Abstract
Circuit mapping requires knowledge of both structural and functional connectivity between cells. While optical tools have been made to assess either the morphology and projections of neurons or their activity and functional connections, few probes integrate this information. We have generated a family of photoactivatable Genetically Encoded Ca2+ Indicators (pa-GECIs) that combines attributes of high-contrast photo-labeling with high-sensitivity Ca2+ detection in a single-color, protein-sensor. We demonstrate the utility of pa-GECIs in cultured neurons and in vivo in Drosophila and zebrafish larvae. We show how single cells can be selected out of dense populations for Golgi-like visualization of morphology and high signal-to-noise measurements of activity, synaptic transmission and connectivity. Our design strategy is readily transferrable to other sensors based on circularly permutated GFP (cpGFP).
Introduction
An important part of understanding brain function lies in deciphering how individual neurons integrate synaptic inputs to function together in circuits. This requires detailed information of structural and functional connectivity. Since Golgi staining was first discovered to fill sparsely distributed neurons, enabling morphological tracing and physical connectivity to be assessed by contemporaries like Ramόn y Cajal1, structural mapping of neuronal circuits has progressed with development of optical highlighting tools2-8. In particular, photoactivatable fluorescent proteins (PA-FP) have provided a non-invasive way to highlight individual cells in situ in live tissues, facilitating tracking fine neuronal processes with a clarity that cannot be achieved with dense expression of standard FPs9-13. Genetically Encoded Ca2+ Indicators (GECIs14-16) have, likewise, revolutionized modern neurobiology by enabling non-invasive, rapid measurements of changes in intracellular Ca2+, an important correlate of neuronal activity and excitatory synaptic transmission17,18. It would be therefore a major benefit for mapping of neuronal circuits with a single protein reporter with the dual capability of photo-activated morphological highlighting and reporting Ca2+ activity, which could also be genetically targeted.
Here, we present a set of novel photoactivatable GECIs (pa-GECIs) that combine attributes of high-contrast in situ highlighting with high-sensitivity Ca2+ detection. Out of the GECI superfamily19-22, we focused on the commonly used GCaMP indicators because they have single emission color, express efficiently in a large number of preparations and have high sensitivity16,23-25. We arrive at a rational design in which mutations of a small set of key residues in the FP domain confers high contrast photoactivation onto GCaMPs with retention of Ca2+ sensitivity. Our pa-GECIs enable resolution of Ca2+ activity in processes and dendritic spines in individual neurons, selected in situ in culture and in vivo. We show this engineering strategy to be transferable among different GCaMP versions and related GECIs, suggesting that this method will be applicable to future generations of GECIs and potentially to other sensors, based on circularly permuted GFP20,26-29.
Results
Engineering photoactivatable GCaMPs
We designed a photoactivatable GECI based on the structure and function of the photoactivatable green fluorescent protein (PA-GFP)11,30 (see Supplementary Notes). Briefly, we rationally introduced mutations within the fluorescent protein of various GCaMPs to yield superfolder photoactivatable GCaMPs (spa-GCaMPs) (Fig. 1a) that show strong, near-UV-dependent photoactivation and large Ca2+ responses in several cell lines, primary cultured hippocampal neurons and cultured glia (Supplementary Figs. 1-4 and Video 1). From in vitro measurements, we found that spa-GCaMP6f has an apparent Ca2+ affinity (Kd of ~650 nM) similar to the parent, GCaMP6f-WT23 (Supplementary Fig. 5).
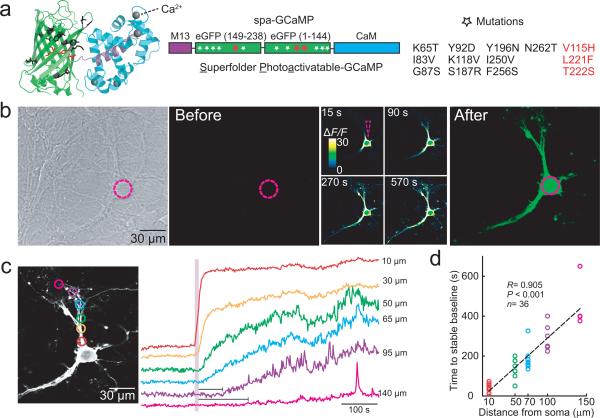
Figure 1. Structure and activity highlighted simultaneously by spa-GCaMP6f in cultured hippocampal neurons.
a, Cartoon (left, PDB: 3SG4) and schematic of spa-GCaMP (middle), consisting of M13 (violet), a circularly permutated GFP (cpGFP, green) and calmodulin (CaM, cyan), indicating three necessary and sufficient mutations for photoactivation (red stars) and superfolding (white stars), denoted on the right. Note that all mutations are within the cpGFP, leaving the Ca2+ transduction domains intact. b, Representative transmitted-light (left) and 3D fluorescence projection micrographs showing densely cultured neurons expressing spa-GCaMP6f, before and after photoactivation by 405 nm illumination of the soma (dashed violet circle)(n>50). (middle) Single plane, time-lapse, color-coded fluorescence micrographs after photoactivation, normalized to the basal fluorescence at 0 s (see Online Methods). c, Dendritic highlighting following somatic photoactivation occurs rapidly in proximal, thick processes and more slowly, following a delay (horizontal bars below traces) in distal, thinner processes (distances are relative to center of soma.) d, Summary of distance-dependent filling, fitted with a linear regression (Pearson correlation, P<0.001, n = 36).
spa-GCaMP6 highlights morphology and reports on Ca2+ activity
We assessed the functionality of the spa-GCaMP6 family in cultured hippocampal neurons. Prior to photoactivation, neurons expressing spa-GCaMP6 variants exhibit very little fluorescence when imaged with 488 nm laser excitation (Fig. 1b, Supplementary Videos 2-4). To identify cells that express spa-GCaMP, we used brief, low intensity illumination at 405 nm (5 μW, <0.64 μs dwell/pixel) that generated sufficient green emission for visualization but, importantly, caused no persistent photoactivation (Supplementary Figure 6). Higher intensity 405 nm illumination (2.1 mW, 0.64-1 μs dwell/pixel) on selected soma induced large and persistent increases in somatic fluorescence that spread throughout the cell within minutes (Fig. 1b). As observed with other cytosolic photoactivatable FPs12,31, spa-GCaMP6f highlighted neuronal processes by diffusion, with distant processes displaying a short delay in filling (Fig. 1c, d). Repeated bouts of photoactivation caused no deleterious effect on cell health, as measured by membrane potential or evoked currents (Supplementary Figure 7a, b).
We next assessed the sensitivity with which spa-GCaMP6 variants detect action potentials (APs) by combining loose-patching and confocal imaging. Electrical and fluorescent signals simultaneously recorded from cultured neurons expressing either spa-GCaMP6f or spa-GCaMP6s showed that fluorescence transients closely followed APs (Fig. 2a, b), with larger ΔF/Fs in spa-GCaMP6s (Fig. 2c). Single APs were also detected by both spa-GCaMP6f and spa-GCaMP6s (Fig. 2a-c), but with signals that were 4-5 fold smaller than the parent GCaMPs, consistent with differences in basal brightness (Supplementary Figure 7c-e).
