Abstract
High-throughput somatic mutation screening using FFPE tissues is a major challenge because of a lack of established methods and validated variant calling algorithms. We aimed to develop a targeted sequencing protocol by Fluidigm multiplex PCR and Illumina sequencing and to establish a companion variant calling algorithm. The experimental protocol and variant calling algorithm were first developed and optimized against a series of somatic mutations (147 substitutions, 12 indels ranging from 1 to 33 bp) in seven genes, previously detected by Sanger sequencing of DNA from 163 FFPE lymphoma biopsy specimens. The optimized experimental protocol and variant calling algorithm were further ascertained in two separate experiments by including the seven genes as a part of larger gene panels (22 or 13 genes) using FFPE and high-molecular-weight lymphoma DNAs, respectively. We found that most false-positive variants were due to DNA degradation, deamination, and Taq polymerase errors, but they were nonreproducible and could be efficiently eliminated by duplicate experiments. A small fraction of false-positive variants appeared in duplicate, but they were at low alternative allele frequencies and could be separated from mutations when appropriate threshold value was used. In conclusion, we established a robust practical approach for high-throughput mutation screening using archival FFPE tissues.
The advent of next-generation sequencing (NGS) technology has transformed the landscape of life science research and has led to unprecedented discoveries. In the field of cancer research, NGS has already uncovered a catalog of extensive somatic mutations and continues to extend this ever-growing list of genetic changes in human cancer. In most human malignant tumors, somatic mutations are observed in a wide spectrum of diverse oncogenes and tumor suppressor genes at variable frequencies. For example, in diffuse large B-cell lymphoma (DLBCL), somatic mutations are observed in >300 cancer genes, and on average each lymphoma harbors approximately 30 pathogenic mutations.1–4 Most of these pathogenic mutations occur in <20% of cases, but different somatic mutations may affect a common molecular pathway.1–4 One of the major challenges is to investigate somatic mutations in these newly identified cancer genes; investigate their potential value in diagnosis, prognosis, and treatment stratification; and translate the relevant research findings into clinical practice using routine formalin-fixed, paraffin-embedded (FFPE) diagnostic tissue biopsies.
There are several target enrichment approaches for high-throughput mutation screening by NGS, for example, hybrid capture with Agilent SureSelect (Agilent Technologies, Santa Clara, CA) or NimbleGen SeqCap (Roche, Basel, Switzerland) products and PCR using HaloPlex (Agilent Technologies) or RainDance technology (RainDance Technologies, Cambridge, UK). These targeted resequencing approaches were originally developed based on high-molecular-weight (HMW) DNA samples and have now been successfully applied to those from FFPE tissues and circulating cell-free tumor DNA.5–7 Several commercial NGS-based assays have been developed for detection of well-characterized somatic alterations, particularly the hotspot mutations, in cancer genes, and these assays, particularly those by Ion Torrent (Life Technologies, Carlsbad, CA), can be applied to a minute amount of DNA extracted from FFPE tissue biopsies.8–10 Nonetheless, there is no established protocol for discovery research (ie, detecting unknown mutations in the newly identified cancer genes using FFPE tissue specimens, particularly small biopsy specimens). Many of the caveats for NGS-based mutation screening using FFPE tissue DNA, such as artifacts due to poor DNA quality and sequencing errors, false-negative variants due to inadequate target enrichment, suboptimal performance of variant calling algorithm, the cutoff value of variant allele frequency for diagnosis of somatic mutation, minimal DNA quantity, and quality required for successful NGS, have not been properly investigated.
Among the various target enrichment methods currently available, PCR using the microfluidic technology (Fluidigm Access Array System; Fluidigm, South San Francisco, CA) represents a practical alternative for high-throughput mutation screening using routine FFPE tissue biopsies. The Fluidigm Access Array System offers several distinct advantages, including i) being amenable to small amounts (50 ng) of DNA samples, ii) allowing parallel amplification of 48 samples with 48 pairs of PCR primers, and iii) offering great flexibility in choice of primers and genetic targets. The system has been successfully used for targeted sequencing using HMW DNA, plasma DNA, and very recently FFPE tissue DNA.11–13 However, all these caveats, including the strategies to eliminate false-positive variants and the cutoff value of variant allele frequency for diagnosis of somatic mutation, remain to be established. In the present study, we developed a protocol for high-throughput mutation analysis by multiplex PCR with Fluidigm Access Array System using DNA samples from FFPE tissues, followed by Illumina MiSeq sequencing. We also developed and validated an in-house variant calling algorithm against a wide range of known mutations. More importantly, we have addressed the above issues through a series of designed experiments and data analysis.
Materials and Methods
Tumor Materials and DNA Extraction
FFPE lymphoma specimens from 163 cases of DLBCL were retrieved from the Haematological Malignancy Diagnostic Service at St James's University Hospital, Leeds, and Addenbrooke's Hospital, Cambridge. Local ethical guidelines were followed for the use of archival tissues for research with the approval of the ethics committees of the involved institutions.
Hematoxylin and eosin slides were reviewed, and crude microdissection was performed in each case to enrich tumor cells, ensuring that a tissue area that contained >60% of tumor cells was used for DNA extraction. DNA was extracted using the QIAamp DNA Micro Kit (Qiagen, Crawley, UK) and quantified using Qubit Fluorometer (Life Technologies). In addition, DNA was extracted from FFPE reactive tonsils and used for validation of various PCR conditions.
Assessment of DNA Quality by Conventional PCR
