Notch signalling is a key intercellular communication mechanism that is essential for cell specification and tissue patterning, and which coordinates critical steps of blood vessel growth1–3. Although subtle alterations in Notch activity suffice to elicit profound differences in endothelial behaviour and blood vessel formation2,3, little is known about the regulation and adaptation of endothelial Notch responses. Here we report that the NAD+-dependent deacetylase SIRT1 acts as an intrinsic negative modulator of Notch signalling in endothelial cells. We show that acetylation of the Notch1 intracellular domain (NICD) on conserved lysines controls the amplitude and duration of Notch responses by altering NICD protein turnover. SIRT1 associates with NICD and functions as a NICD deacetylase, which opposes the acetylation-induced NICD stabilization. Consequently, endothelial cells lacking SIRT1 activity are sensitized to Notch signalling, resulting in impaired growth, sprout elongation and enhanced Notch target gene expression in response to DLL4 stimulation, thereby promoting a non-sprouting, stalk-cell-like phenotype. In vivo, inactivation of Sirt1 in zebrafish and mice causes reduced vascular branching and density as a consequence of enhanced Notch signalling. Our findings identify reversible acetylation of the NICD as a molecular mechanism to adapt the dynamics of Notch signalling, and indicate that SIRT1 acts as rheostat to fine-tune endothelial Notch responses.
We investigated the role of SIRT1 in the regulation of endothelial Notch signalling. Notch activity was analysed in response to DLL4 stimulation in scrambled or SIRT1 short interfering (si)RNA-transfected human umbilical vein endothelial cells (HUVECs) by assessing the activity of the Notch reporter genes TP1-luciferase and 4×CBF1-luciferase as well as the expression of the endogenous Notch target genes NRARP and HEY2. Knockdown of SIRT1 by siRNA did not alter basal Notch activity, but significantly enhanced Notch reporter gene activity and target gene expression after DLL4 stimulation (Fig. 1a–c and Supplementary Fig. 2a, b). Comparable results were obtained with a non-related pool of SIRT1 siRNAs (Supplementary Fig. 2b, c). Importantly, enhanced Notch responsiveness to DLL4 in SIRT1-deficient endothelial cells was abrogated by the γ-secretase inhibitor dibenzazepine (DBZ) or by mutation of the CBF1-binding sites in the Notch reporter gene (Fig. 1a, b). Conversely, activation of SIRT1 signalling using the small molecule SRT2183 (ref. 4) inhibited DLL4-mediated induction of Notch reporter activity and target gene expression, indicating that SIRT1 negatively modulates DLL4/Notch signalling in endothelial cells (Fig. 1d, e).
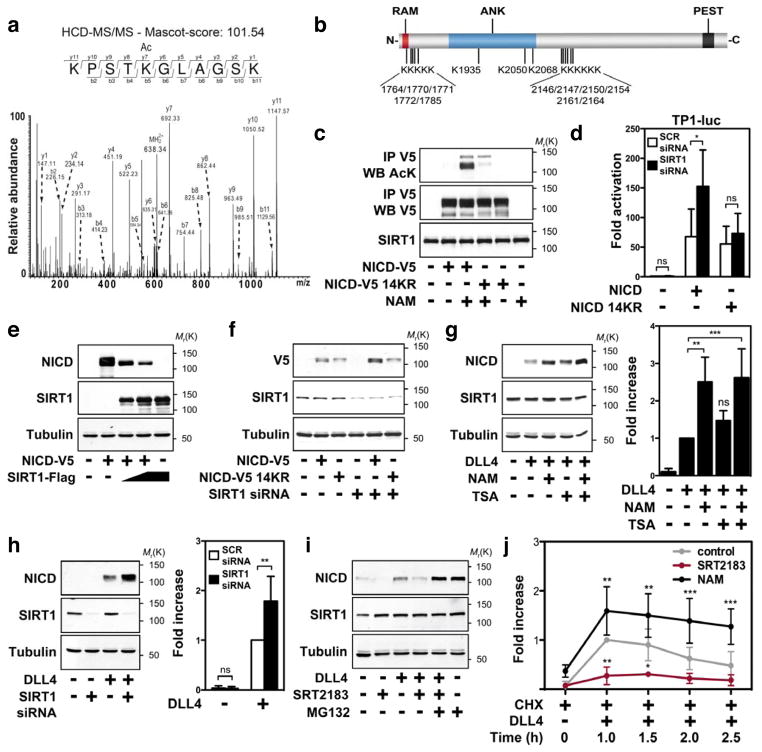
Figure 1. SIRT1 limits endothelial DLL4/Notch signalling and targets NICD for deacetylation.
a, b, TP1- (a), 4×CBF1- or CBF1 mutant- (mt) luciferase activity (b) in control (scrambled) or SIRT1-siRNA-transfected endothelial cells. c, NRARP and HEY2 mRNA levels in SCR or SIRT1 siRNA-transfected endothelial cells. d, e, TP1-luciferase activity (d) and NRARP and HEY2 mRNA levels (e) in endothelial cells stimulated with SRT2183. f, TP1-luciferase activity in control and SIRT1-deficient endothelial cells co-transfected with NICD. g, TP1-luciferase activity in endothelial cells transfected with combinations of NICD, SIRT1 and SIRT1 H363Y. h, i, Co-immunoprecipitations from SIRT1–Flag and NICD–V5 or NICD(ΔC)–Myc co-transfected 293 cells. IP, immunoprecipitation; WB, western blot. j, Co-immunoprecipitation of NICD and SIRT1 in endothelial cells treated with or without MG132. k, Acetylation of NICD–V5 in 293 cells treated with NAM, TSA, or combinations thereof. l, Acetylation of NICD in endothelial cells treated with NAM. m, Acetylation of NICD–V5 in SIRT1-siRNA-transfected 293 cells. n, Deacetylation assay with NICD(ΔC)–Myc, recombinant wild-type or H363Y SIRT1. Relative quantifications of NICD acetylation are shown on the right of the panel in l, m. DLL4 was used to stimulate Notch signalling in endothelial cells in a–e, j and l. All experiments n ≥ 3; error bars, mean ± s.d. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
Analysis of the basal messenger RNA expression of critical Notch pathway components such as NOTCH1, NOTCH4, MAML1 and NCOR1 in control and SIRT1-deficient endothelial cells revealed no significant differences (Supplementary Fig. 2d). However, SIRT1-deficient endothelial cells displayed markedly enhanced Notch activity after overexpression of NICD, as assessed using different Notch reporter genes (Fig. 1f and Supplementary Fig. 2e, f). On the other hand, co-transfection of SIRT1 with NICD inhibited Notch activity in a dose-dependent manner (Supplementary Fig. 2g). The inhibitory effect of SIRT1 required its deacetylase activity, as a catalytically inactive SIRT1 mutant (SIRT1 H363Y) was unable to inhibit Notch responses (Fig. 1g). Taken together, these results demonstrate that inhibition of Notch signalling by SIRT1 occurs in endothelial cells that receive activating DLL4/Notch signals and indicate a regulatory mechanism involving NICD as well as the deacetylase activity of SIRT1.
