Abstract
The nucleosomal subunit organization of chromatin provides a multitude of functions. Nucleosomes elicit an initial ~7-fold linear compaction of genomic DNA. They provide a critical mechanism for stable repression of genes and other DNA-dependent activities by restricting binding of trans-acting factors to cognate DNA sequences. Conversely they are engineered to be nearly meta-stable and disassembled (and reassembled) in a facile manner to allow rapid access to the underlying DNA during processes such as transcription, replication and DNA repair. Nucleosomes protect the genome from DNA damaging agents and provide a lattice onto which a myriad of epigenetic signals are deposited. Moreover, vast strings of nucleosomes provide a framework for assembly of the chromatin fiber and higher-order chromatin structures. Thus, in order to provide a foundation for understanding these functions, we present a review of the basic elements of nucleosome structure and stability, including the association of linker histones.
The Nucleosome Core
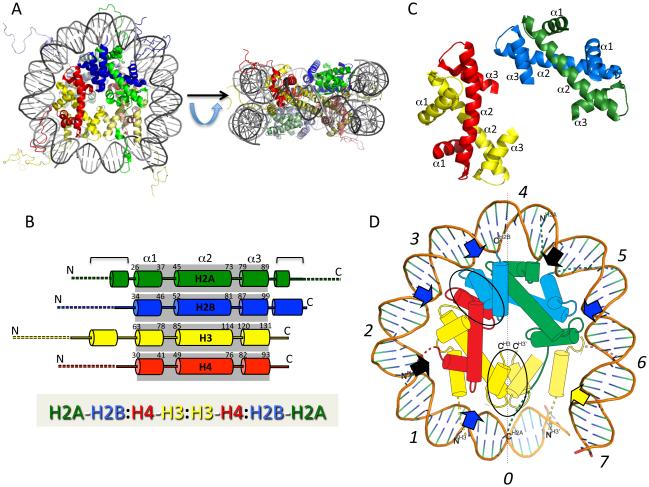
Nucleosomes constitute the basic repeating subunit of chromatin. Each nucleosome can be considered as composed of a nucleosome ‘core’, linker DNA, and in most instances, a linker histone. We will first consider the components and structure of the nucleosome core, and then the linker DNA and linker histone [chromatin structures beyond the nucleosome level will be covered in subsequent chapters in this volume]. The structure of the nucleosome core is relatively invariant from yeast to metazoans [1,2], and includes a 147 bp segment of DNA and two copies each of four core histone proteins. The core histones assemble into a spool-like structure onto which the core DNA is wrapped, in about 1¾ left-handed superhelical turns, forming a squat disc-like structure about 5.5 nm in height and 11 nm in diameter [3] (Fig. 1A). The core DNA is in tight association with the core histones and is protected from nuclease digestion whereas the linker DNA is rapidly digested. Indeed the term “nucleosome core particle” was originally defined as the product of extensive micrococcal nuclease digestion of native chromatin [4].
Fig. 1. Structural Details of a Nucleosome Core.
A. Model of a nucleosome core (PDB: 1KX5, ref: [20]). Shown are a view down the superhelical axis, and a view rotated 90°about a horizontal axis, as shown, looking down the dyad axis of the nucleosome. H2A, green, H2B, blue, H3, yellow, H4, red. Proteins in lower half of nucleosome are lighter in color. B. Top: Schematic showing secondary structure of the core histone proteins, with α-helices represented by columns. Dashed lines indicate approximate residues within ‘tail’ domains; shaded boxes indicate the 3-helix histone fold domains within each protein, with first and last residues within α1, α2 and α3 helices indicated. Additional helices outside the histone fold domain are indicated by brackets, Bottom: Linear representation of primary contacts between the core histone proteins in the nucleosome core. Core histone dimerization partners are separated by dashes; dimer-dimer interactions via 4-helix bundles are indicated by colons. C. H2A-H2B (green/blue) and H3-H4 (yellow/red) histone fold domain dimers. α1, α2 and α3 helices indicated, corresponding to B. D. Schematic showing one-half of nucleosome core, looking down the DNA superhelix axis. Superhelix sites are indicated by italicized numerals; 4-helix bundles between H3:H3 and H4:H2B are highlighted by ovals; blue and black arrows indicate paired loop and paired-end-of-helix DNA interaction sites. Yellow arrow indicates site of interaction centered on the N-helix in H3. Note, a small amount of DNA and H3 from the non-depicted half of the nucleosome core are shown for clarity, lighter in color.
The Core Histones
The four core histones are relatively small (11-15 kDa), very basic proteins that are highly conserved among eukaryotic species. About 25% of the mass of each core histone is contained within an N-terminal ‘tail’ domain that is unstructured in the absence of DNA or other macromolecular interactions (discussed below). The bulk of the histone protein mass is comprised by a largely α-helical C-terminal domain that provides for histone-histone interactions to form the octameric column-like structure onto which the DNA is wrapped. A ‘histone fold’ motif comprises the majority of each of the C-terminal domains, which has a nearly identical structure in all four core histones, despite little primary sequence homology between the proteins [5]. This motif is comprised of two shorter α-helices (α1 and α3) 9 to 14 residues in length, that bracket a relatively long (~29 residue) central α-helix (α2), with the α-helices connected by short loop/β-segments [1] (see Fig. 1B). Note α-helical regions are also present outside of the histone fold domain (Fig. 1B, brackets, see below). The histone fold forms an extensive protein-protein interface described as a ‘handshake’ interaction that directs heterodimerization of histones H2A with H2B and H3 with H4 [5,6] (Fig. 1C). Interestingly, the H3/H4 dimer has a modest stability of about 6-7 kcal/mol under physiological conditions, compared to 11-12 kcal/mol for the H2A/H2B dimer [7-9]. Notably, the histone fold motif has come to be recognized as a ubiquitous dimerization interface conserved throughout evolution and found in a wide array of protein complexes, including those that do not associate with DNA [10].