This was performed by PCR of variably sized genomic fragments (Table 1),14 using a 2-ng template DNA in a 10-μL reaction mixture. PCR conditions were 95°C for 10 minutes, 40 cycles of 95°C for 20 seconds, 60°C for 20 seconds, 72°C for 1 minute, and a final extension at 72°C for 5 minutes.
Table 1.
Primers Used for Conventional PCR and Sanger Sequencing
| Genes | Exon | Primer name | Sequence (5′-3′) | Amplicon size (bp) | Ta (°C) |
|---|---|---|---|---|---|
| Primers for Assessment of DNA Quality by Conventional Multiplex PCR | |||||
| RAG1 | E2 | Forward | 5′-TGTTGACTCGATCCACCCCA-3′ | 200 | 60 |
| Reverse | 5′-TGAGCTGCAAGTTTGGCTGAA-3′ | ||||
| PLZF | E1 | Forward | 5′-TGCGATGTGGTCATCATGGTG-3′ | 300 | 60 |
| Reverse | 5′-CGTGTCATTGTCGTCTGAGGC-3′ | ||||
| AF4 | E11 | Forward | 5′-CCGCAGCAAGCAACGAACC-3′ | 400 | 60 |
| Reverse | 5′-GCTTTCCTCTGGCGGCTCC-3′ | ||||
| AF4 | E3 | Forward | 5′-GGAGCAGCATTCCATCCAGC-3′ | 600 | 60 |
| Reverse | 5′-CATCCATGGGCCGGACATAA-3′ | ||||
| Primers for Investigation of Mutations in Oncogenes and Tumor Suppressor Genes Involved in DLBCL by PCR and Sanger Sequencing | |||||
| CARD11 | E5-1 | Forward | 5′-GTGCCCCCTCTCCACAGT-3′ | 200 | 62 |
| Reverse | 5′-AGTACCGCTCCTGGAAGGTT-3′ | ||||
| E5-2 | Forward | 5′-GAAGAAGCAGATGACGCTGA-3′ | 235 | 62 | |
| Reverse | 5′-GTCACCCTGGCGGAGTAG-3′ | ||||
| E6 | Forward | 5′-CACCCTTGGGGTATTTCAGA-3′ | 210 | 59 | |
| Reverse | 5′-CAGGCCCTCACCTGGATG-3′ | ||||
| E7 | Forward | 5′-CCTGACCCTCTGAAACCTCCT-3′ | 204 | 62 | |
| Reverse | 5′-GCGATCCCCACTCCCAC-3′ | ||||
| E8 | Forward | 5′-TCGATGCGCATATTGATTTC-3′ | 181 | 62 | |
| Reverse | 5′-CTGCAGGTGGTGCCTGTA-3′ | ||||
| E9 | Forward | 5′-CCCAAAGCAGCCTTCGTC-3′ | 234 | 62 | |
| Reverse | 5′-cCTGGTCCAGGTTGTTGCTGTCC-3′ | ||||
| MYD88 | E1-1 | Forward | 5′-CTCGGGGCTCCAGATTGTA-3′ | 327 | 58 |
| Reverse | 5′-GCCGGATCTCCAAGTACTCA-3′ | ||||
| E1-2 | Forward | 5′-GCTGCTCTCAACATGCGAGT-3′ | 317 | 62 | |
| Reverse | 5′-GGAAAGTCAGCCTCCTCACC-3′ | ||||
| E2 | Forward | 5′-CTGGATCCTGACTGTGGGTAA-3′ | 281 | 62 | |
| Reverse | 5′-GCTTCAAACACCCATGCTCT-3′ | ||||
| E3 | Forward | 5′-TCTGACCACCACCCTTGTG-3′ | 264 | 62 | |
| Reverse | 5′-CAGGGCAGGGCTTCATGC-3′ | ||||
| E4 | Forward | 5′-GGCCCTTCCTGAAGCTATTC-3′ | 270 | 62 | |
| Reverse | 5′-TGGTACTGCATCCACAGTCC-3′ | ||||
| E5 | Forward | 5′-GTTGAAGACTGGGCTTGTCC-3′ | 292 | 59 | |
| Reverse | 5′-AGGAGGCAGGGCAGAAGTA-3′ | ||||
| CD79A | E5 | Forward | 5′-ATGAAGTGAGTGAAGGGTGGG-3′ | 326 | 58 |
| Reverse | 5′-AGAATGTCCCAGGGAAGTGAG-3′ | ||||
| CD79B | E5 | Forward | 5′-TAGGTGGCTGTCTGGTCAATG-3′ | 306 | 58 |
| Reverse | 5′-TGTTCTTGCAGAATGCACCTC-3′ | ||||
| E6 | Forward | 5′-CTGGAGACAAATGGCAGCTC-3′ | 362 | 58 | |
| Reverse | 5′-CACCTACGAGGTAAGGAGAGGG-3′ | ||||
| PRDM1 | E1 | Forward | 5′-GGAGAATGTGGACTGGGTAG-3′ | 220 | 55 |
| Reverse | 5′-TAAGTGCACTAAAGCAGAAGC-3′ | ||||
| E2-1 | Forward | 5′-TGTATTAGTCATAGCCTCTCA-3′ | 367 | 55 | |
| Reverse | 5′-CTCTTCAAACTCAGCCTCTGT-3′ | ||||
| E2-2 | Forward | 5′-TGTGAGGTTTCAGGGATTGGCAGA-3′ | 261 | 55 | |
| Reverse | 5′-AGGTCCCCAATCTTCTTGTC-3′ | ||||
| E3 | Forward | 5′-TTGTATTACTACTTGACAGTCC-3′ | 258 | 55 | |
| Reverse | 5′-TTCCTAACATTTAATGGGTCTG-3′ | ||||
| E4-1 | Forward | 5′-GTTTATTCTGAGAGGTGCTGG-3′ | 253 | 55 | |
| Reverse | 5′-CAAGAAGTTCCTGGTTGGCA-3′ | ||||
| E4-2 | Forward | 5′-ATGTGAATCCAGCACACTCTC-3′ | 289 | 55 | |
| Reverse | 5′-TCCAACACATGACAAAGCCGT-3′ | ||||
| E5-1 | Forward | 5′-TTGCTTCAGTTCTCTCTAGCC-3′ | 243 | 55 | |
| Reverse | 5′-TTCTAAAGTCATCGAGGTCC-3′ | ||||
| E5-2 | Forward | 5′-TCCTAAAATTGGACTCCAACC-3′ | 294 | 55 | |
| Reverse | 5′-TGGAGCTCTTGAGGCTTTGGT-3′ | ||||
| E5-3 | Forward | 5′-AGACTTTTTGAAAGCTTCC-3′ | 300 | 55 | |
| Reverse | 5′-TTGTACGAGGGGATGAAAGCTG-3′ | ||||
| E5-4 | Forward | 5′-TACGCTTACTTGAACGCGTC-3′ | 278 | 55 | |
| Reverse | 5′-AGAGAAGTGGGGTTGAGCAT-3′ | ||||
| E5-5 | Forward | 5′-ATCAACAACTTTGGCCTCTTC-3′ | 296 | 55 | |
| Reverse | 5′-TTGGGCTGCACCACATGTTCT-3′ | ||||
| E5-6 | Forward | 5′-ATGAAGGACAAGGCCTGTAG-3′ | 314 | 55 | |
| Reverse | 5′-CAAACACAAGCATGCACTCAC-3′ | ||||
| E6 | Forward | 5′-AACACTTGAGTCTTGGAGCAG-3′ | 267 | 55 | |
| Reverse | 5′-GTGACTCACAGACATTTTCTA-3′ | ||||
| E7-1 | Forward | 5′-TTGGCAACTCTTAATCTTCTGG-3′ | 284 | 55 | |
| Reverse | 5′-TCAGGTGAACCTTGAGGCTACAG-3′ | ||||
| E7-2 | Forward | 5′-CCAAGTTCACCCAGTTTGTG-3′ | 297 | 55 | |
| Reverse | 5′-TTCTGACCACGGCCAGAATTTC-3′ | ||||
| E7-3 | Forward | 5′-TGACATCAGTGACAATGCTGAC-3′ | 313 | 55 | |
| Reverse | 5′-ACTCACCAAGTCATAACTTA-3′ | ||||
| TP53 | E5 | Forward | 5′-CACTTGTGCCCTGACTTTCA-3′ | 267 | 64 |
| Reverse | 5′-AACCAGCCCTGTCGTCTCT-3′ | ||||
| E6 | Forward | 5′-GAGAGACGACAGGGCTGGT-3′ | 231 | 62 | |
| Reverse | 5′-CACTGACAACCACCCTTAACC-3′ | ||||
| E7 | Forward | 5′-CCTGCTTGCCACAGGTCT-3′ | 295 | 62 | |
| Reverse | 5′-GTGATGAGAGGTGGATGGGTAG-3′ | ||||
| E8 | Forward | 5′-GGACAGGTAGGACCTGATTTCC-3′ | 257 | 64 | |
| Reverse | 5′-TGAATCTGAGGCATAACTGCAC-3′ | ||||
| E9 | Forward | 5′-GACCAAGGGTGCAGTTATGC-3′ | 277 | 62 | |
| Reverse | 5′-ACCAGGAGCCATTGTCTTTG-3′ | ||||
| E10 | Forward | 5′-AACTTGAACCATCTTTTAACTCAGG-3′ | 242 | 62 | |
| Reverse | 5′-GAATCCTATGGCTTTCCAACCT-3′ | ||||
| TNFAIP3 | E2a | Forward | 5′-CTGCAGGCAGCTATAGAGGAG-3′ | 272 | 58 |
| Reverse | 5′-CGAAACTGAGGACAAAACTGG-3′ | ||||
| E2b | Forward | 5′-GCAATATGCGGAAAGCTGTG-3′ | 300 | 58 | |
| Reverse | 5′-GCTATCACCCAGGCAAAAGA-3′ | ||||
| E3 | Forward | 5′-TTGCTGGGTCTTACATGCAG-3′ | 378 | 58 | |
| Reverse | 5′-GCTTCGCTTAGCCAAATTCA-3′ | ||||
| E4 | Forward | 5′-GGGAGTACAGGATACATTCAAGC-3′ | 251 | 58 | |
| Reverse | 5′-AAGGCATAAGGCTGAAAGCA-3′ | ||||
| E5 | Forward | 5′-ACCTAAGGGCCTCATTTTCC-3′ | 275 | 58 | |
| Reverse | 5′-GCAAAAAGGAAAACCCTGATG-3′ | ||||
| E6 | Forward | 5′-TGAGATCTACTTACCTATGGCCTTG-3′ | 315 | 58 | |
| Reverse | 5′-TCAGGTGGCTGAGGTTAAAGA-3′ | ||||
| E7a | Forward | 5′-ACAGGCCTGCATTTCAGTG-3′ | 282 | 58 | |
| Reverse | 5′-GGAAGGTTCCATGGGATTC-3′ | ||||
| E7b | Forward | 5′-GCAGGAAAACAGCGAGCA-3′ | 272 | 58 | |
| Reverse | 5′-CCAAGGGCTCATAGGCTTCT-3′ | ||||
| E7c | Forward | 5′-ACTCCCAAAGCTGAACTCCA-3′ | 304 | 58 | |
| Reverse | 5′-GGGATCCAAGTGCCTTGT-3′ | ||||
| E7d | Forward | 5′-ACTGCCATGAAGTGCAGGAG-3′ | 279 | 58 | |
| Reverse | 5′-ATCTGACTTGGAACGCTGGT-3′ | ||||
| E7e | Forward | 5′-TGCAGTACTTGCTTCAAAAGGA-3′ | 313 | 58 | |
| Reverse | 5′-CCACTTCACTCACGTTTGTTTT-3′ | ||||
| E8 | Forward | 5′-GGGGTGACCCCTATGTGGTACT-3′ | 293 | 58 | |
| Reverse | 5′-CCAGTTGCTCTTCTGTCCTTTT-3′ | ||||
| E9a | Forward | 5′-GTGCTCTCCCTAAGAAATGTGAG-3′ | 205 | 58 | |
| Reverse | 5′-CTGGTTGGGATGCTGACACT-3′ | ||||
| E9b | Forward | 5′-CTCTGCATGGAGTGTCAGCAT-3′ | 257 | 58 | |
| Reverse | 5′-GGGTTCAGAGGATAGCACCA-3′ | ||||
DLBCL, diffuse large B-cell lymphoma; E, exon.
