Abstract
Medicinal plants have long been utilized in traditional medicine and ethnomedicine worldwide. This review presents a glimpse of the current status of and future trends in medicinal plant genomics, evolution, and phylogeny. These dynamic fields are at the intersection of phytochemistry and plant biology and are concerned with the evolution mechanisms and systematics of medicinal plant genomes, origin and evolution of the plant genotype and metabolic phenotype, interaction between medicinal plant genomes and their environment, the correlation between genomic diversity and metabolite diversity, and so on. Use of the emerging high-end genomic technologies can be expanded from crop plants to traditional medicinal plants, in order to expedite medicinal plant breeding and transform them into living factories of medicinal compounds. The utility of molecular phylogeny and phylogenomics in predicting chemodiversity and bioprospecting is also highlighted within the context of natural-product-based drug discovery and development. Representative case studies of medicinal plant genome, phylogeny, and evolution are summarized to exemplify the expansion of knowledge pedigree and the paradigm shift to the omics-based approaches, which update our awareness about plant genome evolution and enable the molecular breeding of medicinal plants and the sustainable utilization of plant pharmaceutical resources.
Keywords: genome evolution, medicinal plants, phylogeny, phylogenomics, drug discovery and development
Introduction
There are over 300,000 species of extant seed plants around the globe.1 About 60% of plants have found medicinal use in the post-Neolithic human history. Nowadays, people collect plants for medicinal use not only from the wild but also through artificial cultivation, which is an indispensable part of human civilization. There are over 10,000 medicinal plant species in China, accounting for ~87% of the Chinese materia medica (CMM).2 Medicinal plants are also essential raw materials of many chemical drugs, eg, the blockbuster drugs for antimalarial and anticancer therapy. Currently, more than one-third of clinical drugs are derived from botanical extracts and/or their derivatives. Unfortunately, most medicinal plants have not been domesticated, and currently there is no toolkit to improve their medicinal attributes for better clinical efficacy. Immoderate harvesting has led to supply crisis of phytomedicine, exemplified by the taxane-producing Taxus plants.3 On the other hand, successful domestication and improvement are not realistic without deeper insights into the evolutionary pattern of medicinal plant genomes. Artificial selection can be regarded as an accelerated and targeted natural selection. Studies of medicinal plant genome evolution are crucial not only for the understanding of the ubiquitous mechanisms of plant evolution and phylogeny but also for plant-based drug discovery and development, as well as the sustainable utilization of plant pharmaceutical resources. This review gives a preliminary examination of the recent developments in medicinal plant genome evolution research and summarizes the benefits, gaps, and prospects of the current research topics.
Evolution of Genome, Gene, and Genotype
Genome sequencing
The genomic studies of medicinal plants lag behind those of model plants and important crop plants. The genome sequences encompass essential information of plant origin, evolution, development, physiology, inheritable traits, epigenomic regulation, etc., which are the premise and foundation of deciphering genome diversity and chemodiversity (especially various secondary metabolites with potential bioactivities) at the molecular level. High-throughput sequencing of medicinal plants could not only shed light on the biosynthetic pathways of medicinal compounds, especially secondary metabolites,4 and their regulation mechanisms but also play a major role in the molecular breeding of high-yielding medicinal cultivars and molecular farming of transgenic medicinal strains.
A few principles should be considered when selecting medicinal plants for whole-genome sequencing projects: first, source plants of well-known and expensive CMMs or important chemical drugs that are in heavy demand, eg, Panax ginseng5,6 and Artemisia annua7; second, representative plants whose pharmaceutical components are relatively unambiguous and that have typical secondary metabolism pathways, eg, Salvia medicinal plants8,9; third, characteristic plants that are in a large medicinal genus/family, such as Glycyrrhiza uralensis (Chinese liquorice; Fabaceae)10,11 and Lycium chinense (Chinese boxthorn; Solanaceae)12; fourth, medicinal plants that are potential model plants and have considerable biological data; and last, medicinal plants whose genetic backgrounds are known, with reasonably small diploid genome and relatively straightforward genome structure, should be given priority.
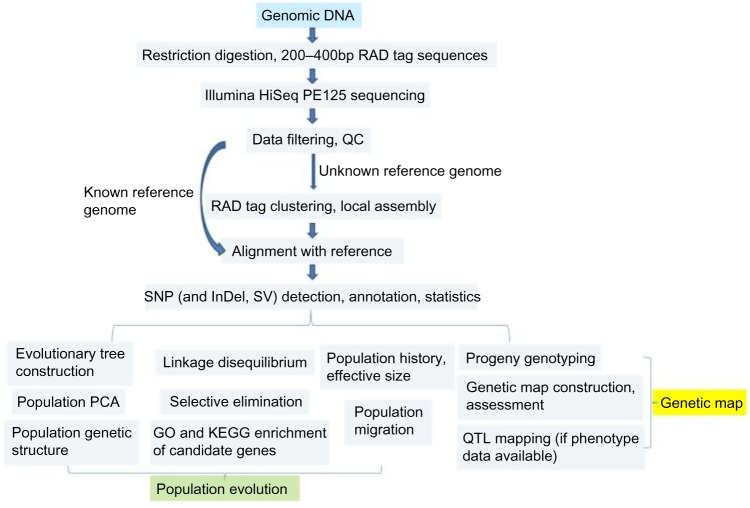
As there is a lack of comprehensive molecular genetic studies on most medicinal plants, it is vital to have some preliminary genome evaluations done before whole-genome sequencing. First, DNA barcoding techniques13 could be used to authenticate the candidate species; second, karyotypes should be determined by observing metaphase chromosomes; last, flow cytometry and pulsed-field gel electrophoresis (PFGE)9,14 could be used to determine the ploidy level and genome size. For example, flow cytometry was used to determine the genome size of four Panax species15 with Oryza sativa as the internal standard. P. notoginseng (San Qi in traditional Chinese medicine) has the largest genome (2454.38 Mb), followed by P. pseudoginseng (2432.72 Mb), P. vietnamensis (2018.02 Mb), and P. stipuleanatus (1947.06 Mb), but their genomes are smaller than the P. ginseng genome (~3.2 Gb). A more reliable approach for species identification without the reference genome is the genome survey via the whole-genome shotgun sequencing.16 Such non-deep sequencing (30×coverage), followed by the bioinformatics analysis, is highly valuable in assessing the genome size, heterozygosity, repeat sequence, GC content, etc, facilitating decision making on the whole-genome sequencing approaches. In addition, RAD-Seq (restriction-site associated DNA sequencing; Fig. 1)17 could be chosen to construct a RAD library and perform the low-coverage genome sequencing of reduced representation, which is an effective approach for assessing the heterozygosity of the candidate genome.
Figure 1.
Technology roadmap of RAD-Seq and its utility in population evolution and genetic map.
Abbreviations: PE, paired end; QC, quality control; InDel, insertion and deletion; SV, splice variant; PCA, principal component analysis; QTL, quantitative trait loci.
The whole-genome sequencing platform is chosen based on the budgetary resources and the preliminary evaluation of candidate genomes.2 The GS FLX or Illumina HiSeq 2500 platform might be suitable for a small, simple genome. However, the majority of the plant genomes are complex, which means they are diploid/polyploidy genomes with >50% repeat sequences and >0.5% heterozygosity. Two or more sequencing platforms could be combined for shotgun and paired-end sequencing, while large insert libraries, eg, BAC (bacterial artificial chromosome),9 YAC (yeast artificial chromosome),18 and Fosmid,14 can be constructed for sequencing; then a sophisticated bioinformatics software19–23 can be used for sequence quality control and assembly. For instance, GS FLX and shotgun sequencing can be used for the initial genome assembly to generate 454 contigs, and then the paired-end sequencing data from the Illumina HiSeq or SOLiD platform used to determine the order and orientation of 454 contigs, thus generating scaffolds. Next, Illumina HiSeq or SOLiD data are used to fill the gap between some contigs. These steps streamline the genome sequencing pipeline as a whole.
The genetic map and physical map are fundamental tools for the assembly of the complex plant genomes and functional genomics research. The genetic linkage map of Bupleurum chinense (Bei Chai Hu in traditional Chinese medicine, TCM) was constructed using 28 ISSR (inter-simple sequence repeat) and 44 SSR (microsatellite) markers24; 29 ISSRs and 170 SRAPs (sequence-related amplified polymorphisms) were mapped to 25 linkage groups of Siraitia grosvenorii (Luo Han Guo in TCM).25 These preliminary results are useful in metabolic gene mapping, map-based cloning, and marker-assisted selection of medicinal traits. The high-throughput physical map could be anchored via the BAC-pool sequencing,26 which, along with its integration with high-density genetic maps, could benefit from next-generation sequencing (NGS) and high-throughput array platforms.27 The development of dense genetic maps of medicinal plants is still challenging, as the parental lines and their progenies with the unambiguous genetic link are not available for most medicinal plants.
Chloroplast genome evolution
Chloroplast (cp) is responsible for photosynthesis, and its genome sequences have versatile utility in evolution, adaptation, and robust growth of most medicinal plants. The substitution rate of the cp nucleotide sequence is 3–4 times faster than that of the mitochondria (mt) sequence,5 implicating more uses of the former in inferring both interspecific and intraspecific evolutionary relationships.5,28–33
P. ginseng is a “crown” TCM plant and frequently used in health-promoting food and clinical therapy. NGS technology provides insight into the evolution and polymorphism of P. ginseng cp genome.5 The cp genome length of Chinese P. ginseng cultivars Damaya (DMY), Ermaya (EMY), and Gaolishen (GLS) was 156,354 bp, while it was 156,355 bp for wild ginseng (YSS), which are smaller than Omani lime (C. aurantiifolia;159,893 bp)29 and 12 Gossypium cp genomes (159,959–160,433 bp)32 but bigger than Rhazya stricta cp genome (154,841 bp).34 Gene content, GC content, and gene order in DMY are quite similar to those of other strains, and nucleotide sequence diversity of the inverted repeat region (IR) is lower than that of large single-copy region (LSC) and small single-copy region (SSC). The high-resolution reads were mapped to the genome sequences to investigate the differences of the minor allele, which showed that the cp genome attained heterogeneity during domestication; 208 minor allele sites with minor allele frequencies (MAFs) of ≥0.05 were identified. The polymorphism site numbers per kb of the cp genome of DMY, EMY, GLS, and YSS were 0.74, 0.59, 0.97, and 1.23, respectively. All minor allele sites were in the LSC and IR regions, and the four strains showed the same variation types (substitution base or indel) at all identified polymorphism sites. The minor allele sites of the cp genome underwent purifying selection to adapt to the changing environment during domestication. The study of the cp genome of medicinal plants with particular focus on minor allele sites would be valuable in probing the dynamics of the cp genomes and authenticating different strains and cultivars.
The genus Citrus contains many economically important fruits that are grown worldwide for their high nutritional and medicinal value. Due to frequent hybridizations among species and cultivars, the exact number of natural species and the evolutionary relationships within this genus are blurred. It is essential to compare the Citrus cp genomes and to develop suitable genetic markers for both basic research and practical use. A reference-assisted approach was adopted to assemble the complete cp genome of Omani lime,29 whose organization and gene content are similar to those of most rosids lineages characterized to date. By comparing with the sweet orange (C. sinensis), 3 intergenic regions and 94 SSRs were identified as potentially informative markers for resolving interspecific relationships, which can be harnessed to better understand the origin of domesticated Citrus and foster germplasm conservation. A comparison among 72 species belonging to 10 families of representative rosids lineages also provides new insights into their cp genome evolution.
