Abstract
The objective of this study was to evaluate the effects of NaCl on the biofilm formations of the isolate from Staphylococcus aureus outbreaks linked to ham. The S. aureus ATCC13565 isolated from ham was exposed to NaCl concentrations of 0%, 2%, 4%, and 6% supplemented in tryptic soy broth (TSB) for 24 h at 35℃, followed by plating 0.1 mL of the culture on tryptic soy agar containing 0%, 2%, 4%, and 6% NaCl, respectively. After incubating at 35℃ for 24 h, the colonies on the plates were collected and diluted to OD600 = 0.1. The diluents of S. aureus were incubated on a 96-well flat bottom plate containing TSB plus the appropriate NaCl concentrations, and the biofilm formation was quantified by crystal violet staining after being incubated at 35℃ for 9 h. Confocal laser scanning microscope (CLSM) was also used for visualizing the biofilm formation of S. aureus at NaCl concentrations of 0%, 2%, 4%, and 6%. The transcriptional analysis of biofilm-related genes, such as icaA, atl, clfA, fnbA, sarA, and rbf, was conducted by quantitative real-time PCR. Crystal violet staining and CLSM showed that the biofilm formations of S. aureus increased (p<0.05) along with the NaCl concentrations. Moreover, the expression of the icaA genes was higher at the NaCl concentrations of 4% and 6% as compared with 0% of NaCl by approximately 9-folds and 20-folds, respectively. These results indicated that the NaCl formulated in processed food may increase the biofilm formations of S. aureus by increasing the icaA gene expressions.
Keywords: ham, Staphylococcus aureus, biofilm, NaCl
Introduction
A biofilm is considered a community of microorganisms surrounded by an extracellular matrix composed of polymeric substances, DNA, and proteins (Cue et al., 2009; Donlan, 2002). Bacteria may adhere to food or food-contact surfaces and form a biofilm as a response to stresses (Rode et al., 2012). Biofilms can be found on food and food-contact surfaces, and the bacterial cells in biofilms can be protected from various environmental stresses such as disinfectants, antibiotics, nutrient limitation, osmotic stress, and other stresses, resulting in sources of crosscontamination (Bower et al., 1999; Poulsen, 1999; Vázquez-Sánchez et al., 2013).
Staphylococcus aureus is a gram-positive bacterium, and the pathogen causes nosocomial and community-acquired infections, including septic arthritis, endocarditis, and osteomyelitis (Sambanthamoorthy et al., 2008). This pathogen is known to be resistant to antibiotics and, particularly, methicillin-resistant S. aureus (MRSA) is considered a critical problem in controlling S. aureus and treating infections (Livermore, 2000; Zapun et al., 2008). S. aureus is also a common causative agent for foodborne diseases, which are caused by its enterotoxins (Le Loir et al., 2003; Lowy, 1998). S. aureus usually exists on the skin and nasal mucosa of humans (Otto, 2008). Thus, S. aureus is often isolated in the foods directly handled by humans. Because S. aureus is poorly competitive in microbiota and has salt-tolerance (10-15%) (Bhunia, 2008), the foodborne outbreaks are usually associated with salted foods, such as dried fermented sausages and hams, rather than raw foods (USFDA, 2004).
NaCl has been used for food preservation by increasing the osmotic pressure of foods, but many foodborne pathogens have shown tolerance to NaCl, which alternates their physiological characteristics (Rode et al., 2012). Yoon et al. (2013) showed that exposure of Salmonella to NaCl increased its heat resistance and Caco-2 cell invasion. S. aureus also can tolerate high NaCl concentrations in foods and regulate global gene expression, such as sigma factor, to survive, and subsequently, the global gene expression can be related to increased resistance to sublethal stresses (Huang et al., 2009; Rode et al., 2012). Additionally, the heat resistance of S. aureus was increased by the regulation of various genes after the pathogen was exposed to high NaCl concentrations (Smith et al., 1982).
Although NaCl is included in many foods including meat products, and only a few studies that NaCl may influence biofilm formation of S. aureus have been reported. Therefore, the objective of this study was to evaluate the effect of NaCl on the biofilm formation of the isolate from S. aureus outbreak linked to ham.