Quantification of DNA Copy Number by qPCR
Quantification of DNA copy number by real-time quantitative PCR (qPCR) was performed on a Quantstudio 6 instrument (Life Technologies) using a custom TaqMan assay of a 195-bp fragment of the PPIA gene, which was chosen because there is no evidence of PPIA gene copy number change in lymphoma. Primers and probe were designed using the Primer Express software version 3.0.1 (Life Technologies). The sequences of the primers (Thermo Fisher Scientific, Ulm, Germany) and the probe (Life Technologies) are as follows: forward primer 5′-TATGGCTGTCAGGAGCAGTTCTT-3′, reverse primer 5′-AAATGGACCAACCTGCTGTCTT-3′, and probe 5′-ACTAAGCAACAAAATAAGCA-VIC-3′. The qPCR conditions and performance were systematically tested and validated before data collection. The qPCR was performed in 10-μL reaction that contained 5 μL of TaqMan gene expression master mix (Life Technologies), 0.9 μL of each primer (final concentration of 900 nmol/L), 0.25 μL of probe (final concentration of 250 nmol/L), 1.95 μL of PCR-certified water (Teknova, Hollister, CA), and 1 μL of template genomic DNA. PCR cycling conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. For each sample to be quantified, DNA concentration was measured by Qubit double-stranded DNA HS assay (Life Technologies), and serial dilutions were performed to give 10-ng/μL, 5-ng/μL, and 2.5-ng/μL solutions. A 10-point standard curve with DNA quantity ranging from 10 to 0.020 ng was prepared using high-quality human genomic DNA (Promega, Madison, WI) (Supplemental Figure S1). TaqMan qPCR was performed in a batch of 38 DNA samples together with negative control and standard curve in triplicate. The estimated copy number was then calculated and expressed as the percentage of functional DNA copies relative to the standard curve, with a mean of the three dilutions taken as the final result (Supplemental Figure S1).
Sanger Sequencing
Mutations in seven genes, including CARD11, CD79A, CD79B, MYD88, TNFAIP3, PRDM1, and TP53, were first investigated by PCR and Sanger sequencing in 163 DLBCLs using the primers detailed in Table 1. PCR was performed in a 10-μL reaction mixture with 5- to 10-ng template DNA and AmpliTaq Gold 360 (Life Technologies) master mix plus GC-enhancer according to the manufacturer's instructions. The PCR conditions were 95°C for 10 minutes to activate the enzyme, followed by 40 cycles of denaturation at 95°C for 20 to 30 seconds, annealing at 55°C to 64°C (depending on the primer set) for 20 seconds, extension at 72°C for 30 to 45 seconds (depending on the amplicon size), and a final 5-minute extension at 72°C. PCR products were routinely sequenced using the BigDye Terminator 3.1 System (Applied Biosystems) on an ABI 3730 instrument (Applied Biosystems). In each case, sequence change was confirmed by at least two independent PCR and sequencing experiments. The somatic nature of mutations was ascertained by excluding germline changes through single-nucleotide (SNP) database search and sequencing DNA samples from the microdissected normal cells.
PCR Product Cloning and Sequencing
To confirm mutations that were detected by Illumina MiSeq but not by conventional Sanger sequencing, the relevant PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) and then transformed into TOP10 competent cells. Colonies were screened by PCR using vector primers, and up to 30 positive clones were routinely sequenced by the Sanger method as above.
Primer Design and Validation for PCR with Fluidigm Access Array
PCR primer pairs were redesigned for the above seven genes and a further 15 genes using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0, last accessed May 10, 2011) based on hg19 of the human genome. A set of criteria were followed for the primer design: i) targeting a small segment of the coding sequence with all amplicons in the range of 144 to 213 bp, thus amenable to DNA samples from FFPE tissues; ii) covering the entire coding sequence or all the regions known to be mutated in human malignant tumors; iii) giving a means ± SD Tm value at 60°C ± 3°C; and iv) where possible avoiding any known SNPs and GC-rich sequence region. The specificity of the primers designed and their potential formation of primer dimers were checked with Primer Blast (www.ncbi.nlm.nih.gov/tools/primer-blast, last accessed May 10, 2011), then further assessed by In-Silico PCR (http://genome.ucsc.edu/cgi-bin/hgPcr?command=start, last accessed May 10, 2011) and the AutoDimer program (http://www.cstl.nist.gov/strbase/AutoDimerHomepage/AutoDimerProgramHomepage.htm, last accessed November 7, 2012) (Supplemental Figure S2).
For each primer pair designed, the forward and reverse primers were tagged with a common sequence 1 (CS1: 5′-ACACTGACGACATGGTTCTACA-3′) and common sequence 2 (CS2: 5′-TACGGTAGCAGAGACTTGGTCT-3′), respectively. All primer pairs were purchased from Thermo Fisher Scientific GmbH and then experimentally validated by PCR using DNA samples from FFPE tonsils. Any primer pairs that failed to yield satisfactory amplification of the expected PCR product or gave rise to a nonspecific product were redesigned. In total, 343 primer pairs for 22 genes were successfully designed and validated and used for PCR with Fluidigm Access Array (Supplemental Table S1).
Preamplification to Enrich Template Target
For PCR with the Fluidigm Access Array using DNA samples from FFPE tissues, it was necessary to perform a preamplification with gene-specific primers to enrich the template targets before PCR with the Fluidigm Access Array (Figure 1). Our initial experiments revealed that it was not feasible to include all primer pairs in a single preamplification reaction and achieve a uniform amplification of all of the targets due to overlapping primers and primer dimer interactions. We then separated the primer pairs that might potentially give rise to the above issues based on In-Silico PCR and AutoDimer analyses and performed two separate preamplifications for each sample accordingly.
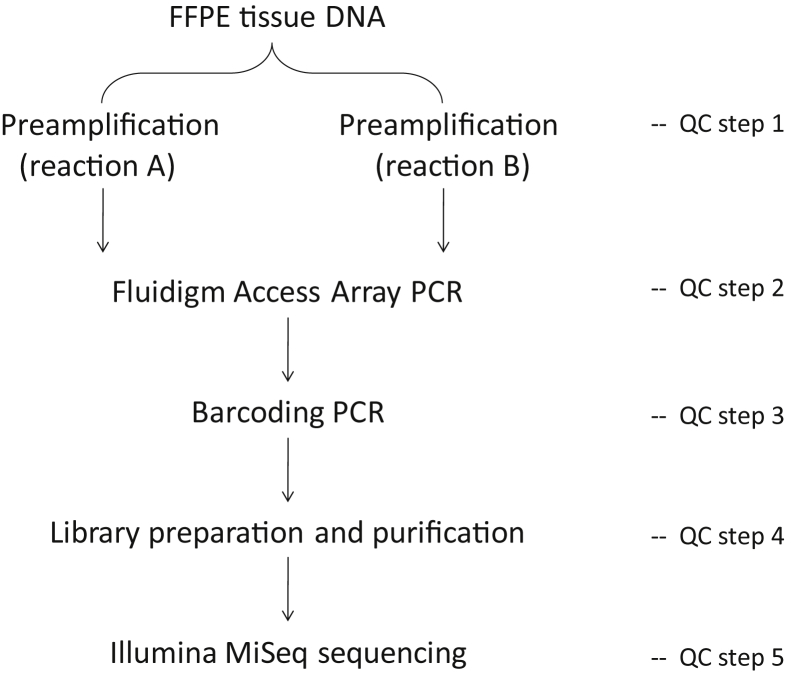
Figure 1.
Outline of experimental design for mutation screening using Fluidigm Access Array PCR and Illumina MiSeq sequencing. FFPE, formalin-fixed, paraffin-embedded; QC, quality control.
For each DNA sample, the preamplification and Fluidigm Access Array PCR were performed in duplicate. The preamplification was performed in a 10-μL FastStart High Fidelity Reaction mixture (Roche, Basel, Switzerland) that contained 50 ng of genomic DNA from FFPE tissues (or 20 ng of HMW DNA from fresh frozen tissues), 50 nmol/L of each primer, 4.5 mmol/L MgCl2, 5% dimethyl sulfoxide (DMSO), 200 μmol/L dNTPs, 1× FastStart High Fidelity Reaction Buffer with MgCI2, and 1 U of FastStart High Fidelity Enzyme, under the following conditions: 95°C for 10 minutes, 2 cycles of 95°C for 15 seconds, 60°C for 4 minutes, 13 cycles of 95°C for 15 seconds, and 72°C for 4 minutes. The preamplified products were routinely treated with 4 μL of ExoSAP-IT enzyme (Affymetrix, Santa Clara, CA) to eliminate the unincorporated primers and dNTPs. The efficacy of preamplification was then validated by conventional PCR (Supplemental Figure S3A).