Co-immunoprecipitation experiments illustrated that overexpressed NICD associates with SIRT1–Flag and endogenous SIRT1, but not with other tested sirtuins (Fig. 1h and Supplementary Figs 3a–c). Intriguingly, NICD–V5 levels were consistently lower in SIRT1–Flag overexpressing lysates (Fig. 1h and Supplementary Fig. 3a). This reduction in NICD protein levels was not observed when SIRT1 was co-expressed with a NICD mutant lacking the carboxy-terminal PEST domain important for proteasomal degradation (NICD(ΔC)), although its binding to SIRT1 was retained (Fig. 1i). These data indicate that SIRT1 might negatively regulate Notch signalling by promoting NICD degradation. Consistent with this model, the interaction between endogenous NICD and SIRT1 was enhanced when endothelial cells were treated with the proteasomal inhibitor MG132 (Fig. 1j). Further characterization of this protein interaction revealed that deletion of the SIRT1 catalytic domain abolished the binding of SIRT1 to NICD, indicating that NICD might be a substrate for the deacetylase SIRT1 (Supplementary Fig. 3d, e).
To assess whether SIRT1 acts as a NICD deacetylase, we asked whether NICD is an acetylated protein. We transfected 293 cells with NICD–V5 and the acetyltransferases PCAF and p300, which bind to NICD and function as coactivators at Notch-regulated promoters5,6. NICD was robustly acetylated in cells co-expressing PCAF or p300 (Supplementary Fig. 4a, b). In contrast, Tip60, which has been reported to acetylate NICD after ultraviolet radiation7, was unable to acetylate NICD under our experimental conditions (Supplementary Fig. 4c). Blocking deacetylase activity with the sirtuin inhibitor nicotinamide (NAM) induced acetylation of overexpressed as well as endogenous NICD in endothelial cells, whereas the class I/II histone deacetylase inhibitor trichostatin A (TSA) had only a minor effect (Fig. 1k, l). Notably, knockdown of SIRT1 increased basal and p300-induced NICD acetylation, whereas overexpression prevented its acetylation (Fig. 1m and Supplementary Fig. 4d, e). Moreover, wild type but not the inactive H363Y SIRT1 mutant deacetylated NICD (NICD(ΔC)) in an in vitro deacetylation assay in a NAD+-dependent manner (Fig. 1n). These data demonstrate that NICD is reversibly acetylated and identify SIRT1 as a bona fide NICD deacetylase.
Using liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS) we found 14 acetylation sites in the NICD targeting conserved lysine residues (Fig. 2a, b and Supplementary Fig. 5a, b). Mutation of these lysines to arginine (NICD(14KR)) abolished NICD acetylation induced by PCAF, p300, or upon NAM treatment or SIRT1 knockdown (Fig. 2c and Supplementary Fig. 5c–e). Compared to wild-type NICD, the NICD(14KR) mutant had a similar ability to activate Notch reporter genes, but was resistant to changes in the level of SIRT1 (Fig. 2d and Supplementary Fig. 6a, b). These data indicate that a set of NICD lysine residues confer negative regulation of Notch signalling by SIRT1.
Figure 2. Destabilization of NICD by SIRT1.
a, HCD (higher energy collisionally activated dissociation) MS/MS spectrum showing acetylation of NICD lysine residue 2154. b, Overview of acetylated lysines in the NICD. RAM, RBPj-associated molecule; ANK, ankyrin repeats; PEST, proline (P), glutamic acid (E), serine (S) and threonine (T) enriched sequence. c, Acetylation of NICD–V5 wild type or 14KR in transfected 293 cells treated with NAM. d, TP1-luciferase activity in scrambled and SIRT1-siRNA-transfected endothelial cells co-transfected with wild-type or NICD(14KR). e, NICD protein levels in 293 cells expressing NICD–V5 and SIRT1–Flag. f, NICD–V5 protein levels in 293 cells transfected with scrambled or SIRT1 siRNA and NICD–V5 wild type or 14KR. g, h, NICD protein expression in endothelial cells treated with NAM and/or TSA (g) or transfected with SIRT1 siRNA (h). Relative quantifications of NICD protein levels are shown on the right of the panel. i, NICD protein levels in endothelial cells pre-treated with or without MG132 and/or SRT2183. j, Endothelial cells were pre-treated with CHX and NAM or SRT2183 and NICD protein levels analysed at the indicated time points. DLL4 was used to stimulate Notch signalling in endothelial cells in g–j. NICD protein levels in g–i were assessed after 6 h of DLL4 stimulation. All experiments n ≥ 3; error bars, mean ± s.d. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
Intriguingly, conditions that favour NICD acetylation increased NICD protein levels (Fig. 1k, l and Supplementary Figs 4a, b and 7a). Because NICD undergoes rapid ubiquitin-mediated degradation1, and acetylation can impair ubiquitination, we explored the effects of SIRT1 on NICD protein degradation and stability. Knockdown of SIRT1 increased, whereas overexpression decreased, NICD protein levels (Fig. 2e, f and Supplementary Fig. 7b). Importantly, protein levels of the NICD(14KR) mutant were not enhanced in SIRT1-deficient or NAM-treated cells (Fig. 2f and Supplementary Fig. 7c). Similarly, blocking SIRT1 activity in endothelial cells by NAM or siRNA increased endogenous NICD protein levels, whereas activation of SIRT1 by SRT2183 reduced endogenous NICD protein levels (Fig. 2g, h and Supplementary Fig. 7d). Treatment of endothelial cells with MG132 led to an increase in NICD protein levels, which were not further enhanced by NAM treatment (Supplementary Fig. 7e). MG132 also prevented downregulation of NICD after stimulation with SRT2183 (Fig. 2i). Together with the unaltered NOTCH1 mRNA expression these data indicate that SIRT1 affects the proteasomal degradation of NICD (Supplementary Fig. 7f). Next, we examined the effect of SIRT1 on the decay of NICD protein levels in response to DLL4 stimulation in cycloheximide-treated endothelial cells. Blocking SIRT1 activity by NAM or SIRT1 siRNA increased, whereas activation by SRT2183 decreased, the amplitude and duration of endogenous NICD protein levels (Fig. 2j and Supplementary Fig. 8a–d). Concomitantly, SIRT1 inhibition by NAM or siRNA-mediated knockdown reduced levels of ubiquitinated NICD in cells that co-expressed NICD–V5 and HA-tagged ubiquitin (Supplementary Fig. 9a–c). Together, these data support a model in which acetylation interferes with ubiquitin-dependent proteasomal degradation of NICD, hence enhancing Notch responses. The deacetylase activity of SIRT1 counteracts the stabilization of NICD by priming it for ubiquitin-mediated proteolysis (Supplementary Fig. 1).