Figure 2. spa-GCaMP6s and spa-GCaMP6f report on action potentials and local calcium in dendritic spines of cultured hippocampal neurons.
a-b, APs recorded in loose patch (bottom black traces) are reliably followed by fluorescence transients in spa-GCaMP6s (red) and spa-GcaMP6f (cyan) expressing cells. Number of action potentials per burst noted above fluorescence traces. c, Summary of ΔF/F of responses to different numbers of APs (n; spa-GCaMP6s (red)= 18, 8, 8, 5, 7, 12, spa-GCaMP6f (cyan)-9, 3, 4, 3, 4, 10; *, p<0.05, ± s.e.m., one way ANOVA). d-e, Calcium imaging in individual dendritic spine heads and shafts following photoactivation of spa-GCaMP6f. d, Highlighting of small compartments and individual spines following somatic photoactivation (as shown in Fig. 1). Prior to photoactivation (before), activity is undetectable, but becomes increasingly apparent once filling occurs (after violet bar). e, (top image and traces) A spine with a thin, long (2.5 μm) neck exhibits Ca2+ activity in the head (green arrowhead and corresponding trace) that is irregularly associated with Ca2+ activity in the shaft (magenta arrowhead and trace; black arrowheads atop traces denote concurrent activity in head and shaft compartments). (bottom image and traces) A stubby spine with a fat, short (0.4 μm) neck, exhibits a high correlation between Ca2+ activity in head and shaft. f, Inverse dependence of spine neck-length and the cross-correlation factor (CCF) of Ca2+ activity in head and shaft. Spines in (e) represented by magenta-green inverted triangle and square, respectively. (Pearson correlation, n = 10 cells, n = 35 spines, dashed red line, linear fit).
Somatic photoactivation in hippocampal neurons enabled APs to be detected continuously for over 30 minutes, and importantly, continuous imaging with 488 nm for such extended periods of time did not itself induce further photoactivation (Supplementary Figures 8a-c). Dendritic processes filled with photoactivated spa-GCaMP6f displayed large Ca2+-transients (Fig. 2d), substantially larger than the somatic responses; spa-GCaMP6s- soma: 1.84 ± 0.26 (±s.e.m., n = 12) vs. processes: 5.22 ± 0.95 ( n = 9) (Supplementary Figure 8b), as previously reported for the parent GCaMPs32,33. spa-GCaMP6f filling of dendritic spines was bright enough to visualize fine structure and provide a robust readout of Ca2+ activity in spine heads (ΔF/F = 3.7 ± 1.5, n = 35) (Fig. 2e). As observed with conventional calcium indicators34,35, spines with shorter (typically thicker) necks displayed a considerably higher coupling to the shaft (i.e. higher cross correlation factor [CCF]) than mushroom-type spines (neck length ~1-2.5 μm) or filopodia (>2.5 μm), which typically had longer, thinner necks (Fig. 2f and Supplementary Video 5). Highlighted spines could be imaged at high speed (line-scans at 1 kHz) with minimal signal loss or increase in baseline fluorescence (Supplementary Figure 9a). Centripetal highlighting (from processes to soma) could also be achieved by photoactivation in distal dendrites that filled the dendrite, secondary dendrites and soma, where Ca2+ activity could then be imaged (Supplementary Figure 9b-d). Importantly, photoactivation in dendrites and dendritic spines did not affect spontaneous synaptic transmission (Supplementary Figure 10).
Finally, we found that 2-photon (2P) photoactivation was efficiently achieved with 750-76036 nm excitation (Supplementary Figure 11a-c). Imaging at 940 nm37 did not produce photoactivation in cultured neurons or astrocytes, as seen by the stable baseline fluorescence (Supplementary Figs. 11c-f). Detecting activity in soma, but also in small structures was also achievable using 2P imaging (following 2P photoactivation), such as synaptic contacts (Supplementary Figure 11g-j). These results show that spa-GCaMPs are photostable, efficiently photoactivated (1 or 2P) without affecting cell health, and subsequently sensitive enough to report on neuronal activity for extended periods without additional photoactivation by the imaging laser (either 1P or 2P imaging).
Mapping cellular and synaptic connectivity
Efforts to map functional connections and analyze dendritic computations are often hampered by the difficulty of tracing morphology, and detecting activity, in individual processes under dense GECI expression38. On the other hand, sparse expression limits the likelihood of finding connected cells. To model these challenges, we transfected high density dissociated neurons to easily find multiple spa-GCaMP6-expressing cells in the field of view of a 20x objective. To note, aside using low intensity 405 nm illumination for cell detection, because spa-GCaMP displays weak, albeit detectable, basal fluorescence (i.e. prior photoactivation), this feature could also be used to detect highly expressing cells. Not only did this enable us to identify cells, it also permitted the detection of neuronal activity and connectivity among cells (Supplementary Figure 12).
To map synaptic connectivity, we photoactivated neurons sequentially, waiting 2-10 minutes for each cell to fill before the next cell was photoactivated (Fig. 3a, b). In this way, we traced the morphology of each cell independently and pseudo-colored it to identify sites of apparent contact where synapses might be located (Fig. 3c). We then imaged Ca2+ activity in the full field of view. APs, recorded in loose-patch or whole-cell mode, were consistently followed by large Ca2+ transients in the soma and proximal axon, which then spread into dendrites (Fig. 2 and Supplementary Figures 12c and 13f-g). These somatic Ca2+ transients in one cell were often associated with local Ca2+ transients at dendritic sites of other highlighted cells at locations of contact with the axon of the first cell (Fig. 3c), suggesting functional synapses. These were visualized as activity correlation maps, and revealed functional connections with a wide range of correlation strengths, from very reliable contacts with an almost 1:1 correspondence, and large contact area, to smaller contacts with weak correlation (Fig. 3d, e and Supplementary Figure 13, but also additional examples in Supplementary Figure 14 and Video 11). These experiments demonstrate the potential of spa-GCaMP6 for functional circuit mapping in dense preparations.
