Abstract
The aim of this study was to identify the synergistic effect of microRNA expression with classical risk factors of coronary heart disease (CHD) and to explore their diagnostic value for coronary stenotic lesions in subjects with CHD. Plasma samples were obtained from 66 subjects with CHD and from 58 control individuals. A quantitative reverse-transcription PCR (RT-qPCR) assay was conducted to confirm the relative expressions of the known CHD-related miRNAs. The severity of coronary atherosclerosis was based on the Gensini scoring system. The expression of miR-125b in plasma of the CHD group was lower than that of the non-CHD group (0.14 ± 0.09 vs. 0.18 ± 0.10, p = 0.055), and the miR-125b levels significantly decreased following an increasing Gensini score (P = 0.037). Spearman correlation analyses indicated the Gensini score was negatively associated with miR-125b (r = −0.215, p = 0.017). Of all the miRNAs, miR-125b showed the lowest AUC (0.405; 95% CI: 0.305 ~ 0.506, p = 0.070). We found several synergistic effects between miR-125b and classical risk factors, such as age, sex, CR, FBG and HDL-C; the proportion of CHD attributable to the interaction of miR-125b and age was as high as 80%. Therefore, miR-125b was shown to play an important role in individual’s susceptibility to developing CHD.
Cardiovascular diseases (CVDs) are the leading causes of morbidity and mortality worldwide. Over the past few decades, the diagnosis, treatment, and prognosis of coronary heart disease (CHD) has dramatically changed and has led to a decrease in the mortality risk. However, there is still a clinical need for new diagnostic and prognostic biomarkers and novel modern interventions to reduce the incidence of CHD1. Recently, microRNAs (miRNAs) have been shown to be associated with CHD2,3,4. MiRNAs are small noncoding RNA molecules that negatively regulate gene expression by triggering either translation repression or RNA degradation on the post-transcriptional level5,6. Approximately 1000 miRNAs have been found in humans to date. Bioinformatics and cloning studies have estimated that miRNAs regulate 30% of all human genes7,8. Among them, there are approximately 50 circulating miRNAs believed to be associated with cardiovascular diseases1. By analysing the profiles of miRNAs released into the blood plasma or serum from cells and tissues of the cardiovascular system in CHD patients, it has been shown that some miRNAs are down-regulated while others are up-regulated9. Many studies have demonstrated the role of cardiomyocyte-enriched miRNAs (miR-1, miR133a, miR-133b, miR-208a, miR-208b, and miR-499) in cardiac damage and myocardial infarction; moreover, some studies have highlighted the underlying effect of circulating miRNAs as diagnostic and prognostic biomarkers in AMI10.
Although coronary angiography has significantly improved the early diagnosis of CHD, it remains a major clinical challenge. Genetic epidemiology is increasingly focused on the study of common diseases with both genetic and environmental determinants. The concept of a gene-environment interaction is becoming a central theme in epidemiologic studies that assess the causes of human disease in populations11. However, the microRNA-environment interaction on CHD has not yet been reported. Therefore, we identified the synergistic effect of microRNA expression with classical risk factors on CHD and explored their respective diagnostic value for coronary stenotic lesions in subjects with CHD.
Subject and Methods
Study subjects
From 2012 to 2013, 124 consecutive subjects (91 males and 33 females), aged 37–77 years, who underwent coronary angiography for suspected or known coronary atherosclerosis at the Friendship Hospital of Ili Kazakh Autonomous Prefecture in China were enrolled in this study. Among them, 66 subjects with CHD (54 males and 12 females, age 39–77 years) and 58 subjects (37 males and 21 females, age 37–73 years) with angiographic exclusion of CHD served as the control group. General exclusion criteria included the following: subjects with spastic angina pectoris, infectious processes within 2 weeks, heart failure, adrenal dysfunction, and thyroid dysfunction. The methods were performed in accordance with the approved guidelines, and all experimental protocols were approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University and the Friendship Hospital of Ili Kazakh Autonomous Prefecture in China. All subjects provided written informed consent.
Coronary Angiography
Coronary arteries were cannulated using either the Judkins technique12 or through a radial artery approach with 6F catheters and recorded at a rate of 30 frames/s. The presence of coronary artery stenosis was evaluated after the direct intracoronary injection of isosorbide dinitrate (ISDN; 2.5 mg/5 ml solution over 20 s). One minute after the injection of ISDN through the Judkins catheter, coronary angiography was performed from several projections. Coronary angiograms were evaluated independently by operators who made visual estimations of the luminal narrowing in multiple segments, based on the AHA/ACC classification, of the coronary tree. Significant CHD was defined as at least one major epicardial vessel with >50% stenosis; the control was defined as all of the major epicardial vessels with <50% stenosis13. The severity of the coronary atherosclerosis was based on the Gensini scoring system. The Gensini score was computed by assigning a severity score in each coronary stenosis according to the degree of luminal narrowing and its geographic importance. Reduction in the lumen diameter and the roentgenographic appearance of concentric lesions and eccentric plaques were evaluated (reductions of 25%, 50%, 75%, 90%, 99% and complete occlusion were given Gensini scores of 1, 2, 4, 8, 16 and 32, respectively). Each principal vascular segment was assigned a multiplier according to the functional significance of the myocardial area supplied by that segment: the left main coronary artery, ×5; the proximal segment of left anterior descending coronary artery (LAD), ×2.5; the proximal segment of the circumflex artery, ×2.5; the mid-segment of the LAD, ×1.5; the right coronary artery, the distal segment of the LAD, the posterolateral artery, and the obtuse marginal artery, ×1; and all others, ×0.514.
Laboratory measurements
Four millilitres of venous blood was drawn after 12 hours of fasting to perform biochemical assays in the routine laboratory on the second day of hospitalization; the total cholesterol (TCH, mmol/L), triglyceride (TG, mmol/L), fasting blood glucose (FBG, mmol/L), creatinine (CR, umol/L), fasting high-density lipoprotein cholesterol (HDL-C, mmol/L) and fasting low-density lipoprotein cholesterol (LDL-C, mmol/L) were determined by enzymatic procedures on an automated autoanalyzer (AU 2700 Olympus, 1st Chemical Ltd, Japan). Excellent intra-assay and inter-assay CVs of <5% were obtained with our assay method.
Selection of miRNAs
Based on previous studies, 20 CHD-related miRNAs were employed as candidates in the study: miR-433, miR-485-3p, miR-1, miR-122, miR-133a, miR-133b, miR-14515, miR-21416, miR-21, miR-25, miR-20a, miR-106a, miR-92a17, miR-130a, miR-155, miR-221, miR-208a, miR-208b, miR-49918, and miR-125b19,20,21,22.
Plasma preparation and RNA isolation
All samples (5 ml) were collected in EDTA plasma tubes on the morning following arrival. Samples were processed within 4 hours and stored at 4 °C. Plasma was collected after centrifugation (15 min at 1000 × g) and was stored at −80 °C until further analysis was performed.
For the RT-qPCR assay of plasma, total RNA was extracted using a one-step phenol/chloroform purification protocol, as Cheng Wang et al. reported23 in 2011. Briefly, 100 μl of plasma was mixed with 300 μl of RNase-free water, 200 μl of phenol and 200 μl of chloroform. After 10 min at room temperature, the mixture was centrifuged at 16,000 × g for 20 min. After phase separation, the upper aqueous layer was mixed with 2 volumes of isopropyl alcohol and 0.1 volumes of 3 M sodium acetate (pH = 5.3). The total RNA was precipitated at −20 °C for 1 hour. The RNA pellet was collected by centrifugation at 16,000 × g for 20 min. The pellet was subsequently washed with 75% ethyl alcohol and dried for 10 min at room temperature. Finally, the RNA was dissolved in 20 ml of RNase-free water and stored at −80 °C until further analysis was performed.
