Abstract
Influenza A is a negative-sense RNA virus with an eight-segment genome. Some segments code for more than one polypeptide product, but how the virus accesses alternate internal open reading frames (ORFs) is not completely understood. In segment 2, ribosomal scanning produces two internal ORFs, PB1-F2 and N40. Here, chemical mapping reveals a Mg2+ dependent pseudoknot structure that includes the PB1-F2 and N40 start codons. The results suggest that ribosome interactions with the pseudoknot may affect the level of translation for PB1-F2 and N40.
Influenza A is a negative-sense RNA virus composed of eight discrete segments. Influenza infections are predicted to cause 25,000 deaths annually in the United States.1 The possibility of pandemic influenza strains, illustrated by the recent H5N1 and H7N9 outbreaks, makes this virus a public health concern.2,3 While each segment of influenza A virus codes for a main protein product, segments 2, 3, 7, and 8 also code, respectively, for additional polypeptide products by ribosomal scanning, ribosomal frame shifting, and, for 7 and 8, alternative splicing.4–7 Segment 2 (PB1) codes for the alternative polypeptide products PB1-F2 and N40, the former acting as a pro-apoptotic factor and the latter of unknown function.8,9 While ribosomal scanning past the main PB1 start codon is responsible for accessing the internal PB1-F2 and N40 open reading frames (ORFs), there may be other factors that control initiation at the proper start codon.
RNA secondary structure is a well-known mechanism for facilitating non-canonical translation. For example, frameshifting pseudoknots and internal ribosome entry sites (IRES) represent structured RNA domains that provide alternative ORFs by causing, respectively, the translating ribosome to “slip” backwards or bypass standard cap dependent translation initiation by serving as an internal ribosome binding domain.10,11 To search for potential secondary structures containing the start codons for PB1-F2 and N40, a consensus sequence was derived for the local region. When the program, DotKnot,12 was used to scan the consensus sequence, a conserved pseudoknot was predicted. Base pairing frequencies determined from an alignment of all available unique influenza sequences for this region are consistent with this folding, but revealed no compensatory changes (see Figure 11 in ref 16). Computational predictions of pseudoknots in biologically relevant sized sequences without any chemical mapping or phylogenetic data predicts as few as 5% of pseudoknotted base pairs correctly.13 Here, chemical mapping in vitro with NMIA, DMS, and CMCT14 is shown to be consistent with the possible pseudoknot, and the folding is found to depend on Mg2+ (Figure 1). As a reminder, NMIA modifies the ribose 2´-OH group of flexible (single-stranded) nucleotides, DMS modifies the pairing faces of single-stranded A’s and C’s, while CMCT similarly modifies U’s. All chemical mapping data can be found in the SNRNASM database (see supplementary Materials & Methods section).
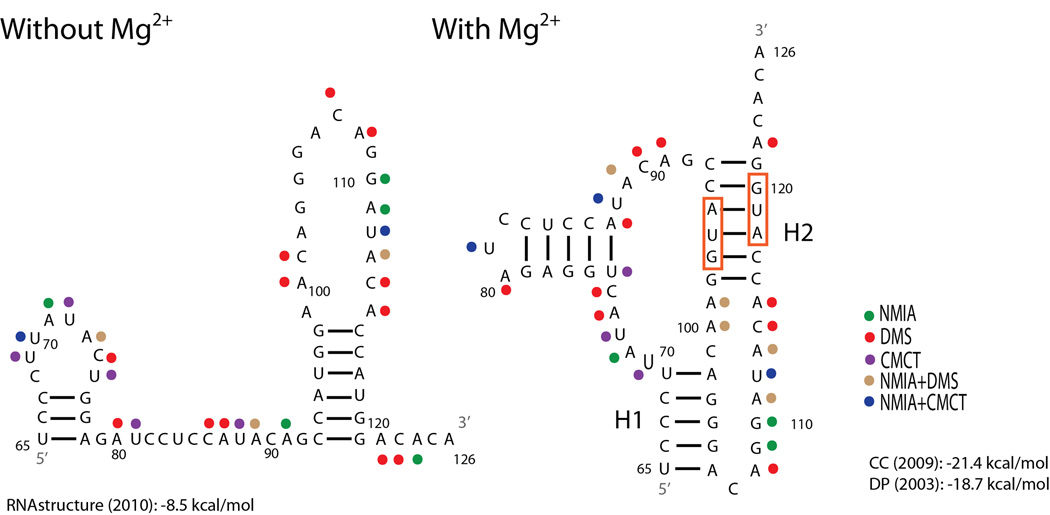
Figure 1.
