Abstract
Background
Glutathione (GSH) is an important molecule involved in cell detoxification and antioxidation and may affect cancer development or outcome. We hypothesized that genetic variation in the GSH pathway might influence the clinical outcome of patients who have non-muscle invasive bladder cancer (NMIBC).
Methods
A total of 114 single nucleotide polymorphisms (SNPs) in 21 GSH pathway genes were genotyped in 414 NMIBC patients treated with transurethral resection alone (TUR) and both TUR and intravesical bacillus Calmette–Guérin instillation (BCG) therapy. The effect of each SNP on time to recurrence was estimated using the multivariate Cox proportional hazards model. Cumulative effect and survival tree analyses were performed to determine the joint effects of unfavorable genotypes and gene–gene interactions on bladder cancer prognosis.
Results
Seven SNPs showed significant associations with cancer recurrence in the TUR group and 15 SNPs showed significant associations with recurrence in the BCG group. The most significant SNP in the TUR group was rs3746162 in GPX4, whose variant genotype conferred a 5.4-fold increased risk of recurrence compared with wild-type (hazard ratio [HR] = 5.43, 95 % confidence interval [CI] = 2.19–13.46), whereas the most significant SNP in the BCG group was rs7265992 in GSS (HR 3.43, 95 % CI 1.56–7.56). The risk of recurrence increased with the number of unfavorable genotypes in both groups. Within treatment group, stratified analyses by tumor grade also indicated predictive variants.
Conclusions
Genetic variants in GSH pathway may influence cancer recurrence in NMIBC patients receiving curative treatment.
Bladder cancer accounts for approximately 4.4 % of new primary cancers and 2.6 % of cancer deaths in the United States, with an estimated 72,570 new cases and 15,210 deaths in 2013.1 Most bladder cancers (75–85 %) are non-muscle invasive at diagnosis.2 Although multimodal treatments have been standard treatment for many years, the prognosis for non-muscle invasive bladder cancers (NMIBCs) remains variable, as seen in the 30–80 % recurrence rate and 1–45 % progression rate within 5 years.2 Tumor cell implantation after transurethral resection (TUR), the standard treatment for NMIBC, is attributed to many early recurrences.3 To reduce the risk of recurrence, clinicians use perioperative intravesical therapy to prevent tumor reimplantation.4 The major choice for intravesical therapy is immunotherapy with bacillus Calmette–Gueérin (BCG), which results in a massive local immune response characterized by the induced expression of cytokines in the urine and bladder wall and the accumulation of granulocytes and mononuclear cells.5 BCG is considered the best adjuvant therapy after TUR for preventing recurrence and delaying progression in NMIBC; however, clinical response is variable ranging from 36–71 %.6–8 Moreover, intravesical BCG administration commonly causes side effects that affect the patient’s quality of life and thus the success of treatment. Most patients experience urinary frequency and urgency, and serious sequelae and rare deaths have occurred.9 Therefore, it is imperative to identify new biomarkers that accurately predict a patient’s response to BCG therapy and post-treatment prognosis.
Glutathione (GSH) is the most abundant low-molecular-weight peptide present in eukaryotic cells and is one of the major compounds involved in cellular antioxidation and detoxification.10 It is synthesized intracellularly from glutamic acid, cysteine, and glycine in two steps; this process is catalyzed by γ-glutamylcysteine synthetase and GSH synthetase (GSS). In antioxidation, GSH, a reduced compound, is oxidized to GSH disulfide by GSH peroxidase (GPX), by GSH-dependent transhydrogenases, or by direct interaction with free radicals. GSH disulfide is then reduced to GSH by NADPH-dependent GSH reductase (GSR). Through this oxidation process, GSH can effectively scavenge free radicals and other reactive oxygen species.11 The detoxification role of GSH involves GSH-S-transferases (GSTs), a family of phase II detoxification enzymes. These enzymes conjugate GSH with various electrophiles, physiological metabolites, and xenobiotics to form mercapturates, which can then be eliminated via an ATP-dependent GSH S-conjugate export pump.12 GSH also is required in many aspects of the immune response. It is essential for cytotoxic T lymphocyte activation, proliferation, and differentiation and also is involved in the activation of polymorphonuclear leukocytes as well as cytokine production.11
It is clear that GSH plays important role in antioxidation and detoxification and that there is strong evidence linking bladder cancer development with exposure to chemical carcinogens. Because intravesical immunotherapy with BCG instillation is the standard treatment for NMIBC, we performed this study to test the hypothesis that genetic variants in the GSH pathway may affect patients’ clinical response to BCG instillation therapy.