The monocot family Orchidaceae, which is evolutionarily more ancient than asterids and rosids, is one of the largest angiosperm families, including many medicinal, horticultural, and ornamental species. Orchid phytometabolites display antinociceptive,35 antiangiogenic,36 and antimycobacterial37 activities, among others. In south Asia, orchid bulb is used for the treatment of asthma, bronchitis, throat infections, dermatological infections, and also as a blood purifier.38 Sequencing the complete cp genomes of the medicinal plant Dendrobium officinale (Tie Pi Shi Hu in TCM) and the ornamental orchid Cypripedium macranthos revealed their gene content and order, as well as potential RNA editing sites.39 The cp genomes of these two species and those of five known photosynthetic orchids are similar in structure as well as gene order and content, but the organization of the IR/SSC junction and ndh genes is distinct. IRs flanking the SSC region underwent expansion or contraction in different Orchidaceae species. Fifteen highly divergent protein-coding genes were identified, which are useful in phylogenetic inference of orchids. Phylogenomic analysis of cp can be used to resolve the interspecific relationship, which cannot be inferred by a few cp markers. Bamboo leaves are used as a component in TCM for the anti-inflammatory function.40 Medicinal bamboo cupping therapy is applied to reduce fibromyalgia symptoms.41 Bamboo extracts exhibit antioxidant effects42 and are used to treat chronic fever and infectious diseases.43 The whole cp genome datasets of 22 temperate bamboos considerably increased resolution along the backbone of tribe Arundinarieae (temperate woody bamboo) and afforded solid support for most relationships regardless of the very short internodes and long branches in the tree.33 An additional cp phylogenomic study, involving the full cp genome sequences of eight Olyreae (herbaceous bamboo) and 10 Arundinarieae species, strengthened the soundness of the above study and recovered monophyletic relationship between Bambuseae (tropical woody bamboo) and Olyreae.44
The monocot genus Fritillaria (Liliaceae) consists of nearly 140 species of bulbous perennial plants and includes the taxa of both horticultural and medicinal importance. The bulbs of plants belonging to the Fritillaria cirrhosa group have been used as antitussive and expectorant herbs in TCM for thousands of years.45 The anticancer activity and cardiovascular effects of Fritillaria phytometabolites are well documented.10 Fritillaria species have attracted attention also because of their remarkably large genome sizes, with all values recorded to date above 30 Gb.46 A phylogenetic reconstruction including most currently recognized species diversity of the genus was performed.46 Three regions of the cp genome were sequenced in 92 species (~66% of the genus) and in representatives of nine other genera of Liliaceae. Eleven low-copy nuclear genes were screened in selected species, but they had limited utility in phylogenetic reconstruction. Phylogenetic analysis of a combined plastid dataset supported the monophyly of the majority of presently identified subgenera. However, the subgenus Fritillaria, which is by far the largest and includes the most important species used in TCM, is found to be polyphyletic. Clade containing the source plants of Chuan Bei Mu, Hubei Bei Mu, and Anhui Bei Mu might be treated as a separate subgenus.47 The Japanese endemic subgenus Japonica, which contains the species with the largest recorded genome size for any diploid plant, is sister to the largely Middle Eastern and Central Asian subgenus Rhinopetalum, which is significantly incongruent with the nuclear ITS tree. Convergent or parallel evolution of phenotypic traits may be a common cause of incongruence between morphology-based classifications and the results of molecular phylogeny. While the relationships between most major Fritillaria lineages can be resolved, these results also highlight the need for data from more independently evolving loci, which is quite perplexing given the huge nuclear genomes found in these plants.
Medicinal plant diversity, comprising genetic diversity, medicinal species diversity, ecological system diversity, and so on,48 results from the intricate interactions between the plant and its environment, and thus is profoundly influenced by the ecological complex and the relevant versatile ecological processes. The effects of the evolutionary processes have to be taken into full consideration when explaining the link between climatic/ecological factors and medicinal plant diversity, especially that in a region where there is strong, uneven differentiation of species. A distinguished example is the “sky islands” of southwest China,49 where the extraordinarily rich resources of medicinal plants rose and thrived during the Quaternary Period. To date, many medicinal tribes and genera, eg, Pedicularis,50 Clematis,51 Aconitum,52,53 and Delphinium,54 are still in the process of rapid radiation and dynamic differentiation. The cp genome sequence can be regarded as the super-barcode of the organelle scale, and thus can be used to probe the intraspecific variations55 and phylogeographic patterns of the same species in disparate geographic locations (eg, geoherb or Daodi medicinal materials).56 The application of cp genome sequencing at the population level may provide clues for the timing and degree of intraspecific differentiation. Deriving the inter-population relationship from cp dataset can be considered as the more detailed phylogenetic reconstruction.
Mitochondria genome evolution
Some fundamental evolution concepts, such as lateral gene transfer, are bolstered by the inquiry of the origin of mt, while plants are especially useful in elucidating the mechanisms of cytonuclear coevolution. Although the gene order of the mt genome might evolve relatively fast in land plants, the substitution rate of its nucleotide sequence is merely 1/100 of that of animal sequence.48 Therefore, the mt genome sequence is less useful than the cp genome in inferring the phylogenetic relationship of medicinal species.57 Notwithstanding, analysis of the genome sequence is still able to contribute to the knowledge on the evolution of the mt genome. Moreover, the terpene synthase has been found in mt,58 highlighting its utility in secondary metabolism.
Rhazya stricta (Apocynaceae) is native to arid regions in South Asia and the Middle East and is used extensively in folk medicine. Analyses of the complete cp and mt genomes and a nuclear (nr) transcriptome of Rhazya shed light on intercompartmental transfers between genomes and the patterns of evolution among eight asterid mt genomes.34 The Rhazya genome is highly conserved, with gene content and order identical to the ancestral organization of angiosperms. The 548,608 bp mt genome contains recombination-derived repeats that generate a compound organization; transferred DNA from the cp and nr genomes, and bidirectional DNA transfers between the mt and the nucleus are also disclosed. The mt genes sdh3 and rps14 have been transferred to the nucleus and have acquired targeting transit peptides. Two copies of rps14 are present in the nucleus; only one has the mt targeting transit peptide and may be functional. Phylogenetic analyses suggest that Rhazya has experienced a single transfer of this gene to the nucleus, followed by a duplication event. The phylogenetic distribution of gene losses and the high level of sequence divergence in targeting transit peptides suggest multiple, independent transfers of both sdh3 and rps14 across asterids. Comparative analyses of mt genomes of eight asterids indicates a complicated evolutionary history in this thriving eudicot clade with substantial diversity in genome organization and size, repeat, gene and intron content, and the amount of alien DNA from the cp and nr genomes. The genomic data enable a rigorous inspection of the gene transfer events.
Nuclear genome evolution
The whole cp genome data-set is not enough to elucidate the phylogenetic relationship of groups undergoing rapid radiation, eg, Zingiberales.59 The cp genome is equivalent to one gene locus, thus it only represents one fulfillment to the coalescent random processes and cannot be used with confidence to reconstruct the evolution history of the populations. Most genetic history of any medicinal plant hides in the nr genome.
High-throughput sequencing and the relevant bioinformatics advances have revolutionized contemporary thinking on nuclear genome/transcriptome evolution and provided basic data for further breeding endeavor. Coix (Poaceae), a closely related genus of Sorghum and Zea, has 9–11 species with different ploidy levels. The exclusively cultivated C. lacrymajobi (2n = 20) is widely used in East and Southeast Asia as food and traditional medicine. C. aquatica has three fertile cytotypes (2n = 10, 20, and 40) and one sterile cytotype (2n = 30), C. aquatica HG, which is found in Guangxi, China.60 Low-coverage genome sequencing (genome survey) showed that ~76% of the C. lacrymajobi genome and 73% of the C. aquatica HG genome are repetitive sequences, among which the long terminal repeat (LTR) retrotransposable elements dominate, but the proportions of many repeat sequences vary greatly between the two species, suggesting their evolutionary divergence. A novel 102 bp variant of centromeric satellite repeat CentX and two other satellites are exclusively found in C. aquatica HG. Fluorescence in situ hybridization (FISH) analysis and fine karyotyping showed that C. lacrymajobi is likely a diploidized paleotetraploid species and C. aquatica HG is possibly from a recent hybridization. These Coix taxa share more coexisting repeat families and higher sequence similarity with Sorghum than with Zea, which agrees with the phylogenetic relationship.
Whole-genome sequencing has been implemented in the representative species of some plant families/genera (Fig. 2), eg, Capsicum annuum,19,20 Coffea canephora,21 Brassica napus,22 Phalaenopsis equestris,23 etc. The genome sequences of the cultivated pepper Zunla-1 (Capsicum annuum) and its wild progenitor Chiltepin (Capsicum annuum var. glabriusculum) were compared to provide insights into Capsicum domestication and specialization. The pepper genome expanded ~0.3 Mya by a rapid amplification of retrotransposon elements, resulting in a genome containing ~81% repetitive sequences and 34,476 protein-coding genes. Comparison of cultivated and wild pepper genomes with 20 resequencing accessions revealed molecular signature of artificial selection, providing a list of candidate domestication genes.19 Dosage compensation effect of tandem duplication genes might contribute to the pungency divergence in pepper.19 The Capsicum reference genome, along with tomato and potato genomes, provides critical information for the study of the evolution of other Solanaceae species, including the well-known Atropa medicinal plants.
Figure 2.
Examples of the phylogeny and genome duplication history of core eudicots.
Notes: Arrowheads indicate hexaploidization; triangles indicate tetraploidization. The current evidence does not suggest further polyploidization after speciation in the genomes of potato, eggplant, chili pepper, tobacco, coffee, grape, papaya, cacao, strawberry, and peach. Few genomic data are available in pepino, tomatillo, and many other species.
One of the milestone breakthroughs is the successful sequencing and assembly of the complex heterozygous genome. The heterozygous genome of C. canephora has been deciphered,21 which displays a conserved chromosomal gene order among asteroid angiosperms. Although it shows no sign of the whole-genome triplication identified in Solanaceae species, the genome includes several species-specific gene family expansions, eg, N-methyltransferases (NMTs) involved in caffeine biosynthesis, defense-related genes, and alkaloid and flavonoid enzymes involved in secondary metabolite production. Caffeine NMTs expanded through sequential tandem duplications independently and are distinct from those of cacao and tea, suggesting that caffeine in eudicots is of polyphyletic origin and its biosynthesis underwent convergent evolution. The heterozygous genome sequences of the tropical epiphytic orchid P. equestris provide insights into the unique crassulacean acid metabolism (CAM).23 The assembled genome contains 29,431 predicted protein-coding genes and is rich in genes that might be involved in self-incompatibility pathways, which ensures genetic diversity and enhances fitness and survival. An orchid-specific paleopolyploidy event is disclosed, which preceded the radiation of most orchid clades, and gene duplication might have contributed to the evolution of CAM photosynthesis in P. equestris. The expanded and diversified families of MADS-box C/D-class, B-class AP3, and AGL6-class genes might contribute to the highly specialized morphology of orchid flowers. LTRs are the most abundant transposable element (Fig. 3), followed by long interspersed elements (LINEs).