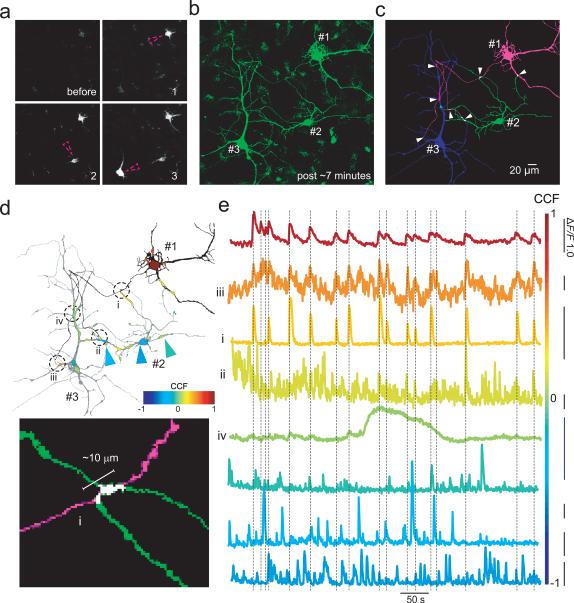
Figure 3. Functional highlighting of functionally connected cultured hippocampal neurons.
a, Time lapse images of sequential somatic photoactivation (violet circle) of three closely situated sspa-GCaMP6f expressing neurons (1, 2 and 3) with ~2-5 minutes apart between photoactivation (n = 7). b, 3D projection image showing filling of the three neurons 7 min after photoactivation of cell #3. c, Sequential photoactivation enables neurons 1-3 to be traced with high fidelity, then color-coded to help identify crossing points between processes (white arrowheads) that could form potential synaptic contacts. d, (top) Black and white image (as shown in c) overlaid with a cross-correlation map (see Online Methods). Selected points of contact shown in c (arrowheads) are underscored by dashed circles and roman numerals. (bottom) A high power image of a potential contact between the long and thin axon-like process of neuron #1 (magenta) and a thick dendritic segment of neuron #2 (region “i”, green), spanning over 10 μm. e, Time resolved fluorescent traces of Ca2+ activity (color coded based on the correlation map shown in d as well as enumerated with roman numerals as shown in d) for the different points of contact (size of region on correlation map reflects the region of interest from which fluorescence was collected. Hot colors (yellow to red) indicate strong correlated activity with neuron #1 (top, dark red trace), whereas cold colors (light green to blue) indicate less of a correlation. Bottom three traces depict regions (arrowheads in d) outside the selected regions in neuron #2.
spa-GCaMPs in Drosophila in vivo
We next tested functional highlighting in vivo in Drosophila melanogaster. We generated multiple transgenic lines with spa-GCaMP6f or sspa-GCaMP6f inserted in various chromosomes (Supplementary Figure 15) under the control of the Gal4-UAS expression system39. First, we focused on the larval ventral nerve cord (VNC) of third instar larvae expressing UAS-spa-GCaMP6f under the control of the motor neuron-specific Gal4, OK6-Gal4 (ref. 40)(Fig. 4a). Illumination of the VNC (Fig. 4b), triggered robust photoactivation, yielding bright fluorescently fills of somata and fine processes (ΔF/F= 8.5 ± 0.6, n = 27, 4 larvae, Supplementary Figure 15b), which diffused out of the region of photoactivation (Supplementary Video 8). Spontaneous, widespread and directional Ca2+ activity was frequently observed in the VNC, as observed when using conventional Ca2+ indicators41,42 (Supplementary Figure 15c). Antidromic stimulation of motor neuron axons evoked large fluorescence signals by spa-GCaMP6f, that were larger in neuropil than in somata (Fig. 4c, d), with similar evoked activity patterns as observed for GCaMP6f-WT23 (Supplementary Figure 15d). The similarities between the patterns of activity further suggest that the photoactivation process did not damage or change the activity of the neurons (Supplementary Fig. 15d).
Figure 4. In vivo highlighting and imaging neural activity in Drosophila.
a, Cartoon and transmitted-light micrograph of Drosophila larval nervous system, including the ventral nerve cord (VNC), with motor nerves extending to muscle and stimulation electrode. b, 3D projection images of VNC (outlined) from a transgenic larva expressing spa-GCaMP6f in motor neurons, before (left) and 5 minutes after (right) photoactivation (dashed pink region). Note the spread of fluorescence outside the region of photoactivation. c, Photoactivation of a large posterior region of the VNC enables imaging structure and activity of individual somata and neuropil. (right) Color-coded traces of time resolved fluorescence intensity following antidromic electrical stimulation (1 s at 20 Hz, every 20 s) yields large fluorescence transients from neuropil and individual somata, summarized in d (±s.e.m., n = 3 animals per group, t-test, P<0.05). Fluorescence baseline and transients are stable over 300 seconds of chronic imaging. e, Schematic of the in vivo experimental imaging set-up of adult Drosophila flies where female flies are mounted and head cuticle is opened to reveal the brain. A glass capillary filled with 2 M sucrose is placed next to the extended proboscis, from which sucrose is presented. f, Contrast images showing the subesophogeal zone (SEZ) region of a transgenic fly expressing spa-GCaMP6f in sweet taste cells (Gr5a-Gal4>UAS-spa-GCaMP6f) before (left, 0 s) and after (right, 150 s) 2P-photoactivation (red dashed area). g, (top trace) 2 M Sucrose, but not 100 mM denatonium (bottom trace), repeatedly elicits responses in nerve terminals following photoactivation. h, Summary of first responses to 2 M sucrose (n = 3 females flies, 5 SEZs).
Backward highlighting from processes to soma was also possible with photoactivation in the peripheral nerve (n = 5 animals). Photoactivation at the presynaptic motor neuron terminals of the neuromuscular junction allowed detection of presynaptic Ca2+ transients following motor nerve stimulation (Supplementary Figure 16a-c). Photoactivation at the axon terminal did not induce a detectable morphological change, or affect muscle membrane potential or resistance, EPSP or miniature EPSP amplitude, or miniature EPSP frequency (Supplementary Figure 16d-i).
As in cultured cells, 2P photoactivation was also achieved using 2P illumination of single motor neurons in the VNC (Supplementary Figure 17a). Ca2+ responses to electrical stimulation could then be imaged by 1- or 2P excitation (at 940 nm) without further photoactivation (Supplementary Figure 17b-e), indicating that pa-GCaMP is fully compatible with 2P microscopy, and that neither 1P imaging of the pa-GECI at 488 nm, nor 2P imaging at 940 nm, causes substantial photoactivation (Supplementary Figures 11, 12 and 17).
To test spa-GCaMP6 in vivo in a light scattering, dense tissue, we turned to brain imaging of transgenic adult flies. Flies were mounted and the head cuticle was opened to reveal the brain43 (Fig. 4e; Supplementary Figure 18a). 2P photoactivation of spa-GCaMP6f in the antennal lobe (AL) and mushroom body (MB) was efficient (Supplementary Figure 18b, c). To image Ca2+ activity in response to a physiological stimulus, we focused on the gustatory nerve axons and terminals in the subesophogeal zone (SEZ) by crossing UAS-spa-GCaMP6f flies with Gr5a-Gal4 flies to drive expression only in sweet sensory cells13. We photoactivated the axon terminals at the SEZ with 2P illumination at 760 nm light (600-700 ms bouts at 5-7 mW/pixel), at depths of 25-50 μm and imaged with 488 nm (Fig. 4f). Application of sucrose to the proboscis, but not of the bitter-tastant denatonium, triggered robust, progressively desensitizing Ca2+ transients in the highlighted axon terminals (Fig. 4f-h). Following more than 30 minutes, we observed that the fluorescence signal spread from axon endings towards the somata in the proboscis that enabled tracing parts of these neuronal projections (n = 3)(Supplementary Figure 18d). Ca2+ transients measured with photoactivated spa-GCaMP6f were similar in shape and kinetics to those obtained with the parent GCaMP, GCaMP6f-WT (Supplementary Figure 18e, f), although they were ~5-fold smaller in amplitude (Supplementary Figure 18g). To further test the ability to confine photoactivation to individual cells in a very dense preparation, we turned to image Kenyon cells in the mushroom body of the Drosophila larva. To identify individual cells amongst the hundreds of cells located at the Calyx44, we expressed a nuclear red marker (nls-mCherry), which allowed us to target and photoactivate individual cells by 760 nm (Supplementary Figure 18h). Together, these observationsdemonstrate that spa-GCaMP6f can provide an in vivo readout of neuronal activity as well as highlighting from optically selected cells in the scattering medium of the young and adult Drosophila brain, and is compatible with two color imaging.