The histone dimers associate with each other primarily via 4-helix bundles at the dimer-dimer interfaces (Fig. 1D, highlighted by black ovals). The H3/H4 dimer self-associates via an H3:H3 interface to form a stable tetramer in solutions of physiological ionic strengths. A single H2A/H2B dimer associates with either end of the H4/H3:H3/H4 tetramer via a H4:H2B 4-helix bundle to form a symmetrical string of four tetramers that generate a helical ramp of DNA contact sites (Fig. 1B, bottom, and D). Of note, the H2B:H4 interface is significantly less stable than the H3:H3 interface such that the entire histone octamer is formed only when wrapped by DNA or in solutions of high (~2 M) salt due to the high positive charge of the component proteins. The H2A/H2B and H3/H4 dimers form a symmetrical arc-shaped structure in which three distinct DNA binding sites are arranged along the outer curved edge (Fig. 1D, blue and black arrows). Each of the three DNA contacts are composed of paired structural elements from each dimer and include two paired β-loops (blue arrows) and a central set of paired α-helical ends (black arrows), each spaced to contact the DNA backbone where it faces the proteins, approximately once every helical turn of DNA [1,6]. DNA contacts within the nucleosome are discussed further below.
Nucleosome Core DNA
The DNA associates with the histone octamer such that nucleosome dyad axis passes through a single base pair at the center of the structure. The DNA helix at the dyad straddles the H3:H3 interface with the minor groove oriented directly away from the histone surface (Fig. 1 D). This central (dyad) position is typically demarcated as position 0 in the DNA superhelix, with other super helix positions (+/−1, +/−2, etc,) located at outward-facing minor grooves found at successive helical turns extending in either direction from super helix location (SHL) 0 (Luger et al., 1997) (Fig 1D). Given the formal 2-fold symmetry in the nucleosome, the location of a single base pair on the dyad indicates that an odd number of base pairs, nominally 147, should be considered as the length of DNA within the nucleosome core. However, it is important to keep in mind that micrococcal nuclease excision of nucleosome core particles from chromatin results in a wide distribution of DNA lengths due to the sequence preference of the enzyme [11].
DNA is highly contorted by assembly into nucleosome cores. First, the DNA, which has a persistence length of about 150 bp, must be bent into ~1¾ left-handed superhelical turns of about 80 bp/turn. This bending is largely accomplished through base pair roll into the minor and major grooves where these grooves are oriented toward the histone octamer surface, with very little contribution of tilt [1,12,13]. The nucleosomal DNA superhelix is relatively flat, with a rise of about 30 Å per ~80 bp superhelical turn and a diameter of about 42 Å, although the curvature is not uniform, with sharpest bends occurring at about 1½ and 4½ turns from the center (dyad position) of the nucleosome [1] (Fig. 1D). Interestingly, the superhelical rise is accomplished primarily through base pair slide that occurs primarily at the aforementioned periodic sites of roll-based bending, where the minor and major grooves face the histone octamer [13]. Thus dinucleotide base pair steps with roll-based bending and slide occur every 5 bp and separate short stretches of relatively straighter DNA within the nucleosome.
Although some features of sequence-dependent variation in B-DNA structure found in naked DNA persist in nucleosomes, and affect the overall stability of DNA wrapping in the structure, clearly the histones proteins are dominant in defining the overall shape of the DNA [14,15]. The significant and well-defined deformation of the nucleosome core DNA and the sequence-dependent structure and anisotropic deformability of DNA provides for the important phenomenon of sequence-dependent nucleosome positioning(s) [16].
Furthermore, association with the core histone octamer constrains the helical twist of the DNA, decreasing the average # bp/helical turn from about ~10.5 to ~10.2 within the nucleosome [1,12,17,18]. In addition to providing an additional structural feature that might be exploited for translational positioning, the change in twist accounts for the so-called nucleosomal linking number paradox whereby the nucleosome contains an apparent 1¾ superhelical turns but constrains only ~1 negative supercoil of DNA [19]. However, it is important to note that the DNA twist is constrained by localized histone-DNA contacts that precisely position DNA backbone positions along one face of the DNA and thus the total twist between contact points formally may vary by integral base pair units [12]. Indeed, the initial high-resolution crystal structure analysis of a nucleosome core containing 146 base pairs of DNA showed an asymmetric disposition of 72 and 73 base pairs in each of the two halves of the structure (with a single base pair at the dyad) resulting in a ‘twist defect’, in which the DNA was overwound by 1 base pair between sets of contact points [1] to account for the ‘missing’ base pair. Further analyses indicated that the site of over-winding was partially delocalized throughout the nucleosome core [20,21], and that such ‘twist defects’ may be a common feature of most nucleosomes. This suggests a mechanism whereby the DNA may be moved with respect to the histone octamer without the need for concerted disruption of all histone-DNA interactions by diffusion of overwound regions directionally through the nucleosome, [1,12,20].
Histone-DNA contacts within the nucleosome core
Investigations of the salt-dependence of heat-induced DNA end fraying in nucleosome core particles led McGhee and Felsenfeld to the conclusion that a surprising small number (15%) of DNA phosphates were involved in direct charge-charge interactions with the core histone octamer at the nucleosome periphery [22]. Indeed the initial high-resolution X-ray crystal structure of a nucleosome core showed that only 2 phosphates per strand per helical turn are involved in direct interactions with the core histones [1]. Contacts occur primarily between the paired-loop and paired-end-of-helix elements in the histone fold domains and the DNA backbone where the minor groove is oriented toward the protein surface (Fig. 1D, see above). These contacts to the DNA include interactions between the phosphate residues and lysine and arginine side chains as well as main chain amide nitrogens [1,20]. Importantly, at each of the fourteen main backbone contact points over the length of the 147 bp of nucleosome core DNA, a highly conserved arginine interacts with the minor groove, to help precisely position the DNA, facilitate overall DNA bending, and the superhelical shape [1,20,23]). Most of these arginines insert into the minor groove and also form hydrogen-bonds and non-polar interactions with deoxyriboses [20]. Eight of these arginines (H3 R83, H4 R45, H2A R42 and H2A R77) emanate from paired loop elements and are especially conserved in position among available crystal structures of nucleosome cores and precisely position a phosphate in relation to the protein [12,23]. Indeed mutation of at least some of these ‘special’ positions are associated with SIN mutations in yeast, and lead to increased nucleosome mobility and accessibility.