Figure 3.
Categories of transposable elements predicted in the orchid genome (according to Ref. 23).
Abbreviations: DNA, DNA transposon; LINE, long interspersed element (retrotransposon); SINE, short interspersed nuclear element (retrotransposon); LTR, long terminal repeat (retrotransposon).
More than 40 plant genomes have been sequenced, representing a diverse set of taxa of agricultural, energy, medicinal, and ecological importance.19–23 Gene family members are often inferred from DNA sequence homology, but deeper insights into evolutionary processes contributing to gene family dynamics are imperative. In a comparative genomics framework, multiple lines of evidence can be generated by gene synteny, sequence homology, and protein-based hidden Markov modeling (HMM) to extract homologous super-clusters composed of multi-domain resistance (R)-proteins of the NB-LRR (nucleotide binding-leucine rich repeat) type, which are involved in plant innate immunity.61 Twelve eudicot plant genomes were screened to assess the intra and interspecific diversity of R-proteins, where 2,363 NB-LRR genes were found. Half of the R-proteins have tandem duplicates, and 22% of gene copies are left from ancient polyploidy events (ohnologs, whole-genome duplication duplicates). The positive Darwinian selection and major differences in molecular evolution rates (Ka/Ks) were detected among tandem (mean = 1.59), ohnolog (1.36), and singleton (1.22) R-gene duplicates. The distribution pattern of all 140 NB-LRR genes present in the model plant Arabidopsis is species-specific, and four distinct clusters of NB-LRR “gatekeeper” loci sharing syntenic orthologues across all analyzed genomes were identified, which could be useful for the gene-edited plant breeding. The near-complete set of multidomain R-protein clusters in a eudicot-wide scale could shed light on the evolutionary dynamics underlying diversification of innate immune system of the plant. More functional NB-LRR genes could be identified from more sequenced plant species.
The estimated upper limit of extant plants is ~450,000, indicating the potentially enormous biological space. Multiple and recurrent genome duplications during plant evolution result in the generation of novel biosynthetic pathways of diverse medicinal compounds, which are frequently involved in plant defense and disease resistance, and, more importantly, create a huge chemical space for drug discovery and development. The duplicated gene copies could explain the diversification processes of the multigene secondary metabolism pathways, such as those involved in the biosynthesis of terpenoids,4 benzoxazinoids,62 steroidal glycoalkaloids,63 and glucosinolates.64 More than 200,000 secondary metabolites have been found in angiosperms, many of which could stem from the genome-duplication-based rapid innovation of the complex traits.48
Single-copy genes are common across angiosperm genomes. Based on 29 sufficiently high quality sequenced genomes, the large-scale identification and evolutionary characterization of single-copy genes from among multiple species is possible.65 A significant negative correlation was found between the number of duplicate blocks and the number of single-copy genes. Only 17% of the single-copy genes are located in organelles, most of which are involved in binding and catalytic activity. Most single-copy genes are in nuclear genomes. Single-copy genes have a stronger codon bias than non-single-copy genes in eudicots.65 The relatively high expression level of single-copy genes was partially confirmed by the RNA-Seq (transcriptome sequencing) data. Unlike in most other species, there is a strong negative correlation between Nc (effective number of codons) and GC3 (G+C content at third codon position) of single-copy genes in grass genomes. Compared to non-single-copy genes, single-copy genes are more conserved, as indicated by Ka and Ks values. Selective constraints on alternative splicing are weaker in single-copy genes than in low-copy family genes (1–10 paralogues) and stronger than high-copy family genes (>10 paralogues). Using concatenated, shared single-copy genes, a well-resolved phylogenetic tree can be obtained. Addition of intron sequences improved the branch support, but striking incongruences were also obvious. Inclusion of intron sequences might be more appropriate for the phylogenetic reconstruction at lower taxonomic levels. Evolutionary constraints between single-copy genes and non-single-copy genes are distinct, and are somewhat species-specific, especially between eudicots and monocots.
Transcriptome
The high cost of the whole-genome sequencing is still formidable. Accurate sequence assembly is still challenging, especially when the genome contains a high proportion of repeat sequences, high heterozygosity, and non-diploids.2 RNA-Seq is a powerful tool for the assessment of gene expression and the identification and characterization of molecular markers in non-model organisms.11 Unlike genome sequences, the intron sequences are not included in the RNA-Seq dataset, and the Unigene (contig) assembly is not disturbed by the repeat sequences and the ploidy level. A global view of the ethnomedicine resources and accurate delimitation of the novel medicinal taxa cannot be achieved without the molecular phylogeny based on the complete taxon sampling of the relevant tribes/genera. Because of the plummeting cost of RNA-Seq, dense taxon sampling is now possible in phylotran-scriptomic studies. It is obvious that large-scale comparative transcriptome studies, including those of medicinal plants, are more feasible than comparative genomics based on the whole-genome sequencing. As shown in the NCBI PubMed, SRA, and GEO databases, transcriptomes of hundreds of medicinal plants have been sequenced, eg, Caryophyllales,66 Fabaceae,67 Oenothera (Onagraceae),68 Rhodiola algida (Crassulaceae),69 Salvia sclarea (Lamiaceae),8 Polygonum cuspidatum (Polygonaceae),70 and Taxus mairei (Taxaceae).71 The single-copy orthologous gene sequences could be extracted from the Unigene datasets of multiple medicinal plants,70 which can be used in phylogenetic reconstruction and evolutionary analyses.66,70 The information uncovered in transcriptome studies could serve in the characterization of important traits related to secondary metabolite formation and for probing the relevant molecular mechanisms.8,69–71
Reconstructing the origin and evolution of land plants and their algal relatives is a vital problem in plant phylogenetics and is essential for understanding how novel adaptive traits, eg, secondary metabolites, arose. Despite advances in molecular systematics, some evolutionary relationships remain poorly resolved. Inferring deep phylogenies with rapid diversification is often tricky, and genome-scale data significantly increase the number of informative characters for analyses. Since sparse taxon sampling could result in inconsistent results, transcriptome data of 92 streptophyte taxa were generated and analyzed along with 11 published plant genome sequences.72 Phylogenetic reconstructions were conducted using 852 nuclear genes and 1,701,170 aligned sites. Robust support for a sister-group relationship between land plants and one streptophyte green algae, the Zygnematophyceae, was obtained. Strong and robust support for a clade comprising liverworts and mosses contradicts the widely accepted view of early land plant evolution. Phylogenetic hypotheses could be tested using the phylotranscriptomic approach to give deeper insights and novel arguments into the evolution of fundamental plant traits, including their fascinating chemodiversity.
Transcriptome sequencing also sheds light on other untapped issues of plant evolution. Arbuscular mycorrhizae (AM) are symbiotic systems in nature and have great significance in promoting the growth and stress resistance of medicinal plants.73 AM have multifaceted effects on the active ingredients of TCM plants. The transcriptomes of nine phylogenetically divergent non-AM symbiosis plants were analyzed to reveal the correlation between the loss of AM symbiosis and the loss of many symbiotic genes,74 which was found in four additional plant lineages besides the Arabidopsis lineage (Brassicales), implicating the convergent evolution. RNA-Seq was used to outline the gene sequence and expression discrepancy between cultivated tomato and five allied wild species.75 Human handling of the genome has profoundly altered the tomato transcriptome via directed admixture and by secondarily choosing nonsynonymous over synonymous substitutions. A hitherto unidentified paleopolyploidy event that arose 20–40 million years ago was uncovered based on the transcriptomes of 11 Linum species,76 which is specific to a clade enclosing cultivated flax (L. usitatissimum) and other mainly blue-flowered species.
Evolution and population genetics/genomics
SSRs play a major role as molecular markers for genome analysis and plant breeding. The microsatellites existing in the complete genome sequences would have a direct role in the genome organization, recombination, gene regulation, quantitative genetic variation, and evolution of genes. Microsatellite markers have been characterized for many medicinal plant families and genera, eg, Acanthaceae family,77 Artemisia genus,78 Camellia genus,79 and Chinese jujube.80 For instance, 11 nuclear SSR loci were used to reveal the relative low genetic diversity of three Camellia taliensis (Da Li tea) populations, three C. sinensis var. assamica (Pu Er tea in TCM) populations, and two transitional populations of C. taliensis. An important genetic differentiation was found between C. sinensis var. assamica and C. taliensis populations. The transitional populations of C. taliensis stemmed mainly from C. taliensis and underwent genetic differentiation during domestication. Gene introgression was spotted in the cultivated C. sinensis var. assamica and C. taliensis of the same tea garden, and genetic material of C. taliensis seemingly intruded into C. sinensis var. assamica, suggesting that the former was genetically involved in the domestication of the latter. These results are useful in protecting the genetic resources of ancient tea plants. The whole nucleotide sequences, eg, the genomic sequences9 or the transcriptomic sequences,8,11 of plant species can be obtained from NCBI databases and screened for the presence of SSRs.
Both ISSR81 and SRAP markers were suitable for discriminating among the studied individuals, but the SRAP markers were more efficient and preferable.78 Multiple regression analysis revealed statistically significant association between rust resistance and some molecular markers, which can provide clues for the identification of the individuals with higher rust resistance. RAPD (randomly amplified polymorphic DNA) and ISSR markers were used to characterize Schisandra chinensis with white fruit.82 The molecular-marker-based study of genetic diversity helps in assessing the studied germplasm, which would be a valuable genetic resource for future breeding. Based on such a study, in situ conservation measures or other methods could be recommended to preserve the valuable genetic resources of medicinal plants.
Sinopodophyllum hexandrum is an endangered Berberidaceae (Ranunculales) medicinal plant, and its genetic diversity must be protected against habitat loss and anthropogenic factors. The Qinling Mountains is a distribution area for S. hexandrum, where unique environmental features highly affect the evolution of the species. ISSR analysis of 32 natural populations revealed the genetic diversity and population structure of S. hexandrum in Qinling, and provided reference data for evolutionary and conservation studies.83 The 32 populations fell into three major groups, and analysis of their molecular variance confirmed significant variation among populations. The high genetic differentiation may be attributed to the limited gene flow within the species. The spatial pattern and geographic locations of different populations were not correlated. In light of the low within-population genetic diversity, high differentiation among populations, and the increasing anthropogenic pressure on the species, in situ conservation is proposed to preserve S. hexandrum in Qinling. Other populations must be sampled to maintain the genetic diversity of the species for ex situ preservation.