pa-GCaMPs in zebrafish in vivo
To test in vivo functional highlighting in a model vertebrate, we generated a UAS-sspa-GCaMP6f construct to inject into embryos of zebrafish expressing Gal4. In a motor neuron selective Gal4 , 1P photoactivation of somata was sufficient to highlight axon terminals located 180 ± 20 μm (n = 7) away within 10 minutes (Fig. 5a). At approximately 30 hours post-fertilization (hpf), highlighted motor neurons showed large spontaneous fluorescent activity (Fig. 5b), typical of previously described spontaneous activity45,46. In an enhancer trap Gal4 line broadly targeting retinal cells, 2P photoactivation of single Müller glia cells yielded an increase in basal fluorescence by 7-fold, which highlighted distal regions of the cell within 5 minutes (Fig. 5c and Supplementary Figure 19a), and revealed hotspots of fast Ca2+ activity (Supplementary Video 9), as well as fine structural features (Fig. 5d; Supplementary Video 10), similar to observations with conventional indicators47. Photoactivated cells remained visible for long periods of time; for instance, muscle cells photoactivated by 405 nm have 72 ± 8% change in basal fluorescence over 24 hr period (n = 3 animals). Photoactivation was also compatible with two-color in vivo imaging when UAS-sspa-GCaMP6f was co-expressed with a membrane-bound version of the red fluorescent protein KillerRed48: brief bouts of 405 nm light (rastering 2.1 mW, 1.8 s, 0.64 μs dwell/pixel) sufficient to photoactivate sspa-GCaMP6f did not affect the fluorescence a red fluorescent protein (Supplementary Figure 19b, c).
Figure 5. In vivo highlighting and imaging neural and glial activity in larval zebrafish.
a, 3D projection images of photoactivated sspa-GCaMP6f, transiently expressed in zebrafish (UAS-sspa-GCaMP6f injected into mnx-Gal4 embryo), in spinal cord motor neurons in 3 dpf larva before (left) and after (right) photoactivation of the soma (pink circle, which fills the axon and its bifurcations; inset-arrowheads) (n =7). b, Spontaneous activity recorded from the soma of another motor neuron following photoactivation. c, 2P photoactivation of retinal Müller glial cell (UAS-sspa-GCaMP6f injected into 1003-Gal4 embryo) highlighted the structure spanning the retinal layers in (GCL, gangion cell layer; IPL, inner plexiform layer; INL, Inner nuclear layer; ONL, outer nuclear layer) (n = 3). (left) 3D projection image of highlighted Müller cell ~10 minutes after 2P photoactivation. (right) Higher power image of the stalk of the cell showing three regions of interest (arrowheads). d, Compartmentalized Ca2+ activity from regions shown in c. e, 2P photoactivation in retinal ganglion cell axonal arbor in 5 dpf larva. Single plane fluorescent micrographs showing gradual filling of an axon arbor (1 - 20 min) that was repeatedly photoactivated at a single axon terminal (dashed red triangles) using 760 nm. i, 3D projection of highlighted arbor 30 minutes after initial photoactivation. ii, After 1 hr, the arbor has comparable brightness using same imaging settings. iii, After 4 hr, the arbor is half as bright, but still visible with increased imaging power (iv). f, Axon arbor remodeling is highlighted by diffusion of photoactivated molecules. Overlay (black) of arbors imaged at 30 minutes (green) and 4 hrs (magenta) post-photoactivation showed both pruning and growth of axonal branches.
To test whether pa-GECIs would be suitable for chronic imaging in vivo, we photoactivated sspa-GCaMP6f in retinal ganglion cell axons in the tectum at 5 days post-fertilization (dpf). 2P photoactivation at individual axon terminals (5 mW/pixel, 1 sec every 10 frames) yielded robust local photoactivation in the illuminated area (~20-fold). Photoactivated molecules spread to highlight additional branches in the same axonal arbor within 30 minutes (Fig. 5e). The highlighting was long lasting enough to enable chronic imaging to track the dynamic remodeling of immature axonal branches (Fig. 5f). The degree of branch tip remodeling observed with sspa-GCaMP6f highlighting was comparable to control experiments made with a sparsely expressed non-photoactivatable membrane bound GFP (Supplementary Figure 19d, e), suggesting that neither the expression of sspa-GCaMP6f nor 2P photoactivation in the fine processes of the axons affect axonal arbor development.
Engineering other cpGFP-based Ca2+ indicators
There would be a major advantage if the strategy that worked to create photoactivatable GCaMPs could be transferrable to other GECIs. We asked whether the green variant of the GECO family, G-GECO1.2 (ref. 20), could also be made photoactivatable. G-GECO1.2 has a high sequence homology with GCaMPs (>85%), and an even higher homology around the chromophore (Supplementary Figure 20). We introduced the three mutations (Fig. 6a) that made GCaMP photoactivatable along with an additional mutation in the cpGFP next to the linker (Fig. 6a and Supplementary Figure 20c, d) to generate pa-G-GECO1.2*. pa-G-GECO1.2* expressed robustly and showed strong photoactivation in cultured hippocampal neurons (n = 8)(Fig. 6b). Once photoactivated, pa-G-GECO1.2* rapidly filled the cell's processes, including dendrites and dendritic spines (Fig. 6b,c and Supplementary Figure 20a) and robustly reported on Ca2+ activity (Fig. 6d).
Figure 6. Transferability of engineering strategy to other GECIs.
a, Schematic of pa-G-GECO1.2* construction strategy with three mutations required for photoactivation (red stars) along a M84K mutation (yellow star; see Supplementary Notes and Figure 20). b, 3D projection fluorescence micrographs of hippocampal neurons expressing pa-G-GECO1.2* before (0 s) and after (300 s) somatic photoactivation (pink dashed regions). (Right) Contrast image showing the degree of photoactivation at 300 s. (inset) Magnified dendrite with highlighted dendritic spines (white arrowheads). c, Ca2+ activity recorded in soma of a hippocampal neuron expressing pa-G-GECO1.2*, following somatic photoactivation (n = 3). Images display pa-G-GECO1.2* fluorescence of a neuron at trough (left) and peak (right) of fluorescent transients, displayed in d. Note spread of activity throughout the cell.