Materials and Methods
Preparation of inocula
S. aureus ATCC13565 was the strain isolated from S. aureus outbreak linked to ham consumption. The isolate was cultured in tryptic soy broth (TSB; Difco, Becton Dickinson, and Company, USA) at 35℃ for 24 h, and 0.1 mL of the culture was transferred into 10 mL of TSB. After incubation, the cells were harvested by centrifugation (1,912 g, 15 min, 4℃), washed twice, and resuspended in phosphate-buffered saline (PBS, pH 7.4; 0.2 g of KH2PO4, 1.5 g of Na2HPO4·7H2O, 8.0 g of NaCl, and 0.2 g of KCl in 1 L of distilled water). The suspension was serially diluted in PBS to obtain 4 Log CFU/mL. The diluent (0.1 mL) was then inoculated in 10 mL of TSB plus 0%, 2%, 4%, and 6% NaCl and incubated at 35℃ overnight. A portion (0.1 mL) of each culture exposed to 0%, 2%, 4%, and 6% NaCl was then plated on tryptic soy agar (TSA; Difco) added 0%, 2%, 4%, and 6% NaCl, respectively, to obtain only survival cells from NaCl stress, which is found in meat processing environment. After incubating at 35℃ for 24 h, 4 mL of PBS was added over the colonies, and the colonies were scraped with a glass rod. The collected S. aureus cells were centrifuged (1,912 g, 4℃, 15 min), and the pellets were then washed with PBS twice. These bacterial cell suspensions were then adjusted to OD600=0.1 (Yoon et al., 2013).
Crystal violet staining
To quantify the biofilm, 20 μL of the S. aureus cell suspension with OD600=0.1 was inoculated in 230 μL of TSB supplemented with 0%, 2%, 4%, and 6% NaCl in a 96-well flat bottom plate. The plate was then incubated at 35℃ for 9 h. After the incubation, the supernatant was removed, cells were washed three times with sterile distilled water, and 250 μL of methanol (99.9%) was added into each well and left for 15 min to fix the S. aureus cells. After removing the methanol, 250 μL of crystal violet was added to each well, cells were stained for 5 min at room temperature. After staining, the dye was discarded, and cells were washed three times with sterile distilled water. Acetic acid (33%, 250 μL) was then added and left for 5 min, and the OD was determined at 595 nm using a microplate reader (Bio Tek Instruments, USA).
Image analysis of confocal laser scanning microscopy (CLSM)
Pre-sterilized 8-well chamber glass slides (Thermo Fisher Scientific Inc., USA) were used to form the staphylococcal biofilm, and the LIVE/DEAD® Biofilm Viability Kit (Molecular Probes, Netherlands) was applied to staining live and dead cells of S. aureus. Aliquots (30 μL) of the S. aureus cell suspensions (OD60=0.1) were inoculated in 320 μL of TSB plus the appropriate concentration of NaCl (0%, 2%, 4%, and 6% ) in 8-well chamber glass slides and incubated at 35℃ for 9 h. After removing the supernatants and washing the bacterial cells three times with PBS, 200 μL of the fluorescent stains SYTO9 and propidium iodide (PI) were added, and the cells were left for 20 min at room temperature with protection from the light. SYTO9, which is excitated and emitted at 482 and 500 nm, respectively, stains live cells green by penetrating the live bacterial membrane, and PI, which is excitated and emitted at 490 and 635 nm, respectively, stains cells red by penetrating only the cells having damaged membranes (Takenaka et al., 2001). SlowFade Gold Antifade Reagent (4 μL; Molecular Probes) was then added on the samples to prevent quenching of the fluorescent signal. The confocal images were acquired using the OlympusTM FluoView FV300 (Olympus Co., Japan) confocal scanning laser microscope, and the biofilms were observed with a 40× water-immersion lens.
Transcriptional analysis
To evaluate the relative expression levels of the genes listed in Table 1, S. aureus cell suspensions were inoculated in TSB plus 0%, 4%, and 6% NaCl and incubated at 35℃ until reaching approximately OD600=0.5. After centrifuging 1 mL of the culture (5,000 g, room temperature, 5 min), the supernatant was discarded, 200 μL of lysostaphin (200 μg/mL) (Sigma-Aldrich Co., USA) was added to the pellets, and cells were incubated at 37℃ for 20 min to be lysed. RNA was extracted from the lysed cells using the Qiagen RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instruction. The concentration of total RNA was measured using Epoch Microplate Spectrophotometer System with Take3 (Bio Tek Instruments). Reverse transcription of the extracted RNA was performed using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s protocols.