Quantification of miRNAs by RT-qPCR analysis
For miRNA profiling, the RT-qPCR assay was performed using a TaqMan PCR kit according to the manufacturer’s instructions (Applied Biosystems, Foster City, USA); a minor modification was made according to the State Key Laboratory of Pharmaceutical Biotechnology (School of Life Sciences, Nanjing University), reported in 201024. TaqMan miRNA assays are all available through Applied Biosystems. In brief, the reverse transcription reaction was performed with a final volume of 10 μl, which included 2 μl of plasma extract RNA, 0.5 μl of AMV reverse transcriptase (TaKaRa), 1 μl of stem-loop RT primer (Applied Biosystems), 1 μl of 10 mMdNTPs, 2 μl of 5× reverse transcription buffer and 3.5 μl of RNase-free water. For synthesis of cDNA, the reaction solutions were incubated at 16 °C for 30 min, at 42 °C for 30 min, at 85 °C for 5 min, and then held at 4 °C. One reverse transcriptase reaction with no-template control was included. The quantitative PCR reaction was then performed in 20 μl, containing 1 μl of cDNA, 0.3 μl of Taq polymerase, 0.33 μl of TaqMan probe, 0.4 μl of 10 mMdNTPs, 1.2 μl of 25 mM MgCl2, 2 μl of 10 × PCR buffer (MgCl2 free) and 14.77 μl of RNase-free water. Real-time PCR was performed with the Roche Molecular Systems Mastercycler® ep realplex with one cycle of 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 sec and 60 °C for 60 sec. All reactions were run in triplicate, containing three wells of product of reverse transcriptase reaction with no-template control and water blanks as negative controls. Due to the superior performance of a combination of let-7d, let-7g and let-7i, this combination was chosen as a reference for the normalization of plasma miRNAs rather than the commonly used reference genes U6, RNU44, RNU48 and miR-1625. The resulting threshold cycle (CT) values were determined according to the default threshold settings when the reactions were completed. The relative amount of each miRNA was calculated based on the internal control, i.e., the combination of let-7d, let-7g and let-7i, analysed using the 2−Δct method, which is a widely used method for presenting relative gene expression by comparative CT, and the calculation formula was as following: 2exp-(mean Ct target miRNA-mean Ct internal control)26,27. Present the data as 2−Δct and plot the heatmap as described by means of MultiExperiment Viewer v4.928.
Statistical analysis
Data were statistically analysed using Statistics Package for Social Sciences (ver. 16.0; SPSS Incorporated, Chicago, IL, USA). Subjects were classified into 2 groups according to CHD status and 4 groups according to the quartile of the Gensini score. Data for age, TCH, CR, HDL-C, LDL-C, miR-20a, and miR-125b were normally distributed parameters and presented as the mean ± SD; comparisons were performed using the independent-samples T test and one-way ANOVA. Skewed data, including the TG, FBG, Gensini scores, miR-485-3p, miR-133b, miR-221, miR-208a, miR-433, miR-1, miR-122, miR-133a, miR-145, miR-214, miR-21, miR-25, miR-106a, miR-92a, miR-130a, miR-155, miR-208b, and miR-499 were expressed as median and quartile ranges, and comparisons were performed using the Mann-Whitney U test and Kruskal-Wallis H test. Categorical variables of gender were compared between the groups of patients using a chi-squared analysis. The Spearman two-way test was used to assess the relationship between Gensini scores with miRNAs and classical risk factors. As a measure of effect strength and direction when using the Mann–Whitney test, the area under the receiver operating characteristic (ROC) curve was computed29.
To analyse possible positive or negative interactions between microRNA expression with classical risk factors, the 4 × 2 table approach was used to calculate odds ratio (OR), respective 95% confidence intervals (CI) and two-tailed p values; additionally, this approach was used in synergy measures in additive (SI) or multiplicative models (SIM). It was assumed that unexposed individuals without the susceptibility microRNA had a certain background risk for CHD (OR00 is assumed to be 1); OR10 refers to the relative risk for CHD among people without the susceptibility microRNA for CHD but exposed to the environmental risk factor relative to those with neither the susceptibility microRNA nor exposure; OR01 refers to the relative risk among people with the susceptibility microRNA who are not exposed to the risk factor relative to those with neither the susceptibility microRNA nor exposure; and OR11 is the ratio of CHD risk among exposed people with susceptibility microRNA to CHD risk among unexposed people without the susceptibility microRNA. These ORs were then used in the calculation of synergy indices: SI = (OR11 − 1)/(OR10 + OR01 − 2), SIM = OR11/(OR10 × OR01); the relative excess risk due to interaction, RERI = OR11 − OR10 − OR01 + 1; and the attributable proportion of the disease due to interaction, AP = RERI/OR1111,30. The susceptibility microRNAs, environment risk factors and their cut-offs were determined using the area under the receiver operating characteristic (ROC) curve.
Differences were considered to be significant if the null hypothesis could be rejected with >95% confidence. All p-values were two-tailed.
Results
Characteristics of the study population grouped by CHD and controls
Table 1 presents the characteristics of the study population. A total of 66 subjects with CHD and 58 controls were enrolled in the study. As expected, increasing age (p = 0.000) and male (p = 0.023) are risk factors for CHD in this population. Additionally, the level of HDL-C (p = 0.020), FBG (p = 0.006), CR (p = 0.033), and Gensini scores (p = 0.000) were significantly different between CHD and controls.
Table 1. Characteristics of the study population categorized by CHDs and controls.
| Characteristics | CHDs (N = 66) | Controls (N = 58) | Statistical parameter | Pvalue |
|---|---|---|---|---|
| Age (years) | 62.20 ± 8.95 | 55.93 ± 8.68 | −3.946 | 0.000 |
| Gender (F/M) | 12/54 | 21/37 | 5.136 | 0.023 |
| TCH | 4.75 ± 1.04 | 4.64 ± 0.99 | −0.606 | 0.546 |
| HDL-C | 1.35 ± 0.33 | 1.50 ± 0.34 | 2.354 | 0.020 |
| LDL-C | 2.93 ± 0.91 | 2.76 ± 0.85 | −1.045 | 0.298 |
| FBG | 5.44(4.88 ~ 6.34) | 5.04(4.71 ~ 5.50) | 1266.00 | 0.006 |
| CR | 74.10 ± 15.87 | 68.56 ± 11.62 | −2.161 | 0.033 |
| Gensini scores | 35.50(16.75 ~ 64.50) | 0.00(0.00 ~ 2.00) | 51.000 | 0.000 |
| miR-433 | 0.60(0.31 ~ 1.15) | 0.62(0.37 ~ 1.13) | 1868.500 | 0.820 |
| miR-1 | 1.56(0.49 ~ 3.18) | 0.87(0.44 ~ 2.18) | 1651.500 | 0.189 |
| miR-122 | 0.48(0.12 ~ 1.42) | 0.31(0.10 ~ 0.8) | 1733.000 | 0.366 |
| miR-133a | 0.43(0.19 ~ 0.9) | 0.35(0.23 ~ 0.79) | 1810.000 | 0.602 |
| miR-145 | 0.10(0.04 ~ 0.17) | 0.08(0.04 ~ 0.18) | 1829.000 | 0.670 |
| miR-214 | 0.18(0.04 ~ 1) | 0.13(0.03 ~ 0.49) | 1698.500 | 0.280 |
| miR-21 | 0.61(0.38 ~ 1.01) | 0.64(0.42 ~ 1.09) | 1763.500 | 0.451 |
| miR-25 | 0.02(0.01 ~ 0.08) | 0.02(0.01 ~ 0.08) | 1851.000 | 0.747 |
| miR-20a | 0.38 ± 0.26 | 0.39 ± 0.33 | 0.028 | 0.978 |
| miR-106a | 2.41(1.51 ~ 3.99) | 2.69(1.79 ~ 4.31) | 1757.000 | 0.432 |
| miR-92a | 0.19(0.04 ~ 0.78) | 0.20(0.08 ~ 1.16) | 1707.500 | 0.301 |
| miR-130a | 54.35(29.32 ~ 111.82) | 61.82(35.26 ~ 107.49) | 1880.500 | 0.867 |
| miR-155 | 197.19(102.36 ~ 371.59) | 201.56(118.86 ~ 322.18) | 1908.500 | 0.978 |
| miR-208b | 1.13(0.63 ~ 4.13) | 1.02(0.6 ~ 3.56) | 1791.500 | 0.540b |
| miR-499 | 3.5(2.19 ~ 8.62) | 3.09(2.03 ~ 6.66) | 1739.500 | 0.382 |
| miR-485-3p | 1.79(0.97 ~ 5.53) | 1.54(0.76 ~ 3.35) | 1708.000 | 0.302 |
| miR-133b | 0.14(0.06 ~ 0.23) | 0.17(0.11 ~ 0.22) | 1662.000 | 0.207 |
| miR-221 | 0.4(0.19 ~ 0.82) | 0.55(0.22 ~ 0.92) | 1740.000 | 0.384 |
| miR-208a | 1.13(0.63 ~ 4.13) | 1.87(1.19 ~ 3.78) | 1632.500 | 0.159 |
| miR-125b | 0.14 ± 0.09 | 0.18 ± 0.10 | 1.938 | 0.055 |
Data are summarized by either mean ± standard deviation or 50th (25th/75th) percentiles for continuous variables and N1/N2 for binary variables. CHD, coronary heart disease; TCH, total cholesterol; HDL-C, fasting high-density lipoprotein cholesterol; LDL-C, fasting low-density lipoprotein cholesterol; TG, triglyceride; FBG, fasting blood glucose; CR, creatinine. The value of each miRNAs means the relative mount calculated by 2−Δct method.