Discussion
We describe a generalizable strategy for generating photoactivatable GECIs by introducing PA-GFP mutations into the conserved FP domain of GCaMPs. Two analogous approaches have been reported. First, a photoactivated TNXL (PA-TNXL) was made using PA-GFP as a fluorescence resonance energy transfer donor and a non-emitting fluorescent protein acceptor49. While PA-TNXL exhibits small signals, among other limitations50, more sensitive TNXL-based reporters, e.g., Twitch51, may make it possible to engineer a higher sensitivity PA-TNXL. However, it remains to be seen whether photoactivation can be transferred to Twitch, which structurally differs from PA-TNXL in many aspects. The second approach consists of a photoconvertible GECI, GR-GECO, which shifts fluorescence from green to red following near-UV illumination52. The sensitivity of the red form of GR-GECO is considerably higher than with our pa-GECIs, allowing active cells to be readily selected prior photoconversion. However, because GR-GECO occupies both green and red channels, it may be difficult to use in combination with additional fluorescent proteins (see Supplementary Discussion).
With pa-GECI, expressing cells could be identified, and Ca2+ activity detected, under weak illumination that did not produce significant photoactivation (Supplementary Figures 5, 6, 8, 12). Expressing cells could then be targeted for photoactivation with one of four strategies: i) Group targeting by illumination in a continuous region of interest (e.g. Fig. 4; Supplementary Figures 2, 6, 15 and 17), ii) Single cell photoactivation (e.g. Figs. 1, 5, 6; Supplementary Figures 3, 8, 10-12, 14 and 17; Supplementary Videos 2-4), iii) Sequential single cell photoactivation (e.g. Fig. 3; Supplementary Figures 12-14; Supplementary Videos 2 and 7), and iv) Simultaneous photoactivation of individual cells (e.g. Supplementary Figures 11, 15, 17 and Supplementary Video 6).
Once photoactivated, the pa-GECIs diffuse quickly and efficiently throughout the cell to enable the mapping of fine cellular architecture and report on action potential firing or synaptic transmission. Sequential photoactivation enables the morphology and projections of a series of neurons to be reconstructed and this, then, permits a powerful form of functional connectivity mapping using a straightforward cross-correlation analysis. The pa-GECIs can be efficiently photoactivated and imaged with either 1- or 2P illumination on standard microscopes and function in mammalian primary neuronal cell culture, as well as in vivo in Drosophila and zebrafish. The pa-GECIs can function at multiple scales, from composite activity of large groups of neurons to sensory-specific activity in a few projections in a limited subpopulation of neurons. They are bright enough and sufficiently persistent to enable highlighting and tracking of cellular morphology over time scales that are relevant for development. Importantly, neither photoactivation nor imaging has an impact on the electrical properties, synaptic transmission or morphology of the cells. Because the absorbance of the pre-activated spa-GCaMPs peaks at ~400 nm and is negligible at wavelengths >510 nm11 (Supplementary Figure 5d), it should be compatible with red-shifted opsins, such as ReaChR53, Jaws54 or ChRimson55, which are optimally excited with wavelengths >600 nm, which does not photoactivate spa-GCaMPs.
Directed evolution has proven powerful for optimizing the performance of other photoactivatable proteins, but the set of mutations that work in one protein could not be easily transferred to others56,57. In contrast, our rational design strategy was sufficient to rapidly generate multiple single colored, monomeric pa-GECIs from some of the most broadly used GECIs. We show that the three necessary and sufficient residues for efficient photoactivation are transferable from GCaMPs to G-GECOs (Fig. 6), suggesting that it could be extended to other GECIs, such as GCaMP7, 8, GCaMP-fast, and ultra-fast-GCaMP16,23-25,58,59, as well as to future GECIs. The observation that different combinations of mutations and truncations tested here are well tolerated by several cpGFP-based Ca2+-indicators is of significance not only for Ca2+ sensors, but potentially also for other cpGFP-based indicators, such as inositol-sensors28, diacylglycerol-sensors27, glutamate sensors60 and voltage sensors29,61.
Online Methods
Molecular biology
GCaMP3 (plasmid #22692) and GCaMP6s, m and f (#40753, 4, 5, respectively), and G-GECO1.2 (#32446) were purchased from Addgene. All GCaMP indicators are composed of the Methionine-Glycine-Serine sequence, denoted MGS (without the Arginine62), directly followed by an artificial poly Histidine-tag; denoted MGS-HISx6, a transcript stabilizing sequence from gene 10 of phage T7, the Xpress epitope and the enterokinase cleavage recognition sequence, denoted RSET62, a M13 fragment from myosin light chain kinase, a circularly permutated GFP (cpGFP) and calmodulin (CaM)63. G-GECO is of similar design, but does not contain the MGS and RSET domains64. Point mutations were done using standard PCR reactions. For full list of primers see Table 1. Following the introduction of the numerous mutations found in superfolder GFP onto GCaMP3 along with the 3 mutation required for photoactivation (denoted by spa), the modified spa-cpGFP fragment was incorporated into GCaMP6 (s, m and f) in two steps, rather than re-introducing all the mutations anew in GCaMP6. To do so, we only amplified the cpGFP component from GCaMP3, using the forward-5′-GGTCGGCTGAGCTCACTCGAGAACGTCTATATCACCGCCGACAAGCAG and reverse 5′- CTCTTCAGTCAGTTGGTCCGGCAGGTTGTATTCCAGCTTGTGCCC primers, to generate a MEGA primer, which was directly used to amplify and introduce cpSPA into GCaMP6. To generate shorter versions of the various spa-GCaMPs (sspa-GCaMPs), we truncated the MGS and RSET domains. To generate pa-GECO1.0 and 1.2, we used the same set of primers used for pa-GCaMP3. Knowingly, by using the GCAMP3 V115H primer-set, we introduced an additional mutation (M84K, G-GECO numbering) in pa-GECO, denoted pa-GECO* (see Supplementary Note), because this incorporated Lysine lines the hole of the barrel and was suspected to play an important role in solution accessibility, thus fluorescence (see Supplementary Figure 10). pa-G-GECO1.2 (without the K84 mutation) displayed similar properties as pa-G-GECO1.2*, albeit slightly smaller Ca2+-responses. To insert the various clones into pRSETB (Kindly provided by R.Y. Tsien), we introduced a BgLII restriction site before the starting codon upstream to the His(x6)-tag. This was done to remove the entire RSET sequence found in the original vector and to allow insertion of the modified RSET sequence found in the various GCaMPs (see list of primers and Supplementary Figure 6).