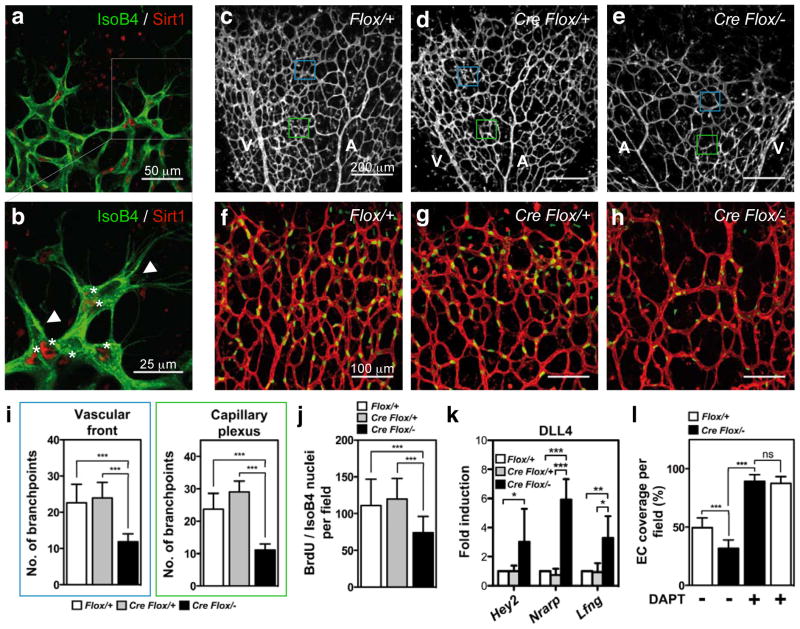
To assess whether SIRT1 regulates Notch responses in vivo, we inactivated the catalytic domain of SIRT1 in endothelial cells (Supplementary Fig. 10a)8 and assessed postnatal retinal angiogenesis in mice. Recent studies showed that DLL4/Notch signalling coordinates retinal angiogenesis by controlling the specification of endothelial cells into tip and stalk cells9–14. Tip cells express high levels of DLL4 and guide nascent sprouts, whereas following stalk cells receive DLL4/Notch signals from tip cells to form the vascular tube. Immunofluorescence staining revealed that Sirt1 is robustly expressed at the angiogenic front of the vasculature, particularly in stalk cells (Fig. 3a, b), in which Notch activity is most prominent9,13. Compared to Sirt1flox/+ and Tie2cre;Sirt1flox/+ control mice, endothelial-cell-restricted Sirt1 mutant mice (Tie2cre;Sirt1flox/−) displayed a delayed expansion of the vascular plexus and a significant reduction in vessel density and endothelial cell proliferation (Fig. 3c–j and Supplementary Fig. 10b–e). These phenotypic changes resemble those caused by enhanced Notch activity in stalk cells13 and were mirrored by enhanced expression of the Notch target genes Nrarp, Hey2 and Lfng in Dll4-treated endothelial cells derived from these mice (Fig. 3k). However, the ability of tip cells to extend filopodia was unaffected in Sirt1 mutant mice (Supplementary Fig. 10f–i). Together with the prominent expression of Sirt1 in stalk cells, these findings indicate that Sirt1 negatively modulates Notch signalling in stalk cells to facilitate endothelial branching and proliferation, thereby controlling vascular growth and density.
Figure 3. Inactivation of SIRT1 enhances endothelial Notch responses in mice.
a, Sirt1 localization (red) in the retinal endothelium (isolectin B4, green). b, Higher magnification of inset in a is shown. Asterisks indicate Sirt1 expression in stalk endothelial cells; arrowheads indicate tip cells with weak or absent Sirt1 expression. c–e, Images of P5 Sirt1 control and mutant retinas stained with isoB4. Blue and green boxes indicate the vascular front and capillary plexus, respectively. A, arteries; V, veins. f–h, IsoB4 (red) and BrdU (green) labelling in P5 retinas of the respective genotypes. i, j, Statistical summary of the number of vessel branch points and the number of BrdU/IsoB4-positive cells of the respective genotypes. k, qPCR of Hey2, Nrarp and Lfng mRNA expression in Dll4-stimulated endothelial cells derived from the respective genotypes. l, Statistical summary of the percentage of IsoB4-positive vessel coverage in vehicle and DAPT-treated retinas of control and Sirt1 mutant mice. All experiments n ≥ 4; error bars, mean ± s.d. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
To understand further the effects of SIRT1 on the dynamics of tip/stalk cell behaviour, we used computational modelling15. Modelling the increase in NICD levels induced by Sirt1-deficiency revealed a delayed negative feedback regulation of the DLL4/Notch pathway, resulting in a slower selection rate of tip and stalk cells. The tip cells were, however, phenotypically normal, producing similar numbers of filopodia (Supplementary Movies 1 and 2). The predicted delay in tip and stalk cell selection would translate into both reduced branching and delayed expansion of the vascular plexus, as observed in vivo.