Massive Parallel PCR with the Fluidigm Access Array
This procedure was performed essentially according to the manufacturer's instructions. Briefly, a sample mixture was prepared by mixing 1 μL of the fivefold diluted preamplified product with 4 μL of FastStart High Fidelity Reaction Buffer containing 4.5 mmol/L MgCl2, 5% DMSO, 200 μmol/L each dNTPs, and 0.25 U of FastStart High Fidelity Enzyme. Separately, a primer mixture was prepared for each primer pair or multiple primer pairs where indicated, with 6 μmol/L of each primer and 1× Access Array Loading Reagent in a final volume of 50 μL. The Fluidigm 48.48 Access Array was loaded with the sample and primer mixtures via the appropriate inlets using an integrated fluidic circuits controller. The array chip was then placed in the Fluidigm Thermal Cycler, and PCR was performed under the default conditions of the manufacturer (Supplemental Table S1). The amplified products for each sample were harvested together using an integrated fluidic circuits controller. Harvested PCR products were assessed by gel electrophoresis (Supplemental Figure S3B).
At the initial stage of the method development, the seven genes were screened for mutations in each of the 163 lymphoma samples by singleplex PCR with the Fluidigm Access Array. While at the late stage of the method validation, the seven genes were included as a part of the 22-gene panel and screened for mutation in 142 cases of the above lymphoma samples where sufficient DNA was available by multiplex PCR with the Fluidigm Access Array. In both experiments, the preamplification and Fluidigm PCR for each DNA sample were performed in duplicate.
In a separate parallel study, the seven genes were included as a part of the 13-gene panel and screened for mutation in 38 cases of splenic marginal zone lymphoma by multiplex PCR with Fluidigm Access Array using HMW DNA.15 This experiment was similarly performed in duplicate. The novel variants identified in these samples were further verified by a totally independent experiment. The sequence data from these HMW DNA were analyzed in parallel as a comparison.
Barcoding and Illumina MiSeq Sequencing
Barcoding was performed in a 20-μL reaction mixture that contained 1 μL of the 100-fold diluted harvested Fluidigm PCR products and 400-nmol/L barcode primers (Fluidigm) in FastStart High Fidelity reaction buffer. The reaction was performed on a conventional PCR thermal cycler under the following conditions: 95°C for 10 minutes, 15 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 1 minute, with a completion step at 72°C for 3 minutes.
The barcoded PCR products from various samples were pooled, assessed by gel electrophoresis (Supplemental Figure S3C), and purified using AMPure XP beads (Beckman Coulter, Pasadena, CA) following the manufacturer's instructions. A ratio of bead to library at 0.8:1 efficiently removed nonspecific products, commonly <200 bp (Supplemental Figure S3D). Purified PCR product library was quantified using a Qubit Fluorometer. Purified libraries were routinely sequenced on an Illumina MiSeq sequencer using 250-bp end sequencing protocol.
MiSeq Sequence Data Analysis
The fastq conversion from BCL and demultiplexing were performed using the MiSeq Reporter software version 2.4 (Illumina, San Diego, CA). The adaptor sequence (TGTAGAACCATGTCGTCAGTGT) was removed using cutadapt.16 The reads were aligned to the target sequences using BWA aln and sampe with the -e 50 parameter for the latter.17 The coordinates of the aligned reads were transposed into GRCh37/HG19 coordinates using an in-house perl program and transformed to a bam file using samtools.18 Variants were identified using an in-house developed variant caller python program, which was specially designed to identify variants by Fluidigm PCR and MiSeq sequencing, and systematically validated against a large number of known mutations from seven genes (Figure 2). The identified variants were annotated using the ensembl human database, using the ensembl Variant Effect Predictor,19 and the result was transformed into an Excel sheet using a bespoke perl script. After filtering baseline sequence errors and germline changes through an SNP database search, novel variants seen in both replicates of the same sample were recorded.
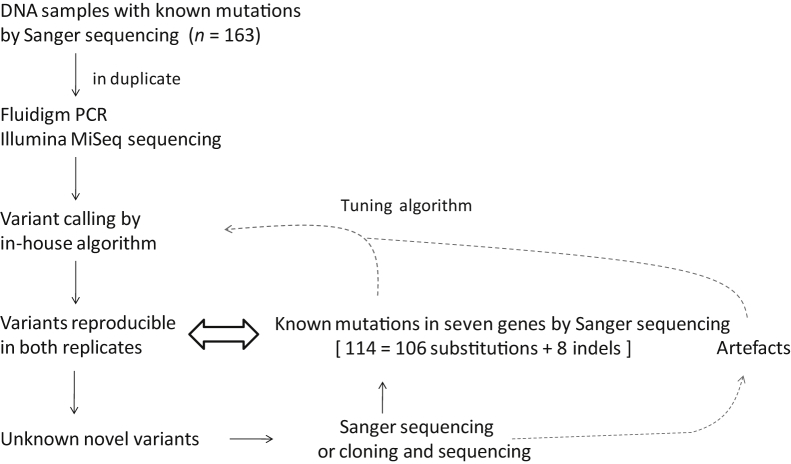
Figure 2.
Strategies for development and improvement of in-house variant calling algorithm. At first, the performance of the in-house variant calling algorithm was assessed and tuned against 114 known mutations by Sanger sequencing. The additional novel variants identified by Fluidigm PCR/MiSeq sequencing were further validated by PCR and Sanger sequencing, where necessary by cloning and sequencing of the PCR products. The resulting sequence data were used to further fine-tune the algorithm. Taken together, a total of 159 Sanger sequencing confirmed somatic mutations, including 147 substitutions and 12 indels (range, 1 to 33 bp) were used to optimize the algorithm.
Sequence Search for Features Potentially Associated with False-Positive Variants
For each type of nucleotide substitutions, the 21-bp sequence flanking the nucleotide change was extracted. These sequences were aligned together, and the position weight matrices were calculated and displayed by WebLogo.20 The de novo enriched motifs were discovered from these sequences using the MEME suite.21
Results
In the initial study, the experimental protocol and variant calling algorithm were developed and validated against the somatic mutations in seven genes detected by Sanger sequencing of DNA samples from a total of 163 FFPE DLBCL biopsy specimens. In the subsequent study, the above optimized experimental protocol and variant calling algorithm were tested in two sets of independent experiments with larger panels of genes.
Development of Multiplex PCR with the Fluidigm Access Array
Because DNA samples from FFPE tissues are highly fragmented and inefficient for direct PCR with Fluidigm Access Array, the template targets were first enriched by preamplificaton of each DNA sample with gene-specific primers. The major challenges for preamplification are to design primers that can work efficiently in the presence of a large number of other primer sets and yield uniform target enrichment with minimal nonspecific products. We started with the seven genes and designed 111 primer pairs, covering a 21-kb sequence. Despite a meticulous effort in primer design, uniform target enrichment could not be achieved in a single preamplification reaction because of undesired amplification by overlapping primers and poor amplification with a small proportion of primer pairs due to primer dimer interaction. To resolve this, we separated the primer pairs that potentially gave rise to these problems into two independent preamplifications based on In-Silico PCR and AutoDimer analyses (Figure 1, Supplemental Figure S2). The preamplified products were first validated by conventional PCR and then by Fluidigm Access Array PCR and Illumina MiSeq sequencing (Supplemental Figure S2). Illumina MiSeq sequencing confirmed adequate depth of read for each of the 111 amplicons.
The standard protocol for the Fluidigm Access Array allows PCR with 48 pairs of primers. To increase capacity, we tested a range of multiplex PCR (2 to 10 primer pairs) with the Fluidigm Access Array. The combination of various primer pairs for multiplex PCR was guided by In-Silico PCR and AutoDimer analyses. Illumina MiSeq sequencing of the Fluidigm amplified products revealed adequate depth of coverage for each of the amplicons by multiplex PCR with up to four primer pairs (Supplemental Figure S4) but unsatisfactory coverage for some of the amplicons by multiplex PCR with five or more primer pairs.
In addition to the strategies outlined above, a series of quality control measures were established at various steps of Fluidigm PCR/MiSeq sequencing, including quality control assessment of template DNA, preamplification, Fluidigm PCR, barcode labeling, and library purification (Supplemental Figure S3).
Development of Strategy and Variant Calling Algorithm for Mutation Detection
PCR using FFPE DNA is prone to generate sequence errors due to a variety of reasons, such as DNA base modification or damage, few copies of intact templates for PCR, and Taq polymerase error. Most of these errors are likely to be random and thus not reproducible and could be efficiently eliminated by performing Fluidigm PCR/MiSeq sequencing analyses in duplicate. As expected, most nonreproducible changes were observed at a lower alternate allele frequency (AAF), particularly <10% (Figure 3A). Nonetheless, a very small fraction of nonreproducible changes were seen at a much higher AAF, even up to 100% of all reads, indicating errors introduced at the very early steps of the amplification procedure. The level of these nonreproducible variants was also dependent on DNA quality (Figure 3A).