Tissue culture and transfection
HEK293T and HeLa cells were purchased from the Molecular and Cell Biology Tissue Culture Facility, University of California, Berkeley. All cells were tested for Mycoplasma and authenticated by DDC Medical. Cells were maintained in DMEM with 5% FBS on poly-L-lysine-coated 12 mm glass coverslips at 37°C, 5% CO2 and transiently co-transfected with 0.5-1 μg of the various pa-GECIs plasmids using Lipofectamine 2000 (Invitrogen). Imaging was performed 10-12h after transfection. Dissociated postnatal rat hippocampal neurons (P0-P5) were prepared and transfected as described previously65, kept in 32-37°C, 5% CO2 and transiently transfected with 0.5-1 μg of cDNA encoding the various pa-GECIs by the calcium phosphate method and imaged at 13-15 DIV.
Calcium Imaging
HEK293T, HeLa cells and cultured hippocampal neurons were imaged in a standard extracellular solution containing (in mM): 138 NaCl, 1.5 KCl, 1.2 MgCl2, w/o 2.5 CaCl2 (unless indicated otherwise), 10 D-glucose, 10 Hepes, 1 EDTA (where indicated), pH 7.4. Imaging was done using an upright confocal microscope (Zeiss, LSM-780/880) with a 20x water-immersion objective, equipped with 405, 488, 561 nm lasers as well as a 690-1040 nm tunable 2P laser, with a spectrally-resolved 32-pixel GaAsP detector array or BIG-1/2 GaAsP detectors. Brief pulses of 405 nm (rastering at 2.1 mW/pixel, dwell time ~0.64-1.23 μs/pixel) were used to illuminate selected regions. To detect cells prior photoactivation, rapid, single line rastering was performed using low intensity 405 nm illumination (5 μW/pixel) with a large increase in gain (30-45% increase). ΔF/F was calculated as fluorescence at time t (Ft) subtracted from initial fluorescence at time 0 (Fi), divided by fluorescence at time 0; ΔF/F=(Ft-Fi)/Fi. For mean ΔF/F, we averaged the fluorescence before photoactivation to reflect Fi and average fluorescence following photoactivation to reflect Ft. Titration curves were fitted with a Hill-type equation; f=axb/(cb+xb), with a=1.
Two-Photon imaging (940 nm) and photoactivation (760 nm) of Drosophila Kenyon cells in vivo (see below) was done on a custom built 3i/Sutter In Vivo SLM system. Spatially targeted photoactivation was performed using a 2P digital holography66,67, using a commercial spatial light modulator system (Phasor; Intelligent Imaging Innovations, Inc.). Holograms were calculated from user-defined ROIs based on images obtained from fluorescence images of the preparation. NIR laser pulses encoding phase holograms were projected onto the back aperture of a 20x/1.0NA water-dipping objective.
Electrophysiology
Patch clamp recordings used an Axopatch 200A amplifier. Recordings were carried out 2-3 days after transfection of 15 days in vitro (DIV) hippocampal neurons. When in whole cell mode, cells were voltage-clamped at −60 mV or Current-clamped at I=0 (to assess resting potential). Pipettes had resistances of 8-12 MΩ and were filled with a solution containing (in mM): 140 K-gluconate, 10 NaCl, 2 MgATP, 1 CaCl(H2O)2, 5 EGTA, 2 MgCl2(H2O)6, 10 Hepes, pH 7.3. Loose patching was performed using a similar internal solution, but with pipettes with lower resistance (3-5 MΩ). After partial seal was formed (30-50 MΩ), recordings were done in current clamp mode (lowpass Bessel Filter at 10 kHz). Whereas most neurons exhibited spontaneous activity that allowed investigating the fluorescence correlates, in some cells we induced action potential firing by brief 0.5 ms voltage triggers (voltage ZAP). For the dissociated rat hippocampal neurons, the extracellular recording solution contained (in mM): 138 NaCl, 1.5 KCl, 1.2 MgCl2, 10 D-glucose, 2.5 CaCl2, 10 Hepes, pH 7.4 (NaOH). Electrophysiological data was recorded with pClamp and analyzed by Clampfit 10.2 software.
Zebrafish
spa-GCaMP6f and sspa-GCaMP6f were cloned with a 10X repeat of an upstream activating sequence (UAS) into a pT2KXIG-delta-IN vector containing Tol2 transposon recognition sites 68 and SV40 polyA, using restriction enzyme sites AscI and MluI (inserted into the vector) for ligation (see Supplementary Table 1). The resulting pT2-UAS-s/spa-GCaMP6f constructs were diluted to 50 ng/uL with 0.25 ng Tol2 transposase and 0.05% Phenol Red (Sigma) and injected into embryos at the 1-2 cell stage. Transiently expressing embryos were raised in 0.05% PTU and embedded in 1.4% agarose without anesthetic for imaging. Eggs were obtained by incrossing transgenic fish expressing Gal4 using the following lines: motor neuron expression, Tg[mnx1-gal4]69 and Tg[s1020t-gal4;UAS-KillerRed]; Muller glia, Tg[s1003t-gal4]; retinal ganglion cells, Tg[ath5-gal4]. Control experiments for retinal ganglion cell axon dynamics were done by injecting an ath5-gap43-GFP plasmid at 50 ng/uL into AB embryos at the 2-4 cell stage.
Transgenic Drosophila melanogaster adults and larvae
spa-GCaMP6f and sspa-GCaMP6f DNA were cloned into the pUAST vector70. UAS-spa-GCaMP6f and UAS-sspa-GCaMP6f transgenic flies were made with P-element transformation (BestGene Inc., 2918 Rustic Bridge, Chino Hills CA) into the w1118 background (Table 2). Expression was driven in larval motor neurons using the OK6-Gal471. Flies were raised on standard corn-meal and molasses media at 25°C. Third instar larvae with the genotype w1118; OK6-Gal4;UAS-spa-GCaMP6f were selected for all experiments unless otherwise noted. Larvae were dissected and imaged in HL3 solution containing (in mM): 70 NaCl, 5 KCl, 0.45 CaCl2, 20 MgCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, 5 HEPES, pH 7.2. For imaging evoked activity (orthodromic and antidromic) larvae were imaged in HL3 solution as mentioned above, but with (in mM): 1.5 CaCl2 and 7 mM Glutamate (to reduce muscle contraction and movements by desensitizing postsynaptic glutamate receptors). Larvae were dissected by making a longitudinal dorsal incision, removing all organs, and pinning the cuticle flat under high tension to prevent significant movement upon body wall muscle contraction. The CNS and peripheral motor neurons were left intact. Targeted photoactivation and recording of spontaneous activity was performed on an upright confocal microscope (Zeiss LSM-780)(as described in Calcium Imaging) or on a spinning disk confocal microscope (3i) equipped with a spatial light modulator (PhasorTM, 3i) to target 405 nm laser. Nerve stimulation was induced with a suction electrode filled with HL3. Imaging of Kenyon cells was done on dissected third instar larvae bearing the genotype: Genotype: w1118; UAS-mCherry-nls/UAS-spa-GCaMP6f; UAS-spa-GCaMP6f/UAS-spa-GCaMP6f; OK107-Gal4/+, and imaged in the HL3 solution mentioned above, but with 0.45 mM Ca2+.