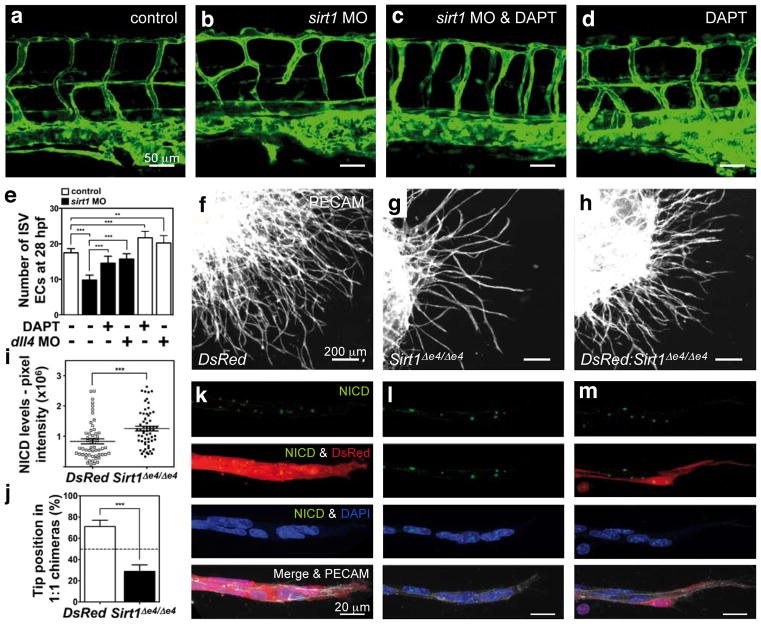
Next, we tested whether Notch inhibition would restore endothelial cell responses in Sirt1 mutants. Treatment of control animals with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) caused a hyperdense and hyperbranched vessel network, as reported9,11,13,14 (Fig. 3l and Supplementary Fig. 10m). In response to DAPT treatment, vessel density in Sirt1 mutants increased to a similar extent as in DAPT-treated controls, indicating that the reduction in vessel density is a consequence of increased Notch activity (Fig. 3l and Supplementary Fig. 10j–m). Likewise, knockdown of sirt1 in zebrafish by antisense morpholino-modified oligonucleotides resulted in defects of the trunk vasculature similar to those caused by increased Notch activity in stalk cells13,16,17 and were characterized by thin, misguided, poorly connected and hypocellular intersomitic vessels8 (Fig. 4a, b, e and Supplementary Fig. 11a, b). Treatment of sirt1 morphants with DAPT or knockdown of the artery-specific Notch ligand dll4, at least in part, normalized the patterning and cellularity of intersomitic vessels (Fig. 4a–e and Supplementary Fig. 11a–c), indicating that misregulation of Notch activity in endothelial cells contributes to the vascular patterning defects in sirt1 morphants.
Figure 4. Inactivation of SIRT1 leads to a cell-autonomous increase in Notch signalling and defective endothelial cell sprouting.
a–d, Lateral trunk views of 48 h post fertilization Tg(fli1a:EGFP)y71 zebrafish embryos. Control embryos or sirt1 morphants were treated with DAPT and DMSO. e, Endothelial cell nuclei counts in the intersomitic vessels (ISVs) of transgenic Tg(fli1a:nEGFP)y7 embryos at 28 h post fertilization (h. p. f.) treated with or without DAPT, or injected with dll4-specific morpholinos. f–h, Overview of vascular sprouts from EBs of DsRed (WT) (f), Sirt1Δe4/Δe4 (g) ES cells and 1:1 chimaeras (h). i, Quantification of total pixel intensity of the NICD immunostaining per nucleus in DsRed and Sirt1 mutant endothelial cells. j, Quantification of tip-cell contribution of each ES cell genotype in vascular sprouts. k–m, NICD immunostaining (green) of individual sprouts of EBs from the respective genotypes. DsRed cells (red), Sirt1 mutant cells (non-labelled), DAPI (blue), PECAM1 (grey). All experiments n ≥ 4. Error bars, mean ± s.d. **, P < 0.01; ***, P < 0.001.
To dissect further the effects of SIRT1 on Notch-controlled cell responses, we monitored sprout formation and proliferation in isolated human endothelial cells. Control endothelial cell spheroids extended multicellular sprouts (Supplementary Fig. 12a, b). In contrast, knockdown of SIRT1 impaired sprout formation and elongation8 (Supplementary Fig. 12a, b). Importantly, these defects were restored by γ-secretase inhibition (DBZ) (Supplementary Fig. 12a, b). Likewise, inhibition of endothelial cell growth by DLL4/Notch signalling was enhanced in SIRT1-deficient endothelial cells and rescued by DBZ (Supplementary Fig. 12c). To test the cell-autonomous effect of Sirt1 in modulating Notch responses, we used clusters of differentiating mouse embryonic stem cells (embryoid bodies (EBs)), which form branched vascular networks in response to VEGF-A stimulation18. Control EBs expressing the fluorophore DsRed-MST (DsRed) developed extensively branched vessel networks, whereas genetic inactivation of Sirt1 (Sirt1Δex4/Δex4) impaired vascular outgrowth and density (Fig. 4f, g). Recent work on genetically mosaic EBs illustrated that the ability of individual cells to gain the tip position is dynamically and competitively regulated by DLL4/Notch signalling19. Adding DsRed cells to Sirt1 mutant cells in a 1:1 ratio partially restored sprouting and branching (Fig. 4f–h), as DsRed cells (wild type for Sirt1) effectively formed tip cells, whereas Sirt1 mutant cells, exhibiting increased NICD levels, preferentially formed stalk cells (Fig. 4i–m). Furthermore, inhibition of Sirt1 by NAM in Dll4 heterozygote EBs (Dll4+/lacZ), which are characterized by reduced Notch signalling and excessive vessel branching19, restored normal branching behaviour (Supplementary Fig. 13), indicating that SIRT1 inhibition can re-adjust adequate levels of Notch signalling in Dll4+/lacZ cells.