Several structural elements of the core histones outside of the histone fold domains, including as the histone tail domains (discussed below) and extra-fold α-helices (Fig 1B) also play important roles in contacting nucleosome DNA. For example, each of the four histone fold dimers spans about 30 bp of contact with nucleosome DNA, such that the 4-dimer unit accounts for about 120 bp of contact with DNA within the nucleosome core [1]. Importantly, the full 147 bp of stable interactions found in the nucleosome core includes an additional strong contact to the edge of the nucleosome core DNA made by the N-terminal non-histone fold α-helix within H3 and the beginning of the H3 N-terminal tail domain [1] (Fig. 1B, brackets and Fig. 1D, yellow arrow). This crucial point of contact inhibits invasion of the nucleosome by processive enzymes and includes a lysine residue (K56) that is acetylated in vivo and regulates the strength of histone-DNA interactions and the probability of DNA unwrapping at the edge of the nucleosome core [24].
Core histone tail domains
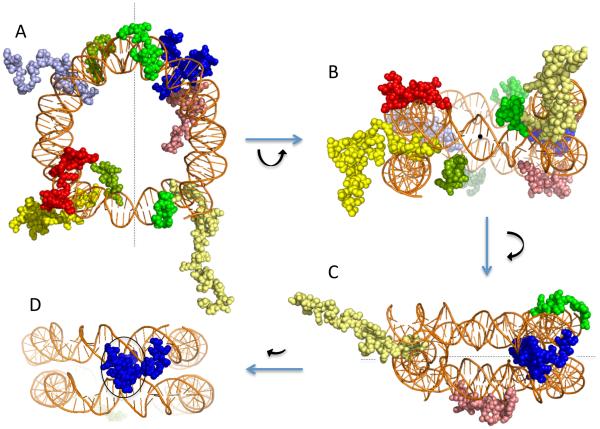
Beyond the structured regions that form the spool onto which the DNA is wrapped, the remaining 25–30% of the mass of the core histones consists of the largely structurally undefined but evolutionarily conserved “tail” domains (Fig. 1B, dashed lines). These domains are located at the N-terminal portion of all four core histone proteins and the C-terminus of histone H2A and were originally defined by their sensitivity to proteases, indicating their exposure to solvent and dynamic nature relative to the so-called ‘structured’ domains [25]. X-ray crystal structures of nucleosome core particles show that the tail domains follow minor grooves of DNA to the exterior of the nucleosome [1,20]. The tails of histones H3 and H2B exit to the exterior of the nucleosome core between adjacent DNA superhelical gyres where the minor grooves of both the upper and lower gyres are aligned to form a channel (Fig. 2, yellow and blue tails, respectively). The H3 tail domains exit through aligned grooves beginning approximately at SHL 6.5/−0.5 and −6.5/0.5, while the H2B tail exits at 4.5/−2.5 and −4.5/2.5 [1,20]. The N-terminal tail domains of H2A and H4 follow minor grooves over/under the top/bottom superhelical turns to the exterior of the nucleosome (Fig. 2, green and red tails, respectively), but alternative paths are observed in some models from crystal structures, highlighting the flexible nature of these domains.
Fig. 2. Core histone tail domains and nucleosome DNA.
Several orientations with only tail domains and nucleosome DNA are shown from an X-ray crystal structure model of a nucleosome core (PDB: 1KX5, ref: [20]). A. View down the nucleosome superhelix axis. B. View rotated 90°, looking down the nucleosome dyad axis. C. View rotated 90°about vertical axis looking orthogonal to dyad axis, showing tails of H3 (yellow) and H2B (blue) exiting through the superhelical gyres while the H4 (red) and H2A (green) tails exit over/under the superhelix. D. View with additional 30°rotation about vertical axis, showing only H2B tail exiting through aligned minor grooves (black oval). Protein colors as in Fig. 1. Nucleosome dyad axis is indicated (dotted line).
The tail domains are often referred to as the “unstructured,” portions of the core histones and indeed these regions adopt random coil conformations when the proteins are free in solution, unassociated with DNA, or released from their nucleosome binding sites in high salt solutions [26,27]. The core histone tail domains contain a preponderance of lysines and arginine residues, with significant amounts of glycine, alanine and threonine in certain tail domains. Indeed, sequence analysis places these domains firmly within the class of intrinsically disordered peptides [28]. However, the majority of the tail domains associate tightly with nucleosome binding sites in nucleosomes in physiological salts [26,27], with conformational equilibrium constants of ~50-100 [29]. The tails contribute marginally to the thermal stability of mononucleosomes [30] and removal of the tail domains increases unwrapping of DNA from the nucleosome surface and accessibility of DNA-binding factors to nucleosome DNA [31-35]. Thus protease sensitivity and reported mobility on the NMR timescale are likely attributable to the ~2-4% of the time the tails spend dissociated from the nucleosome surface and in random coil conformations in physiological salt conditions. Moreover, when bound within chromatin, crosslinking evidence suggests that the tails domains adopt defined structures and make specific interactions when bound within the chromatin fiber [36,37]. Moreover, CD measurements of tail-containing and tail-lacking nucleosome cores suggest that the tail domains adopt a significant amount (about 20%) of α-helical conformation at physiological salts [38]. Interestingly, hyperacetylation of tail domains appears to increase the α-helical content of the tails [38].
The tails are readily accessible to enzymes that carry out posttranslational modifications important for epigenetic signaling. Moreover, these domains are in position to mediate internucleosomal interactions within condensed chromatin structures, and, in fact, this appears to be their primary role in organizing higher-order chromatin structures. The tail domains are essential for condensation of nucleosome arrays into secondary and tertiary chromatin structures [39-41] and interact with multiple protein and DNA targets in chromatin [42]. For example, as originally observed in a crystal structure of a nucleosome core [1], a portion of the H4 tail interacts with an acidic patch on the surface of the H2A/H2B dimer of an adjoining nucleosome, both in solution [43,44] and in cells [45]. Likewise, the H3 tail domain contacts the DNA of its own nucleosome in extended nucleosome arrays, while exclusively contacting the DNA of neighboring nucleosomes in condensed chromatin [46]. Consistent with the proposal that the tail domains fulfill multiple independent functions, both the H3 and H4 tails participate in local inter-nucleosome contacts to facilitate folding of the chromatin fiber and long-range fiber-fiber contacts to mediate formation of higher order chromatin structures [46-49]. Moreover, specific posttranslational modifications within the tail domains likely elicit distinct chromatin states by either directly or indirectly altering tail interactions [42,50,51]. For example the acetylation of lysine 16 within the H4 N-terminal tail domain reduces H4 tail interactions with an acidic pocket on the protein surface of the nucleosome formed by residues from H2A and H2B, an interaction shown to contribute to the stability of condensed chromatin structures [1,43,52-54]. In general, acetylation restricts the ability of nucleosome arrays to undergo salt-dependent folding and oligomerization, presumably by directly modulating tail interactions with protein or DNA targets [41,55,56]. However, the mechanism(s) by which most acetylation alters tail interactions is not known. Thus the core histone tail domains play critical structural and regulatory roles in nucleosomes and higher order chromatin structure.