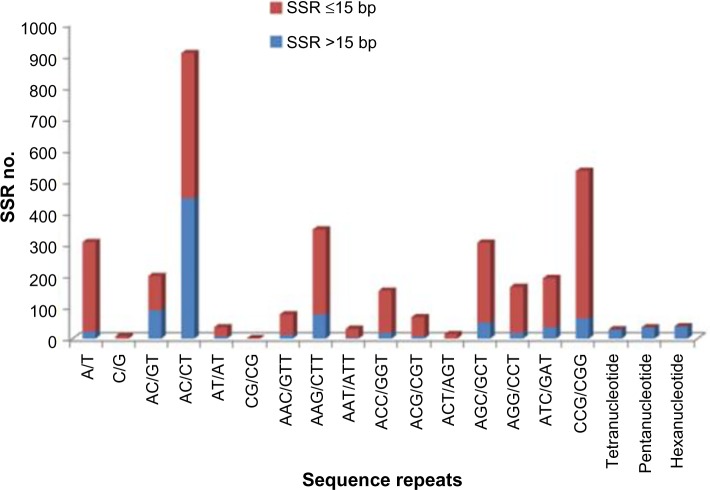
SNPs (single nucleotide polymorphisms) are much more abundant than SSRs in most species,84 including medicinal plants. The mutation rate of SNPs (10−9 per locus per generation) is much lower than that of SSRs (10−3–10−4).85 Generally, there are only two alleles in each SNP site, whereas there can be more than 10 alleles in each SSR. The highly polymorphic SSRs are especially suitable for detecting the hybridization between closely related species and studying the gene flow/introgression.86 SSRs are of lower ascertainment bias and are also good for studying the recent population structure. Mining suitable SSR sites via transcriptome sequencing datasets is fast and affordable. For example, 3,446 microsatellites were identified from 2718 Unigenes (16.8% of 16,142 assembled sequences) of the Salvia sclarea transcriptome (Fig. 4). Tri-nucleotide (1,883) is the predominant microsatellite, followed by dinucleotide (1,144) and mononucleotide (315), indicating that many microsatellites are in the translated regions of the expressed genes. CCG/CGG is the predominant trinucleotide SSR, followed by AAG/CTT and AGC/GCT. AG/CT is the most common dinucleotide SSR. Of the identified repeats, 601 (19.2%) have sufficient flanking sequence information to allow polymerase chain reaction (PCR) primer design. Intriguingly, many SSR motifs are linked with unique sequences encoding enzymes involved in phenylpropanoid/terpenoid metabolism. For instance, SSRs were detected in phenylalanine ammonia-lyase, 4-coumarate-CoA ligase, hydroxyphenylpyruvate dioxygenase, flavonoid 3′-hydroxylase, cinnamyl alcohol dehydrogenase, and lignan glycosyltransferase sequences, which belong to the phenylpropanoid pathway; SSRs were also found in 2-C-methyl-d-erythritol 4-phosphate pathway genes (1-deoxy-d-xylulose 5-phosphate synthase, 1-deoxy-d-xylulose 5-phosphate reductoisomerase, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase), mevalonate pathway genes (mevalonate pyrophosphate decarboxylase) and other terpenoid biosynthesis genes (isopentenyl diphosphate isomerase, cytochrome P450 71D18, pinene synthase, squalene synthase, squalene monooxygenase). These SSRs might be useful in future breeding and ecological studies. One of the major drawbacks of SSRs is the low universality and poor transferability; ie, usually the species-specific SSR primers have to be developed. The other disadvantage of SSRs is their uncertain mutation model, which is often simplified to be the stepwise mutation model, whereas the actual mutation pattern might be more complicated.
Figure 4.
SSRs predicted from the Salvia sclarea transcriptome dataset.8 Msatcommander (http://code.google.com/p/msatcommander/) was used to annotate SSRs. BatchPrimer387 was employed to design PCR primers in the flanking regions of the detected SSRs, setting a minimum product size of 100 bp, a minimum primer length of 18 bp, a minimum GC content of 30%, a melting temperature between 50 and 70 °C, and a maximum melting temperature difference between primers of 8 °C.
NGS toolkits could provide grist for the medicinal plant phylogeography mill. Sufficiently abundant SNPs could be identified directly from the genome sequences of the model plants. Most medicinal plants lack genomic data; therefore two alternative strategies can be adopted. The faster and cheaper one is mining suitable SNP sites via transcriptome sequencing datasets.11 However, the subsequent PCR primer design might not be successful, as no information about the intergenic sequences and the introns is available from the RNA-Seq data. On the other hand, large amounts of SNPs can be obtained by the simplified genome sequencing, mainly referring to RAD-Seq50 and genotyping-by-sequencing (GBS),88 although their reproducibility and reliability need further improvements.
Plants of various evolutionary levels, not only higher plants, are harnessed in TCM and ethnomedicine worldwide. The caterpillar fungus Ophiocordyceps sinensis (Dong Chong Xia Cao in TCM) is one of the most valuable medicinal fungi in the world, and host insects of family Hepialidae (Lepidoptera) are a must to complete its life cycle. The genetic diversity and phylogeographic structures of the host insects are characterized using mt COI (cytochrome oxidase subunit I) sequences.89 Abundant haplotype and nucleotide diversity were mainly found in the east edge of the Qinghai–Tibet Plateau (QTP), which is the diversity center or micro-refuges of the host insects. The genetic variation of the host insects is negligible among 72.1% of all O. sinensis populations. All host insects are monophyletic except for those of four O. sinensis populations around the Qinghai Lake. A significant phylogeographic structure was revealed for the monophyletic host insects, and the three major phylogenetic groups corresponded to specific geographical areas. The divergence of most host insects might have occurred at ~3.7 Ma, shortly before the rapid uplift of the QTP. The geographical distribution and star-like network of the haplotypes implied that most host insects were derived from the relicts of a once-widespread host that subsequently became fragmented. Most host insects underwent recent demographic expansions, which began ~0.118 Ma in the late Pleistocene, suggesting that the genetic diversity and distribution of the present-day insects could be ascribed to effects of the QTP uplift and glacial advance/retreat cycles during the Quaternary ice age. These results provide valuable reference to the conservation and sustainable use of both host insects and O. sinensis.
Population genetics can be upgraded to population genomics using the large dataset of transcriptomes from multiple species.68 The dearth of extant asexual species might be partially caused by the buildup of harmful mutations and intensified elimination risk linked with repressed recombination and segregation in these species, which was tested with a dataset of 62 transcriptomes of 29 Oenothera species.68 Non-synonymous polymorphism is more abundant than the synonymous variation within asexual species, implying relaxed purifying selection. Asexual species also displayed more transcripts with premature stop codons. The increased proportion of nonsynonymous mutations was positively associated with the divergence time between sexual and asexual species. These results suggest that sex enables selection against deleterious alleles.
Mechanisms of Species Evolution and Diversification
The incidence of polyploidy in land plant evolution has led to an acceleration of genome variations compared with other crown eukaryotes and is connected with key innovations in plant evolution.67 Increasing genome resources facilitate linking genomic alterations to the origins of novel phytochemical and physiological features of medicinal plants. Ancestral gene contents for key nodes of the plant family tree are inferred.90 The ancestral WGDs (whole-genome duplications) concentrating ~319 and 192 million years ago expedited the diversification of regulatory genes vital to seed and flower development, and were responsible for key innovations followed by the upsurge and ultimate supremacy of seed plants and flowering plants.1 Widespread polyploidy in angiosperms might be the major factor generating novel genes and expanding some gene families.64 However, most gene families lose most duplicated copies in a nearly neutral process, and a few families are actively selected for single-copy status. It is challenging to link genome modifications to speciation, diversification, and the phytochemical and/or physiological innovations that jointly comprise biodiversity and chemodiversity. Ongoing evolutionary genomics investigation may greatly improve the resolution, enabling the identification of specific genes responsible for particular innovations. More concise understanding of plant evolution may enrich fundamental knowledge of botanical diversity, including medicinally important traits that sustain humanity.
Case studies are important to illustrate the correlation between WGD and the diversification of secondary metabolism pathways. WGD and the tandem duplication facilitated glucosinolate pathway diversification in the mustard family (Brassicaceae) (Fig. 5).64 In Arabidopsis thaliana, at least 52 biosynthetic and regulatory genes are involved in the glucosinolate biosynthesis. Aethionema arabicum, basal to other Brassicales species, harbors 67 glucosinolate biosynthesis genes, most of which have the orthologue in A. thaliana, displaying the syntenic relationship. In A. thaliana, 45% of the protein-coding genes have more than one copy, while 95% of A. thaliana and 97% of Aethionema glucosinolate pathway genes possess multiple copies, suggesting the particular diversification of this defense pathway. Sequence alignment and phylogenetic analysis showed that the significant duplications of glucosinolate pathway genes occurred during the last common WGD event. The tandem duplication and the subsequent subfunctionalization and neofunctionalization further increase the genetic diversity and chemodiversity of the glucosinolate secondary metabolites, thus enhancing the phenotypic plasticity and adaptation. More importantly, the chemical space of the diverse secondary metabolites has great potential in drug discovery. The duplicated gene copies also explain the diversification process of terpenoids,4 the largest class of plant natural products. Tracing the roots of terpene biosynthesis and diversification in plants reveals that distinct genomic mechanisms of pathway assembly have been evolved in eudicots and monocots.
Figure 5.

Duplicate distribution among Arabidopsis thaliana (At) protein-coding genes compared with AtGS (glucosinolate) and Aethionema arabicum (Aab) GS loci, according to Ref. 64. The percentage of genes with retained ohnolog (clusters of dose-sensitive genes organized in functional modules), tandem duplicate (TD), and gene transposition duplicate (GTD) are shown. GS metabolic plasticity during lineage evolution arose from a combination of increased ohnolog retention and TD rates.
Besides polyploidy, allopatric divergence, climatic oscillation-based divergence, hybridization and introgression, and pollination-mediated isolation are also highlighted as the mechanisms of medicinal species evolution, especially in the hotspot area of biodiversity, such as QTP.91 Rapid species diversification followed the extensive uplift of QTP brought about numerous morphologically and phytochemically distinct species. Both morphological and metabolic phenotype innovations are apparently ecologically adaptive, and the underlying molecular mechanisms are still elusive.
Phenotype Evolution and Ecology
Medicinal plants synthesize an arsenal of protective molecules, most of which are secondary metabolites, can be ingested by animals and humans, and then help them antagonize against disadvantageous environmental conditions.92 The epidemiological (parasite prevalence and virulence) and environmental (medicinal plant toxicity and abundance) conditions that predict the evolution of genetically fixed versus phenotypically plastic forms of animal medication could be identified using the tritrophic interaction between the monarch butterfly, its protozoan parasite, and its food plant Asclepias spp. as a test case.93 Analogously, in folk medicine practice people have accumulated knowledge about the relative benefits (the antiparasitic/antimicrobial properties of medicinal plants) and costs (side effects of phytomedicine, the costs of searching for medicine) in ethnomedicine practice, which determine whether medication is for therapeutic use or preventive use.