Our studies unravel a novel role for SIRT1 as a cell-autonomous negative modulator of Notch signalling and highlight reversible acetylation of NICD as a key molecular mechanism to adjust the dynamics of Notch responses. Negative regulation of Notch signalling by SIRT1 might, however, involve additional components of the Notch pathway such as the co-repressor NCOR and transcription factors of the HES/HEY and FOXO family, which have been shown to be regulated by SIRT1 (refs 20–24). By cooperative regulation of these factors that function at different steps in the Notch pathway, SIRT1 might synergistically modulate Notch signalling to achieve robust regulation of Notch-controlled cellular responses. Notably, the regulatory function of SIRT1 in the Notch pathway seems to extend beyond NOTCH1 and to include other family members such as NOTCH4 (Supplementary Fig. 14), indicating a general mechanism for Notch regulation by SIRT1 that may apply to several Notch-controlled biological processes. However, we cannot rule out that SIRT1 might also indirectly affect Notch signalling in other tissues or cellular environments, as shown in ageing neuronal cells, where SIRT1 has been found to increase Notch signalling through a RXR-dependent regulation of ADAM10 (ref. 25). However, in endothelial cells ADAM10 expression was not affected by SIRT1 (Supplementary Fig. 15), indicating that the indirect modulation of Notch signalling through RXR is not operational in endothelial cells. Because SIRT1 requires NAD+ for its catalytic activity, it is responsive to changes in the metabolic and redox state of the cell26 and might therefore function as a cellular sensor coupling energy and oxygen homeostasis to Notch-dependent control of branching vessel morphogenesis.
METHODS
Cells and cell culture
Pooled human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and cultured as described27. HEK293T cells were purchased from ATCC and Invitrogen and cultured as recommended. Mouse lung endothelial cells (MLECs) were isolated, purified and cultured as described8.
DLL4 stimulation of endothelial cells
Lyophilized recombinant human or mouse DLL4 was purchased from R&D Systems and reconstituted at 200 μg ml−1 in PBS containing 0.1% bovine serum albumin. For stimulation of cultured endothelial cells, DLL4 was immobilized by coating culture dishes with 500 ng ml−1 DLL4 in PBS for 1 h at room temperature or overnight at 4 °C.
Reagents and pharmacological in vitro cell treatments
HUVECs or HEK293 cells were pharmacologically treated with 10 μM trichostatin A (TSA; Calbiochem), 20 mM nicotinamide (NAM; Sigma), 10 μM MG132 (Calbiochem) or 50 ng ml−1 cyclohexemide (CHX; Calbiochem). SRT2183 was used at a concentration of 20 μM (Sirtris Pharmaceuticals). Control groups were treated with the respective vehicles. To inhibit Notch signalling in vitro, HUVECs were incubated with 0.08 μM DBZ ((S,S)-2-[2-(3,5-Difluorophenyl)acetylamino]-N-(5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)propionamide). When HUVECs were co-stimulated with DLL4, cells were pre-treated for at least 1 h before being replated on DLL4-coated dishes.
Plasmids and transfections
The intracellular domain of murine Notch1 (bp 5,230 to bp 7,593) and Notch4 (bp 4,417 to bp 5,895) was cloned in-frame into the mammalian expression vectors pcDNA3.1/nV5-DEST or pDEST40 as well as pcDNA3.0 N-Flag and pDEST490 harbouring V5 or Flag tags at the N or C terminus, respectively. Human SIRT1, SIRT2, SIRT3 and SIRT5 were subcloned in a derivative of the pcDNA3.1(+) vector to generate C-terminal Flag-tagged fusions as described28. The serial deletions mutants of SIRT1 were provided by Z. Lou29, the NICD(ΔC)–Myc by R. Kopan, the p300 expression plasmid by R. Eckner, PCAF–Flag by S. L. Berger and TIP60–Flag by S. McMahon. The Notch-regulated luciferase reporter genes TP1 and 4×CBF1 were from U. Zimber-Strobl and D. Hayward, respectively. Transient transfections of HEK293 cells were carried out with Lipofectamine 2000 transfection reagent (Invitrogen). HUVECs were grown to 60–70% confluence and transfected with the Trans Pass V reagents (New England Biolabs) as recommended.
RNA interference
To silence SIRT1 gene expression, cells were transfected with a specific SIRT1 siRNA synthesized by Eurogentec (5′-GAAGTTGACCTCCTCATTG-3′) or a validated pool of siRNA duplexes directed against human SIRT1 (On-Target plus SMART pool), which was purchased from Dharmacon. A scrambled siRNA was used as a control (5′-TTCTCCGAACGTGGCACGA-3′). HUVECs and HEK293 cells were transfected with the indicated siRNAs (50 nM) using the GeneTrans II reagent (MoBiTec) or Lipofectamine 2000 (Invitrogen), respectively.
Luciferase assays
Reporter assays in HUVECs were performed with the Dual-Luciferase Reporter Assay System (Promega) and a LUMAT LB 9507 luminometer (BERTHOLD Technologies). Briefly, 24 h after co-transfection with the Notch luciferase reporters and the constitutive Renilla luciferase reporter pGL4.74hRluc/TK (Promega) HUVECs were lysed and reporter assays performed according to the manufacturers’ protocols. For siRNA experiments, cells were transfected with siRNAs and after 24 h transfected with the Notch luciferase reporters and the constitutive Renilla luciferase reporter. For experiments in which Notch activity was induced by DLL4, transfected HUVECs were replated on DLL4-coated dishes 6 h after plasmid transfections. Luciferase activity was measured after an additional 24 h. Reporter activity was adjusted for the internal Renilla luciferase control and is expressed relative to control.