Nucleosome dynamics
As pointed out above, the assembly of DNA into nucleosomes results in protection from nucleases and greatly restricts the binding of trans-acting factors. While a full treatment of this subject is beyond the scope of this review, it is important to highlight that nucleosomes are dynamic entities that undergo facile excursions to conformational states other than the fairly uniform structures that are observed in X-ray crystal studies. For example, as was first quantified by Widom and colleagues [57] nucleosome DNA transiently unwraps and rebinds the surface of the nucleosome at sufficiently rapid rates and with sufficient probability to allow access to DNA binding factors [58]. This dynamic behavior exposes DNA sites with a probability of about 1 in 103 to 105 as one moves from the periphery of the nucleosome toward the center so the apparent DNA binding affinities of many trans-acting factors for nucleosomal DNA simply will be reduced by 103-105 compared to the affinities of these factors for the same sites in naked DNA. Thus, although the time-averaged fraction of nucleosomes with exposed binding sites is small, the dynamic nature of the unwrapping process allows factors having sufficiently high affinities for naked DNA and/or present in locally high concentrations to thermodynamically compete with the core histones for binding with the DNA on physiologically relevant timescales. Of note, the conformational equilibrium constant describing this ‘unwrapping’ transition is dependent on DNA sequence [58,59] but does not substantially depend on the presence of core histone tail domains or the presence of neighboring nucleosomes [32,60]. These results show that nucleosomes by themselves provide only a surmountable thermodynamic barrier to factor binding, rather than an absolute kinetic block to factor access to nucleosomal DNA.
Moreover, the protein-protein interactions within the nucleosome also appear to undergo excursions to alternative states that may have physiological significance. Prunell and colleagues provided evidence that the H3/H4 tetramer can, under specific conditions of DNA torsional stress, undergo a change to a right-handed conformation involving realignment of the H3-H3 interface [61]. The altered conformation is promoted by removal or acetylation of the H4 and H3 tail domains, [62-64]. This transition requires dissociation of H2A/H2B dimers from the nucleosome and may result from the positive supercoiling stress that accumulates in front of transcribing polymerases in order to generate a more transcriptionally permissible template [65-67]. Evidence has been presented for other conformational excursions within the nucleosome, including opening of the H2A/H2B dimer – H3/H4 tetramer interface that precedes DNA unwrapping [68] and nucleosome ‘gaping’, which may involve increases in the distance between the superhelical turns of DNA in the nucleosome [69].
Linker Histone (H1)
Linker histones (H1s) are a primary component of nucleosomes in higher eukaryotes and thus should be included in any discussion of basic nucleosome structure. The H1 family of proteins is less conserved between species than that of the core histones, varying in both sequence homology and number of non-allelic variants across eukaryotes. Linker histones are present with an abundance of about 1 per nucleosome in vivo [4] and exhibit stoichiometric and preferential binding to nucleosomes in vitro [70]. H1s fulfill numerous functions, both in vitro and in vivo, including stabilizing wrapping of DNA around the nucleosome, promoting folding and assembly of higher order chromatin structures [47,71], influencing nucleosome spacing on DNA [72,73], regulating specific gene expression [74,75], and suppressing transcription of repetitive transposable DNA elements packaged into heterochromatin [76]. Moreover, specific posttranslational modifications of linker histones have been linked to numerous cellular processes, including replication timing and mitosis [77-79]. H1s bind to the nucleosome exterior and can be extracted from native chromatin by lower salt concentrations than required for removal of the core histone proteins [4]. Consistent with their weaker binding, linker histones exhibit much greater mobility about the nucleus than the core histones, as shown by fluorescence recovery after photobleaching (FRAP) experiments with an H1-GFP fusion [80].
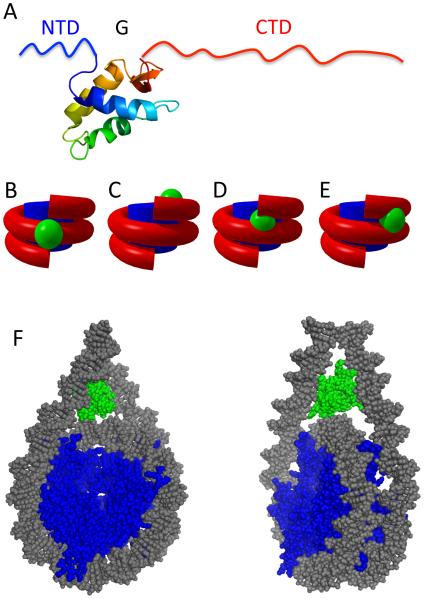
Early sequence comparisons and analysis of trypsin proteolysis indicated metazoan H1s typically have a tripartite structure, with a short (~30 residue), protease sensitive N-terminal tail domain, a central (~80 residue) stably folded and protease resistant globular domain and, a ~100 residue, protease sensitive and highly basic C-terminal domain [25,81] (Fig. 3A). However, variations on this theme can be found, with some H1s exhibiting much longer C-terminal domains and others exhibiting alternative domain structures, typically in lower eukaryotes [4]. For example, S. cerevisiae has a single H1-like protein, Hho1p, with a unique structure containing two globular domains connected by a short C-terminal tail-like domain [82], whereas Tetrahymena H1 lacks a globular region altogether [83,84]. Though in higher eukaryotes the number of H1 variants within an organism is typically much larger, the tripartite domain structure is generally maintained, with differences in sequences primarily found in the C-terminal regions. In mammals there are 11 variant subtypes, five of which are ubiquitously expressed in somatic cells, with additional subtypes with tissue specificity including H1.0 found in terminally differentiated cells [85].
Fig. 3. Model of H1 binding to nucleosome.