Numerous botanical compounds, as the integral part of plant defense mechanisms, also bind and modify fundamental regulators of animal physiological processes in ways that enhance animal adaptation to the ever-changing environments.94 The underlying mechanism might be that animals and fungi, as heterotrophs, are capable of sensing chemical signals produced by plants and responding actively to the biotic/abiotic stress (xenohormesis).95 These plant-derived cues offer early warning about fading ecological circumstances, permitting the heterotrophs to get ready for misfortune when conditions are still favorable. Plant secondary metabolites could activate the evolutionarily conserved cellular stress response and subsequently enhance the cellular adaptation to adversity in both plants themselves and animals that consume them. Xenohormesis could explain TCM pharmacological effects from an evolutionary and ecological perspective.96 Medicinal herb, microbial, and human cellular signal transduction pathways have many conserved similarities, enabling beneficial effects of botanical metabolites in humans via a process of “cross-kingdom” signaling.94
Daodi medicinal material (geoherb) is produced in particular geographic regions, where there is defined ecological environment and cultivation pipeline.56 The clinical efficacy of a geoherb is superior to that of the same medicinal plant growing in other regions. The special medicinal features of a plant are determined by its genome, while the proper ecological conditions have major effects on the formation of a geoherb. For instance, Zhejiang, China, is the best production area of the geoherb Bai Shao (Paeonia lactiflora), where the paeoniflorin content of P. lactiflora roots was positively correlated with soil pH and rhizosphere bacterial diversity97 but negatively correlated with the organic matter content of the rhizosphere. The rhizosphere soil properties have a close relationship with the geoherbalism of F. thunbergii (Zhe Bei Mu in TCM)98 and Panax ginseng.99
The section Moutan of the genus Paeonia consists of eight species that are distributed in a particular area of China, and various secondary metabolites, including monoterpenoid glucosides, flavonoids, tannins, stilbenes, triterpenoids, steroids, paeonols, and phenols, have been found in these species. The metabolic phenotype evolves in the differentiated niche and in response to the plant–insect and plant–microbe interactions,97 which can be used for the chemotaxonomy of the section Moutan.100 Forty-three metabolites were identified from eight species by HPLC-Q-ToF MS (high-performance liquid chromatography-quadrupole time-of-flight-mass spectrometry), including 17 monoterpenoid glucosides, 11 galloyl glucoses, 5 flavonoids, 6 paeonols, and 4 phenols. PCA (principal component analysis) and HCA (hierarchical cluster analysis) showed a clear separation between the species based on metabolomic similarities, and four groups were identified, which is in good agreement with the conventional classification based on the morphology and geographical distributions. P. decomposita, from the geoherb production region Sichuan, China, was found to be a transition species between two subsections. According to the metabolic fingerprints, P. ostii (Feng Dan in TCM) and P. suffruticosa (Mu Dan) could be the same species. The metabolic profiles of P. delavayi (wild Mu Dan) were highly variable, and no significant difference was found between P. delavayi and P. ludlowii (yellow Mu Dan), implying that they either have a close evolutionary relationship or underwent the convergent evolution of the specialized metabolism. The combination of metabolomics and multivariate analyses has great potential for guiding chemotaxonomic studies of other medicinal plants.3
With the surge in NGS technology, it is becoming common to perform the phylogenetic study based on genomic data. However, for most medicinal plants it is not realistic to rely on the whole-genome sequencing data. RAD-Seq is easily applied to non-model plants for which no reference genome is available (Fig. 1),50 and it is promising for reconstructing phylogenetic relationships in evolutionarily younger clades in which sufficient numbers of orthologous restriction sites are retained across species.17 Coincidentally, the younger clades are more capable of harboring a wider variety of secondary metabolites, as chemodiversity often accompanies the rapid radiation and diversification. The evolutionarily young Pedicularis section Cyathophora is a systematically refractory clade of the broomrape family (Orobanchaceae). Phylogenetic inferences were performed based on the datasets of 40,000 RAD loci.50 The maximum likelihood and Bayesian methods generated similar trees that had two major clades: a “rex-thamnophila” clade, comprised of two species and several subspecies with relatively low floral diversity, and geographically widespread distributions at lower elevations; and a “superba” clade, consisting of three species with high floral diversity and isolated geographic distributions at higher altitudes. Levels of molecular divergence between subspecies in the rex-thamnophila clade are similar to those between species in the superba clade. The significant introgression among nearly all taxa in the rex-thamnophila clade was identified, while no gene flow was detected between clades or among taxa within the superba clade. The geographic isolation, following the uplift of QTP in the Quaternary Period, and the emergence of “sky islands”49 might be crucial in the advent of species barriers, by enabling local adaptation and differentiation without the influence of homogenizing gene flow. Pedicularis plants are traditionally used in folk medicine. It will be interesting to study its chemotaxonomy and treat the chemo-diversity and biodiversity data in a holistic approach for drug discovery and development.
Pharmacophylogeny vs Pharmacophylogenomics
Diverse new terms are emerging in the genomic era, such as phylogenomics, pharmacophylogenomics, and phylotranscriptomics, which are somewhat overlapping with pharmaphylogeny (pharmacophylogeny/pharmacophylogenetics).48 Phylogenomics is the crossing of evolutionary biology and genomics, in which genome data are utilized for evolutionary reconstructions. Pharmaphylogeny, advocated by Pei-gen Xiao since the 1980s,101,102 focuses on the phylogenetic relationship of medicinal plants and aims to foster the sustainable utilization of TCM resources, and is thus nurtured by molecular phylogeny, chemotaxonomy, ethnopharmacology, and bioactivity studies (Fig. 6). Phylogenomics can be integrated into the pipeline of drug discovery and development, and extends the field of pharmaphylogeny at the omic level; thus the concept of pharmacophylogenomics, initially emphasizing the genomic analysis of the evolutionary history of drug targets,103 could be redefined as an upgraded version of pharmaphylogeny.
Figure 6.

Omics data that could be used in the pharmacophylogeny inference.
Abbreviations: RAD, restriction site associated DNA; SNP, single nucleotide polymorphism; SSR, simple sequence repeat; EST, expressed sequence tag.
The new conceptual framework of pharmacophylogenomics highlights the comprehensive analysis of the evolutionary history of medicinal organisms (especially the predominant medicinal plants), in particular the congruence and conflict between molecular phylogeny and chemotaxonomy,46,47,51,52,100 the orthology and paralogy relationships,66,104 the degree and landscape of evolutionary transformation they have undergone, and the involved evolving metabolic pathways and regulatory networks. More specifically, first, the tree of life of different scales can be constructed based on the genomic information to determine the phylogenomic relationship of medicinal plant groups, eg, the relationship between the geoherb (higher content of medicinal compounds and better therapeutic efficacy)56 and non-geoherb populations; second, the genomic data, in particular those from the RAD-Seq or GBS, can be exploited to estimate the divergence time, reconstruct the geographic distribution, and infer the origin and the spatial distribution pattern of extant medicinal plants/geoherbs (Fig. 1)48,53; third, within the context of the temporal tree, the ecological factors, environmental attributes, and evolutionarily innovative traits can be combined to dissect the diversification process and mechanism of medicinal plants; fourth, the origin and structure of the phylogenetic diversity of medicinal plants could be revealed; fifth, the diversity of medicinal compounds could be dissected based on biodiversity to promote drug discovery via the high-throughput screening105; last but not least, the dynamic alteration of the medicinal plant diversity can be predicted, and then the appropriate conservation and development strategies can be put forward.
During evolution, plants develop tactics of chemical defenses, leading to the evolution of specialized metabolites with diverse potencies. A correlation between phylogeny and biosynthetic pathways could offer a predictive approach, enabling more efficient selection of alternative and/or complementary plants for guaranteeing clinical use and novel lead discovery. This relationship has been rigorously tested and the potential predictive power subsequently validated.106 A phylogenetic hypothesis was put forward for the medicinal plant subfamily Amaryllidoideae (Amaryllidaceae) based on parsimony and Bayesian analysis of nuclear, cp, and mt DNA sequences of over 100 species.106 It is interesting to test whether alkaloid diversity and activity in bioassays related to the central nervous system are significantly correlated with molecular phylogeny. Evidence for a significant phylogenetic signal in these traits has been found, but the effect is not strong. Several genera are non-monophyletic, highlighting the importance of using phylogeny for understanding character distribution. Lack of congruence between specialized metabolism and molecular phylogeny is not unusual,10,46,47,51,52,100 and the prominent factor is convergent evolution. Alkaloid diversity and in vitro inhibition of acetylcholinesterase and binding to the serotonin reuptake transporter are significantly correlated with phylogeny, illustrating the validity of pharmaphylogeny, which has implications for the use of molecular phylogenies to interpret chemical evolution and biosynthetic pathways, to select candidate taxa for lead discovery, and to make policies regarding therapeutic use and conservation priorities.
The correlation between the plant molecular phylogeny and therapeutic utility has been suggested.107–109 For instance, bulky, juicy leaves representative of medicinal aloes (Aloeaceae, Liliales) rose during the most recent expansion ~10 million years ago and are powerfully associated with the molecular phylogeny and correlated to the probability of a species being used for therapy.109 A noteworthy, though feeble, phylogenetic hint is apparent in the remedial uses of aloes, signifying that their pharmaceutical properties do not arise stochastically across the clades of the evolutionary tree. The taxonomic clades included in native pharmacopoeias are indeed associated with certain disease groups, and ecology and angiosperm phylogeny, which could be the alternative and/or complementary for chemical kinship and convergence, to a certain extent explain the observed preference of the therapeutic use. For instance, evolutionarily related plants from New Zealand, Nepal, and the Cape of South Africa are used to combat diseases of the same therapeutic spaces,108 which powerfully shows the self-determining discovery of the botanical value. A considerably greater fraction of recognized medicinal plants is present in these phylogenetic groups than in haphazard samples, suggesting that screening work should be focused on a subgroup of traditionally used plants that are more affluent in medicinal molecules. The phylogenetic/phylogenomic cross-cultural evaluations would invigorate the use of old-fashioned knowledge in bioprospecting. Statistical analysis of the ethnopharmacology data based on Chinese medicinal plants of Magnoliidae,110 Hamamelidae, and Caryophyllidae111 has been performed to summarize the distribution pattern of ethnomedicine uses across three subclasses. These nearly extinct traditional knowledge, collected nationwide during a TCM resources survey, lay the foundation for further quantitative correlation studies of molecular phylogeny and therapeutic efficacy.
Chinese medicinal material resource is the foundation of the development of TCM. In the study of sustainable utilization of TCM resource, adopting innovative theory and method to find new TCM resource is one of the hotspots and always highlighted.53 Pharmacophylogeny interrogates the phylogenetic relationship of medicinal organisms (especially medicinal plants), as well as the intrinsic correlation of morphological taxonomy, molecular phylogeny, chemical constituents, and therapeutic efficacy (ethnopharmacology and pharmacological activity). This new discipline may have the power to change the way we utilize medicinal plant resources and develop plant-based drugs. Phylogenomics can be integrated into the flowchart of drug discovery and development, and extends the field of pharmacophylogeny at the omic level. Analogously, phyloproteomics can be used in the proteome-based phylogeny study112; it can be used to examine the evolutionary relationship at the epigenomic level.113 phylometagenomics is also applicable in the exploration of medicinal plant-associated microbiota.114
Many medicinally important tribes and genera, such as Clematis,51 Pulsatilla, Anemone, Cimicifugeae,115 Nigella, Delphinieae,54 Adonideae, Aquilegia, Thalictrum,116 and Coptis, belong to the Ranunculaceae family. Chemical components of this family include several representative groups: benzylisoquinoline alkaloid, ranunculin, triterpenoid saponin, and diterpene alkaloid, etc. Ranunculin and magnoflorine were found to coexist in some genera. Other medicinal compounds also show some intriguing distribution patterns in 5 subfamilies and 10 tribes.105,117 Compared to other plant families, Ranunculaceae has the most species that are recorded in China Pharmacopoeia (CP) version 2010. However, many Ranunculaceae species, eg, those that are closely related to CP species, as well as those endemic to China, have not been investigated in depth,105 and their phylogenetic relationship and potential in medicinal use remain elusive. As such, it is proposed to select Ranunculaceae to exemplify the utility of pharmacophylogenomics and to elaborate the new concept empirically. It is argued that phylogenetic and evolutionary relationship of medicinally important tribes and genera within Ranunculaceae could be elucidated at the genomic, transcriptomic, and metabolomic levels, from which the intrinsic correlation between medicinal plant genotype and metabolic phenotype, and between genetic diversity and chemodivesity of closely related taxa, could be revealed. This proof-of-concept study would enrich the intention and spread the extension of pharmacophylogeny, promote the development of TCM genomics, and boost the sustainable development of Chinese medicinal plant resources.