RNA analysis by real-time quantitative PCR
RNA from HUVECs or MLECs was isolated with the RNeasy Kit (Qiagen) as recommended. TaqMan Gene Expression Assays for HEY2/Hey2 (Hs0023262_m1; Mm00469280_m1), NRARP/Nrarp (Hs01104102_m1; Mm00482529_m1), Lfng (Mm00456128_m1), NOTCH1 (Hs01062014_m1) and ADAM10/Adam10 (Hs00153853_m1; Mm00545742_m1) were obtained from Applied Biosystems and qPCR was carried out using the StepOnePlus real-time PCR system (Applied Biosystems).
Co-immunoprecipitations
For co-immunoprecipitations of overexpressed proteins, HEK293 cells were lysed 24 h post-transfection in IPLS buffer (50 mM Tris-HCL pH7.5, 120 mM NaCl, 0.5 mM EDTA and 0,5% Nonidet P-40). After pre-clearing, immunoprecipitations were performed using direct-conjugated immuno-affinity agarose beads against Flag (Sigma), Myc (Sigma) or V5 (Invitrogen) at 4 °C with gentle rotation over night. After western blotting, the presence of the co-immunopurified proteins was analysed with antibodies against Flag (1:1000, Sigma), Myc (1:1000, Santa Cruz) and V5 (1:5000, Invitrogen). For co-immunoprecipitation of endogenous SIRT1 with endogenous NICD, HUVECs were pretreated with 10 μM MG132 (Calbiochem) for 1 h and then re-plated on DLL4-coated culture dishes in the presence of MG132 for 6 h. Cells were lysed in IPLS buffer. After pre-clearing, SIRT1 was immunopurified with a mouse anti-human SIRT1 monoclonal antibody (Santa Cruz) for 12 h at 4 °C followed by incubation with A/G agarose beads (Santa Cruz) for an additional 2 h. Immune complexes were analysed by western blotting using NICD (1:1,000, Cell Signaling Technology) and SIRT1 (1:1,000, Cell Signaling Technology) antibodies.
Detection of NICD acetylation
For detection of acetylated NICD in HEK293 cells, NICD–V5-transfected HEK293 cells were treated with NAM, TSA, the respective vehicles or combinations thereof for 6 or 16 h. Acetylation was also measured in SIRT1 siRNA transfected cells 24 h after NICD–V5 overexpression. For both approaches, cells were lysed in RIPA buffer supplemented with 10 μM TSA and 20 mM NAM (AcRIPA). After preclearing cell extracts were subjected to immunoprecipitation using agarose beads coupled to a V5 antibody. The immune complexes were analysed by western blotting with polyclonal anti-acetylated lysine antibodies (AcK, 1:300, Cell Signaling or 1:1,000, Abcam). For detection of endogenous acetylated NICD, HUVECs were plated on DLL4-coated culture dishes and stimulated for 6 h in the presence or absence of NAM. HUVECs were lysed in AcRIPA buffer. After pre-clearing, NICD was purified from total cell lysates with a Notch1-specific antibody (Cell Signaling) for 12 h at 4 °C followed by incubation with protein A/G agarose beads (Santa Cruz) for an additional 2 h. Western blotting was performed with an antibody targeting acetylated lysines (1:300, Cell Signaling).
In vitro deacetylation assay
NICD(ΔC)–Myc was expressed in HEK293 cells alone or together with p300 and immunopurified using an anti-Myc antibody. Immunoprecipitated NICD(ΔC)–Myc was four times in IPLS buffer and once in ST buffer (50 mM Tris-HCL, pH 9, 4 mM MgCl2 and 0.2 mM DTT). Purified NICD(ΔC)–Myc was then incubated with recombinant wild-type or deacetylase defective (H363Y) SIRT1 in 30 μl of ST buffer containing 800 nM TSA and supplemented or not with 50 μM of NAD+. Reactions were incubated at 30 °C under robust agitation for 2 h and stopped by addition of Leammli buffer. Acetylation of NICD was then assessed by western blotting using an anti-acetylated lysine antibody.
Ubiquitination assay
For detection of ubiquitinated NICD in HEK293 cells, NICD–V5 was overexpressed in HEK293 cells together with HA-tagged ubiquitin. The effect of SIRT1 on NICD ubiquitination was assessed after knockdown of SIRT1 by siRNA or after NAM treatment. Cells were lysed in RIPA buffer and subjected to immunoprecipitation using anti-V5-coupled agarose beads. The immune complexes were analysed by western blotting with anti-HA antibodies (HA, 1:1000, Roche). For quantification of NICD ubiquitination the anti-HA signal was normalized to the NICD–V5 levels.
Western blot analysis and antibodies
SDS–PAGE and western blot analyses were performed according to standard procedures and detected with the ECL detection kit (GE Health Care Bio-Sciences). Quantification of band intensities by densitometry was carried out using the Image J software.
Mass spectrometric analysis
HEK293 cells were transfected with NICD–V5 and PCAF–Flag and 24 h after transfection, treated with nicotinamide for 6 h. After NICD–V5 immunoprecipitation proteins were separated by one-dimensional SDS–PAGE (4–12% Novex-gels, Invitrogen) and stained with colloidal Coomassie. NICD–V5 gel bands were excised and subjected to in-gel digest with trypsin30. The resulting tryptic peptides were extracted with acetonitrile, and desalted with reversed phase C18 STAGE tips31. Mass spectrometric experiments were performed on a nano-flow HPLC system (Proxeon) connected to a LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific) equipped with a nanoelectrospray source (Proxeon). The mass spectrometer was operated in the data dependent mode to monitor MS and MS/MS spectra. Survey full-scan MS spectra (from m/z 300–2,000) were acquired in the Orbitrap with a resolution of R = 60,000 at m/z 400 after accumulation of 1,000,000 ions. The five most intense ions from the preview survey scan delivered by the Orbitrap were sequenced by collision-induced dissociation (CID) in the LTQ. For higher C-trap dissociation (HCD) 30,000 ions were accumulated in the C-trap and MS/MS spectra were detected in the orbitrap at a resolution of 7,500 (ref. 32). Mass spectra were analysed using MaxQuant software33 and automated database searching (Matrix Science). All tandem mass spectra were searched against the mouse International Protein Index protein sequence database (IPI version 3.54) and concatenated with reversed copies of all sequences. The required false positive rate was set to 1% at the protein level, and maximum allowed mass deviation was set to 5 p.p.m. in MS mode and 0.5 Da for MS/MS peaks. Cysteine carbamidomethylation was searched as a fixed modification and N-acetyl protein, oxidized methionine and acetylation of lysine was searched as variable modifications. A maximum of three missed cleavages was allowed.