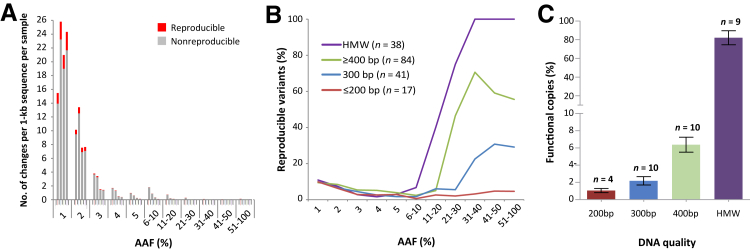
Figure 3.
Impact of DNA quality on baseline sequence errors by Fluidigm Access Array PCR and Illumina MiSeq sequencing. A: The level of both reproducible (seen in both replicates) and nonreproducible (seen only in one replicate) variants according to alternate allele frequency (AAF) and DNA quality. Formalin-fixed, paraffin-embedded (FFPE) DNA samples are further divided into subgroups according to the size of genomic sequence amenable for amplification by conventional multiplex quality control PCR. The data from FFPE tissue DNA are based on 142 cases investigated by two sets of independent experiments (Figure 4) and are shown from one set of the experiments. The levels of both reproducible and nonreproducible variants, particularly the latter, are remarkably high toward lower AAF. Because of small numbers of data points with an AAF >5%, these data are combined and presented in groups as indicated. B: The percentage of reproducible variants of the total (reproducible plus nonreproducible variants) according to AAF and DNA quality as above. Nonreproducible variants are baseline sequence errors. Reproducible variants at high AAFs are likely true mutations, but those at lower AAFs are probably a mixture of false-positive and subclonal genetic changes. Thus, the proportion of reproducible variants could indicate the level of background noise and a putative threshold level of AAF to be used for detection of somatic mutation. The percentage of reproducible variants critically depends on AAF and DNA quality, being minimal at lower AAF, but increasing steadily at >10% AAF in high molecular weight (HMW) and FFPE tissue DNA samples amenable for PCR of ≥300-bp genomic fragments but not in those only supporting PCR of up to 200 bp. C: Quantification of functional copy number of DNA by TaqMan real-time PCR. The level of functional copies amenable to PCR critically depends on DNA quality, being 80% in HMW DNA but only approximately 1% in DNA samples amplifiable for PCR of up to 200 bp.
After elimination of nonreproducible changes and SNPs, the remaining variants represented those seen in both replicates and were designated as reproducible variants. The absolute number of reproducible changes was also much higher at a lower AAF (Figure 3A). However, the percentage of these reproducible variants was minimal at a lower AAF but increased steadily at >10% AAF, particularly for HMW DNA, then followed by FFPE tissue DNA samples amenable to PCR of 400 or 300 bp. In contrast, the percentage of reproducible variants was consistently low in FFPE tissue DNA samples amenable to PCR of up to 200 bp (Figure 3B). To quantify the number of functional copies adequate for PCR, we performed TaqMan qPCR in a series of representative DNA samples (Figure 3C, Supplemental Figure S1). This was successful in all HMW and FFPE tissue DNA samples amenable to PCR of ≥300 bp, but only in four of the seven DNA samples amenable to PCR of up to 200 bp. Of the four samples amenable to PCR of up to 200 bp, which were successfully assayed by TaqMan PCR, the mean percentage of functional copies was only 1.1% (Figure 3C). For these reasons and with further evidence of high baseline sequence errors from later analysis, we excluded the DNA samples amenable for PCR of only up to 200 bp from subsequent mutation analysis.
The reproducible variants at a high AAF were likely true genetic changes, whereas those at a lower AAF were probably a mixture of false-positive variants and subclonal genetic changes. To permit comparison of mutations detected between Fluidigm PCR/MiSeq sequencing and conventional PCR/Sanger sequencing, we thus initially chose an AAF of 10% as a cutoff value because the variants above this value can be validated by conventional PCR and Sanger sequencing or by cloning and sequencing where necessary.
The reproducible variants with an AAF >10% in both replicates were then cross-examined with known somatic mutations detected by Sanger sequencing of the seven genes in a total of 163 DNA samples from FFPE lymphoma tissues (Figure 2, Supplemental Figure S5). At first, the performance of an in-house variant calling algorithm was assessed and tuned against 114 known mutations, including 106 substitutions and eight indels (range, 1 to 33 bp). While the variant calling algorithm was assessed, additional novel variants were identified by Fluidigm PCR/MiSeq sequencing, and these novel variants were further validated by PCR and Sanger sequencing or where indicated by cloning and sequencing of the PCR products (n = 15). The resulting sequence data were used to further fine-tune the algorithm, until the algorithm was able to detect all mutations detected or confirmed by Sanger sequencing (Figure 2), without both false-negative and false-positive variants. Taken together, a total of 159 Sanger sequencing–confirmed somatic mutations, including 147 substitutions and 12 indels (range, 1 to 33 bp), were used to optimize the algorithm.
Testing the Optimized Experimental Protocols and Variant Calling Algorithm and Determining the Cutoff Value of AAF for Somatic Mutation Detection
To further ascertain the performance of the above-optimized experimental protocol and variant calling algorithm, we performed the following two sets of independent experiments (Figure 4A). In one set of experiments, the above 111 PCR primer pairs for the seven genes were further investigated as a part of a total of 343 PCR primer pairs for 22 genes covering 65-kb sequence using the same cohort of FFPE lymphoma DNA samples as above. These independent experiments confirmed the characteristics of nonreproducible and reproducible changes for the seven genes as presented above and also found little difference in these profiles between the seven genes and 15 additional genes. Cross examination of novel reproducible changes in the seven genes between the two sets of independent experiments revealed that the concordance in mutation detection critically depended on the cutoff value of AAF (Figure 4B). With an AAF of 10% as a cutoff value, a 98.8% concordance was observed, whereas with a cutoff AAF value <10%, concordances progressively deteriorated, and therefore this value was unreliable for mutation detection.
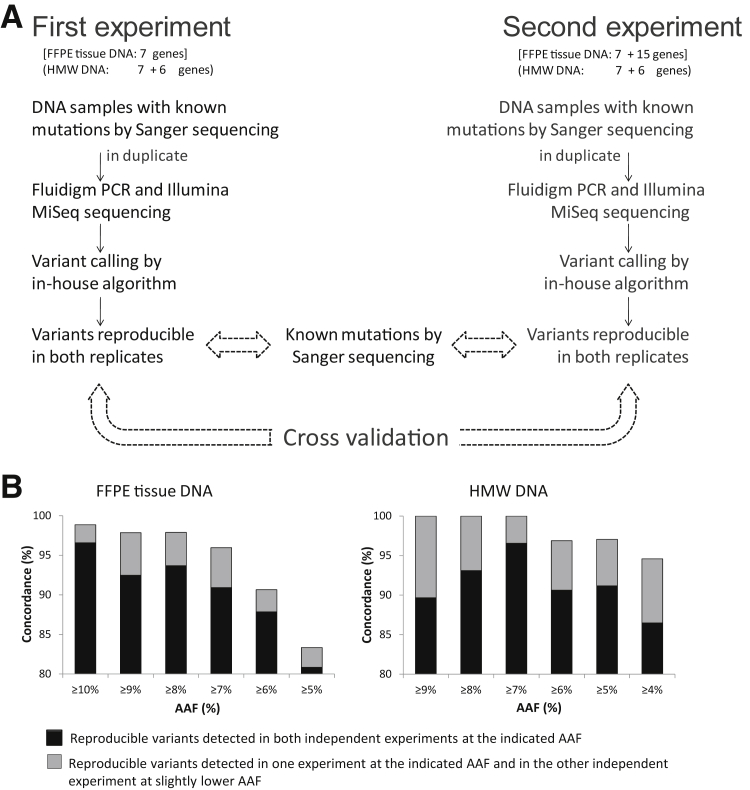
Figure 4.
Determining the cutoff value of alternate allele frequency (AAF) for somatic mutation detection. A: Experimental strategy: two sets of independent experiments were performed, and the reproducible variants detected in the seven genes were cross validated, together with the known mutations by Sanger sequencing. B: Comparison of the reproducible variants from the seven genes between the two independent experiments reveals that the concordances critically depend on the level of AAF. For DNA samples from formalin-fixed, paraffin-embedded (FFPE) tissues, a cutoff value of ≥10% AAF yields 98.8% concordance, whereas for high-molecular-weight (HMW) DNA, the cutoff value can be as low as ≥7% AAF, generating 100% concordance.
In the other set of experiments, the same 111 PCR primer pairs for the seven genes were analyzed as a part of 157 PCR primer pairs for 13 genes in an additional cohort of 38 HMW DNA samples from splenic marginal zone lymphoma,15 and this experiment was performed twice independently. Cross examination of novel reproducible changes from the seven genes between the two sets of independent experiments revealed a 100% concordance at an AAF of ≥7% (Figure 4B). Thus, the best cutoff value for HMW DNA was defined as 7%.