A. Domain structure of metazoan linker histone showing the 25-35 residue ‘unstructured’ N-terminal domain (NTD), ~80 residue globular domain (G), and the 100-120 residue ‘unstructured’ C-terminal domain (CTD). Model of the globular domain from ref [91]. B-E. Simplified models showing approximate sites for interaction of H1 globular domain with the nucleosome from refs. [81,112,113,115], respectively. F. Three-contact model for linker histone globular domain binding within a nucleosome. Two views of a nucleosome with proposed location of a linker histone globular domain (green) are shown, with DNA and core histones colored grey and blue, respectively. Approximately 20 additional bp of DNA are depicted beyond the boundary of the nucleosome core region.
H1 interactions and functions in chromatin
Although H1 is an essential component of higher-order chromatin structure [73], many molecular details of how H1 binds to nucleosomes in chromatin remain unknown. Early studies exploiting sensitivity of chromatin to nucleases revealed an increased rate of micrococcal nuclease (MNase) digestion of the ~50 bp region of DNA between nucleosome cores upon removal of histone H1 [86], indicating H1 is important for protecting linker DNA. Careful analysis of products of MNase I digestion of chicken erythrocyte chromatin showed a defined product larger than a nucleosome core particle, termed the chromatosome, which contains DNA fragments of ~168 bp, the four core histones and linker histone [87]. This work indicated that linker histone protects an additional 20 bp of DNA from nuclease digestion compared to nucleosome core particles, that is, on average, symmetrically disposed to either side of the 147 bp core DNA [87]. Thus high-affinity H1 binding to the nucleosome requires ~10 bp extensions to each of the ends of the nucleosome core DNA and suggests that this single domain contacts 3 distinct DNA segments in chromatin. The appearance of chromatin under electron microscope in various ionic conditions also indicated that H1 is located near the entry/exit points of DNA and stabilizes the wrapping of nucleosome DNA near the edge of the core region, as evident by changes in salt dependent structures in H1-depleted and native chromatin [88]. Though a single H1 was initially presumed to interact with each nucleosome, it was not clear if H1:nucleosome stoichiometry was the same for all cell nuclei. A direct chemical method radiolabeling method used to measure linker histone stoichiometry relative to core histones showed, in vivo, one H1 is generally associated with each nucleosome, although that the ratio of H1:nucleosomes in the nucleus naturally varies over a range of about 0.8-1.4 [4,89].
H1 Domain Structure and Function
Early studies of the re-association of H1 peptides with reconstituted oligonucleosomes or linker histone-depleted chromatin revealed the trypsin resistant globular domain (GH1) is also sufficient to protect ~20 bp of DNA in chromatosome MNase digestion assay, though full length H1 may be required for full compaction of chromatin [81]. These studies revealed that the globular domain is sufficient for structure-specific recognition of nucleosomes, and likely binds along the nucleosome dyad, contacting the central wrap of nucleosome core DNA and the two 10 bp extensions of the linker DNA entering/exiting the structure. Thus these studies suggested the globular domain binds in a pocket formed by three distinct DNA surfaces where the linker DNA exits and enters the nucleosome (Fig. 3B).
Though less conserved than core histones, the linker histone globular domain is more conserved and more hydrophobic than either N- or C-terminal regions (NTD and CTD, respectively). While GH1 can be obtained by trypsin digest, subsequent homogeneous preparation of recombinant GH1 allowed for analysis of secondary structure by homonuclear and 1H-15N heteronuclear 2D NMR, which revealed 3 α–helices and a possible -hairpin [90]. Also using a recombinant polypeptide, Ramakrishnan and colleagues obtained a 2.5 Å resolution X-ray crystal structure that showed the entire fold of the globular domain of the closely related linker histone H5, including the beta hairpin described in the GH1 structure [91] (Fig. 3A). Comparison of the GH5 structure to the tertiary structure assignment of GH1 from a later NMR study revealing a remarkably similar 3D structure of linker histone globular domains, albeit with slight differences in electrostatic potentials, which may relate to differences in function [92]. Both globular domain structures contain a characteristic DNA-binding, winged-helix domain, as well as two additional surfaces with the potential for DNA binding, consistent with the three DNA surface contact model mentioned above (Fig. 3B).
The H1 NTD contains two unique regions, a proline and alanine rich subdomain and a shorter highly basic region near the globular domain [93]. H1 peptides lacking NTD are as effective as full-length H1 in inducing higher-order chromatin structure but do not as efficiently confer protection of 168 bp DNA from MNase digestion [71]. An H1 NTD contribution to binding affinity is also reflected in FRAP analysis of murine H1.0 and H1c NTD domain swapping experiments [94]. These observations suggest that though H1 NTD is not necessary for chromatin condensation, it may serve as an anchor for H1 positioning in such a way that seals off DNA entering and exiting the nucleosome.
The C-terminal domain of linker histones has a sequence loosely based on a repeat of the sequence S/TPXK [95] and typically •100 amino acid residues in length, with lysine constituting ~40% of the residues, evenly distributed throughout the domain [28,96]. Alanine is also highly abundant, representing ~20-35% of the residues, followed by, proline, valine, and serine, while Gln, Asn, Glu, Asp, His, Phe, Ile, Leu, Cys, Trp, and Tyr are either not found or rarely found in H1 CTDs [28]. Interestingly, arginine is rarely found and is restricted to quiescent cell-type specific H1 variants. The large fraction of basic residues found within the CTD is consistent with the primary function of this domain in stabilizing higher order chromatin structure via neutralization of DNA charge, primarily in the linker DNA [71,97,98]. The CTD also greatly stabilizes binding of H1 to chromatin within nuclei [80,99].
The CTD is sensitive to proteases and is known to be disordered when the protein is free in solution [100]. Moreover, the amino acid content of the CTD is characteristic of an intrinsically disordered protein domain [28]. Consistent with this idea, the amino acid composition rather than the primary sequence appears to be more important for the chromatin condensing function of the protein [101,102]. The CTD also interacts with numerous nuclear factors, both in chromatin and in the nucleolus [103,104]. Thus, the function of this intrinsically disordered region may be to accommodate disparate interactions with diverse macromolecular partners in the nucleus.
Consistent with its molecular behavior as an intrinsically discorded domain, a substantial amount of α-helical structure within peptides derived from CTDs can be induced by secondary structure stabilizing solvents [105]. Moreover, FT-IR spectroscopy indicates that diverse secondary structural elements can be induced in the presence of short DNA fragments or in solution conditions leading to substantial neutralization of the negative charge within the domain [106,107]. More recent work employing FRET indicates that the CTD undergoes a transition from a random coil conformation to a much more condensed structure upon H1.0 binding to nucleosomes [108]. Interestingly, while H1.0 binding to naked DNA fragments also induces CTD condensation, the induced structure is distinct from that found when H1 is bound to nucleosomes [108,109]. Future work defining the structure of the CTD in different contexts, with posttranslational modifications such as phosphorylation, and in complex with different macromolecular binding partners will clearly be important in elucidating the molecular mechanisms related to H1 structure and function.