Site-directed mutagenesis
The acetylation-defective NICD(14KR)–V5 mutant was generated by site-directed mutagenesis replacing the acetylated lysines by arginine using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent technologies). Primer sequences are available upon request. In addition, we generated a synthetic gene encoding for the murine NICD sequence harbouring the lysine-to-arginine substitutions at the sites identified by mass spectrometry (Geneart). Using Gateway cloning the synthesized gene was cloned in frame into the mammalian expression vector pDEST490 containing a V5 tag at the C terminus.
Endothelial cell proliferation and three-dimensional sprouting assay
Cell growth was assayed by a colorimetric procedure with the cell counting kit-8 (Dojindo Laboratories), modified from a method that uses 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as recommended. HUVECs were cultured in DLL4 or solvent and treated with DBZ (0.08 μM) or DMSO (solvent). The cell counting kit-8 reagent was added to the cultures according to manufacturer’s instructions and growth assessed after 0, 24, 48, 72 and 96 h in culture. Endothelial cell spheroids of defined cell number were generated as described previously27 and sprouting assessed after 24 h.
Mouse embryonic stem cells and embryoid bodies assay
DsRed-MST ES cells and Dll4+/lacZ ES cells were obtained from A. Nagy and G. Thurston (Regeneron Pharmaceuticals), respectively, and have been described previously34, 19. The generation of Sirt1 mutant ES cells was previously described35. The lack of exon 4 in the Sirt1 gene generates a truncated protein that is catalytically inactive and both cells and mice exhibit the same phenotype as the full Sirt1 knockout, as previously described35. ES cell culture and generation of EBs were performed as described18. The following primary antibodies were used for detection: rat anti-mouse CD31 antibody (BD Biosciences), rabbit anti-mouse cleaved Notch1 (Abcam). Secondary antibodies: Alexa 647 donkey anti-rat IgG, Alexa 488 donkey anti-rabbit, (Molecular Probes), and DAPI (Sigma) for staining of nuclei.
Quantification analysis of tip cells and NICD levels in EB sprouts
Complete high-resolution three-dimensional rendering of EBs was acquired using a Laser Scanning Microscope 710 (Zeiss) equipped with a motorized stage. For quantification of tip cells, 50–200 tip cells per EB were manually scored and marked for genotypic origin using the Imaris 7.0.0 software. Tip regions were defined as the regions from the very tips to the second or third vascular loop. Quantification was done using Prism 5.0c software. For the quantification of NICD protein levels, complete 1,024 × 1,024, 12 bit images of vascular sprouts were collected with ×63 objective lens using a Laser Scanning Microscope 710 (Zeiss), keeping the same settings for all images (Supplementary Fig. 16). Images were processed using the Imaris 7.0.0 software. To identify nuclear NICD spots we used ‘Surface’ Imaris function. Measurements of number of spots, volume and total pixel intensity of each spot were taken and statistics were analysed using Prism 5.0c software.
Zebrafish strains, maintenance and drug treatment
Zebrafish strains were maintained and bred under standard conditions. AB wild type strain, Tg(fli1:nEGFP)y7 and Tg(fli1a:EGFP)y136 lines were used. Fish were raised/maintained according to EU regulations on laboratory animals. DAPT (Sigma) was dissolved in DMSO and added to E3 media to give a final concentration of 100 μM DAPT plus 0.2% DMSO. Embryos were dechorionated and incubated at 28.5 °C in E3 containing 100 μM DAPT and 0.2% DMSO from the end of gastrulation until 28 h.p.f. E3 containing DMSO alone was used as a control.
Antisense morpholino oligonucleotides, fluorescence microscopy and cell counts
Antisense morpholino oligonucleotides against the exon1–intron1 splice site, 5′-CGAACCAAACTCACCAATCTGTGGC-3′, specifically targeting zygotic sirt1 (MO2-sirt1, a splice blocking MO)8, and the 5′-GTTCGAGCTTACCGGCCACCCAAAG-3′, specifically targeting zygotic dll4 (MO1-dll4, a splice blocking MO)16 were used. For cell counting experiments, 28 h.p.f. Tg(fli1a:nEGFP)y7 embryos were fixed in 1% paraformaldehyde in PBS containing CaCl2 0.15 mM and 4% sucrose overnight at 4 °C and were then washed in PBS at room temperature. Embryos were permeabilized for 1–3 h at room temperature and then mounted in 85% glycerol solution. Using confocal microscope, endothelial cells of Tg(fli1a:nEGFP)y7 were counted from projections of Z-series of optical sections. The region counted in each specimen spanned two somites (three intersegmental vessels).