For in vivo imaging of adult fly's SEZ, experiments were performed as described in 72 where 2 day old female flies were placed into a small slit on a plastic mount, such that the head was separated from the rest of the body. Following immobilization of the head using nail-polish, the head cuticle was dissected open, in ice-cold hemolymph-like (AHL) solution devoid of Ca2+ and Mg2+, to reveal the brain. However, legs and proboscis were left intact. Before imaging, the AHL solution was replaced with AHL containing Ca2+ and Mg2+. Application of sucrose to the proboscis was performed as described in 73.
Purified proteins
His(X6)-tagged proteins were expressed and purified as previously described74, with several modifications. Competent E. coli (T7 express Competent, Biolabs) were transformed with different pRSET-spa/WT-GCaMP6 clones, pelleted and lysed. Lysate was clarified by ultracentrifugation and proteins were isolated on a column. Eluted proteins (by imidazole gradient) were dialyzed for 2 days in darkness. Initial dialysis was performed in 20 mM Tris pH 7.8, 100 mM NaCl, 10% glycerol (v/v) and 1 mM EDTA to remove imidazole and (most) residual Ca2+. This was followed by a second dialysis, but without EDTA. Absorption and excitation emission spectrum measurements were done on a Cary spectrophotometer and a Photon Technology International fluorimeter, respectively.
Calcium titrations were done using EGTA-buffered Ca2+ solutions (Calcium Calibration Buffer Kit #1, Life Technologies). Purified GCaMP variants were diluted 1:100 in 30 mM MOPS, pH 7.2, 100 mM KCl containing either 10 mM EGTA ([Ca2+]free= 0 μM) or 10 mM CaEGTA ([Ca2+]free]= 39 μM free calcium). These were mixed in various ratios (as specified by the kit) to give 11 different Calcium concentrations. Green GCaMP fluorescence (excitation 485 nm, 5 nm bandpass; emission 510 nm, 5 nm bandpass) was measured using a fluorimiter. Sigmoidal binding functions (f=axb/(cb+xb), with a=1), were fitted to the calcium titration fluorescence data to extract the Kd for Ca2+ for each GCaMP variant. pH titrations were carried out by diluting concentrated purified proteins into pH buffers containing 50 mM citrate, 50 mM Tris, 50 mM glycine, 100 mM NaCl, and either 5 mM CaCl2 or 5 mM EDTA. pKa values were determined from the inflection point of a sigmoid fit to 520 nm fluorescence versus pH.
Statistical analysis
Results are shown as mean ± SEM. Multiple group comparison was performed using one-way analysis of variance (ANOVA) with post hoc Tukey test (all-pairwise analysis) and two group comparisons were done using two-tailed t-test (SigmaPlot 2011). Correlations between two independent parameters were examined using Spearman correlation. Asterisks indicate statistically significant differences as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Correlation maps
Movies were corrected for exponential bleach (if any) and registered to correct for x-y drift. ROIs were selected semi-automatically using the Analyze Particles plug-in in ImageJ. Zero-lag correlations were computed on ΔF/F traces in Matlab.
Structure rendering
Protein structures were rendered using Pymol (PyMOL™, 2010 Schrodinger, LLC.). Neural tracing was performed using Imaris 8 (Bitplane) and ImageJ (NIH).
Animal Usage
All animal experiments were done under oversight by the university of California institutional review board (Animal Care and Use Committee).
Supplementary Material
Acknowledgments
We thank C. Stanley and Z. Fu for help with molecular biology, H. Aaron for technical help with microscopy and C. Chang for use of fluorimeter. We also want to thank R.Y. Tsien (University of California San Diego) for the pRSETB vector, J.L. Bruses (University of Kansas) for the generous gift of the mnx1-Gal4 construct and D. Friedmann for generating mnx1-Gal4 transgenic zebrafish line. The work was supported by National Science Foundation Graduate Research Fellowship 1106400 (Z.L.N), National Science Foundation MRI grant 1041078 (E.Y.I), NIHGMS R01 GM068552 (J.C.L.) and National Institutes of Health Nanomedicine Development Center for the Optical Control of Biological Function 2PN2EY01824 (E.Y.I).
Footnotes
Authors Contributions
S.B., E.C.C. and E.Y.I. conceived of the project. S.B. designed sequences and constructed clones with help from H.O.O. S.B. performed all experiments in cultured cells and patching experiments. Z.L.N. maintained transgenic Drosophila lines and conducted larval Drosophila experiments with help from S.B. and E.C.C. B.K. conducted adult Drosophila experiments with help from S.B. E.C.C. and Z.L.N. E.C.C. conducted zebrafish experiments with help from C.M.Q. and S.B. S.S.M. purified proteins with supervision of J.C.L and N.C.R. E.C.C. and S.B. characterized the purified proteins. S.B., E.C.C, Z.L.N. and E.Y.I wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Cajal SR. Estructura de los centros nerviosos de las aves. Revista Trimestral de Histología Normal y Patológica. 1888;1:1–10. [Google Scholar]
- 2.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat. Methods. 2013;10:508–13. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 3.Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat. Rev. Neurosci. 2008;9:417–22. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 5.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods. 2007;4:47–9. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alivisatos AP, et al. Nanotools for neuroscience and brain activity mapping. ACS Nano. 2013;7:1850–66. doi: 10.1021/nn4012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underwood E. New Tools Light Up the Intricacies of the Brain. Science. 2013;342:917–918. doi: 10.1126/science.342.6161.917. [DOI] [PubMed] [Google Scholar]
- 8.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014 doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lien AD, Scanziani M. In vivo Labeling of Constellations of Functionally Identified Neurons for Targeted in vitro Recordings. Front. Neural Circuits. 2011;5:16. doi: 10.3389/fncir.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peter M, et al. Transgenic mouse models enabling photolabeling of individual neurons in vivo. PLoS One. 2013;8:e62132. doi: 10.1371/journal.pone.0062132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–7. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 12.Ruta V, et al. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–90. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–91. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 15.Tian L, Looger LL. Genetically encoded fluorescent sensors for studying healthy and diseased nervous systems. Drug Discov. Today Dis. Models. 2008;5:27–35. doi: 10.1016/j.ddmod.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001;19:137–41. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 17.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 18.Zucker RS. Calcium- and activity-dependent synaptic plasticity. Curr. Opin. Neurobiol. 1999;9:305–13. doi: 10.1016/s0959-4388(99)80045-2. [DOI] [PubMed] [Google Scholar]
- 19.McCombs JE, Palmer AE. Measuring calcium dynamics in living cells with genetically encodable calcium indicators. Methods. 2008;46:152–9. doi: 10.1016/j.ymeth.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–91. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian L, Hires SA, Looger LL. Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb. Protoc. 2012;2012:647–56. doi: 10.1101/pdb.top069609. [DOI] [PubMed] [Google Scholar]