Distinct Difference in the Nature of False-Positive Variants between FFPE Tissue and HMW DNA
To understand the potential factors underpinning false-positive variants, we examined the nature of nonreproducible and reproducible variants in both FFPE tissue and HMW DNA. Separate analyses of data from the seven genes and others revealed no apparent difference, and the data were thus combined and presented together.
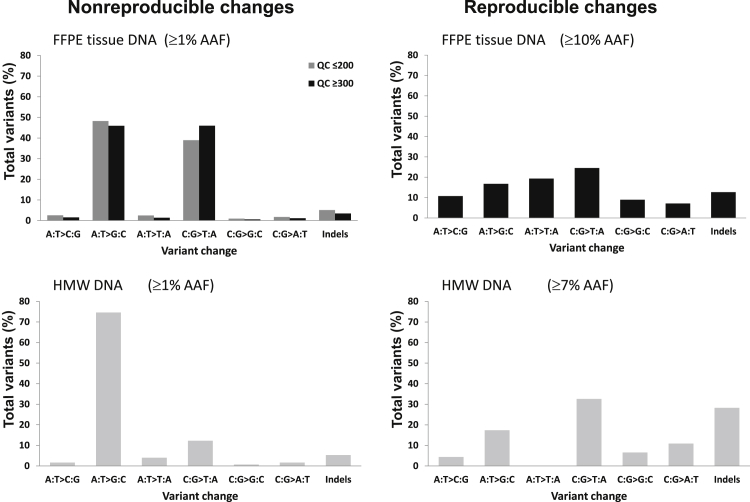
For nonreproducible changes, there was a broad similarity in the pattern of nucleotide changes between FFPE tissue and HMW DNA samples, and both revealed frequent C:G>T:A and A:T>G:C alterations, with other base changes being at relatively low frequencies (Figure 5). However, there were marked differences in the frequencies of these changes between FFPE tissue and HMW DNA samples, with the frequencies of C:G>T:A change being remarkably higher in the FFPE tissue (Figure 5). There was neither an apparent difference in the frequency of indels between FFPE tissue and HMW DNA nor any association between the nature of nonreproducible changes and their AAFs (Supplemental Figure S6).
Figure 5.
The nature of nonreproducible and novel reproducible changes in formalin-fixed, paraffin-embedded (FFPE) tissue and high-molecular-weight (HMW) DNA samples. For nonreproducible changes, there are marked differences in the frequencies of base changes between FFPE tissue and HMW DNA samples, with the frequencies of C:G>T:A change being remarkably higher in the FFPE tissue DNA. For novel reproducible changes, the spectrum of base changes between FFPE tissue and HMW DNA is similar, being broad without major bias toward any particular changes.
For reproducible changes, we further subdivided them according to the cutoff AAF. Those above this value were true genetic changes, whereas those below this value were a mixture of subclonal changes and false-positive variants. In contrast to nonreproducible changes, the spectrum of the reproducible changes above the cutoff value in both FFPE tissue and HMW DNA was broad, without apparent bias toward any particular nucleotide changes (Figure 5). The slightly more variations of the spectrum of the reproducible changes in the HMW group are most likely due to a small number (n = 46) of mutations in this group.
Finally, we also searched for sequence features that might be potentially associated with nonreproducible or reproducible variants in both FFPE tissue and HMW DNA using the MEME suite,21 but the analyses did not identify any sequence features associated with false-positive or true mutations.
Discussion
In this study, we developed and validated a robust high-throughput mutation screen using DNA samples from archival FFPE tissues. Experimentally, we established a practical protocol with various quality control steps for multiplex PCR with the Fluidigm Access Array, providing a uniform amplification of target genes for Illumina MiSeq sequencing. Bioinformatically, we generated an in-house variant calling algorithm and fine-tuned its performance against somatic mutations detected by Sanger sequencing. In addition, we established a strategy to maximally eliminate false-positive variants, enabling detection of known and novel mutations. Our study also highlights several critical issues for application of PCR-based target enrichment and NGS to DNA samples from FFPE tissues.
Potential Sources of False-Positive Sequence Changes
There are many potential causes leading to a false-positive sequence change. Apart from those associated with Illumina sequencing, the major causes for false positivity in the context of the current study are poor quality of DNA and errors of Taq polymerase.
It is known that poor quality of DNA is prone to PCR and sequencing errors. We found in the present study that the extent of false-positive changes as measured by nonreproducible changes between the two replicates of the same DNA sample depended on DNA quality, with the poorer-quality DNA samples having higher rates of false-positive changes. The propensity of the DNA samples from FFPE tissues to generate false-positive changes is most likely due to DNA damage and few copies of intact templates for PCR. In comparison with HMW DNA, those from FFPE tissues had a remarkably high incidence of C:G>T:A, accounting for approximately 40% of nonreproducible changes. This extraordinarily high false-positive rate is most likely due to deamination of cytosine during tissue formalin fixation and storage.13,22,23
Because of degradation, only a small fraction of a DNA sample from FFPE tissue is adequate to serve as templates for PCR despite the fact that primers were designed to amplify short fragments (200 bp) of genomic sequences. By TaqMan qPCR, we found that only 1.1% of genomic DNA from FFPE tissues was adequate for PCR of 200 bp of genomic sequences. For a DNA sample amenable to conventional PCR of up to 200 bp, 50 ng of DNA contains only approximately 170 functional copies adequate for PCR of 200-bp genomic sequences. Because few functional templates are available for PCR, any errors introduced at the early steps of the amplification process would appear in a substantial proportion of the amplified products. Consistently, the other major nonreproducible changes are A:T>G:C alterations, which are likely the result of Taq polymerase errors.24
Despite the fact that HMW DNA samples are far better in quality than those from FFPE tissues, these samples also gave rise to considerable false-positive results at low AAFs. In contrast to FFPE tissue DNA, most false-positive results in the HMW DNA samples as measured by nonreproducible changes were A:T>G:C changes, being far more frequent than C:G>T:A alterations.
Strategies to Eliminate False-Positive Results
We established several practical means to eliminate false-positive results, allowing highly efficient and specific detection of somatic mutations.
Assessing DNA Quality to Select Those with Adequate Quality and Quantity
By quality control PCR and further supported by TaqMan qPCR, we observed that DNA samples amenable to PCR of ≥300 bp were adequate for mutation screening with Fluidigm PCR and Illumina MiSeq sequencing. Under the protocols described in this study, 50 ng of FFPE tissue DNA or 20 ng of HMW DNA yielded excellent results for sequencing of 343 amplicons covering 65 kb. However, the amount of template DNA may be subjected to change, depending on the number of primer pairs used and the size of the amplicons.
Investigating Each DNA Sample in Duplicate
Most false-positive results are not reproducible and thus can be efficiently eliminated by analysis of each DNA sample in duplicate. Under the experimental conditions described, duplicate analyses are sufficient, and there is no need to further increase the number of replicates. Theoretically, this approach is potentially capable of eliminating all types of random false-positive changes resulting from poor quality DNA or Taq polymerase errors. An alternative approach to reduce false-positive results is treatment of FFPE tissue DNA with uracil glycosylase.13,25 This can significantly reduce false-positive changes resulting from deamination of cytosine; however, uracil glycosylase is active only at uracil lesions but not thymine lesions resulting from deamination of 5-methyl cytosine. In addition, the C:G>T:A artifact at CpG dinucleotides is resistant to uracil glycosylase treatment.25 Thus, duplicate experiments offer broader efficacy in elimination of false-positive results, which is very much highlighted by a recent review.26
Choosing Appropriate Cutoff Value of AAF for Reliable Detection of Somatic Mutations
On the basis of the concordance of reproducible variants between two sets of independent experiments, together with known somatic mutations by Sanger sequencing, we suggest using 10% and 7% as the optimal cutoff value of AAF for mutation detection in FFPE tissue and HMW DNA, respectively. For detection of well-characterized hotspot mutations, it is possible to go below these cutoff values with caution. However, for detection of unknown mutations, it is impossible to distinguish somatic mutations from baseline sequence errors when AAF is below the cut-off value. These findings further emphasize the importance of DNA preparation from specimens with high tumor cell content or microdissected tumor cells.
A Fully Validated In-House Variant Calling Algorithm
A fully validated in-house variant calling algorithm was developed and fine-tuned by assessing its performance on detection of a large number of known somatic mutations, including a variety of indels. We also tested this in-house variant calling algorithm in two independent ongoing studies, including one on solid tumors with different gene panels, and confirmed its excellent performance as judged by correlation with known mutations by Sanger sequencing. In comparison with commercial software, the validated in-house variant calling algorithm gave much better performance particularly in indel calling.