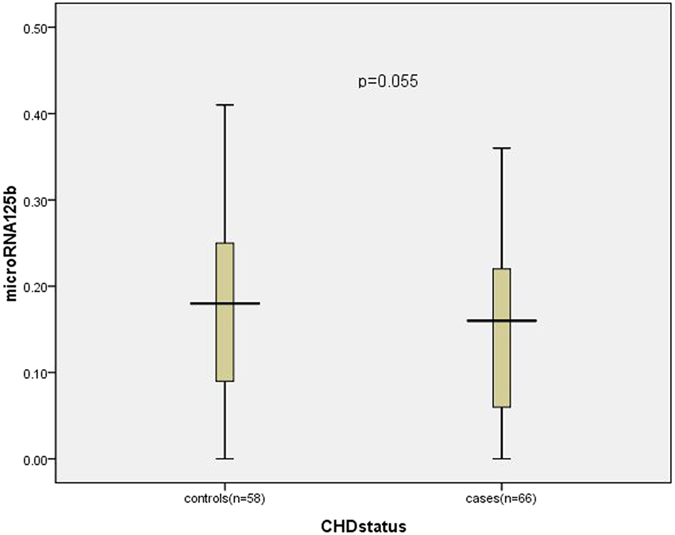
To determine miRNAs in CHD, we compared the levels of 20 circulating miRNAs in plasma samples of CHD subjects to controls. The relative expression of miR-125b in plasma of the CHD group (n = 66) was 0.14 ± 0.09, lower than that of the non-CHD group (n = 58), which was 0.18 ± 0.10 (p = 0.055) (Fig. 1); and the relative expression of miR-133b in plasma of the CHD group lower than that of the non-CHD group (0.14(0.06 ~ 0.23) vs 0.17(0.11 ~ 0.22), p = 0.207); however, no significant difference was reached.
Figure 1. The relative expression of miR-125b in plasma between controls and CHD cases.
Characteristics of the study population categorized by the quartile of Gensini score
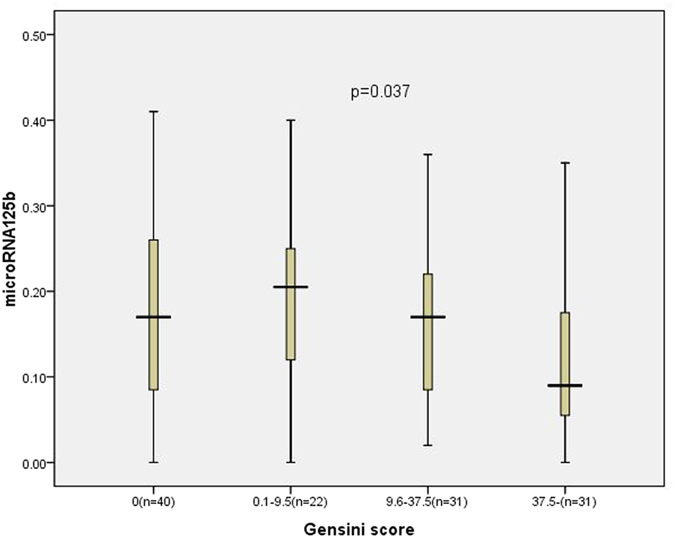
Subjects were divided into 4 groups according to the quartiles of the Gensini score: 0 (first quartile; n = 40 subjects), 0.1 to 9.5 (second quartile; n = 22 subjects), 9.6 to 37.5 (third quartile; n = 31 subjects) and ≥37.5 (fourth quartile; n = 31 subjects). The characteristics of the study population categorized by the quartile of the Gensini score are shown in Table 2. Age (P = 0.001) and serum CR levels (P = 0.003) significantly increased as the involvement of the Gensini score increased. The prevalence of CHD (P = 0.000) and the proportion of male gender (P = 0.009) were also significantly greater in the highest quartile of the Gensini score. Moreover, plasma miR-125b levels significantly decreased following an increasing Gensini score (P = 0.037) (Fig. 2).
Table 2. Characteristics of the study population categorized by the quartile of Gensini score.
| Characteristics | Gensini score |
Statistical parameter | Pvalue | |||
|---|---|---|---|---|---|---|
| 0(n = 40) | 0.1–9.5(n = 22) | 9.6–37.5(n = 31) | 37.5-(n = 31) | |||
| Age (years) | 55.70 ± 8.41 | 56.86 ± 9.50 | 61.68 ± 9.14 | 63.16 ± 8.70 | 5.460 | 0.001 |
| Gender (F/M) | 13/27 | 11/11 | 5/26 | 4/27 | 11.599 | 0.009 |
| TCH | 4.72 ± 0.97 | 4.52 ± 1.07 | 4.79 ± 1.10 | 4.72 ± 0.97 | 0.313 | 0.816 |
| HDL-C | 1.48 ± 0.35 | 1.51 ± 0.33 | 1.33 ± 0.28 | 1.37 ± 0.39 | 1.846 | 0.143 |
| LDL-C | 2.82 ± 0.83 | 2.61 ± 0.86 | 3.01 ± 1.00 | 2.89 ± 0.85 | 0.936 | 0.426 |
| FBG | 5.07(4.64–5.73) | 5.04(4.80–5.49) | 5.08(4.78–6.29) | 5.89(4.95–6.99) | 7.610 | 0.055 |
| CR | 69.73 ± 12.62 | 63.11 ± 7.85 | 74.73 ± 19.35 | 76.71 ± 10.58 | 5.048 | 0.003 |
| CHD (case/control) | 0/40 | 5/17 | 30/1 | 31/0 | 104.600 | 0.000 |
| miR-433 | 0.57(0.32 ~ 1.09) | 0.77(0.47 ~ 1.38) | 0.57(0.31 ~ 1.14) | 0.58(0.28 ~ 1.04) | 2.959 | 0.398 |
| miR-1 | 0.85(0.36 ~ 2.93) | 1.25(0.73 ~ 1.89) | 1.80(0.52 ~ 3.85) | 1.34(0.47 ~ 3.02) | 3.012 | 0.390 |
| miR-122 | 0.28(0.09 ~ 0.56) | 0.62(0.15 ~ 1.67) | 0.46(0.09 ~ 1.41) | 0.33(0.10 ~ 1.32) | 3.012 | 0.390 |
| miR-133a | 0.33(0.19 ~ 0.77) | 0.58(0.31 ~ 0.92) | 0.38(0.23 ~ 0.89) | 0.32(0.16 ~ 0.97) | 3.797 | 0.284 |
| miR-145 | 0.08(0.04 ~ 0.18) | 0.07(0.03 ~ 0.18) | 0.11(0.04 ~ 0.22) | 0.09(0.05 ~ 0.16) | 1.308 | 0.727 |
| miR-214 | 0.15(0.02 ~ 0.67) | 0.11(0.02 ~ 0.34) | 0.17(0.04 ~ 0.72) | 0.20(0.03 ~ 1.08) | 1.436 | 0.697 |
| miR-21 | 0.59(0.42 ~ 1.00) | 0.82(0.42 ~ 1.55) | 0.62(0.44 ~ 0.96) | 0.60(0.32 ~ 1.07) | 2.457 | 0.483 |
| miR-25 | 0.02(0.01 ~ 0.07) | 0.03(0.01 ~ 0.11) | 0.01(0.01 ~ 0.07) | 0.02(0.01 ~ 0.09) | 0.950 | 0.813 |
| miR-20a | 0.35(0.14 ~ 0.54) | 0.33(0.19 ~ 0.64) | 0.34(0.14 ~ 0.58) | 0.38(0.18 ~ 0.44) | 0.595 | 0.898 |
| miR-106a | 2.69(1.65 ~ 4.31) | 2.58(1.92 ~ 4.36) | 2.60(1.85 ~ 4.59) | 1.92(1.48 ~ 3.81) | 1.628 | 0.653 |
| miR-92a | 0.65 ± 0.95 | 1.29 ± 1.82 | 1.91 ± 6.21 | 0.26 ± 0.42 | 1.544 | 0.207 |
| miR-130a | 59.52(35.26 ~ 88.03) | 96.18(38.95 ~ 30.52) | 76.90(28.34 ~ 235.57) | 46.53(29.24 ~ 97.34) | 4.440 | 0.218 |
| miR-155 | 159.51(106.22 ~ 337.12) | 246.85(154.05 ~ 331.43) | 204.36(127.56 ~ 384.01) | 158.68(62.25 ~ 314.08) | 3.045 | 0.385 |
| miR-208b | 1.05(0.56 ~ 3.90) | 0.75(0.59 ~ 1.61) | 1.15(0.67 ~ 4.91) | 1.30(0.65 ~ 3.38) | 1.859 | 0.602 |