- 22.Whitaker M. Genetically encoded probes for measurement of intracellular calcium. Methods Cell Biol. 2010;99:153–82. doi: 10.1016/B978-0-12-374841-6.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, et al. Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement. Protein Cell. 2013;4:299–309. doi: 10.1007/s13238-013-2103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, et al. Improved orange and red Ca2+ indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci. 2013;4:963–72. doi: 10.1021/cn400012b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tewson P, et al. Simultaneous detection of Ca2+ and diacylglycerol signaling in living cells. PLoS One. 2012;7:e42791. doi: 10.1371/journal.pone.0042791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaguchi R, et al. A single circularly permuted GFP sensor for inositol-1,3,4,5-tetrakisphosphate based on a split PH domain. Bioorg. Med. Chem. 2009;17:7381–6. doi: 10.1016/j.bmc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 29.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014;17:884–9. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson JN, et al. Structure and mechanism of the photoactivatable green fluorescent protein. J. Am. Chem. Soc. 2009;131:4176–7. doi: 10.1021/ja808851n. [DOI] [PubMed] [Google Scholar]
- 31.Ruthazer ES, et al. Bulk electroporation of retinal ganglion cells in live Xenopus tadpoles. Cold Spring Harb. Protoc. 2013;2013:771–5. doi: 10.1101/pdb.prot076471. [DOI] [PubMed] [Google Scholar]
- 32.Yamada Y, Mikoshiba K. Quantitative comparison of novel GCaMP-type genetically encoded Ca(2+) indicators in mammalian neurons. Front. Cell. Neurosci. 2012;6:41. doi: 10.3389/fncel.2012.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, et al. A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging. Nat. Commun. 2014;5:5262. doi: 10.1038/ncomms6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noguchi J, Matsuzaki M, Ellis-Davies GC, Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron. 2005;46:609–22. doi: 10.1016/j.neuron.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araya R, Jiang J, Eisenthal KB, Yuste R. The spine neck filters membrane potentials. Proc. Natl. Acad. Sci. U S A. 2006;103:17961–6. doi: 10.1073/pnas.0608755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider M, Barozzi S, Testa I, Faretta M, Diaspro A. Two-photon activation and excitation properties of PA-GFP in the 720-920-nm region. Biophys. J. 2005;89:1346–52. doi: 10.1529/biophysj.104.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A. Two-photon absorption properties of fluorescent proteins. Nat. Methods. 2011;8:393–9. doi: 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branco T, Staras K, Darcy KJ, Goda Y. Local dendritic activity sets release probability at hippocampal synapses. Neuron. 2008;59:475–85. doi: 10.1016/j.neuron.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 40.Aberle H, et al. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–58. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- 41.Fox LE, Soll DR, Wu CF. Coordination and modulation of locomotion pattern generators in Drosophila larvae: effects of altered biogenic amine levels by the tyramine beta hydroxlyase mutation. J. Neurosci. 2006;26:1486–98. doi: 10.1523/JNEUROSCI.4749-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohsaka H, Okusawa S, Itakura Y, Fushiki A, Nose A. Development of larval motor circuits in Drosophila. Dev. Growth. Differ. 2012;54:408–19. doi: 10.1111/j.1440-169X.2012.01347.x. [DOI] [PubMed] [Google Scholar]
- 43.Marella S, et al. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49:285–95. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 44.Kurusu M, et al. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc. Natl. Acad. Sci. U S A. 2000;97:2140–4. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saint-Amant L, Drapeau P. Motoneuron activity patterns related to the earliest behavior of the zebrafish embryo. J. Neurosci. 2000;20:3964–72. doi: 10.1523/JNEUROSCI.20-11-03964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muto A, Kawakami K. Imaging functional neural circuits in zebrafish with a new GCaMP and the Gal4FF-UAS system. Commun. Integr. Biol. 2011;4:566–8. doi: 10.4161/cib.4.5.15848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shigetomi E, Kracun S, Sofroniew MV, Khakh BS. A genetically targeted optical sensor to monitor calcium signals in astrocyte processes. Nat. Neurosci. 2010;13:759–66. doi: 10.1038/nn.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulina ME, et al. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24:95–9. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda T, Horikawa K, Saito K, Nagai T. Highlighted Ca2+ imaging with a genetically encoded 'caged' indicator. Sci. Rep. 2013;3:1398. doi: 10.1038/srep01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mank M, et al. A FRET-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys. J. 2006;90:1790–6. doi: 10.1529/biophysj.105.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thestrup T, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods. 2014;11:175–82. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- 52.Hoi H, Matsuda T, Nagai T, Campbell RE. Highlightable Ca2+ indicators for live cell imaging. J. Am. Chem. Soc. 2013;135:46–9. doi: 10.1021/ja310184a. [DOI] [PubMed] [Google Scholar]
- 53.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16:1499–508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci. 2014;17:1123–9. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–46. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods. 2009;6:153–9. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J. Am. Chem. Soc. 2010;132:6481–91. doi: 10.1021/ja100906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muto A, Ohkura M, Abe G, Nakai J, Kawakami K. Real-time visualization of neuronal activity during perception. Curr. Biol. 2013;23:307–11. doi: 10.1016/j.cub.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 59.Nordine Helassa EE, Carter Tom, Bradley Jonathan, Ogden David, Török Katalin. Ultrafast Genetically Encoded Calcium Indicators for visualizing Calcium Flux and Action Potentials. Biophysical Society Meeting. 2014 [Google Scholar]
- 60.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods. 2013;10:162–70. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker BJ, et al. Genetically encoded fluorescent sensors of membrane potential. Brain Cell Biol. 2008;36:53–67. doi: 10.1007/s11068-008-9026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001;19:137–41. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 64.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–91. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szobota S, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–45. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 66.Oron D, Papagiakoumou E, Anselmi F, Emiliani V. Two-photon optogenetics. Prog. Brain Res. 2012;196:119–43. doi: 10.1016/B978-0-444-59426-6.00007-0. [DOI] [PubMed] [Google Scholar]
- 67.Carroll EC, et al. Two-photon brightness of azobenzene photoswitches designed for glutamate receptor optogenetics. Proc. Natl. Acad. Sci. U S A. 2015;112:E776–85. doi: 10.1073/pnas.1416942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 69.Zelenchuk TA, Bruses JL. In vivo labeling of zebrafish motor neurons using an mnx1 enhancer and Gal4/UAS. Genesis. 2011;49:546–54. doi: 10.1002/dvg.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 71.Aberle H, et al. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–58. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- 72.Marella S, Mann K, Scott K. Dopaminergic Modulation of Sucrose Acceptance Behavior in Drosophila. Neuron. 2012;73:941–950. doi: 10.1016/j.neuron.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marella S, et al. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49:285–95. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 74.Hirose M, et al. Structure of the human-heart fatty-acid-binding protein 3 in complex with the fluorescent probe 1-anilinonaphthalene-8-sulphonic acid. J Synchrotron Radiat. 2013;20:923–8. doi: 10.1107/S0909049513021298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.