Structural models of the influenza A segment 2 (PB1) 65–126 nucleotide region. To optimize SP6 polymerase efficiency the identities of nucleotides 63 and 64 (not shown) are both A’s, however in the wild-type sequence the nucleotides are A and U, respectively. The structure without Mg2+ is predicted with RNA structure15 where strongly reactive nucleotides were forced to be single-stranded. The structure shown in the presence of Mg2+ is the one most consistent with the model based on sequence comparison.16 This structure is also predicted by ShapeKnots.17 Predicted free energy at 37 °C for the structure without Mg2+ is reported at the bottom left.15 Predicted free energies at 37 °C from two pseudoknot prediction models are reported at the bottom right.18,19 Red boxes indicate the start codons for PB1-F2 and N40 ORFs, respectively. Colored dots represent strong chemical mapping hits for the reagent identified in the key. H1: Helix 1, H2: Helix 2.
Absence of significant chemical modification at positions 65–69 and 102–106 in the presence of Mg2+ is consistent with a pseudoknotted helix at these positions. Moreover, when the NMIA data were incorporated into the ShapeKnots program, the model in Figure 1 was predicted to be the most stable conformation. The topology of this pseudoknot is unusual because the pseudoknotted helices are not coaxially stacked on each other.
An important characteristic of pseudoknots is their dependence on multivalent cations, such as Mg2+, to stabilize their formation.20 As shown in Figure 1, there are important differences between chemical reactivity in the presence and absence of Mg2+. In particular, increased chemical reactivity at positions 69, 86, 101, and 107 in the absence of Mg2+ indicates destabilization of the folded RNA. Approximate initial melting temperatures determined with UV absorbance were 53 °C and 72 °C in the absence and presence of Mg2+, respectively, and the melting curve in the presence of Mg2+ suggests additional transition above 95 °C (Figure S1). These results further indicate that Mg2+ stabilizes the pseudoknot. No cooperative structural changes were observable at 37 °C, thus indicating that the structure is completely folded under the conditions used for chemical mapping experiments.
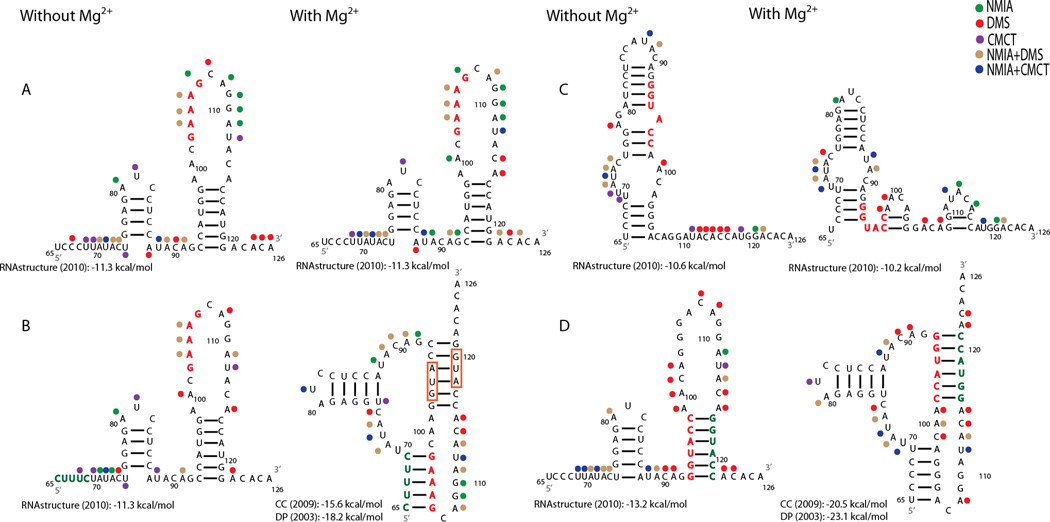
To further investigate the folding of this RNA, two sets of mutant sequences predicted to disrupt and rescue either of the pseudoknotted helices were synthesized. Chemical mapping reveals that the sequence in Figure 2A disrupts Helix 1, as demonstrated by increased chemical reactivity at nucleotides 69 and 103–106 in the presence of Mg2+. Compensatory mutations to restore Helix 1 provide protection to nucleotides 69 and 103–106, but only in the presence of Mg2+ (Figure 2B). These results are consistent with formation of a pseudoknot.