Aconitum (Delphinieae, Ranunculaceae) has more than 300 species in the temperate regions of the Northern Hemisphere, over half of which are distributed in China. This genus has two subgenera, Lycoctonum and Aconitum (Fig. 7).48 Southwest China, particularly the Hengduan Mountains, is the most important center of origin and diversity of the genus. Many Aconitum species are used as poisonous and medicinal plants. Their anticancer activity, cardioactive effect, analgesic activity, anti-inflammatory activity, effect on energy metabolism, and antimicrobial and pesticidal activities, which are mainly due to the abundant diterpenoid alkaloids, are well archived.52 The correlation between molecular phylogeny, chemical components, and medicinal uses in Aconitum is notable.52,118 Diterpenoid alkaloids belong to four skeletal types: C18, C19, C20 and bisditerpenoid alkaloids. The subgenus Lycoctonum contains mainly C18 (lappaconine-type and ranaconine-type) and C19 (lycoctonine-type). Roots of the Lycoctonum plants exhibit relatively low toxicity and have been used to combat rheumatism, pains, irregular menstruation, and so on. This subgenus is worth a more detailed phytochemical investigation for new lead discovery and development. The Chinese taxa of section Aconitum (predominant in subgen. Aconitum) are morphologically divided into 11 series.
Figure 7.

Cladogram of the Ranunculaceae tribe Delphinieae, according to Refs 52, 119. Gymnaconitum and Staphisagria were regarded as the subgenus of Aconitum and Delphinium, respectively. Consolida, usually treated as an independent genus, could belong to the genus Delphinium. 19
Series Tangutica and Rotundifolia have abundant lactone-type C19-diterpenoid alkaloids,118 which can be considered as the chemical markers of these two series. The toxicity of their roots is much lower than those of series Bullatifolia and Brachypoda, and the whole plants are traditionally used in western China for high fever. The highly toxic aconitine-type diester C19 dominate in series Stylosa (Da Wu Tou in TCM). Series Ambigua contains mainly the aconitine-type C19 with anisoyloxy residues, indicating its close affinity to series Stylosa. Several species of series Volubilia have the highly advanced 15-hydroxyl aconitine-type C19, indicating their possible kinship to series Inflata, which harbors two most widely used TCM/CP aconite species A. carmichaeli (Wu Tou in TCM) and A. kusnezoffii (Bei Wu Tou in TCM). A. hemsleyanum of series Volubilia as well as many other Aconitum herbs are morphologically polymorphic and display substantial inter-populational phytochemical variation. Series Grandituberosa is more toxic than series Inflata, Volubilia, and Ambigua.
The morphology-based 11-series classification of section Aconitum, subgenus Aconitum, is not supported by chemotaxonomy and molecular phylogeny. Molecular phylogeny based on nr and cp DNA sequences divided the nine morphologically similar series into two clusters, which is supported by chemotaxonomic data. Series Rotundifolia and Brachypoda, as well as Tangutica and Bullatifolia, are not monophyletic groups and cluster together. Series Ambigua, Stylosa, Volubilia, and Inflata are also not monophyletic but are intermingled on the phylogenetic tree.10,52 Series Grandituberosa is closer to Volubilia than to other series. A. brunneum and A. racemulosum are distinct in both molecular phylogeny and chemotaxonomy. Gymnaconitum, previously regarded as a subgenus of Aconitum but distinct phytochemically from Aconitum, is between Aconitum and the genus Delphinium in molecular phylogeny (Fig. 7) and now treated as an independent genus.119 A high possibility of deriving novel chemical entities from untapped species in traditionally used drug-productive genera/families has been suggested.120 New genomic technologies that discover hidden gene clusters [4], pathways, and interspecific crosstalk allow the unearthing of innovative natural products.121 It is critical to assimilate the omic platforms into Aconitum studies for both the sustainable utilization of Aconitum pharmaceutical resources and finding novel compounds with potential clinical utility and less toxicity.
Conclusion and Prospects
The trend of integrating genomics and evolution into studies of medicinal plants is perceivable, and therefore it is time to summarize the current progress in the relevant fields in order to make full use of evolutionary biology/genomics and revolutionize the roadmap of medicinal plant inquiries. This review gave a brief analysis of the association and the distinguishing features of the multifaceted medicinal plant evolution and genomics studies, in the context of the plant-based drug discovery and the sustainable utilization of traditional pharmaceutical resources. A phylogenetic approach along with transcriptomics and other omics has value for understanding the evolution of medicinal plants, and a stronger case for the utility of these methods for future identification of useful genes and/or taxa for medicinal use is warranted.
The research paradigm of medicinal plant genome and evolution is evolving, and the use of omics techniques is reshaping the landscape of this dynamic field. Genomics, transcriptomics, proteomics, metabolomics, and other omics platforms generate formidably large data, which cannot be used efficiently in probing plant genome and evolution without the aid of advancing bioinformatics. Medicinal plants evolve new traits to adapt to the changing environments and pave the road toward a better life for themselves, while both hypothesis-driven and big data-driven studies integrate herbal technology, biotechnology, and information technology to pave the road toward a healthier life for humans.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,675 words, excluding any confidential comments to the academic editor.
FUNDING: This work is supported by the Scientific Research Foundation for ROCS, Ministry of Education, China, and Natural science fund of Liaoning Province. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived the review: DCH, PGX. Analyzed data and wrote the manuscript: DCH. Provided some reference information and edited the manuscript: PGX. Both authors reviewed and approved the final manuscript.
REFERENCES
- 1.Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 2.Chen SL, Sun YZ, Xu J, et al. Strategies of the study on herb genome program. Yao Xue Xue Bao. 2010;45(7):807–12. [PubMed] [Google Scholar]
- 3.Hao DC, Xiao PG, Ge GB, Liu M. Biological, chemical, and omics researchof Taxus medicinal resources. Drug Dev Res. 2012;73:477–86. [Google Scholar]
- 4.Boutanaev AM, Moses T, Zi J, et al. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci U S A. 2015;112(1):E81–8. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Yin J, Guo H, et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci. 2015;5:696. doi: 10.3389/fpls.2014.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Luo H, Li Y, et al. 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep. 2011;30(9):1593–601. doi: 10.1007/s00299-011-1070-6. [DOI] [PubMed] [Google Scholar]
- 7.Moses T, Pollier J, Shen Q, et al. OSC2 and CYP716A14v2 catalyze the biosynthesis of triterpenoids for the cuticle of aerial organs of Artemisia annua. Plant Cell. 2015;27(1):286–301. doi: 10.1105/tpc.114.134486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao da C, Chen SL, Osbourn A, Kontogianni VG, Liu LW, Jordán MJ. Temporal transcriptome changes induced by methyl jasmonate in Salvia sclarea. Gene. 2015;558(1):41–53. doi: 10.1016/j.gene.2014.12.043. [DOI] [PubMed] [Google Scholar]
- 9.Hao DC, Vautrin S, Song C, et al. The first insight into the Salvia (Lamiaceae) genome via BAC library construction and high-throughput sequencing of target BAC clones. Pak J Bot. 2015;47(4):1347–57. [Google Scholar]
- 10.Hao DC, Gu XJ, Xiao PG. Medicinal Plants: Chemistry, Biology and Omics. 1st ed. Oxford: Elsevier-Woodhead; 2015. [Google Scholar]
- 11.Hao DC, Chen SL, Xiao PG, Liu M. Application of high-throughput sequencingin medicinal plant transcriptome studies. Drug Dev Res. 2012;73:487–98. [Google Scholar]
- 12.Yao X, Peng Y, Xu LJ, Li L, Wu QL, Xiao PG. Phytochemical and biological studies of Lycium medicinal plants. Chem Biodivers. 2011;8(6):976–1010. doi: 10.1002/cbdv.201000018. [DOI] [PubMed] [Google Scholar]
- 13.Hao DC, Xiao PG, Peng Y, Dong J, Liu W. Evaluation of the chloroplast barcoding markers by mean and smallest interspecific distances. Pak J Bot. 2012;44(4):1271–4. [Google Scholar]
- 14.Hao DC, Yang L, Xiao PG. The first insight into the Taxus genome via fosmid library construction and end sequencing. Mol Genet Genomics. 2011;285(3):197–205. doi: 10.1007/s00438-010-0598-4. [DOI] [PubMed] [Google Scholar]
- 15.Pan YZ, Zhang YC, Gong X, Li FS. Estimation of genome size of four Panax species by flow cytometry. Plant Diversity Resour. 2014;36(2):233–6. [Google Scholar]
- 16.Polashock J, Zelzion E, Fajardo D, et al. The American cranberry: first insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 2014;14:165. doi: 10.1186/1471-2229-14-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin BE, Ree RH, Moreau CS. Inferring phylogenies from RAD sequence data. PLoS One. 2012;7(4):e33394. doi: 10.1371/journal.pone.0033394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noskov VN, Chuang RY, Gibson DG, Leem SH, Larionov V, Kouprina N. Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat Protoc. 2011;6(1):89–96. doi: 10.1038/nprot.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin C, Yu C, Shen Y, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci U S A. 2014;111(14):5135–40. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Park M, Yeom SI, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46(3):270–8. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 21.Denoeud F, Carretero-Paulet L, Dereeper A, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–4. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 22.Chalhoub B, Denoeud F, Liu S, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–3. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 23.Cai J, Liu X, Vanneste K, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 2015;47(1):65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 24.Zhan QQ, Sui C, Wei JH, Fan SC, Zhang J. Construction of genetic linkage map of Bupleurum chinense DC. using ISSR and SSR markers. Yao Xue Xue Bao. 2010;45(4):517–23. [PubMed] [Google Scholar]
- 25.Liu L, Ma X, Wei J, Qin J, Mo C. The first genetic linkage map of Luohanguo (Siraitia grosvenorii) based on ISSR and SRAP markers. Genome. 2011;54(1):19–25. doi: 10.1139/G10-084. [DOI] [PubMed] [Google Scholar]