Specific H1-nucleosome interactions
As mentioned above, MNase protection studies and electron microscopy of native chromatin indicated H1 binding near the center of the nucleosome, stabilizing and orienting the linker DNA entering and exiting the nucleosome core [81,88] (Fig. 3B). In addition, DNase I footprinting studies of H1 and H5 within dinucleosomes suggested the globular domain contacts the DNA near nucleosomal DNA entry/exit points, and the dyad axis and that NTD and CTD regions may account for partial protections of adjacent regions of linker DNA [110]. In the following decades, many attempts to better characterize H1- nucleosome interactions resulted in several models [111,112]. Nuclease protection, directed DNA cleavage, and chemical crosslinking studies of H1 and H5 bound nucleosomes reconstituted on 5S RNA gene templates indicated linker histones do not substantially alter the organization of DNA in the core, but did lead to protection of an additional 20 bp of DNA, asymmetrically distributed relative to the core particle, contacting the inner surface of the DNA superhelix, near one end of the 5S nucleosome [70,113,114]. This lead to a model in which the globular domain inserts inside the superhelical DNA wrap, extending the ramp of charge provided by the core histones at one end of the nucleosome [113,114] (Fig. 3C). Site-directed protein-DNA photo-crosslinking to map GH5 on native chicken nucleosomes lead to a model whereby the GH5 is positioned between the central turn of DNA near nucleosome dyad and one of the linker arms [115] (Fig. 3D). Using FRAP to probe effects of mutations within the H1.0 globular domain on the stability of binding to nucleosomes in living cells, Brown et al. defined two principle DNA binding sites on the GH1 surface comprised of clusters of multiple residues, supporting a model whereby GH1 DNA binding domain residues interact with a major groove near the dyad and the minor groove of one linker DNA [112] (Fig. 3E). Computational analysis of GH5-DNA interactions by Fan and Roberts provided evidence that GH5 electrostatic potential is unique from that of other winged-helix proteins, identifying three distinct DNA binding sites where the primary DNA binding site is divided into two separate DNA interaction sites [116]. A model resulting from this work indicated GH5 binding at the nucleosome dyad, contacting three distinct DNA surfaces, consistent with previous models of binding [81]. A recent comprehensive study combining electron cryomicroscopy (cryoEM), and hydroxyl radical footprinting of H1 bound to mono-, di- and tri-, nucleosomes has provided further evidence for the globular domain located at the dyad and contacting 3 DNA surfaces, similar to the original model [117] (Fig. 3B and F). Hydroxyl radical footprinting provides single base resolution and indicated that H1 binding results in the protection of the outward-facing DNA minor groove at the dyad, as well as DNA beyond edge of nucleosomal core, consistent with H1 organizing the linker DNA into a stem structure [117]. Further attempts to understand the H1 binding and orientation were made via solution NMR of Drosophila H1-nucleosome complex using H1 and CTD truncation mutants. Paramagnetic relaxation enhancement (PRE) experiments suggest GH1 bridges nucleosome core and one of the two linker DNA segments asymmetrically, with the 3 helix of the globular domain facing the DNA near the nucleosome dyad, and implicates both the H1 CTD and H2A CTD in formation of the H1-nucleosome complex [118]. Moreover recent FRAP results suggest the possibility that linker histone variants might have unique modes of association with nucleosomes [119].
Despite numerous studies, several critical aspects of how linker histone bind and orient on the nucleosome remain unclear. In order to advance our understanding of the structural and functional role of linker histones, we must elucidate the molecular details of nucleosome recognition by H1. For example, experimental evidence is still needed to confirm the idea that the globular domain interacts with the wide minor groove at the dyad in a manner similar to that observed for winged-helix proteins binding to cognate major grooves. Moreover, little is known regarding specific sites of interactions of the N- and C-terminal domains of H1 within the nucleosome or how posttranslational modifications may affect the structure and/or interactions of these domains.
Acknowledgement
This work supported by NIH Grants R01GM052426 and 5T32 GM068411.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [2].White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–18. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–7. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- [4].van Holde KE. Chromatin. Springer Verlag; New York: 1989. [Google Scholar]
- [5].Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA. 1991;88:10148–52. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc. Natl. Acad. Sci. USA. 1993;90:10489–93. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Karantza V, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: pH and ionic strength effects on the stability of the (H3-H4)/(H3-H4)2 system. Biochemistry. 1996;35:2037–46. doi: 10.1021/bi9518858. [DOI] [PubMed] [Google Scholar]
- [8].Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–16. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gloss LM, Placek BJ. The effect of salts on the stability of the H2A-H2B histone dimer. Biochemistry. 2002;41:14951–9. doi: 10.1021/bi026282s. [DOI] [PubMed] [Google Scholar]
- [10].Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–91. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nikitina T, Wang D, Gomberg M, Grigoryev SA, Zhurkin VB. Combined micrococcal nuclease and exonuclease III digestion reveals precise positions of the nucleosome core/linker junctions: implications for high-resolution nucleosome mapping. J Mol Biol. 2013;425:1946–60. doi: 10.1016/j.jmb.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- [13].Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J Mol Biol. 2007;371:725–38. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hayes JJ, Bashkin J, Tullius TD, Wolffe AP. The histone core exerts a dominant constraint on the structure of DNA in a nucleosome. Biochemistry. 1991;30:8434–40. doi: 10.1021/bi00098a022. [DOI] [PubMed] [Google Scholar]
- [15].Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361:617–24. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- [16].Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–73. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hayes JJ, Clark DJ, Wolffe AP. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA. 1991;88:6829–33. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hayes JJ, Tullius TD, Wolffe AP. The structure of DNA in a nucleosome. Proc. Natl. Acad. Sci. USA. 1990;87:7405–9. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Travers AA, Klug A. The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond B Biol Sci. 1987;317:537–61. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- [20].Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- [21].Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J Mol Biol. 2005;345:103–14. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- [22].McGhee JD, Felsenfeld G. The number of charge-charge interactions stabilizing the ends of nucleosome DNA. Nucleic Acids Res. 1980;8:2751–69. doi: 10.1093/nar/8.12.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang D, Ulyanov NB, Zhurkin VB. Sequence-dependent Kink- and-Slide Deformations of Nucleosomal DNA Facilitated by Histone Arginines Bound in the Minor Groove. J. Biomol. Struct. Dynam. 2010;27:843–859. doi: 10.1080/07391102.2010.10508586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–63. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bohm L, Crane-Robinson C. Proteases as structural probes for chromatin: the domain structure of histones. Biosci. Rep. 1984;4:365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- [26].Walker IO. Differential Dissociation of Histone Tails from Core Chromatin. Biochemistry. 1984;23:5622–5628. doi: 10.1021/bi00318a037. [DOI] [PubMed] [Google Scholar]