| miR-499 | 3.10(2.09 ~ 7.26) | 2.87(1.89 ~ 5.04) | 4.38(2.26 ~ 10.06) | 3.75(1.40 ~ 9.29) | 2.260 | 0.520 |
| miR-485-3p | 1.56(0.70 ~ 3.38) | 1.42(0.84 ~ 2.03) | 1.70(0.81 ~ 15.14) | 1.87(1.04 ~ 6.59) | 1.210 | 0.751 |
| miR-133b | 0.16(0.10 ~ 0.25) | 0.18(0.10 ~ 0.22) | 0.14(0.09 ~ 0.23) | 0.14(0.06 ~ 0.24) | 1.251 | 0.741 |
| miR-221 | 0.62(0.21 ~ 0.91) | 0.43(0.21 ~ 1.12) | 0.36(0.21 ~ 0.93) | 0.43(0.18 ~ 0.80) | 0.648 | 0.885 |
| miR-208a | 1.67(1.12 ~ 3.85) | 2.16(1.21 ~ 4.64) | 2.25(1.31 ~ 4.39) | 2.89(1.69 ~ 4.29) | 3.318 | 0.345 |
| miR-125b | 0.17 ± 0.10 | 0.19 ± 0.10 | 0.16 ± 0.09 | 0.12 ± 0.09 | 2.912 | 0.037 |
Data are summarized by either mean ± standard deviation or 50th (25th/75th) percentiles for continuous variables and N1/N2 for binary variables. CHD, coronary heart disease; TCH, total cholesterol; HDL-C, fasting high-density lipoprotein cholesterol; LDL-C, fasting low-density lipoprotein cholesterol; TG, triglyceride; FBG, fasting blood glucose; CR, creatinine. The value of each miRNAs means the relative mount calculated by 2−Δct method.
Figure 2. The relative expression of miR-125b in plasma among the subjects categorized by the quartile of the Gensini score.
Spearman correlations between Gensini score with clinical characteristics and miRNAs
Table 3 shows the results of Spearman correlation analyses between Gensini score, clinical characteristics, and miRNAs. The results indicate that the Gensini score was positively associated with age (r = 0.339, p = 0.000), FBG (r = 0.223, p = 0.014), and CR (r = 0.276, p = 0.002). A negative association was found between the Gensini score and HDL-C (r = −0.211, p = 0.021), and miR-125b (r = −0.215, p = 0.017). And, the heatmap was generated by transformation of the real-time PCR data presented as 2−Δct and was shown in Fig. 3.
Table 3. Spearman correlations between Gensini score and features of the study population.
| Variables | Relationship coefficient | P value |
|---|---|---|
| Age (years) | 0.339 | 0.000 |
| TCH | 0.019 | 0.841 |
| HDL-C | −0.211 | 0.021 |
| LDL-C | 0.052 | 0.570 |
| FBG | 0.223 | 0.014 |
| CR | 0.276 | 0.002 |
| miR-433 | 0.001 | 0.993 |
| miR-1 | 0.066 | 0.469 |
| miR-122 | 0.071 | 0.431 |
| miR-133a | 0.037 | 0.681 |
| miR-145 | 0.043 | 0.636 |
| miR-214 | 0.089 | 0.328 |
| miR-21 | −0.057 | 0.531 |
| miR-25 | 0.006 | 0.943 |
| miR-20a | 0.007 | 0.934 |
| miR-106a | −0.079 | 0.381 |
| miR-92a | −0.151 | 0.094 |
| miR-130a | −0.066 | 0.466 |
| miR-155 | −0.040 | 0.660 |
| miR-208b | 0.04 | 0.663 |
| miR-499 | 0.06 | 0.507 |
| miR-485-3p | 0.084 | 0.356 |
| miR-133b | −0.083 | 0.352 |
| miR-221 | −0.073 | 0.422 |
| miR-208a | 0.134 | 0.137 |
| miR-125b | −0.215 | 0.017 |
TCH, total cholesterol; HDL-C, fasting high-density lipoprotein cholesterol; LDL-C, fasting low-density lipoprotein cholesterol; TG, triglyceride; FBG, fasting blood glucose; CR, creatinine. The value of each miRNAs means the relative mount calculated by 2−Δct method.
Figure 3. The heatmap of real-time PCR expression profiling of microRNA in subjects with CHD and controls.
Receiver operating characteristic curve analyses in subjects with CHD and controls
To further explore the applicability of circulating miRNAs and classical risk factors as potential diagnostic biomarkers of CHD, subsequent ROC analyses were performed, and the results are shown in Table 4. The AUC was 0.695 for age (95% CI: 0.601 ~ 0.788, p = 0.000) (Fig. 4); 0.354 for HDL-C (95% CI: 0.255 ~ 0.454, p = 0.006) (Fig. 5); 0.647 for FBG (95% CI: 0.549 ~ 0.745, p = 0.006) (Fig. 6); and 0.622 for CR (95% CI: 0.522 ~ 0.723, p = 0.021) (Fig. 7). Of all the miRNAs, miR-125b showed the lowest AUC (0.405; 95% CI: 0.305 ~ 0.506, p = 0.070) (Fig. 8). Due to the levels of HDL-C, miR-133b and miR-125b were lower in patients with CHD than in the controls; the AUC of the above three variables were less than 0.5. The optimal cut-off value, the sensitivity, the specificity and Youden index of age, CR, FBG, HDL-C, and miR-125b are shown in Table 5.