Table 1. Oligonucleotide primers used in the quantitative real-time PCR analysis.
| Gene | Primer | Sequence (5' → 3') | Assession No. | Reference |
|---|---|---|---|---|
| 16s | 16s RNA-F | GAG GGT CAT TGG AAA CTG GA | L37597 | This study |
| rRNA | 16s RNA-R | CAT TTC ACC GCT ACA CAT GG | ||
| icaA | icaA-F | CTG GCG CAG TCA ATA CTA TTT CGG GTG TCT | NC003923 | Cue et al. (2009) |
| icaA-R | GAC CTC CCA ATG TTT CTG GAA CCA ACA TCC | |||
| atl | atl-F | ATA ACC GCA CTG GTT GGG TA | AJ567417 | This study |
| atl-R | TTG GCA GCT GAT GTA GTT GG | |||
| clfA | clfA-F | TGC TGC ACC TAA AAC AGA CG | DQ435612. | This study |
| clfA-R | TGT GTC GTT TCC TGT TGT GC | |||
| sarA | sarA-F | TCG AGC AAG ATG CAT CAA AT | NC002745 | This study |
| sarA-R | TGT CAG CAT AAG TGA CCA TTG A | |||
| fnbA | fnbA-F | GAA GAG CAT GGT CAA GCA CA | NC002951 | This study |
| fnbA-R | ACG TCA TAA TTC CCG TGA CC | |||
| rbf | rbf-F | ACG CGT TGC CAA GAT GGC ATA GTC TT | - | Cue et al. (2009) |
| rbf-R | AGC CTA ATT CCG CAA ACC AAT CGC TA |
The PCR reaction mixture (25 μL) was prepared with the Rotor-Gene SYBR Green PCR Kit (Qiagen) according to the manufacturer’s instruction. The threshold cycle (CT) values of the genes were determined by quantitative real-time PCR (qRT-PCR) with Rotor-Gene Q (Qiagen), and the CT value of each gene was normalized to a reference gene (16S rRNA). The relative expression levels of the genes were analyzed with Rotor-Gene Q software (Qiagen) to compare the expression levels of the genes related to biofilm formation in S. aureus grown in TSB supplemented with 0% NaCl and increasing NaCl concentration. Analysis of the gene expression levels was performed in duplicate per replication, and more than a twofold increase was considered significant (Sambanthamoorthy et al., 2008).
Statistical analysis
Each experiment was performed twice with four repeat samples per trial (n=8). The experimental data for the quantitative analysis were analyzed with the general linear model procedure of SAS® version 9.2 (SAS Institute Inc., USA). The mean comparison was performed by a pairwise t-test at α=0.05.
Results and Discussion
Quantification of biofilm
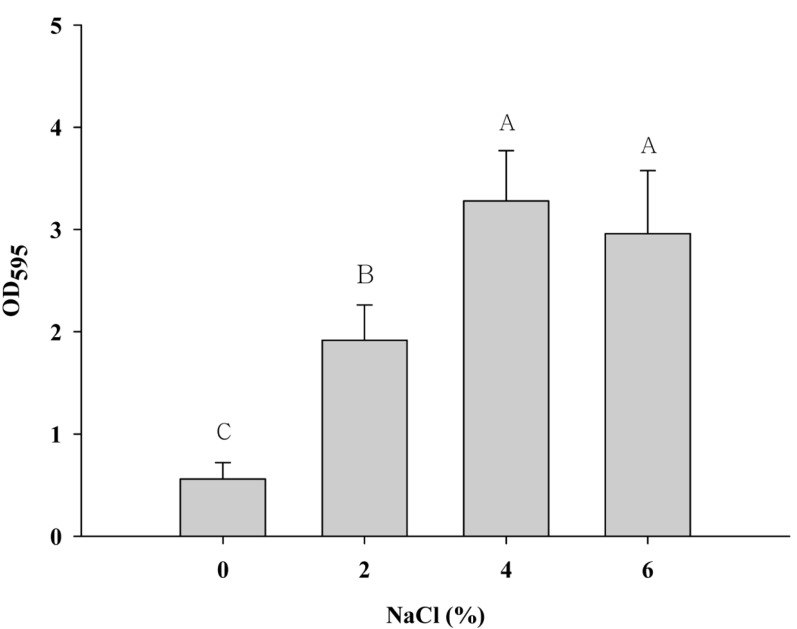
To quantify the biofilm, the biomass of the biofilm formation was measured with increasing NaCl concentrations in a microtiter plate, which is the most common assay to quantify biofilms. The OD595 values of S. aureus were significantly increased at higher NaCl concentrations compared with 0% NaCl (p<0.05) (Fig. 1). This result indicates that the exposure of S. aureus to NaCl may increase the biofilm formation of the pathogen. A study by Xu et al. (2010) showed that the cell aggregation of S. aureus increased when exposed to high NaCl concentrations (up to 10%). Other studies also reported that NaCl affects the increase of biofilm formation in S. aureus (Kennedy and O’Gara, 2004; Rode et al., 2007). However, these studies used the crystal staining method to quantify biofilm formation, which does not distinguish dead cells from live cells. Although there are dead cells caused by stress in certain biofilms, the method shows these cells as part of the biofilm, as suggested by Yoon (2013). Thus, a method, which can distinguish live cells from dead cells in biofilms, is necessary to evaluate appropriately the effect of NaCl on S. aureus biofilms.