Figure 2.
Chemical mapping results for four mutant sequences with and without Mg2+. Red nucleotides indicate predicted destabilizing nucleotide changes and green represents compensatory changes. Predicted non-pseudoknotted structures were modeled with RNA structure as in Figure 1. Predicted pseudoknotted structures (B and D with Mg2+) were previously modeled with ShapeKnots and overlaid with chemical mapping results. (A) Destabilizing mutant sequence for Helix 1. (B) Compensatory mutant sequence for Helix 1. (C) Destabilizing mutant sequence for Helix 2. (D) Compensatory mutant sequence for Helix 2. Other figure annotations are as in Figure 1.
The sequence in Figure 2C is predicted to destabilize Helix 2 by reversing the orientation of nucleotides 93–97. This is confirmed by increased chemical reactivity for nucleotides 116, 117, 119, and 121 in the absence of Mg2+ and 119 and 121 in the presence of Mg2+. When the orientations of nucleotides 115–120 are also reversed to restore base pairing, the nucleotides of Helix 2 are protected from chemical modification (Figure 2D). The resulting reactivity pattern in the presence of Mg2+ is similar to those shown in Figures 1 and 2B, consistent with the proposed pseudoknot model.
Chemical mapping experiments of the native, destabilized, and compensatory mutant sequences strongly favor formation of a pseudoknot at nucleotides 65–126 of influenza A segment 2, which contains the PB1-F2 and N40 start sites. The results suggest a structural role of this pseudoknot in regulating translation initiation and therefore expression of PB1-F2 and N40. In light of PB1-F2 pro-pathogenic characteristics, this pseudoknot may provide a target for rational drug design.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Stanislav Bellaousov for his assistance with ShapeKnots.
Funding Sources
This work was supported by NIH grant R01 GM022939. SFP is a trainee in the MSTP program supported by grant T32 GM007356.
ABBREVIATIONS
- DMS
Dimethyl sulfate
- CMCT
1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene
- NMIA
N-methylisotoic anhydride
- SHAPE
Selective 2´-hydroxyl acylation analyzed by primer extension
Footnotes
ASSOCIATED CONTENT
A detailed Materials and Methods section can be found in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
No competing financial interests have been declared.
REFERENCES
- 1.Estimates of deaths associated with seasonal influenza --- United States, 1976–2007. MMWR Morb Mortal Wkly Rep. 2010;59:1057–1062. [PubMed] [Google Scholar]
- 2.Li FC, Choi BC, Sly T, Pak AW. Finding the real case-fatality rate of H5N1 avian influenza. J Epidemiol Community Health. 2008;62:555–559. doi: 10.1136/jech.2007.064030. [DOI] [PubMed] [Google Scholar]
- 3.Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, Zhang H, Li M, Zeng L, Xu Z, Li Z, Luo H, Li Q, Feng Z, Cao B, Yang W, Wu JT, Wang Y, Leung GM. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet. 2013;382:138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb RA, Choppin PW. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc Natl Acad Sci U S A. 1979;76:4908–4912. doi: 10.1073/pnas.76.10.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb RA, Lai CJ, Choppin PW. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci U S A. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wise HM, Barbezange C, Jagger BW, Dalton RM, Gog JR, Curran MD, Taubenberger JK, Anderson EC, Digard P. Overlapping signals for translational regulation and packaging of influenza A virus segment 2. Nucleic Acids Res. 2011;39:7775–7790. doi: 10.1093/nar/gkr487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 9.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gesteland RF, Weiss RB, Atkins JF. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- 11.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 12.Sperschneider J, Datta A. DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model. Nucleic Acids Res. 2010;38:e103. doi: 10.1093/nar/gkq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellaousov S, Mathews DH. ProbKnot: fast prediction of RNA secondary structure including pseudoknots. RNA. 2010;16:1870–1880. doi: 10.1261/rna.2125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuter JS, Mathews DH. RNA structure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss WN, Priore SF, Turner DH. Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA. 2011;17:991–1011. doi: 10.1261/rna.2619511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirks RM, Pierce NA. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J Comput Chem. 2004;25:1295–1304. doi: 10.1002/jcc.20057. [DOI] [PubMed] [Google Scholar]
- 19.Cao S, Chen SJ. Predicting structures and stabilities for H-type pseudoknots with interhelix loops. RNA. 2009;15:696–706. doi: 10.1261/rna.1429009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyatt JR, Puglisi JD, Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.