Table 4. Receiver operating characteristic curve analyses in subjects with CHD and controls.
| Variables | AUC(95% CI) | P value |
|---|---|---|
| Age (years) | 0.695(0.601 ~ 0.788) | 0.000 |
| TCH | 0.515(0.411 ~ 0.619) | 0.780 |
| HDL-C | 0.354(0.255 ~ 0.454) | 0.006 |
| LDL-C | 0.542(0.438 ~ 0.645) | 0.432 |
| FBG | 0.647(0.549 ~ 0.745) | 0.006 |
| CR | 0.622(0.522 ~ 0.723) | 0.021 |
| miR-433 | 0.488(0.386 ~ 0.590) | 0.820 |
| miR-1 | 0.569(0.467 ~ 0.670) | 0.189 |
| miR-122 | 0.547(0.445 ~ 0.649) | 0.366 |
| miR-133a | 0.527(0.425 ~ 0.630) | 0.602 |
| miR-145 | 0.522(0.419 ~ 0.625) | 0.670 |
| miR-214 | 0.556(0.455 ~ 0.658) | 0.281 |
| miR-21 | 0.461(0.359 ~ 0.562) | 0.451 |
| miR-25 | 0.484(0.381 ~ 0.586) | 0.752 |
| miR-20a | 0.522(0.419 ~ 0.625) | 0.674 |
| miR-106a | 0.459(0.357 ~ 0.561) | 0.432 |
| miR-92a | 0.446(0.344 ~ 0.548) | 0.301 |
| miR-130a | 0.491(0.388 ~ 0.594) | 0.867 |
| miR-155 | 0.501(0.399 ~ 0.604) | 0.978 |
| miR-208b | 0.532(0.430 ~ 0.634) | 0.540 |
| miR-499 | 0.546(0.444 ~ 0.674) | 0.382 |
| miR-485-3p | 0.554(0.452 ~ 0.656) | 0.302 |
| miR-133b | 0.434(0.333 ~ 0.536) | 0.207 |
| miR-221 | 0.455(0.353 ~ 0.556) | 0.384 |
| miR-208a | 0.574(0.430 ~ 0.675) | 0.159 |
| miR-125b | 0.405(0.305 ~ 0.506) | 0.070 |
CI, confidence interval; AUC, area under the receiver operating characteristic curve; TCH, total cholesterol; HDL-C, fasting high-density lipoprotein cholesterol; LDL-C, fasting low-density lipoprotein cholesterol; TG, triglyceride; FBG, fasting blood glucose; CR, creatinine. The value of each miRNAs means the relative mount calculated by 2−Δct method.
AUC < 0.5 indicates the levels in patients with CHD lower than in controls.
AUC > 0.5 indicates the levels in patients with CHD higher than in controls.
Figure 4. The receiver operating characteristic curve of age for the ability to differentiate the CHD cases from the control individuals, AUC (95% CI) was 0.695(0.601 ~ 0.788).
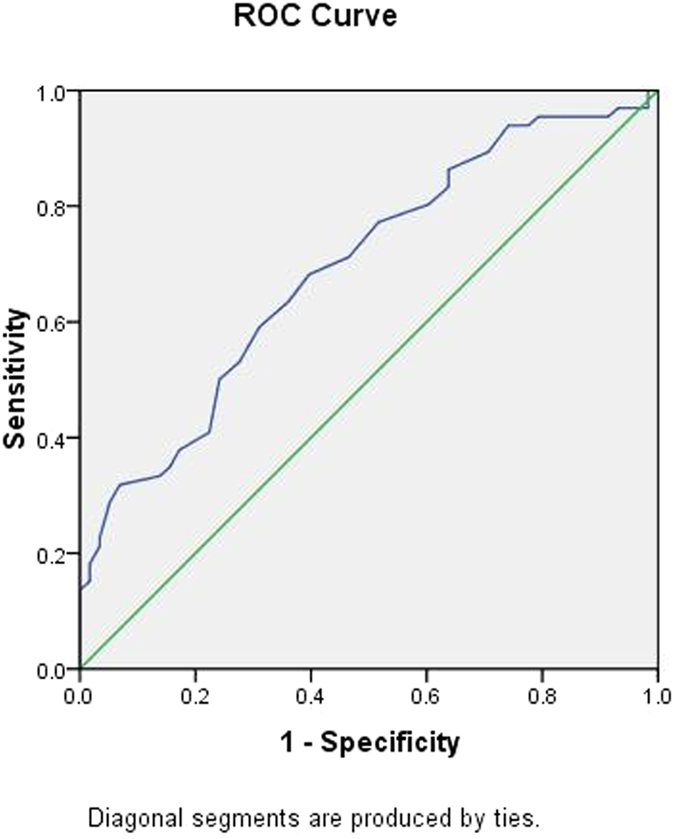
Figure 5. The receiver operating characteristic curve of HDL-C for the ability to differentiate the CHD cases from the control individuals, AUC (95% CI) was 0.354(0.255 ~ 0.454).
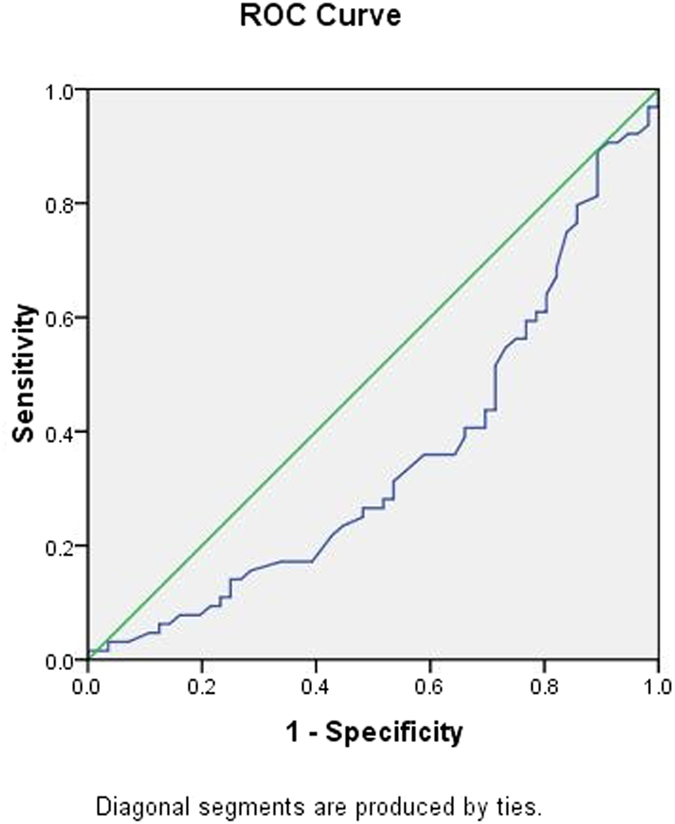
Figure 6. The receiver operating characteristic curve of FBG for the ability to differentiate the CHD cases from the control individuals, AUC (95% CI) was 0.647(0.549 ~ 0.745).
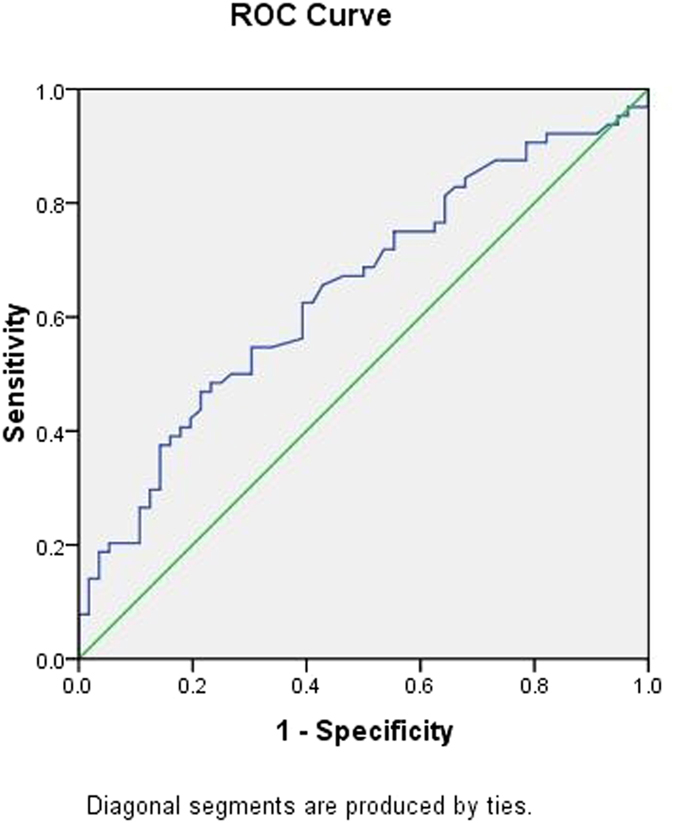
Figure 7. The receiver operating characteristic curve of CR for the ability to differentiate the CHD cases from the control individuals, AUC (95% CI) was 0.622(0.522 ~ 0.723).