- 26.Cviková K, Cattonaro F, Alaux M, et al. High-throughput physical map anchoring via BAC-pool sequencing. BMC Plant Biol. 2015;15(1):99. doi: 10.1186/s12870-015-0429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ariyadasa R, Stein N. Advances in BAC-based physical mapping and map integration strategies in plants. J Biomed Biotechnol. 2012;2012:184854. doi: 10.1155/2012/184854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian J, Song J, Gao H, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS One. 2013;8(2):e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su HJ, Hogenhout SA, Al-Sadi AM, Kuo CH. Complete chloroplast genome sequence of Omani lime (Citrus aurantiifolia) and comparative analysis within the rosids. PLoS One. 2014;9(11):e113049. doi: 10.1371/journal.pone.0113049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu CS, Chaw SM, Huang YY. Chloroplast phylogenomics indicates that Ginkgo biloba is sister to cycads. Genome Biol Evol. 2013;5:243–54. doi: 10.1093/gbe/evt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malé PJ, Bardon L, Besnard G, et al. Phylogenomics and a posteriori data partitioning resolve the cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci U S A. 2012;109:17519–24. doi: 10.1073/pnas.1205818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Q, Xiong G, Li P, et al. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. PLoS One. 2012;7(8):e37128. doi: 10.1371/journal.pone.0037128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma PF, Zhang YX, Zeng CX, Guo ZH, Li DZ. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (poaceae) Syst Biol. 2014;63(6):933–50. doi: 10.1093/sysbio/syu054. [DOI] [PubMed] [Google Scholar]
- 34.Park S, Ruhlman TA, Sabir JS, et al. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution acrossasterids. BMC Genomics. 2014;15:405. doi: 10.1186/1471-2164-15-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales-Sánchez V, Rivero-Cruz I, Laguna-Hernández G, Salazar-Chávez G, Mata R. Chemical composition, potential toxicity, and quality control procedures of the crude drug of Cyrtopodium macrobulbon. J Ethnopharmacol. 2014;154(3):790–7. doi: 10.1016/j.jep.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Basavarajappa HD, Lee B, Fei X, et al. Synthesis and mechanistic studies of a novel homoisoflavanone inhibitor of endothelial cell growth. PLoS One. 2014;9(4):e95694. doi: 10.1371/journal.pone.0095694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponnuchamy S, Kanchithalaivan S, Ranjith Kumar R, Ali MA, Choon TS. Antimycobacterial evaluation of novel hybrid arylidene thiazolidine-2,4-diones. Bioorg Med Chem Lett. 2014;24(4):1089–93. doi: 10.1016/j.bmcl.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Nagananda GS, Satishchandra N. Antimicrobial activity of cold and hot successive pseudobulb extracts of Flickingeria nodosa (Dalz.) Seidenf. Pak J Biol Sci. 2013;16(20):1189–93. doi: 10.3923/pjbs.2013.1189.1193. [DOI] [PubMed] [Google Scholar]
- 39.Luo J, Hou BW, Niu ZT, Liu W, Xue QY, Ding XY. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS One. 2014;9(6):e99016. doi: 10.1371/journal.pone.0099016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koide CL, Collier AC, Berry MJ, Panee J. The effect of bamboo extract on hepatic biotransforming enzymes – findings from an obese-diabetic mouse model. J Ethnopharmacol. 2011;133(1):37–45. doi: 10.1016/j.jep.2010.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao H, Hu H, Colagiuri B, Liu J. Medicinal cupping therapy in 30 patients with fibromyalgia: a case series observation. Forsch Komplementmed. 2011;18(3):122–6. doi: 10.1159/000329329. [DOI] [PubMed] [Google Scholar]
- 42.Jiao J, Lü G, Liu X, Zhu H, Zhang Y. Reduction of blood lead levels in lead-exposed mice by dietary supplements and natural antioxidants. J Sci Food Agric. 2011;91(3):485–91. doi: 10.1002/jsfa.4210. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Yue YD, Tang F, Sun J. Screening and analysis of the potential bioactive components in rabbit plasma after oral administration of hot-water extracts from leaves of Bambusa textilis McClure. Molecules. 2012;17(8):8872–85. doi: 10.3390/molecules17088872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wysocki WP, Clark LG, Attigala L, Ruiz-Sanchez E, Duvall MR. Evolution of the bamboos (Bambusoideae; Poaceae): a full plastome phylogenomic analysis. BMC Evol Biol. 2015;15:50. doi: 10.1186/s12862-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu K, Mo C, Xiao H, Jiang Y, Ye B, Wang S. Imperialine and verticinone from bulbs of Fritillaria wabuensis inhibit pro-inflammatory mediators in LPS- stimulated RAW 264.7 macrophages. Planta Med. 2015;81(10):821–9. doi: 10.1055/s-0035-1546170. [DOI] [PubMed] [Google Scholar]
- 46.Day PD, Berger M, Hill L, et al. Evolutionary relationships in the medicinally important genus Fritillaria L. (Liliaceae) Mol Phylogenet Evol. 2014;80:11–9. doi: 10.1016/j.ympev.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 47.Hao DC, Gu XJ, Xiao PG, Peng Y. Phytochemical and biological research of Fritillaria medicine resources. Chin J Nat Med. 2013;11(4):330–44. doi: 10.1016/S1875-5364(13)60050-3. [DOI] [PubMed] [Google Scholar]
- 48.Hao DC, Xiao PG, Liu M, Peng Y, He CN. Pharmaphylogeny vs. pharmacophylogenomics: molecular phylogeny, evolution and drug discovery. Yao Xue Xue Bao. 2014;49(10):1387–94. [PubMed] [Google Scholar]
- 49.He K, Jiang XL. Sky islands of southwest China. I. An overview of phylogeographic patterns. Chin Sci Bull. 2014;59:585–97. [Google Scholar]
- 50.Eaton DA, Ree RH. Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae) Syst Biol. 2013;62(5):689–706. doi: 10.1093/sysbio/syt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao DC, Gu XJ, Xiao PG. Chemical and biological research of Clematis medicinal resources. Chin Sci Bull. 2013;58:1120–9. [Google Scholar]
- 52.Hao DC, Gu XJ, Xiao PG. Recent advances in the chemical and biological studies of Aconitum pharmaceutical resources. J Chin Pharm Sci. 2013;22(3):209–21. [Google Scholar]
- 53.Hao DC, Xiao PG, Liu LW, et al. Essentials of pharmacophylogeny: knowledge pedigree, epistemology and paradigm shift. China J Chin Mat Med. 2015;40(13):1–8. [PubMed] [Google Scholar]
- 54.Jabbour F, Renner SS. A phylogeny of Delphinieae (Ranunculaceae) shows that Aconitum is nested within Delphinium and that late miocene transitions to long life cycles in the Himalayas and Southwest China coincide with bursts in diversification. Mol Phylogenet Evol. 2012;62(3):928–42. doi: 10.1016/j.ympev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Whittall JB, Syring J, Parks M, et al. Finding a (pine) needle in a haystack: chloroplast genome sequence divergence in rare and widespread pines. Mol Ecol. 2010;19(S1):100–14. doi: 10.1111/j.1365-294X.2009.04474.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhao ZZ, Guo P, Brand E. The formation of daodi medicinal materials. J Ethnopharmacol. 2012;140(3):476–81. doi: 10.1016/j.jep.2012.01.048. [DOI] [PubMed] [Google Scholar]
- 57.Henriquez CL, Arias T, Pires JC, Croat TB, Schaal BA. Phylogenomics of the plant family Araceae. Mol Phylogenet Evol. 2014;75:91–102. doi: 10.1016/j.ympev.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Hsu CY, Huang PL, Chen CM, Mao CT, Chaw SM. Tangy scent in Toona sinensis (Meliaceae) leaflets: isolation, functional characterization, and regulation of TsTPS1 and TsTPS2, two key terpene synthase genes in the biosynthesis of the scent compound. Curr Pharm Biotechnol. 2012;13(15):2721–32. doi: 10.2174/138920112804724864. [DOI] [PubMed] [Google Scholar]
- 59.Barrett CF, Specht CD, Leebens-Mack J, Stevenson DW, Zomlefer WB, Davis JI. Resolving ancient radiations: can complete plastid gene sets elucidate deep relationships among the tropical gingers (Zingiberales) [J]? Ann Bot. 2014;113:119–33. doi: 10.1093/aob/mct264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Z, Liu H, He Q, et al. Differential genome evolution and speciation of Coix lacryma-jobi L. and Coix aquatica Roxb. hybrid guangxi revealed by repetitive sequence analysis and fine karyotyping. BMC Genomics. 2014;15:1025. doi: 10.1186/1471-2164-15-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hofberger JA, Zhou B, Tang H, Jones JD, Schranz ME. A novel approach for multi-domain and multi-gene family identification provides insights into evolutionary dynamics of disease resistance genes in core eudicot plants. BMC Genomics. 2014;15:966. doi: 10.1186/1471-2164-15-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dutartre L, Hilliou F, Feyereisen R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: gene duplications and origin of the Bx cluster. BMC Evol Biol. 2012;12:64. doi: 10.1186/1471-2148-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manrique-Carpintero NC, Tokuhisa JG, Ginzberg I, Holliday JA, Veilleux RE. Sequence diversity in coding regions of candidate genes in the glycoalkaloid biosynthetic pathway of wild potato species. G 3 (Bethesda) 2013;3:1467–79. doi: 10.1534/g3.113.007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hofberger JA, Lyons E, Edger PP, Chris Pires J, Eric Schranz M. Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol Evol. 2013;5:2155–73. doi: 10.1093/gbe/evt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han FM, Peng Y, Xu L, Xiao PG. Identification, characterization, and utilization of single copy genes in 29 angiosperm genomes. BMC Genomics. 2014;15:504. doi: 10.1186/1471-2164-15-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y, Moore MJ, Brockington SF, et al. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Mol Biol Evol. 2015:pii–msv081. doi: 10.1093/molbev/msv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cannon SB, McKain MR, Harkess A, et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 2015;32(1):193–210. doi: 10.1093/molbev/msu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hollister JD, Greiner S, Wang W, et al. Recurrent loss of sex is associated with accumulation of deleterious mutations in oenothera. Mol Biol Evol. 2015;32(4):896–905. doi: 10.1093/molbev/msu345. [DOI] [PubMed] [Google Scholar]
- 69.Zhang F, Gao Q, Khan G, Luo K, Chen S. Comparative transcriptome analysis of aboveground and underground tissues of Rhodiola algida, an important ethno-medicinal herb endemic to the Qinghai-Tibetan Plateau. Gene. 2014;553(2):90–7. doi: 10.1016/j.gene.2014.09.063. [DOI] [PubMed] [Google Scholar]
- 70.Hao D, Ma P, Mu J, et al. De novo characterization of the root transcriptome of a traditional Chinese medicinal plant Polygonum cuspidatum. Sci China Life Sci. 2012;55(5):452–66. doi: 10.1007/s11427-012-4319-6. [DOI] [PubMed] [Google Scholar]