- [27].Smith RM, Rill RL. Mobile histone tails in nucleosomes. Assignments of mobile segments and investigations of their role in chromatin folding. J. Biol. Chem. 1989;264:10574–81. [PubMed] [Google Scholar]
- [28].Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–6. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- [29].Wang X, Hayes JJ. Site-specific binding affinities within the H2B tail domain indicate specific effects of lysine acetylation. J Biol Chem. 2007;282:32867–76. doi: 10.1074/jbc.M706035200. [DOI] [PubMed] [Google Scholar]
- [30].Ausio J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J. Mol. Biol. 1989;206:451–63. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- [31].Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- [32].Polach KJ, Lowary PT, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 2000;298:211–233. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- [33].Vettese-Dadey M, Walter P, Chen H, Juan LJ, Workman JL. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol. Cell. Biol. 1994;14:970–81. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Walter PP, Owen-Hughes TA, Cote J, Workman JL. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 1995;15:6178–87. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang Z, Zheng C, Thiriet C, Hayes JJ. The core histone N-terminal tail domains negatively regulate binding of transcription factor IIIA to a nucleosome containing a 5S RNA gene via a novel mechanism. Mol Cell Biol. 2005;25:241–9. doi: 10.1128/MCB.25.1.241-249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee KM, Hayes JJ. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl. Acad. Sci. USA. 1997;94:8959–64. doi: 10.1073/pnas.94.17.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee KM, Hayes JJ. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry. 1998;37:8622–8. doi: 10.1021/bi980499y. [DOI] [PubMed] [Google Scholar]
- [38].Wang X, Moore SC, Laszckzak M, Ausio J. Acetylation increases the alpha-helical content of the histone tails of the nucleosome. J. Biol. Chem. 2000;275:35013–20. doi: 10.1074/jbc.M004998200. [DOI] [PubMed] [Google Scholar]
- [39].Allan J, Harborne N, Rau DC, Gould H. Participation of the core histone tails in the stabilization of the chromatin solenoid. J. Cell Biol. 1982;93:285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schwarz PM, Hansen JC. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J Biol Chem. 1994;269:16284–9. [PubMed] [Google Scholar]
- [41].Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 1995;270:17923–8. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- [42].Zheng C, Hayes JJ. Structures and interactions of the core histone tail domains. Biopolymers. 2003;68:539–46. doi: 10.1002/bip.10303. [DOI] [PubMed] [Google Scholar]
- [43].Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin Fiber Folding: Requirement for the Histone H4 N-terminal Tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- [44].Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- [45].Wilkins BJ, et al. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343:77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- [46].Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem. 2005;280:33552–7. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- [47].Hansen JC. Conformational Dynamics of the Chromatin Fiber in Solution: Determinants, Mechanisms, and Functions. Annu Rev Biophys Biomol Struct. 2002;31:361–92. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- [48].Kan PY, Caterino TL, Hayes JJ. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2009;29:538–46. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pepenella S, Murphy KJ, Hayes JJ. A distinct switch in interactions of the histone H4 tail domain upon salt-dependent folding of nucleosome arrays. J Biol Chem. 2014;289:27342–51. doi: 10.1074/jbc.M114.595140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- [51].Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–96. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- [52].Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- [53].Sinha D, Shogren-Knaak MA. Role of direct interactions between the histone H4 Tail and the H2A core in long range nucleosome contacts. J Biol Chem. 2010;285:16572–81. doi: 10.1074/jbc.M109.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, Nordenskiold L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–91. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fletcher TM, Hansen JC. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J. Biol. Chem. 1995;270:25359–62. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- [56].Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–38. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–49. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- [58].Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- [59].Ngo TT, Zhang Q, Zhou R, Yodh JG, Ha T. Asymmetric Unwrapping of Nucleosomes under Tension Directed by DNA Local Flexibility. Cell. 2015;160:1135–44. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol. 2008;379:772–86. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hamiche A, Carot V, Alilat M, De Lucia F, O'Donohue MF, Revet B, Prunell A. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left- to a right-handed superhelical form. Proc Natl Acad Sci U S A. 1996;93:7588–93. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sivolob A, De Lucia F, Alilat M, Prunell A. Nucleosome dynamics. VI. Histone tail regulation of tetrasome chiral transition. A relaxation study of tetrasomes on DNA minicircles. J Mol Biol. 2000;295:55–69. doi: 10.1006/jmbi.1999.3302. [DOI] [PubMed] [Google Scholar]
- [63].Peterson S, Jackson V. Acetylation of H4 suppresses the repressive effects of the N-termini of histones H3/H4 and facilitates the formation of positively coiled DNA. Biochemistry. 2008;47:7053–65. doi: 10.1021/bi8004945. [DOI] [PubMed] [Google Scholar]
- [64].White RH, Keberlein M, Jackson V. A mutational mimic analysis of histone H3 post-translational modifications: specific sites influence the conformational state of H3/H4, causing either positive or negative supercoiling of DNA. Biochemistry. 2012;51:8173–88. doi: 10.1021/bi300872t. [DOI] [PubMed] [Google Scholar]
- [65].Levchenko V, Jackson B, Jackson V. Histone release during transcription: displacement of the two H2A-H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry. 2005;44:5357–72. doi: 10.1021/bi047786o. [DOI] [PubMed] [Google Scholar]
- [66].Bancaud A, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–47. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- [67].Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ngo TT, Ha T. Nucleosomes undergo slow spontaneous gaping. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv276. Advance Access published March 30, 2015 online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hayes JJ, Wolffe AP. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc. Natl. Acad. Sci. USA. 1993;90:6415–9. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- [72].Blank TA, Becker PB. Electrostatic mechanism of nucleosome spacing. J Mol Biol. 1995;252:305–13. doi: 10.1006/jmbi.1995.0498. [DOI] [PubMed] [Google Scholar]