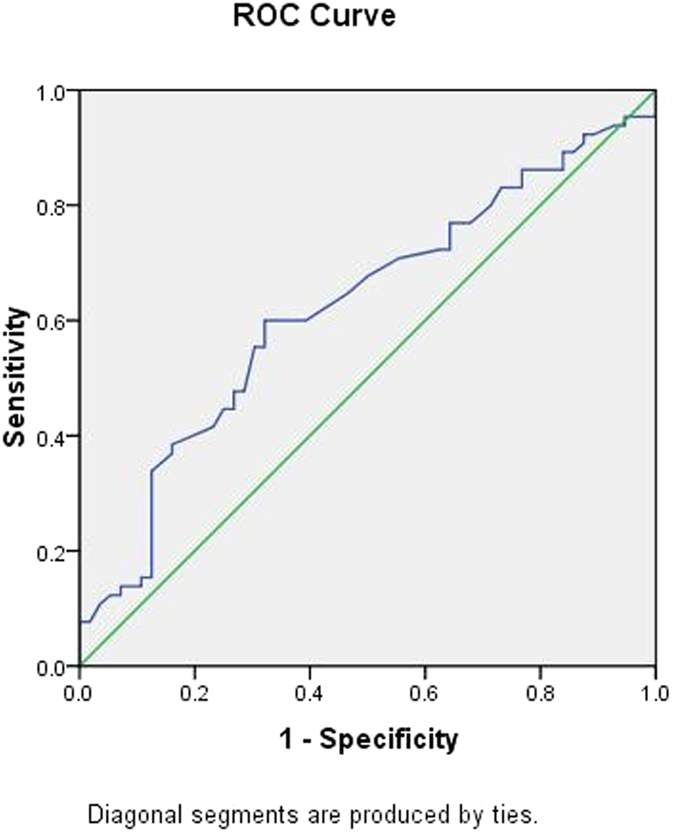
Figure 8. The receiver operating characteristic curve of miR-125b for the ability to differentiate the CHD cases from the control individuals, AUC (95% CI) was 0.405(0.305 ~ 0.506).
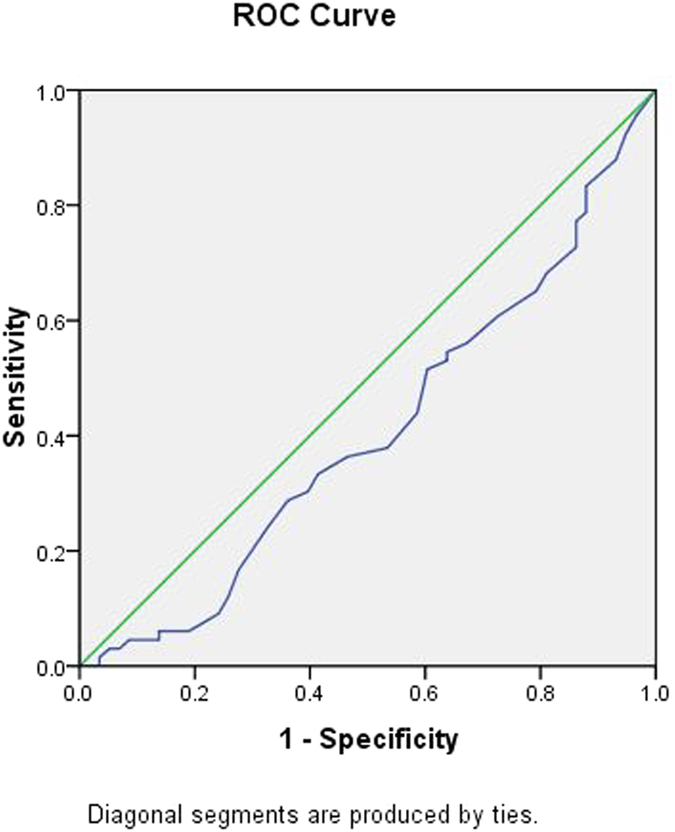
Table 5. The optimal cut-off and the Youden index.
| Variables | Cut-off | Sensitivity | Specificity | Youden index |
|---|---|---|---|---|
| Age | 58.500 | 0.682 | 0.603 | 0.285 |
| CR | 70.700 | 0.600 | 0.679 | 0.279 |
| FBG | 5.590 | 0.469 | 0.786 | 0.255 |
| HDL-C | 1.325 | 0.696 | 0.594 | 0.290 |
| miR-125b | 0.175 | 0.534 | 0.621 | 0.155 |
CR, creatinine; FBG, fasting blood glucose; HDL-C, fasting high-density lipoprotein cholesterol; TG, triglyceride. The value of each miRNA-125b means the relative mount calculated by 2−Δct method.
Interaction between miR-125b and classical risk factors
The results from the analysis of the possible positive/negative association between miR-125b and classical risk factors are presented in Tables 6 and 7. According to the ROC results, the cut-off of 0.175 for expression of miR-125b was the optimal value, accordingly, subjects with expression of miR-125b less than 0.175 were employed as subjects with susceptibility microRNA. Regarding the conventional risk of subjects unexposed to both classical risk factor and miR-125b risk (reference category) as being 1.0, the ORs estimating the effect of joint exposure to miR-125b and age, sex (male was defined as risk gender), CR, FBG, and HDL-C were significantly higher than the ORs estimating the effect of each factor in the absence of the other. A negative association between miR-125b and age was found (SI = −50.99, SIM = 5.48, AP = 0.80), the proportion of CHD attributable to the interaction of miR-125b and age in this group was as high as 80%. After performing the same analysis regarding gender, significant results was found in male subjects with susceptibility microRNA of miR-125b (SI = 1.18, SIM = 0.78, AP = 0.12). The susceptibility microRNA of miR-125b (SI = 1.46, SIM = 1.00, AP = 0.25) interact with CR to develop CHD. The risk provided by FBG was found to be positively reinforced by susceptibility microRNA of miR-125b (SI = 1.25, SIM = 0.77, AP = 0.16) in this study. The HDL-C and miR-125b significantly interact during the onset of CHD; this combination was shown to be accountable for 28% of the disease (SI = 1.50, SIM = 0.92, AP = 0.28).
Table 6. Synergistic effect of miR-125b and classical risk factors in CHD patients and controls.
| Classical risk | miR-125b | CHD | Controls | OR (95% CI) | Pvalue |
|---|---|---|---|---|---|
| Age | |||||
| 0 | 0 | 13 | 18 | 1 | 0.002 |
| 0 | 1 | 8 | 17 | 0.652(0.216 ~ 1.962) | 0.446 |
| 1 | 0 | 12 | 13 | 1.278(0.443 ~ 3.691) | 0.650 |
| 1 | 1 | 33 | 10 | 4.569(1.673 ~ 12.479) | 0.003 |
| Sex | |||||
| 0 | 0 | 6 | 14 | 1 | 0.085 |
| 0 | 1 | 6 | 7 | 2.00(0.469 ~ 8.530) | 0.349 |
| 1 | 0 | 19 | 17 | 2.608(0.819 ~ 8.309) | 0.105 |
| 1 | 1 | 35 | 20 | 4.083(1.355 ~ 12.303) | 0.012 |
| CR | |||||
| 0 | 0 | 12 | 22 | 1 | 0.016 |
| 0 | 1 | 14 | 16 | 1.604(0.587 ~ 4.381) | 0.357 |
| 1 | 0 | 13 | 8 | 2.979(0.965 ~ 9.196) | 0.058 |
| 1 | 1 | 26 | 10 | 4.767(1.731 ~ 13.130) | 0.003 |
| FBG | |||||
| 0 | 0 | 13 | 24 | 1 | 0.017 |
| 0 | 1 | 21 | 20 | 1.938(0.779 ~ 4.822) | 0.155 |
| 1 | 0 | 12 | 6 | 3.692(1.123 ~ 12.136) | 0.031 |
| 1 | 1 | 18 | 6 | 5.538(1.764 ~ 17.391) | 0.003 |
| HDL-C | |||||
| 0 | 0 | 10 | 21 | 1 | 0.009 |
| 0 | 1 | 16 | 18 | 1.867(0.680 ~ 5.126) | 0.226 |
| 1 | 0 | 15 | 9 | 3.500(1.144 ~ 10.706) | 0.028 |
| 1 | 1 | 23 | 8 | 6.037(2.006 ~ 18.173) | 0.001 |
CR, creatinine; FBG, fasting blood glucose; HDL-C, fasting high-density lipoprotein cholesterol.
