PCR product was inserted on pGEM vector and sequenced (Supplementary files 1-3). The sequence of our strains was blasted on NCBI for identification.
Discussion
In recent researches, resistance to antibiotics in the treatment of bacterial infections has been increasingly observed, creating serious problems in treatment of infectious diseases. The resistance has been reported against almost any antibiotic discovered, and hence this issue has turned into one of the most urgent public health concerns and future medicinal challenges. In this regard, probiotics could be considered as living drugs that reduce antibiotic consumption. The aim of this study was the isolation of lactobacilli against human enteropathogenic bacteria for future designing of starter cultures, its application for medical purposes and perhaps the extraction of bacteriocin that is a safe natural antimicrobial for food preservation21 and an alternative substance to antibiotics.22,23 Bacteriocins, which are antimicrobial peptides produced by probiotic bacteria, might seriously be considered as alternatives to antibiotics. These proteins have significant exhibit potency against other human enteropathogenic bacteria as well as antibiotic-resistant strains that are stable and manipulate the future medicine.23 Homemade tvorog curd cheese in Azerbaijan that is consumed as a health promoting food, could also be a natural resource for other probiotic organisms such as yeasts.
Conclusion
In conclusion, screening of the acid- and bile-resistant lactobacilli strains from tvorog curd cheese resulted in the isolation of antibacterial samples which were identified by 16s rDNA as L. plantarum, L. casei and L. rhomnos. These bacteria that were isolated from traditional regions as native microbiome, were preserved in a biobank for future studies for medicinal applications and food industry, especially in culture starter designing.
Acknowledgments
Authors are very grateful to Baku State University for hosting of this Research.
Ethical issues
There is none to be declared.
Competing interests
There is none to be disclosed.
Supplementary materials
consist of sequenced data.
Research Highlights
What is current knowledge?
√ Probiotic candidate bacteria were isolated from traditionally fermented foods.
√ Antibiotic resistance has turned into one of the most urgent public health issues.
√ Probiotic therapy could be used instead of antibiotic therapy.
What is new here?
√ Homemade tvorog curd cheese in Azerbaijan harbor a variety of probiotics.
√ The isolated probiotics have antagonistic activity.
√ The isolated probiotics could be conserved in a biobank for medical applications.
References
- 1. Who website. http://www.who.int/en/.
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neu HC. The crisis in antibiotic resistance. Science. 1992;257:1064–73. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 5. Mancini A, Franciosi E, Carafa I, Fava F, Viola R, Tuohy K, editors. Probiotic potential of a BSH positive, high GABA producing strain, Lactobacillus brevis FEM 1874, isolated from traditional "wild" Alpine cheese. Rowett-INRA 2014: Gut Microbiology: from sequence to function; 2014.
- 6.Houshang J, Hassan SM, Allah DK. Isolation and Identification of Lactobacilli Found in Nomads Traditional Yogurt in the City of Jahrom Using PCR Method and, the Study of Their Interactional Effects on Streptococcus mutans as Cause of Tooth Decay Using Disc and Auger Hole Methods. Advances in Environmental Biology. 2014;8:421–7. [Google Scholar]
- 7.Jayashree S, Pooja S, Pushpanathan M, Vishnu U, Sankarasubramanian J, Rajendhran J. et al. Genome sequence of Lactobacillus fermentum strain MTCC 8711, a probiotic bacterium isolated from yogurt. Genome Announcements. 2013;1:e00770–13. doi: 10.1128/genomea.00770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blana VA, Grounta A, Tassou CC, Nychas G-JE, Panagou EZ. Inoculated fermentation of green olives with potential probiotic Lactobacillus pentosus and Lactobacillus plantarum starter cultures isolated from industrially fermented olives. Food Microbiol. 2014;38:208–18. doi: 10.1016/j.fm.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 9.García-Ruiz A, González de Llano D, Esteban-Fernández A, Requena T, Bartolomé B, Moreno-Arribas M. Assessment of probiotic properties in lactic acid bacteria isolated from wine. Food Microbiol. 2014;44:220–225. doi: 10.1016/j.fm.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Chiu Y-H, Tsai J-J, Lin S-L, Chotirosvakin C, Lin M-Y. Characterisation of bifidobacteria with immunomodulatory properties isolated from human breast milk. Journal of Functional Foods. 2014;7:700–8. doi: 10.1016/j.jff.2013.12.015. [DOI] [Google Scholar]
- 11.Cárdenas N, Calzada J, Peirotén Á, Jiménez E, Escudero R, Rodríguez JM. et al. Development of a Potential Probiotic Fresh Cheese Using Two Lactobacillus salivarius Strains Isolated from Human Milk. BioMed Research International. 2014;2014:801918. doi: 10.1155/2014/801918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubio R, Jofré A, Martín B, Aymerich T, Garriga M. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 2014;38:303–11. doi: 10.1016/j.fm.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Al Kassaa I, Hamze M, Hober D, Chihib N-E, Drider D. Identification of vaginal lactobacilli with potential probiotic properties isolated from women in North Lebanon. Microbial Ecology. 2014;67:722–34. doi: 10.1007/s00248-014-0384-7. [DOI] [PubMed] [Google Scholar]
- 14.Barzegari A, Eslami S, Ghabeli E, Omidi Y. Imposition of encapsulated non-indigenous probiotics into intestine may disturbs human core microbiome. Evolutionary and Genomic Microbiology. 2014;5:393. doi: 10.3389/fmicb.2014.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Zhu H, He F, Luo Y, Kang Z, Lu C. et al. Whole genome sequence of the probiotic strain Lactobacillus paracasei N1115, isolated from traditional chinese fermented milk. Genome Announcements. 2014;2:e00059–14. doi: 10.1128/genomea.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanghah SM, Ganbarov K. Lactobacillus with probiotic potential from homemade cheese in Azerbijan. BioImpacts : BI. 2014;4:49. doi: 10.5681/bi.2014.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hütt P, Shchepetova J, Loivukene K, Kullisaar T, Mikelsaar M. Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero‐and uropathogens. Journal of Applied Microbiology. 2006;100:1324–32. doi: 10.1111/j.1365-2672.2006.02857.x. [DOI] [PubMed] [Google Scholar]
- 18.Atashpaz S, Khani S, Barzegari A, Barar J, Vahed SZ, Azarbaijani R. et al. A robust universal method for extraction of genomic DNA from bacterial species. Microbiology. 2010;79:538–42. [PubMed] [Google Scholar]
- 19.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proceedings of the National Academy of Sciences. 1985;82:6955–9. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanehbandi D, Saei AA, Zarredar H, Barzegari A. Vibration and glycerol‐mediated plasmid DNA transformation for Escherichia coli. FEMS Microbiology Letters. 2013;348:74–8. doi: 10.1111/1574-6968.12247. [DOI] [PubMed] [Google Scholar]
- 21.Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. International Journal of Food Microbiology. 2001;71:1–20. doi: 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 22.Joerger R. Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poultry Science. 2003;82:640–7. doi: 10.1093/ps/82.4.640. [DOI] [PubMed] [Google Scholar]
- 23.Cotter PD, Ross RP, Hill C. Bacteriocins—a viable alternative to antibiotics? Nature Reviews Microbiology. 2012;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
consist of sequenced data.