- [73].Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–72. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–83. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- [75].Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- [76].Lu X, et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Roth SY, Allis CD. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992;17:93–8. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- [78].Thiriet C, Hayes JJ. Linker histone phosphorylation regulates global timing of replication origin firing. J Biol Chem. 2009;284:2823–9. doi: 10.1074/jbc.M805617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Raghuram N, et al. Pin1 promotes histone H1 dephosphorylation and stabilizes its binding to chromatin. J Cell Biol. 2013;203:57–71. doi: 10.1083/jcb.201305159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–81. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- [81].Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- [82].Landsman D. Histone H1 inSaccharomyces cerevisiae: a double mystery solved? Trends Biochem Sci. 1996;21:287–288. [PubMed] [Google Scholar]
- [83].Hayashi T, Hayashi H, Iwai K. Tetrahymena histone H1. Isolation and amino acid sequence lacking the central hydrophobic domain conserved in other H1 histones. J Biochem. 1987;102:369–76. doi: 10.1093/oxfordjournals.jbchem.a122063. [DOI] [PubMed] [Google Scholar]
- [84].Wu M, Allis CD, Richman R, Cook RG, Gorovsky MA. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986;83:8674–8. doi: 10.1073/pnas.83.22.8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- [86].Whitlock JP, Jr., Simpson RT. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976;15:3307–14. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- [87].Simpson RT. Structure of the chromosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- [88].Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–27. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bates DL, Thomas JO. Histones H1 and H5: One Or Two Molecules Per Nucleosome? Nucleic Acids Res. 1981;2:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cerf C, Lippens G, Muyldermans S, Segers A, Ramakrishnan V, Wodak SJ, Hallenga K, Wyns L. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: sequential assignment and secondary structure. Biochemistry. 1993;32:11345–51. doi: 10.1021/bi00093a011. [DOI] [PubMed] [Google Scholar]
- [91].Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–224. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- [92].Cerf C, Lippens G, Ramakrishnan V, Muyldermans S, Segers A, Wyns L, Wodak SJ, Hallenga K. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5. Biochemistry. 1994;33:11079–86. doi: 10.1021/bi00203a004. [DOI] [PubMed] [Google Scholar]
- [93].Bohm L, Mitchell TC. Sequence conservation in the N-terminal domain of histone H1. FEBS Lett. 1985;193:1–4. doi: 10.1016/0014-5793(85)80067-3. [DOI] [PubMed] [Google Scholar]
- [94].Vyas P, Brown DT. N- and C-terminal domains determine differential nucleosomal binding geometry and affinity of linker histone isotypes H1(0) and H1c. J Biol Chem. 2012;287:11778–87. doi: 10.1074/jbc.M111.312819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Subirana JA. Analysis of the charge distribution in the C-terminal region of histone H1 as related to its interaction with DNA. Biopolymers. 1990;29:1351–7. doi: 10.1002/bip.360291003. [DOI] [PubMed] [Google Scholar]
- [97].Clark DJ, Kimura T. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 1990;211:883–96. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- [98].Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–87. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- [99].Hendzel MJ, Lever MA, Crawford E, Th'ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–34. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- [100].Bradbury EM, et al. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The conformation of histone H1. Eur J Biochem. 1975;52:605–13. doi: 10.1111/j.1432-1033.1975.tb04032.x. [DOI] [PubMed] [Google Scholar]
- [101].Lu X, Hamkalo B, Parseghian MH, Hansen JC. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry. 2009;48:164–72. doi: 10.1021/bi801636y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem. 2004;279:8701–7. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- [103].Kalashnikova AA, et al. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013;41:4026–35. doi: 10.1093/nar/gkt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Szerlong HJ, Herman JA, Krause CM, DeLuca JG, Skoultchi A, Winger QA, Prenni JE, Hansen JC. Proteomic Characterization of the Nucleolar Linker Histone H1 Interaction Network. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Clark DJ, Hill CS, Martin SR, Thomas JO. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988;7:69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Roque A, Iloro I, Ponte I, Arrondo JL, Suau P. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol Chem. 2005;280:32141–7. doi: 10.1074/jbc.M505636200. [DOI] [PubMed] [Google Scholar]
- [107].Vila R, Ponte I, Collado M, Arrondo JL, Suau P. Induction of secondary structure in a COOH-terminal peptide of histone H1 by interaction with the DNA: an infrared spectroscopy study. J Biol Chem. 2001;276:30898–903. doi: 10.1074/jbc.M104189200. [DOI] [PubMed] [Google Scholar]
- [108].Caterino TL, Fang H, Hayes JJ. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered h1 carboxyl-terminal domain. Mol Cell Biol. 2011;31:2341–8. doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Fang H, Clark DJ, Hayes JJ. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 2011;40:1475–1484. doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Staynov DZ, Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988;7:3685–91. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Crane-Robinson C. Where is the globular domain of linker histone located on the nucleosome? Trends Biochem Sci. 1997;22:75–7. doi: 10.1016/s0968-0004(97)01013-x. [DOI] [PubMed] [Google Scholar]
- [112].Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–5. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis EN, Wolffe AP. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science. 1996;274:614–7. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- [114].Hayes JJ. Site-directed cleavage of DNA by a linker histone--Fe(II) EDTA conjugate: localization of a globular domain binding site within a nucleosome. Biochemistry. 1996;35:11931–7. doi: 10.1021/bi961590+. [DOI] [PubMed] [Google Scholar]
- [115].Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–5. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- [116].Fan L, Roberts VA. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc Natl Acad Sci U S A. 2006;103:8384–9. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Syed SH, et al. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci U S A. 2010;107:9620–5. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhou BR, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y. Structural insights into the histone H1-nucleosome complex. Proc Natl Acad Sci U S A. 2013;110:19390–5. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].George EM, Izard T, Anderson SD, Brown DT. Nucleosome interaction surface of linker histone H1c is distinct from that of H1(0) J Biol Chem. 2010;285:20891–6. doi: 10.1074/jbc.M110.108639. [DOI] [PMC free article] [PubMed] [Google Scholar]