Detection of Subclonal Mutations
On the basis of the above established protocols, Fluidigm PCR and Illumina MiSeq sequencing are much more sensitive in somatic mutation screening than conventional Sanger sequencing. Nearly one-third of the mutations (45/159 = 28%) detected by Fluidigm PCR and Illumina MiSeq sequencing were missed by original PCR and Sanger sequencing, albeit confirmed by further Sanger sequencing or cloning and sequencing of the PCR products. Consistent with our findings, Bodor et al27 also found an improvement of 39% in mutation detection by amplicon-based NGS in comparison with conventional PCR and Sanger sequencing. A proportion of the somatic mutations additionally detected by Fluidigm/MiSeq sequencing may represent subclonal genetic changes. Nonetheless, subclonal somatic mutations, particularly uncharacterized changes at a frequency below the cutoff value, cannot be reliably identified because these changes are not distinguishable from baseline sequence errors despite being technically detectable by the method. Importantly, it is the cutoff value of AAF, rather than the technical sensitivity of the NGS, which determines how low a subclonal mutation can be reliably detected.
In conclusion, we established a practical protocol for high-throughput mutation analysis using DNA samples from archival FFPE tissues by Fluidigm multiplex PCR/Illumina MiSeq sequencing and an in-house variant calling algorithm. The strategies used to eliminate false-positive results and identify somatic mutations provide a practical solution for high-throughput mutation screening using routine FFPE tissue biopsies.
Acknowledgments
We thank David Withers for his help in DNA sequencing; Ruth Littleboy, Fay Rodger, and Antje Schulze Selting for technical assistance; Mark Ross, Stefano Berri, and Anthony Rogers for helpful discussion on data analysis; and Shubha Anand for critical reading of the manuscript.
M.W., L.E.-I., S.M., N.Z., A.C., Y.H., N.F.G., T.F., J.S., and H.M. designed experiments and collected and analyzed data. S.B., L.W., and A.J. contributed case reports. K.B., X.X., and A.F. analyzed Illumina sequencing and variant calling. M.-Q.D., M.W., L.E.-I., S.M., and A.C. wrote the manuscript. M.-Q.D. designed and coordinated the study. All authors commented on the manuscript and approved its submission for publication.
Footnotes
Supported by Leukaemia & Lymphoma Research grants LLR10006 and LLR13006 (M.-Q.D.) and the Kay Kendal Leukaemia Fund grants KKL649 and KKL582 (M.-Q.D.). S.M. is a Ph.D. student supported by Medical Research Council, Department of Pathology, University of Cambridge, and Addenbrooke's Charitable Trust. L.E.-I. is a Ph.D. student supported by the Pathological Society of UK & Ireland. X.X. was supported by a visiting fellowship from the China Scholarship Council, Ministry of Education, China. N.F.G. was supported by grant KKL649 from the Kay Kendal Leukemia Fund and an Addenbrooke's Charitable Trust fellowship.
M.W. and L.E.-I. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2015.04.008.
Supplemental Data
Quantification of functional copies of DNA by TaqMan real-time quantitative PCR (qPCR). A: Standard curve. High-quality human genomic DNA (Promega) was serially diluted, and input DNA ranging from 10 to 0.020 ng was quantified in triplicate by TaqMan qPCR of a 195-bp fragment of the PPIA gene to generate a standard curve. B: Examples of TaqMan qPCR. For each sample, input DNA of 10, 5, and 2.5 was quantified in triplicate as above. The mean estimated percentage of functional copy number relative to the standard curve was then calculated.
Stratification of PCR primer pairs for different preamplification reactions and multiplex PCR by In-Silico PCR and AutoDimer analysis. A: In-Silico PCR analysis allows identification of primer pairs that give rise to undesired and nonspecific amplification and their stratification in separate preamplification and multiplex PCR reactions. For example, analysis of the 111 primer pairs for the seven-gene panel identifies a number of putative undesired amplifications largely due to overlapping primers. The large putative undesired amplifications (>300 bp) are normally not amplified under the experimental conditions used and thus do not impose any problem. However, the small undesired amplifications are seen experimentally, and these undesired amplifications cause failure of amplification of a proportion of primer pairs in Fluidigm Access Array PCR. These undesired amplifications can be efficiently prevented by placing the relevant primer pairs in different preamplification reactions as guided by In-Silico PCR analysis. After purification, the two separate preamplified products can then be pooled and used as template for multiplex PCR with Fluidigm Access Array. B: AutoDimer analysis identifies primers potentially interacting with each other. Any primers with an AutoDimer score >12 should be placed in separate reactions. As shown here, PRDM1_ex6r_3 and PRDM1_ex6r_1 have a high degree of complementary sequences and prevent amplification of the targeted sequences when present together. By separating them into two separate reactions, both targeted genomic fragments can be efficiently amplified. PCR products were run on 10% PAGE gel.
Quality control (QC) at various steps of Fluidigm Access Array PCR and Illumina MiSeq sequencing. A: QC step 1: Assessment of preamplified products by conventional multiplex PCR and 10% polyacrylamide gel electrophoresis. One microliter of fivefold diluted preamplified product was amplified with three to four primer pairs grouped based on In-Silico PCR and AutoDimer analyses. PCR was performed in a 5-μL reaction mixture containing 4.5 mmol/L MgCl2, 5% dimethyl sulfoxide, 200 μmol/L of each dNTP, 1.5 μmol/L of each primer, and 0.25 U FastStart High Fidelity Enzyme Blend. The PCR conditions are identical to those used for Fluidigm Access Array. Lanes 1 through 7: representative samples; lane 8: negative control from preamplification reaction without template DNA. B: QC step 2: Assessment of Fluidigm Access Array PCR products by 2% agarose gel electrophoresis. Lane 1: a negative control gone through preamplification and Fluidigm PCR; lanes 2 through 8: individual samples. C: QC step 3: Assessment of barcoded PCR products by 2% agarose gel electrophoresis. Lanes 1 through 7: individual samples; lane 8: a negative control gone through preamplification, Fluidigm PCR, and barcoding. D: QC step 4: Assessment of library purification. Left panel: optimization of the ratio of AMPure XP beads to sample volume using a DNA ladder; the ratio at 0.8 gives the best purification, effectively removing DNA fragments <200 bp. Right panel: The barcoded PCR products from various samples were pooled and purified as above. E: QC step 5: Assessment of library quality by Bioanalyser. The above purified library was routinely assessed using Agilent DNA 1000 Bioanalyser. The 300-bp peak represents the purified library products, whereas the 15-bp and 1500-bp peaks are ladder marker. FU, fluorescence units; P, purified product; U, unpurified product.
Examples of depth of coverage by Fluidigm Access Array PCR and Illumina MiSeq sequencing. Adequate depth of coverage can be achieved for each of the amplicons by multiplex Fluidigm Access Array PCR with up to four primer pairs. AVG, average.
Examples of somatic mutations detected by Fluidigm Access Array PCR and Illumina MiSeq sequencing. The aligned sequence reads are visualized using the Integrative Genomics Viewer (IGV). The reads sequenced from the same direction are shown in the same color (light blue or light red). The green box shows representative reads, with annotations on the right detailing the total number of reads and the counts for each of the four nucleotides. The details of the mutation are indicated at the left of each IGV image.
The nature of nonreproducible variants from formalin-fixed, paraffin-embedded tissue DNA is independent ofthe alternate allele frequency (AAF). The nonreproducible variants C:G>T:A and A:T>G:C changes account for most false-positive results, and in general, the extent of these false-positive results is independent of their AAF.