Fig. 1. Biofilm formation of Staphylococcus aureus ATCC 13565 exposed to 0%, 2%, 4%, and 6% NaCl supplemented in tryptic soy broth. Error bars indicate the standard error of the mean. Optical density (OD) was measured at 595 nm.
Image analysis of CLSM
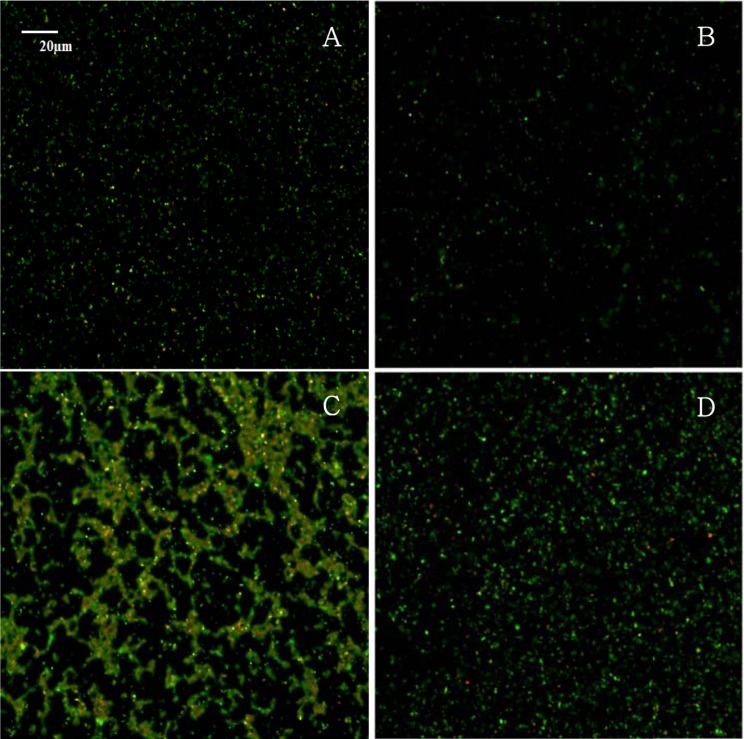
CLSM has the advantage of selectively staining individual live/dead cells with fluorescent stains, and, therefore, is useful in determining S. aureus biofilm formation. Fig. 2 shows merged images with live cells (green) and dead cells (red) of S. aureus after the S. aureus cells were exposed to NaCl concentrations of 0%, 2%, 4%, and 6% supplemented in TSB. The live cells in the biofilm of S. aureus increased at the higher NaCl concentrations rather than at the 0% or 2% NaCl concentrations (Fig. 2), which is in an agreement with the results of the crystal violet staining method. This result suggests that high concentrations of NaCl in food-related conditions increase the biofilm formation of S. aureus. Yoon (2013) evaluated the effect of NaCl on Salmonella biofilm. In the study, crystal violet staining revealed that the biofilm of Salmonella increased as NaCl concentration increased, but the CLSM image showed that most of the Salmonella cells at 4% NaCl were dead. Thus, these data suggest that the NaCl effect on biofilms is dependent on bacteria and that the CLSM image and crystal violet staining methods should be used together in biofilm studies rather than using only the crystal violet staining method.
Fig. 2. Confocal laser scanning microscopic images of a 9 hold biofilm formation of Staphylococcus aureus ATCC 13565 exposed to 0% (A), 2% (B), 4% (C), and 6% (D) NaCl supplemented in tryptic soy broth.