- 71.Hao da C, Ge G, Xiao P, Zhang Y, Yang L. The first insight into the tissue specific taxus transcriptome via illumina second generation sequencing. PLoS One. 2011;6(6):e21220. doi: 10.1371/journal.pone.0021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickett NJ, Mirarab S, Nguyen N, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 2014;111(45):E4859–68. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng Y, Guo LP, Chen BD, et al. Arbuscular mycorrhizal symbiosis for sustainable cultivation of Chinese medicinal plants: a promising research direction. Am J Chin Med. 2013;41(6):1199–221. doi: 10.1142/S0192415X1350081X. [DOI] [PubMed] [Google Scholar]
- 74.Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 2014;10(7):e1004487. doi: 10.1371/journal.pgen.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koenig D, Jiménez-Gómez JM, Kimura S, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci U S A. 2013;110(28):E2655–62. doi: 10.1073/pnas.1309606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sveinsson S, McDill J, Wong GK, et al. Phylogenetic pinpointing of a paleopolyploidy event within the flax genus (Linum) using transcriptomics. Ann Bot. 2014;113(5):753–61. doi: 10.1093/aob/mct306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaliswamy P, Vellingiri S, Nathan B, Selvaraj S. Microsatellite analysis in the genome of Acanthaceae: an in silico approach. Pharmacogn Mag. 2015;11(41):152–6. doi: 10.4103/0973-1296.149731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karimi A, Hadian J, Farzaneh M, Khadivi-Khub A. Evaluation of genetic variability, rust resistance and marker-detection in cultivated Artemisia dracunculus from Iran. Gene. 2015;554(2):224–32. doi: 10.1016/j.gene.2014.10.057. [DOI] [PubMed] [Google Scholar]
- 79.Li MM, Kasun M, Yan L, et al. Genetic involvement of Camellia taliensis in the domestication of C. sinensis var. assamica (assimica tea) revealed by nuclear microsatellite markers. Plant Diversity Resour. 2015;37(1):29–37. [Google Scholar]
- 80.Wang S, Liu Y, Ma L, et al. Isolation and characterization of microsatellite markers and analysis of genetic diversity in Chinese jujube (Ziziphus jujuba Mill.) PLoS One. 2014;9(6):e99842. doi: 10.1371/journal.pone.0099842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hao DC, Chen SL, Xiao PG, Peng Y. Authentication of medicinal plants by DNA-based markers and genomics. Chin Herb Med. 2010;2(4):250–61. [Google Scholar]
- 82.Li XK, Wang B, Zheng YC, Song X, Wu YN, Chen L. Molecular characters of Schisandra chinensis with white fruit by RAPD and ISSR makers. Zhong Yao Cai. 2014;37(4):568–72. [PubMed] [Google Scholar]
- 83.Liu W, Yin D, Liu J, Li N. Genetic diversity and structure of Sinopodophyllum hexandrum (Royle) Ying in the Qinling Mountains, China. PLoS One. 2014;9(10):e110500. doi: 10.1371/journal.pone.0110500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clevenger J, Chavarro C, Pearl SA, Ozias-Akins P, Jackson SA. Single nucleotide polymorphism identification in polyploids: a review, example, and recommendations. Mol Plant. 2015;8(6):831–46. doi: 10.1016/j.molp.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 85.Guichoux E, Lagache L, Wagner S, et al. Current trends in microsatellite genotyping. Mol Ecol Resour. 2011;11(4):591–611. doi: 10.1111/j.1755-0998.2011.03014.x. [DOI] [PubMed] [Google Scholar]
- 86.Wee AK, Takayama K, Chua JL, et al. Genetic differentiation and phylogeography of partially sympatric species complex Rhizophora mucronata Lam. and R. stylosa Griff. using SSR markers. BMC Evol Biol. 2015;15:57. doi: 10.1186/s12862-015-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.You FM, Huo N, Gu YQ, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He J, Zhao X, Laroche A, Lu ZX, Liu H, Li Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. 2014;5:484. doi: 10.3389/fpls.2014.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quan QM, Chen LL, Wang X, et al. Genetic diversity and distribution patterns of host insects of caterpillar fungus Ophiocordyceps sinensis in the Qinghai-Tibet Plateau. PLoS One. 2014;9(3):e92293. doi: 10.1371/journal.pone.0092293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiao Y, Paterson AH. Polyploidy-associated genome modifications during land plant evolution. Philos Trans R Soc Lond B Biol Sci. 2014;369(1648):0355. doi: 10.1098/rstb.2013.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front Genet. 2014;5:4. doi: 10.3389/fgene.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sternberg ED, de Roode JC, Hunter MD. Trans-generational parasite protection associated with paternal diet. J Anim Ecol. 2015;84(1):310–21. doi: 10.1111/1365-2656.12289. [DOI] [PubMed] [Google Scholar]
- 93.Choisy M, de Roode JC. The ecology and evolution of animal medication: genetically fixed response versus phenotypic plasticity. Am Nat. 2014;184(S1):S31–46. doi: 10.1086/676928. [DOI] [PubMed] [Google Scholar]
- 94.Kennedy DO. Polyphenols and the human brain: plant “secondary metabolite” ecologic roles and endogenous signaling functions drive benefits. Adv Nutr. 2014;5(5):515–33. doi: 10.3945/an.114.006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howitz KT, Sinclair DA. Xenohormesis: sensing the chemical cues of other species. Cell. 2008;133(3):387–91. doi: 10.1016/j.cell.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi HY, Li L, Yu J. Xenohormesis: understanding biological effects of traditional Chinese medicine from an evolutionary and ecological perspective. Zhongguo Zhong Yao Za Zhi. 2013;38(19):3388–94. [PubMed] [Google Scholar]
- 97.Yuan XF, Peng SM, Wang BL, Ding ZS. Effects of growth years of Paeonia lactiflora on bacterial community in rhizosphere soil and paeoniflorin content. Zhongguo Zhong Yao Za Zhi. 2014;39(15):2886–92. [PubMed] [Google Scholar]
- 98.Shi JY, Yuan XF, Lin HR, Yang YQ, Li ZY. Differences in soil properties and bacterial communities between the rhizosphere and bulk soil and among different production areas of the medicinal plant Fritillaria thunbergii. Int J Mol Sci. 2011;12(6):3770–85. doi: 10.3390/ijms12063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ying YX, Ding WL, Li Y. Characterization of soil bacterial communities in rhizospheric and nonrhizospheric soil of Panax ginseng. Biochem Genet. 2012;50(11–12):848–59. doi: 10.1007/s10528-012-9525-1. [DOI] [PubMed] [Google Scholar]
- 100.He C, Peng B, Dan Y, Peng Y, Xiao P. Chemical taxonomy of tree peony species from China based on root cortex metabolic fingerprinting. Phytochemistry. 2014;107:69–79. doi: 10.1016/j.phytochem.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 101.Xiao PG. A preliminary study of the correlation between phylogeny, chemical constituents and pharmaceutical aspects in the taxa of Chinese Ranunculaceae. Acta Phytotaxo Sin. 1980;18(2):142–53. [PubMed] [Google Scholar]
- 102.Peng Y, Chen SB, Liu Y, Chen SL, Xiao PG. A pharmacophylogenetic study of the Berberidaceae (s.l.) Acta Phytotaxo Sin. 2006;44(3):241–57. [Google Scholar]
- 103.Searls DB. Pharmacophylogenomics: genes, evolution and drug targets. Nat Rev Drug Discov. 2003;2(8):613–23. doi: 10.1038/nrd1152. [DOI] [PubMed] [Google Scholar]
- 104.Yang Y, Smith SA. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol Biol Evol. 2014;31(11):3081–92. doi: 10.1093/molbev/msu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hao DC, Xiao PG, Ma HY, Peng Y, He CN. Mining chemodiversity from biodiversity: pharmacophylogeny of medicinal plants of the Ranunculaceae. Chin J Nat Med. 2015;13(7):507–20. doi: 10.1016/S1875-5364(15)30045-5. [DOI] [PubMed] [Google Scholar]
- 106.Rønsted N, Symonds MR, Birkholm T, et al. Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol Biol. 2012;12:182. doi: 10.1186/1471-2148-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leonti M, Cabras S, Castellanos ME, Challenger A, Gertsch J, Casu L. Bioprospecting: evolutionary implications from a post-olmec pharmacopoeia and the relevance of widespread taxa. J Ethnopharmacol. 2013;147(1):92–107. doi: 10.1016/j.jep.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 108.Saslis-Lagoudakis CH, Savolainen V, Williamson EM, et al. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc Natl Acad Sci U S A. 2012;109(39):15835–40. doi: 10.1073/pnas.1202242109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grace OM, Buerki S, Symonds MR, et al. Evolutionary history and leaf succulence as explanations for medicinal use in aloes and the global popularity of Aloe vera. BMC Evol Biol. 2015;15:29. doi: 10.1186/s12862-015-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao PG, Wang LW, Lv SJ. Statistical analysis of the ethnopharmacologic data based on Chinese medicinal plants by electronic computer I. Magnoliidae. Zhong Xi Yi Jie He Za Zhi. 1986;6(4):253–6. [PubMed] [Google Scholar]
- 111.Xiao PG, Wang LW, Qiu GS, Sun J. Statistical analysis of the ethnopharmacologic data based on Chinese medicinal plants by electronic computer II. Hamamelidaean d Caryophyllidae. Zhong Xi Yi Jie He Za Zhi. 1989;9(7):429–32. [PubMed] [Google Scholar]
- 112.Villar M, Popara M, Mangold AJ, de la Fuente J. Comparative proteomics for the characterization of the most relevant Amblyomma tick species as vectors of zoonotic pathogens worldwide. J Proteomics. 2013;105:204–16. doi: 10.1016/j.jprot.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 113.Martin DI, Singer M, Dhahbi J, et al. Phyloepigenomic comparison of great apes reveals a correlation between somatic and germline methylation states. Genome Res. 2011;21:2049–57. doi: 10.1101/gr.122721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brindefalk B, Ettema TJ, Viklund J, Thollesson M, Andersson SG. A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS One. 2011;6:e24457. doi: 10.1371/journal.pone.0024457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hao DC, Gu XJ, Xiao PG. Recent advance in chemical and biological studies on Cimicifugeae pharmaceutical resources. Chin Herb Med. 2013;5(2):81–95. [Google Scholar]
- 116.Zhu M, Xiao PG. Chemosystematic studies on Thalictrum L. in China. Acta Phytotaxon Sin. 1991;29(4):358–69. [Google Scholar]
- 117.Wang W, Lu AM, Ren Y, Endressc ME, Chen ZD. Phylogeny and classification of Ranunculales evidence from four molecular loci and morphological data. Perspect Plant Ecol Evol Systemat. 2009;11:81–110. [Google Scholar]
- 118.Xiao PG, Wang FP, Gao F, et al. A pharmacophylogenetic study of Aconitum L. (Ranunculaceae) from China. Acta Phytotaxon Sin. 2006;44(1):1–46. [Google Scholar]
- 119.Wang W, Liu Y, Yu SX, Gao T-G, Chen ZD. Gymnaconitum, a new genus of Ranunculaceae endemic to the Qinghai-Tibetan Plateau. Taxon. 2013;62:713–22. [Google Scholar]
- 120.Zhu F, Ma XH, Qin C, et al. Drug discovery prospect from untapped species: indications from approved natural product drugs. PLoS One. 2012;7(7):e39782. doi: 10.1371/journal.pone.0039782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu F, Qin C, Tao L, et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Natl Acad Sci U S A. 2011;108(31):12943–8. doi: 10.1073/pnas.1107336108. [DOI] [PMC free article] [PubMed] [Google Scholar]