References
- 1.Morin R.D., Mendez-Lago M., Mungall A.J., Goya R., Mungall K.L., Corbett R.D. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pasqualucci L., Trifonov V., Fabbri G., Ma J., Rossi D., Chiarenza A., Wells V.A., Grunn A., Messina M., Elliot O., Chan J., Bhagat G., Chadburn A., Gaidano G., Mullighan C.G., Rabadan R., Dalla-Favera R. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohr J.G., Stojanov P., Lawrence M.S., Auclair D., Chapuy B., Sougnez C., Cruz-Gordillo P., Knoechel B., Asmann Y.W., Slager S.L., Novak A.J., Dogan A., Ansell S.M., Link B.K., Zou L., Gould J., Saksena G., Stransky N., Rangel-Escareno C., Fernandez-Lopez J.C., Hidalgo-Miranda A., Melendez-Zajgla J., Hernandez-Lemus E., Schwarz C., Imaz-Rosshandler I., Ojesina A.I., Jung J., Pedamallu C.S., Lander E.S., Habermann T.M., Cerhan J.R., Shipp M.A., Getz G., Golub T.R. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Grubor V., Love C.L., Banerjee A., Richards K.L., Mieczkowski P.A. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teer J.K., Bonnycastle L.L., Chines P.S., Hansen N.F., Aoyama N., Swift A.J., Abaan H.O., Albert T.J., Margulies E.H., Green E.D., Collins F.S., Mullikin J.C., Biesecker L.G. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 7.Murtaza M., Dawson S.J., Tsui D.W., Gale D., Forshew T., Piskorz A.M., Parkinson C., Chin S.F., Kingsbury Z., Wong A.S., Marass F., Humphray S., Hadfield J., Bentley D., Chin T.M., Brenton J.D., Caldas C., Rosenfeld N. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 8.Kanagal-Shamanna R., Portier B.P., Singh R.R., Routbort M.J., Aldape K.D., Handal B.A., Rahimi H., Reddy N.G., Barkoh B.A., Mishra B.M., Paladugu A.V., Manekia J.H., Kalhor N., Chowdhuri S.R., Staerkel G.A., Medeiros L.J., Luthra R., Patel K.P. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol. 2014;27:314–327. doi: 10.1038/modpathol.2013.122. [DOI] [PubMed] [Google Scholar]
- 9.Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B., Kanagal-Shamanna R., Greaves W.O., Medeiros L.J., Aldape K.D., Luthra R. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Tsongalis G.J., Peterson J.D., de Abreu F.B., Tunkey C.D., Gallagher T.L., Strausbaugh L.D., Wells W.A., Amos C.I. Routine use of the Ion Torrent AmpliSeq Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2014;52:707–714. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 11.Forshew T., Murtaza M., Parkinson C., Gale D., Tsui D.W., Kaper F., Dawson S.J., Piskorz A.M., Jimenez-Linan M., Bentley D., Hadfield J., May A.P., Caldas C., Brenton J.D., Rosenfeld N. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 12.Halbritter J., Diaz K., Chaki M., Porath J.D., Tarrier B., Fu C., Innis J.L., Allen S.J., Lyons R.H., Stefanidis C.J., Omran H., Soliman N.A., Otto E.A. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet. 2012;49:756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 13.Bourgon R., Lu S., Yan Y., Lackner M.R., Wang W., Weigman V., Wang D., Guan Y., Ryner L., Koeppen H., Patel R., Hampton G.M., Amler L.C., Wang Y. High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed PCR and next-generation sequencing. Clin Cancer Res. 2014;20:2080–2091. doi: 10.1158/1078-0432.CCR-13-3114. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Bench A.J., Bacon C.M., Payne K., Huang Y., Scott M.A., Erber W.N., Grant J.W., Du M.Q. A practical strategy for the routine use of BIOMED-2 PCR assays for detection of B- and T-cell clonality in diagnostic haematopathology. Br J Haematol. 2007;138:31–43. doi: 10.1111/j.1365-2141.2007.06618.x. [DOI] [PubMed] [Google Scholar]
- 15.Clipson A., Wang M., de Leval L., Ashton-Key M., Wotherspoon A., Vassiliou G., Bolli N., Grove C., Moody S., Escudero-Ibarz L., Gundem G., Brugger K., Xue X., Mi E., Bench A., Scott M., Liu H., Follows G., Robles E.F., Martinez-Climent J.A., Oscier D., Watkins A.J., Du M. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29:1177–1185. doi: 10.1038/leu.2014.330. [DOI] [PubMed] [Google Scholar]
- 16.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do H., Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase. Oncotarget. 2012;3:546–558. doi: 10.18632/oncotarget.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofreiter M., Jaenicke V., Serre D., von Haeseler A., Paabo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bracho M.A., Moya A., Barrio E. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J Gen Virol. 1998;79(Pt 12):2921–2928. doi: 10.1099/0022-1317-79-12-2921. [DOI] [PubMed] [Google Scholar]
- 25.Do H., Wong S.Q., Li J., Dobrovic A. Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin Chem. 2013;59:1376–1383. doi: 10.1373/clinchem.2012.202390. [DOI] [PubMed] [Google Scholar]
- 26.Robasky K., Lewis N.E., Church G.M. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet. 2014;15:56–62. doi: 10.1038/nrg3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodor C., Grossmann V., Popov N., Okosun J., O'Riain C., Tan K., Marzec J., Araf S., Wang J., Lee A.M., Clear A., Montoto S., Matthews J., Iqbal S., Rajnai H., Rosenwald A., Ott G., Campo E., Rimsza L.M., Smeland E.B., Chan W.C., Braziel R.M., Staudt L.M., Wright G., Lister T.A., Elemento O., Hills R., Gribben J.G., Chelala C., Matolcsy A., Kohlmann A., Haferlach T., Gascoyne R.D., Fitzgibbon J. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013;122:3165–3168. doi: 10.1182/blood-2013-04-496893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of functional copies of DNA by TaqMan real-time quantitative PCR (qPCR). A: Standard curve. High-quality human genomic DNA (Promega) was serially diluted, and input DNA ranging from 10 to 0.020 ng was quantified in triplicate by TaqMan qPCR of a 195-bp fragment of the PPIA gene to generate a standard curve. B: Examples of TaqMan qPCR. For each sample, input DNA of 10, 5, and 2.5 was quantified in triplicate as above. The mean estimated percentage of functional copy number relative to the standard curve was then calculated.
Stratification of PCR primer pairs for different preamplification reactions and multiplex PCR by In-Silico PCR and AutoDimer analysis. A: In-Silico PCR analysis allows identification of primer pairs that give rise to undesired and nonspecific amplification and their stratification in separate preamplification and multiplex PCR reactions. For example, analysis of the 111 primer pairs for the seven-gene panel identifies a number of putative undesired amplifications largely due to overlapping primers. The large putative undesired amplifications (>300 bp) are normally not amplified under the experimental conditions used and thus do not impose any problem. However, the small undesired amplifications are seen experimentally, and these undesired amplifications cause failure of amplification of a proportion of primer pairs in Fluidigm Access Array PCR. These undesired amplifications can be efficiently prevented by placing the relevant primer pairs in different preamplification reactions as guided by In-Silico PCR analysis. After purification, the two separate preamplified products can then be pooled and used as template for multiplex PCR with Fluidigm Access Array. B: AutoDimer analysis identifies primers potentially interacting with each other. Any primers with an AutoDimer score >12 should be placed in separate reactions. As shown here, PRDM1_ex6r_3 and PRDM1_ex6r_1 have a high degree of complementary sequences and prevent amplification of the targeted sequences when present together. By separating them into two separate reactions, both targeted genomic fragments can be efficiently amplified. PCR products were run on 10% PAGE gel.
Quality control (QC) at various steps of Fluidigm Access Array PCR and Illumina MiSeq sequencing. A: QC step 1: Assessment of preamplified products by conventional multiplex PCR and 10% polyacrylamide gel electrophoresis. One microliter of fivefold diluted preamplified product was amplified with three to four primer pairs grouped based on In-Silico PCR and AutoDimer analyses. PCR was performed in a 5-μL reaction mixture containing 4.5 mmol/L MgCl2, 5% dimethyl sulfoxide, 200 μmol/L of each dNTP, 1.5 μmol/L of each primer, and 0.25 U FastStart High Fidelity Enzyme Blend. The PCR conditions are identical to those used for Fluidigm Access Array. Lanes 1 through 7: representative samples; lane 8: negative control from preamplification reaction without template DNA. B: QC step 2: Assessment of Fluidigm Access Array PCR products by 2% agarose gel electrophoresis. Lane 1: a negative control gone through preamplification and Fluidigm PCR; lanes 2 through 8: individual samples. C: QC step 3: Assessment of barcoded PCR products by 2% agarose gel electrophoresis. Lanes 1 through 7: individual samples; lane 8: a negative control gone through preamplification, Fluidigm PCR, and barcoding. D: QC step 4: Assessment of library purification. Left panel: optimization of the ratio of AMPure XP beads to sample volume using a DNA ladder; the ratio at 0.8 gives the best purification, effectively removing DNA fragments <200 bp. Right panel: The barcoded PCR products from various samples were pooled and purified as above. E: QC step 5: Assessment of library quality by Bioanalyser. The above purified library was routinely assessed using Agilent DNA 1000 Bioanalyser. The 300-bp peak represents the purified library products, whereas the 15-bp and 1500-bp peaks are ladder marker. FU, fluorescence units; P, purified product; U, unpurified product.
Examples of depth of coverage by Fluidigm Access Array PCR and Illumina MiSeq sequencing. Adequate depth of coverage can be achieved for each of the amplicons by multiplex Fluidigm Access Array PCR with up to four primer pairs. AVG, average.
Examples of somatic mutations detected by Fluidigm Access Array PCR and Illumina MiSeq sequencing. The aligned sequence reads are visualized using the Integrative Genomics Viewer (IGV). The reads sequenced from the same direction are shown in the same color (light blue or light red). The green box shows representative reads, with annotations on the right detailing the total number of reads and the counts for each of the four nucleotides. The details of the mutation are indicated at the left of each IGV image.
The nature of nonreproducible variants from formalin-fixed, paraffin-embedded tissue DNA is independent ofthe alternate allele frequency (AAF). The nonreproducible variants C:G>T:A and A:T>G:C changes account for most false-positive results, and in general, the extent of these false-positive results is independent of their AAF.