Transcriptional analysis
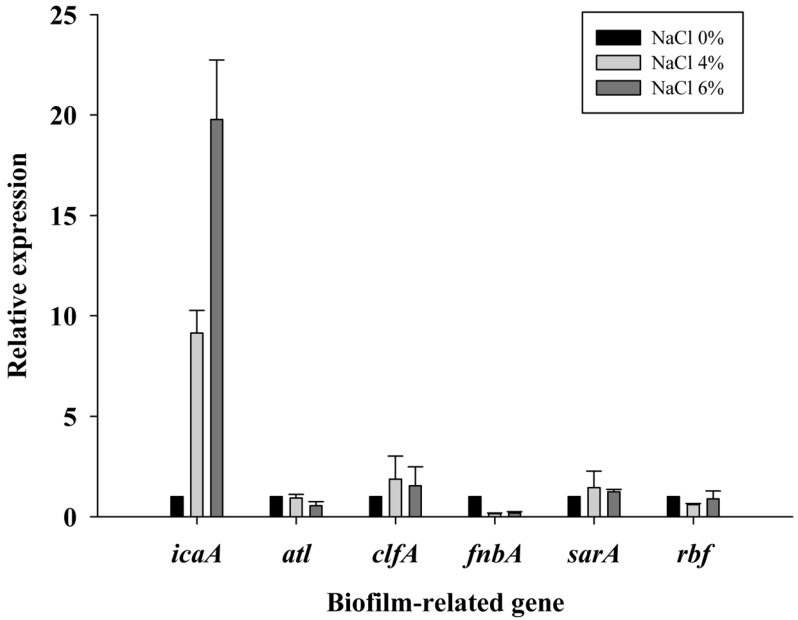
To address the reason for the NaCl-induced increase of S. aureus biofilm formation, the expression levels of the genes related to adherence and biofilm formation were analyzed at various NaCl concentrations by qRT-PCR. The genes investigated in this study are the genes directly or indirectly involved in biofilm formation. The icaA encodes the protein involved in the synthesis in poly N-acetyl glucosamine for intercellular adhesion (Cue et al., 2012), atl is related to promoting the adherence of bacterial cells to surfaces (Biswas et al., 2006), and clfA (clumping factor A) and fnbA (fibronectin-binding protein) encode proteins to mediate bacterial adherence, sarA (staphylococcal accessory regulator) is a global regulator and essential for biofilm formation (Beenken et al., 2004). Among these genes, the expression levels of icaA was increased approximately 9-fold and 20-fold at the 4% and 6% NaCl concentrations, respectively, compared with the 0% NaCl concentration, but the expression levels of the other genes (alt, clfA, fnbA, sarA, and rbf) were not increased. This result suggests that NaCl upregulated icaA and increased intercellular adhesion, which might result in increased biofilm formation of S. aureus at the 4% and 6% NaCl concentrations. Other studies reported that the rbf gene promotes S. aureus biofilm formation by repressing icaR, which is a negative regulator of icaA and that rbf positively regulates biofilm formation in response to high concentrations of NaCl ( Cue et al., 2009; Lim et al., 2004). However, although the expression level of icaA increased, the expression levels of the rbf gene were not different at the 4% and 6% NaCl concentrations compared with the 0% NaCl concentration (Fig. 3). Therefore, the mechanism of the induction of icaA in high osmolarity conditions should be investigated further.
Fig. 3. Relative expression of biofilm-related genes at NaCl concentrations of 0%, 4%, and 6% in Staphylococcus aureus ATCC13565.
In conclusion, NaCl in food-related conditions may increase biofilm formation of S. aureus involved in outbreak linked to ham by upregulating the icaA gene, which is responsible for intercellular adhesion.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2012R1A1A1014562).