The value of each miRNA-125b means the relative mount calculated by 2−Δct method.
Table 7. The indexes of synergistic effect between miRNAs and classical risk factors.
| Variables | SI | SIM | RERI | AP |
|---|---|---|---|---|
| Age-miR-125b | −50.99 | 5.48 | 3.64 | 0.80 |
| Sex-miR-125b | 1.18 | 0.78 | 0.48 | 0.12 |
| CR-miR-125b | 1.46 | 1.00 | 1.18 | 0.25 |
| FBG-miR-125b | 1.25 | 0.77 | 0.91 | 0.16 |
| HDL-miR-125b | 1.50 | 0.92 | 1.67 | 0.28 |
SI, synergy measures in additive models; SIM, synergy measures in multiplicative models; RERI, relative excess risk due to interaction; AP, proportion of disease attributable to interaction; CR, creatinine; FBG, fasting blood glucose; HDL-C, fasting high-density lipoprotein cholesterol.
Discussion
To the best of our knowledge, this is the first study to explore the interaction between microRNA expressions with classical risk factors on coronary heart disease throughout the world. Based on the data of 66 subjects with CHD and 58 subjects with angiographic exclusion of CHD, interactions between miR-125b and classical risk factors were found; furthermore, the proportion of CHD attributable to the interaction of miR-125b and age was as high as 80%.
Cardiovascular diseases are the major cause of death worldwide, particularly in the elderly population who have an increasing rate of mortality and morbidity; cardiovascular diseases are a consequence of genetic and epigenetic interactions31,32,33. The most important epigenetic modifications of mammalian cells are associated with DNA methylation, posttranslational histone modifications, and a class of short noncoding RNAs, the microRNAs (miRNAs or miRs)34,35. MicroRNAs are endogenous, small, non-coding RNAs involved in the regulation of gene expression36. They control expression on a posttranscriptional level as intracellular RNAs and have been discussed as potential therapeutic targets37. MicroRNAs are involved in the pathogenesis of various cardiovascular conditions, such as CHD. We hypothesize that CHD has a multifactorial genetic basis involving a number of microRNAs and interacting environmental factors to determine the potential development of disease. The present study has proven this hypothesis. Therefore, the expression of microRNAs generally predisposes to a greater or lesser extent of CHD, but it is the environmental factors interacting with the individual's expression of microRNAs that determine whether CHD will develop.
The muscle-enriched miRNA, miR-133, is almost the most abundant of the miRNAs present in the normal heart38,39; the gene MIR133b encodes miR-133b. There is evidence that muscle- and/or cardiac-specific miRNA-133b is involved in heart development and some cardiovascular diseases, including myocardial infarction40,41, myocardial injury after operation42,43 and cardiomyopathy44,45. In the present study, the circulating miRNA expression level of miR-133b was lower in subjects with CHD than those in controls, these observations are in line with the results of recently published studies40,41,42,43, showing lower miR-133b levels in patients with CHD than in controls, however, no significant difference was reached regarding as the relative expression of miR-133b between the cases and controls in the present study.
Mature miR-125b originates from two precursors: pre-miR-125b-1 (located at chr11q24.1) and pre-miRNA-125b-2 (located at chr21q21.1)46. A number of studies have investigated the relationships between the members of the miR-125 family, particularly miR-125b, and various diseases, including solid tumours47,48,49, hepatological malignancies50,51, autoimmune diseases52,53, lung disease54,55, chronic kidney disease56, acute stroke57 and obesity58. However, there is little data to support the involvement of miR-125b in human CHD. Suli Huang et al.59 reported that the plasma levels of miR-320b and miR-125b were significantly lower in patients with AMI than in controls. Because miR-320b and miR-125b were expressed in vascular endothelial cells (VECs), the authors hypothesized that the down-regulated miRNAs in AMI might be related to the development and progression of atherosclerosis; this may then trigger myocardial infarction by regulating gene expressions in VECs. The preliminary functional study60 suggested that miR-125a-5p and miR-125b-5p were highly expressed in VECs and that both miR-125a/b-5p could suppress oxLDL-induced endothelin-1 (ET-1) expression by directly targeting the 3′untranslated region of preproET-1 mRNA. Accumulating evidence has suggested that ET-1 plays a significant role in many physiologic and pathophysiological processes, including heart development61, cardiac hypertrophy62, and atherosclerosis63. These functional analyses could further the understanding of the role of circulating miRNAs in atherosclerosis processes and pave the way for further studies. Of all the miRNAs in the present study, miR-125b showed the lowest AUC (0.405; 95% CI: 0.305 ~ 0.506, p = 0.070). To analyse possible positive or negative interactions between miR-125b expressions with classical risk factors, a 4 × 2 table approach was used; the synergy measures in additive (SI) or multiplicative models (SIM) of miR-125b with age, male gender, CR, FBG, and HDL-C were −50.99 or 5.48, 1.18 or 0.78, 1.46 or 1.00, 1.25 or 0.77, and 1.50 or 0.92, respectively. Therefore, the positive interactions between miR-125b with male gender, CR, FBG, and HDL-C were found, and the proportion of CAD attributable to the interaction between HDL-C and susceptibility miR-125b was approximately 28% for the study. Nevertheless, we found a negative interaction between miR-125b expressions with age; the proportion of CHD attributable to the interaction of age and miR-125b was as high as 80%.
In summary, the present study provides the first insights into the interaction between microRNA expressions with classical risk factors of coronary heart disease around the world. Based on the data of 66 subjects with CHD and 58 control individuals, interactions between miR-125b and classical risk factors were found; the proportion of CHD attributable to the interaction of miR-125b and age was as high as 80%. The results of the present study may have important implications for better understanding of the aetiology and pathogenesis of the disease through quantitative assessment of disease risks in some populations. Considering that CHD is a multifactorial and multigenic disease driven by various environmental and genetic components interacting together, our results appear to support the hypothesis that the susceptibility microRNA may indeed enhance the influence of classical risk factors in the development of CHD. However, this was a single-centre study conducted in China, and the conclusions drawn based on this study’s results may not be applicable to other populations. Furthermore, the mechanisms underlying the dysregulation remain to be determined. Therefore, further prospective large-scale studies are required to determine the potential role of circulating miRNAs in the microRNA-environment, interaction, and diagnostic value of coronary stenotic lesions in subjects with CHD.
Additional Information
How to cite this article: Ding, X.-Q. et al. Interaction between microRNA expression and classical risk factors in the risk of coronary heart disease. Sci. Rep. 5, 14925; doi: 10.1038/srep14925 (2015).
Acknowledgments
This study was supported by the National Natural Science Foundations of China (grants 81170180, 30400173 and 30971257), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and a science and technology project funded by the Ili Kazakh Autonomous Prefecture (YZ201401031). Dr. En-Zhi Jia is an Assistant Fellow at the Collaborative Innovation Center for Cardiovascular Disease Translational Medicine.
Footnotes
Author Contributions As the guarantor, E.Z.J. conceived the study. X.Q.D. and Z.L. initially drafted the paper. F.H.A., L.H.L., Z.H.C., H.W.M. and H.J. enrolled participants and collected the data under the supervision of T.B.Z., C.J.L., P.C.G., Z.Y.L. and Y.G. cleaned and analysed the data. X.C. coordinated the study. Z.J.Y., W.Z.M. and L.S.W. monitored the conduct of the study and reviewed the safety and effectiveness data. All authors reviewed the manuscript.