Immunofluorescence and analysis of postnatal angiogenesis in the mouse retina
Endothelial-restricted Sirt1 mutant mice were on a C57BL/6 genetic background and generated as described8. Mice were killed at postnatal day 5 (P5) and their eyes collected for analysis. To inhibit Notch signalling in mice, DAPT (Calbiochem) was dissolved in 10% ethanol and 90% corn oil. The DAPT solution was then injected intraperitoneally at a dose of 100 mg kg−1 per mouse. Control mice were injected with the same amount of vehicle. Mice were treated for 48 h. Injections were performed once a day, at P3 and P4 and eyes harvested at P5. For whole-mount analysis of retinal blood vessels, retinas were fixed in 4% PFA for 2 h at 4 °C, or in MeOH at −20 °C. Antibodies were diluted in PBS containing 0.5% BSA and 0.25% Tween 20 except for isolectin-B4, which was diluted in PBlec (PBS (pH 6.8), 1% Triton X-100, 0.1 mM CaCl2, 0.1 mM MgCl2, 0.1 mM MnCl2). Primary antibodies used: biotinylated-isolectin-B4 (Vector Laboratories and Molecular Probes), BrdU (Molecular Probes) and SIRT1 (Upstate). Alexa-488- or 568-conjugated secondary antibodies were used (Molecular Probes). For labelling of proliferating cells, 50 mg kg−1 of BrdU (BD Biosciences) per pup was injected intraperitoneally 3 h before they were killed. Retinas were fixed for 2 h in 4% PFA and than incubated for 2 h in 2N HCl. Retinas were blocked with 1% BSA/0.5% Tween 20 in PBS and incubated overnight at 4 °C with a mouse monoclonal anti-BrdU antibody. Secondary detection was performed with goat anti-mouse Alexa Fluor-coupled secondary antibody. After BrdU staining, retinas were fixed for 30 min in 4% PFA, washed and than stained with directly conjugated Isolectin-B4. Retinas were flat mounted (Dako fluorescent mounting medium) and confocal laser scanning microscopy was performed using Zeiss LSM 510 microscope. All quantifications were done with highresolution confocal images using Z-sections of the sample. For quantifying vessel migration, the distance of vessel growth from the optic nerve to the periphery was measured using the ImageJ software. For vessel density, the number of vessel branch points was counted from five fields per retina sized 100 × 100 μm, from at least 10 retina samples per group. The number of filopodial extensions were quantified at the angiogenic front and counted in at least 18 fields (sized 132 × 132 μm, 6 retinas per group). The total number of filopodia was normalized to 100 μm of vessel length at the angiogenic front. BrdU-labelled isolectin B4-positive endothelial cells were counted in 30 fields (sized 450 × 450 μm, 6 retinas per group).
Computational modelling of endothelial tip-cell selection
A vessel segment comprising seven cells, with one cell per vessel cross-section, was used with all parameters set as described15. Sirt1 loss was modelled by extending the number of time steps that the ‘current active Notch’ level, which represents NICD in the model, affected downstream targets. In the selection model, Notch activity leads to downregulation of VEGFR2 receptors. It was only effective for one time step when modelling wild-type vessels (one time step representing 15 s in the model). To match the observed 2.5-fold increase in NICD due to the loss of Sirt1 (referred to as Sirt1 null vessels), the time the current active Notch level was effective for was extended to three time steps; however, on the third time step only half of that NICD level continued to affect VEGFR2 downregulation. To investigate the sensitivity and effect of increasing the NICD lifetime, further simulations were performed where it was set to a hypothetical 4.5- and then 9.5-fold increase, using a similar approach.
Statistical analysis
Statistical analysis was performed by ANOVA followed by Bonferroni’s multiple comparison test, or Student’s t-test using Prism 5 (GraphPAD Software Inc.). P values of <0.05 were considered significant.
Supplementary Material
Acknowledgments
We are thankful to F. W. Alt, R. Kopan, Z. Lou, E. Seto, S. L. Berger, S. Diane Hayward, S. McMahon, G. Thurston and N. D. Lawson for reagents and to I. Dikic for comments. This work was supported by grants from the DFG (PO1306/1-1, SFB 834/A6 and Exc 147/1). F.D. was supported by the Interuniversity Attraction Poles Program–Belgian Science Policy (IUAP-BELSPO PVI/28). R.M. is supported by the Sidney Kimmel Cancer Research Foundation, a New Investigator Grant from the Massachusetts Life Sciences Center, an AFAR Research Grant and NIH grants (R01DK088190-01A1 and R01GM093072-01). H.G. is supported by Cancer Research UK, the European Molecular Biology Organisation Young Investigator Programme, and The Lister Institute of Preventive Medicine. H.G. and K.B. are supported by the Fondation Leducq Transatlantic Network of Excellence ARTEMIS. C.A.F. is supported by the Marie Curie FP7 People initiative. G.D. and M.M. thank F. Pezzimenti for fish care and technical help, and AIRC (Associazione Italiana per la Ricerca sul Cancro) for financial support.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Author Contributions V.G. and M.P. designed and guided research. V.G., G.D., C.A.F., M.K., L.-K.P., K.B., L.T., F.D., M.H.H.S., B.Z., R.P.B., M.M., H.G. and M.P. performed research. V.G., G.D., C.A.F., M.K., L.-K.P., K.B., L.T., F.D., M.M., H.G. and M.P. analysed data. R.M., C.H.W. and T.B. provided reagents and/or technical support. T.B., A.M.Z. and S.D. gave conceptual advice. V.G., H.G. and M.P. wrote the paper. All authors commented on the manuscript.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21:2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- 3.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurooka H, Honjo T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 6.Oswald F, et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim MY, et al. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol Cell Biol. 2007;27:6506–6519. doi: 10.1128/MCB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potente M, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 10.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 11.Suchting S, et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci USA. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci USA. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phng LK, et al. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev Cell. 2009;16:70–82. doi: 10.1016/j.devcel.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benedito R, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 15.Bentley K, Mariggi G, Gerhardt H, Bates PA. Tipping the balance: robustness of tip cell selection, migration and fusion in angiogenesis. PLoS Comput Biol. 2009;5:e1000549. doi: 10.1371/journal.pcbi.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- 17.Leslie JD, et al. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- 18.Jakobsson L, et al. Heparan sulfate in trans potentiates VEGFR-mediated angiogenesis. Dev Cell. 2006;10:625–634. doi: 10.1016/j.devcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Jakobsson L, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nature Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- 20.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hisahara S, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci USA. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 23.Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nature Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 24.Kitamura T, et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses β-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potente M, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.North BJ, et al. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 29.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 30.Shevchenko A, et al. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 31.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 32.Olsen JV, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 34.Vintersten K, et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- 35.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.