References
- 1.Beenken K. E., Dunman P. M., McAleese F., Macapagal D., Murphy E., Projan S. J., Blevins J. S., Smeltzer M. S. Global gene expression in Staphylococcus aureus limits biofilm formation. Infect.Immun. (2004);71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhunia A. K. Staphylococcus aureus. In: Bhunia A. K., editor. Foodborne Microbial Pathogens. Springer; NY: (2008). pp. 125–134. [Google Scholar]
- 3.Biswas R., Voffu L., Simon U. K., Hentschel P., Thumm G., Gotz F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. (2006);259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 4.Bower C. K., Daeschel M. A. Resistance responses of microorganisms in food environments. Int. J. Food Microbiol. (1999);50:33–44. doi: 10.1016/S0168-1605(99)00075-6. [DOI] [PubMed] [Google Scholar]
- 5.Cue D., Lei M. G., Lee C. Y. Genetic regulation of the intercellular adhesion locus in staphylococci. Front. Cell Infect. Microbiol. (2012);2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cue D., Lei M. G., Luong T. T., Kuechenmeister L., Dunman P. M., O’Donnell S., Rowe S., O’Gara J. P., Lee C. Y. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J. Bacteriol. (2009);191:6363–6373. doi: 10.1128/JB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donlan R. M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. (2002);8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y., Kan B., Lu. Y., Szeto S. The effect of osmotic shock on RpoS expression and antibiotic resistance in Escherichia coli. J. Exp. Microbiol. Immun. (2009);13:13–17. [Google Scholar]
- 9.Kennedy C. A., O’Gara J. P. Contribution of culture media and chemical properties of polystyrene tissue culture plates to biofilm development by Staphylococcus aureus. J. Med. Microbiol. (2004);53:1171–1173. doi: 10.1099/jmm.0.45764-0. [DOI] [PubMed] [Google Scholar]
- 10.Le Loir Y., Baron F., Gautier M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. (2003);2:63–76. [PubMed] [Google Scholar]
- 11.Livermore D. M. Antibiotic resistance in staphylococci. Int. J. Antimicrob. Agents. (2000);16:S3–S10. doi: 10.1016/s0924-8579(00)00299-5. [DOI] [PubMed] [Google Scholar]
- 12.Lim Y., Jana M., Luong T. T., Lee C. Y. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. (2004);186:722–729. doi: 10.1128/JB.186.3.722-729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowy F. D. Staphylococcus aureus infections. New Eng. J. Med. (1998);339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 14.Otto M. Staphylococcal biofilms. Curr. Top. Microbiol. Immun. (2008);322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulsen L. V. Microbial biofilm in food processing. LWT-Food Sci. Technol. (1999);32:321–326. doi: 10.1006/fstl.1999.0561. [DOI] [Google Scholar]
- 16.Rode T. M., Langsrud S., Holck A., Moretro T. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. Int. J. Food Microbiol. (2007);116:372–383. doi: 10.1016/j.ijfoodmicro.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Rode T. M., Moretro T., Langsrud S., Holck A. Responses of Staphylococcus aureus to environmental stresses. In: Wong H. C., editor. Stress Response of Foodborne Microorganisms. Nova Science Publishers; NY: (2012). pp. 509–546. [Google Scholar]
- 18.Sambanthamoorthy K., Schwartz A., Nagarajan V., Elasri M. O. The role of msa in Staphylococcus aureus biofilm formation. BMC Microbiol. (2008);8:221. doi: 10.1186/1471-2180-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith J. L., Benedict R. C., Palumbo S. A. Protection against heat-injury in Staphylococcus aureus by solutes. J. Food Prot. (1982);45:54. doi: 10.4315/0362-028X-45.1.54. [DOI] [PubMed] [Google Scholar]
- 20.Takenaka S., Iwaku M., Hoshino E. Artificial Pseudomonas aeruginosa biofilms and confocal laser scanning microscopic analysis. J. Infect. Chemothe. (2001);7:87–93. doi: 10.1007/s101560100014. [DOI] [PubMed] [Google Scholar]
- 21.U. S. Food and Drug Administration (FDA) Bad bug book: Foodborne pathogenic microorganisms and natural toxins handbook. International Medical Publishing; McLean, Virginia: (2004). [Google Scholar]
- 22.Vázquez-Sánchez D., Habimana O., Holck A. Impact of food-related environmental factors on the adherence and biofilm formation of natural Staphylococcus aureus isolates. Curr. Microbiol. (2013);66:110–120. doi: 10.1007/s00284-012-0247-8. [DOI] [PubMed] [Google Scholar]
- 23.Xu H., Zou Y., Lee H. Y., Ahn J. Effect of NaCl on the biofilm formation by foodborne pathogens. J. Food Sci. (2010);75:M580–585. doi: 10.1111/j.1750-3841.2010.01865.x. [DOI] [PubMed] [Google Scholar]
- 24.Yoon H. Influence of NaCl on pathogenicity and biofilm formation of Salmonella. Master’s thesis. Sookmyung Women’s Univ.; Seoul, Korea: (2013). [Google Scholar]
- 25.Yoon H., Park B. Y., Oh M. H., Choi K. H., Yoon Y. Effect of NaCl on heat resistance, antibiotic susceptibility, and Caco-2 cell invasion of Salmonella. Biomed Res. Int. (2013) doi: 10.1155/2013/274096. Article ID 274096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zapun A., Contreras-Martel C., Vernet T. Penicillin- binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. (2008);32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]