References
- Sayed A. S., Xia K., Salma U., Yang T. & Peng J. Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ. 23, 503–10 (2014). [DOI] [PubMed] [Google Scholar]
- Schober A., Thum T. & Zernecke A. MicroRNAs in vascular biology–metabolism and atherosclerosis. Thromb Haemost. 107, 603–604 (2012). [DOI] [PubMed] [Google Scholar]
- Schroen B. & Heymans S. Small but smart—microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res. 93, 605–613 (2012). [DOI] [PubMed] [Google Scholar]
- Urbich C., Kuehbacher A. & Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 79, 581–588 (2008). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Sonkoly E., Stahle M. & Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin CancerBiol. 18, 131–140 (2008). [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P. & Burge C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003). [DOI] [PubMed] [Google Scholar]
- Lim L. P. et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773 (2005). [DOI] [PubMed] [Google Scholar]
- Bronze-da-Rocha E. MicroRNAs Expression Profiles in Cardiovascular Diseases. Biomed Res Int. 2014, 985408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T. et al. Blankenberg S. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J. 35, 2106–14 (2014). [DOI] [PubMed] [Google Scholar]
- Yang Q. & Khoury M. J. Evolving methods in genetic epidemiology. III. Gene-environment interaction in epidemiologic research. Epidemiol Rev. 19, 33–43 (1997). [DOI] [PubMed] [Google Scholar]
- Judkins M. P. A percutaneous transfemoral technique. Radiology 89, 815–821 (1967). [DOI] [PubMed] [Google Scholar]
- Sun X. et al. Circulating microRNA-126 in patients with coronary artery disease: correlation with LDL cholesterol. Thromb J. 10, 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensini G. G. A more meaningful scoring system for determinating the severity of coronary heart disease. Am J Cardiol. 51, 606 (1983). [DOI] [PubMed] [Google Scholar]
- D'Alessandra Y. et al. Diagnostic Potential of Plasmatic MicroRNA Signatures in Stable and Unstable Angina. PloS One 8, e80345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Q., He Z. Q. , Fan M. & Wu Z. G. Circulating miR-214 is associated with the severity of coronary artery disease. J Geriatr Cardiol. 10, 34–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. et al. Signature of Circulating MicroRNAs as Potential Biomarkers in Vulnerable Coronary Artery Disease. PLoS One. 8, e80738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtlscherer S. et al. Circulating microRNAs in patients with coronary artery disease. Circulation research 107, 677–84 (2010). [DOI] [PubMed] [Google Scholar]
- Goettsch C. et al. miR-125b Regulates Calcification of Vascular Smooth Muscle Cells. The American Journal of Pathology 179, 1594–1600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. M., Lin K. Y. & Chen Y. Q. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 6, 6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. MicroRNA-125b protects against myocardial ischaemia/reperfusion injury via targeting p53-mediated apoptotic signalling and TRAF6. Cardiovascular research 102, 385–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D. et al. SR-A deficiency reduces myocardial ischemia/reperfusion injury; involvement of increased microRNA-125b expression in macrophages. Biochimica et biophysica acta 1832, 336–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. Altered Profile of Seminal Plasma MicroRNAs in the Molecular Diagnosis of Male Infertility. Clin Chem. 57, 1722–3 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. Expression Profile of MicroRNAs in Serum: A Fingerprint for Esophageal Squamous Cell Carcinoma. Clin Chem. 56, 1871–9 (2010). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. A Combination of Let-7d, Let-7g and Let-7i Serves as a Stable Reference for Normalization of Serum MicroRNAs. PLoS One 8, e79652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y. et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem. 58, 559–67 (2012). [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 3, 1101–8 (2008). [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O. & Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 95, 14863–8 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J. A. & McNeil B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982). [DOI] [PubMed] [Google Scholar]
- Assman S. F., Hosmer D. W., Lemeshow S. & Mundt K. A. Confidence intervals for measures of interaction. Epidemiology 7, 286–290 (1996). [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rodero S., Fernández-Morera J. L., Fernandez A. F., Menéndez-Torre E. & Fraga M. F. Epigenetic regulation of aging. Discov Med. 10, 225–33 (2010). [PubMed] [Google Scholar]
- D'Aquila P., Rose G., Bellizzi D. & Passarino G. Epigenetics and aging. Maturitas. 74, 130–6 (2013). [DOI] [PubMed] [Google Scholar]
- Dimmeler S. & Nicotera P. MicroRNAs in age-related diseases. EMBO Mol Med. 5, 180–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M. A., Partridge L., Serrano M. & Kroemer G. The hallmarks of aging. Cell. 153, 1194–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai S. & Slack F. MicroRNAs and the genetic network in aging. J Mol Biol. 425, 3601–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooij E. The art of microRNA research. Circ Res 108, 219–234 (2011). [DOI] [PubMed] [Google Scholar]
- Small E. M., Frost R. J. & Olson E. N. MicroRNAs add a new dimension to cardiovascular disease. Circulation 121, 1022–1032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L. & Chen C. Characterization of microRNA expression in normal human tissues. BMC Genomics 8, 166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Zhang H., Xiao J. & Wang Z. Regulation of human cardiac ion channel genes by microRNAs: theoretical perspective and pathophysiological implications. Cell Physiol Biochem. 25, 571–586 (2010). [DOI] [PubMed] [Google Scholar]
- Bostjancic E., Zidar N., Stajer D. & Glavac D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology 115, 163–9 (2010). [DOI] [PubMed] [Google Scholar]
- D’Alessandra Y. et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 31, 2765–2773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. et al. Circulating miRNAs reflect early myocardial injury and recovery after heart transplantation. J Cardiothorac Surg. 8, 165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y. et al. Plasma Levels of MicroRNA-499 Provide an early indication of perioperative myocardial infarction in coronary artery bypass graft patients. PLoS One. 9, e104618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucharov C., Bristow M. R. & Port J. D. miRNA expression in the failing human heart functional correlates. J Mol Cell Cardiol. 45, 185–92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. R. et al. MicroRNAs miR-1, miR-133a, miR-133b, miR-208a and miR-208b are dysregulated in Chronic Chagas disease Cardiomyopathy. Int J Cardiol. 175, 409–17 (2014). [DOI] [PubMed] [Google Scholar]
- Pogue A. I. et al. MicroRNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 476, 18–22 (2010). [DOI] [PubMed] [Google Scholar]
- Guan Y., Yao H., Zheng Z., Qiu G. & Sun K. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer 128, 2274–2283 (2010). [DOI] [PubMed] [Google Scholar]
- Huang L. et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer 128, 1758–1769 (2011). [DOI] [PubMed] [Google Scholar]
- Mar-Aguilar F. et al. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 9, 53–9 (2013). [DOI] [PubMed] [Google Scholar]
- Sonoki T., Iwanaga E., Mitsuya H. & Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobul in heavy chain gene locus in a patient with precursor B-cell acute lymphoblasticleukemia. Leukemia 19, 2009–2010 (2005). [DOI] [PubMed] [Google Scholar]
- Chapiro E. et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia 24, 1362–1364 (2010). [DOI] [PubMed] [Google Scholar]
- Lukiw W. J. & Alexandrov P. N. Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer's disease (AD) brain. Mol Neurobiol 46, 11–19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S. et al. Deregulated microRNAs in myotonic dystrophy type 2. PLoS One. 7, e39732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. Circulating microRNAs as potential biomarkers for smoking-related interstitial fibrosis. Biomarkers 17, 435–440 (2012). [DOI] [PubMed] [Google Scholar]
- Van Pottelberge G. R. et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 183, 898–906 (2011). [DOI] [PubMed] [Google Scholar]
- Chen N. X. et al. Decreased microRNA is involved in the vascular remodeling abnormalities in chronic kidney disease(CKD). PLoS One. 8, e64558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepramaniam S. et al. Circulating microRNAs as biomarkers of acute stroke. Int J Mol Sci 15, 1418–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega F. J. et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 59, 781–92 (2013). [DOI] [PubMed] [Google Scholar]
- Huang S. et al. Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ Cardiovasc Genet. 7, 189–98 (2014). [DOI] [PubMed] [Google Scholar]
- Li D. et al. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertens. 28, 1646–54 (2010). [DOI] [PubMed] [Google Scholar]
- Kurihara Y. et al. Aortic arch malformations and ventricular septal defect in mice deficient inendothelin-1. J Clin Invest. 96, 293–300 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H. et al. Endothelin-1is an autocrine/paracrine factor in the mechanism of angiotensinII-induced hypertrophy in cultured rat cardiomyocytes. J Clin Invest. 92, 398–403 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. et al. Endothelin ETA receptor blockade restores NO-mediated endothelial function and inhibits atherosclerosis in apolipoprotein E-deficient mice. Proc Natl Acad Sci USA. 95, 14367–14372 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]