MATERIALS AND METHODS
Details of the methodologies used for this study are described online in Supplementary Methods.
Study Population and Epidemiologic Data
The study population and enrollment procedures were depicted previously.13 Briefly, a total of 414 patients with incident and histologically confirmed NMIBC were recruited. The primary study end point was time to cancer recurrence, defined as a newly diagnosed intravesical tumor subsequent to a previous negative cystoscopy follow-up or a newly found extravesical metastasis. Median follow-up time of the cohort is 48 months (53.3 months for recurrent cases and 38.6 months for nonrecurrent cases). The standard surveillance procedure may vary but usually consists of cystoscopic screening every 3 months for 2 years and then every 6 months for 3 years followed by yearly cytology and imaging screening. This study was approved by the Institutional Review Boards of MD Anderson Cancer Center and Baylor College of Medicine (online Method S1).
Gene/SNP Selection and Genotyping
A total of 114 SNPs from 21 genes (Supplementary Table S1) were identified by data mining the International HapMap Project, dbSNP, and the UCSC Genome Browser. DNA was extracted from peripheral blood, and SNP genotyping was performed using Illumina iSelect custom array platform (online Method S2).
Statistical Analysis
The main effect of a SNP on time to recurrence was estimated using the multivariate Cox proportional hazards model adjusted for potential covariates. Statistical analyses were performed using Stata 10.0 (StataCorp LP, College Station, TX) and STREE program (online Method S3).14
RESULTS
As shown in Table 1, no significant differences were identified between subjects with and without recurrence based on age (P = 0.45), sex (P = 0.15), smoking status (P = 0.44), cancer stage (P = 0.13), cancer grade (P = 0.19), or focality (P = 0.25); however, significant differences were found for treatment (P = 3.42 × 10−16).
TABLE 1.
Demographic and clinical characteristics of the NMIBC patients
| Variables | Recurrence, N
(%) |
No recurrence, N
(%) |
P |
|---|---|---|---|
| Age, mean (SD) | 62.63 (11.04) | 63.49 (11.57) | 0.445 |
| Sex | 0.153 | ||
| Male | 193 (85.0) | 149 (79.7) | |
| Female | 34 (15.0) | 38 (20.3) | |
| Smoking statusa | 0.444 | ||
| Never | 63 (27.8) | 54 (28.9) | |
| Former | 116 (51.1) | 85 (45.5) | |
| Current | 48 (21.1) | 48 (25.7) | |
| Stage | 0.131 | ||
| Tis | 17 (7.5) | 6 (3.2) | |
| Ta | 104 (46.0) | 84 (44.9) | |
| T1 | 105 (46.5) | 97 (51.9) | |
| Gradeb | 0.192 | ||
| G1 | 5 (2.3) | 11 (5.9) | |
| G2 | 77 (36.2) | 66 (35.5) | |
| G3 | 131 (61.5) | 109 (58.6) | |
| Focalityc | 0.254 | ||
| 1 | 76 (52.1) | 86 (31.1) | |
| 2 | 19 (13.0) | 11 (7.4) | |
| Multiple | 51 (34.9) | 51 (34.5) | |
| Treatment | 3.42 × 10−16 | ||
| TUR only | 91 (40.1) | 45 (24.1) | |
| TUR + iBCG | 90 (39.6) | 30 (16.0) | |
| TUR + iBCG + mBCG |
31 (13.7) | 51 (27.3) | |
| Othersd | 15 (6.6) | 61 (32.6) | |
| Total | 227 | 187 |
SD standard deviation, TUR transurethral resection, iBCG induction bacillus Calmette–Guérin, mBCG maintenance bacillus Calmette-Guérin
Individuals who had smoked less than 100 cigarettes in their lifetime were defined as having never smoked. Individuals who had quit smoking at least 1 year before diagnosis were categorized as former smokers
For equivalence with two-grade system, G1 and G2 are considered low grade and G3 high grade
Not counting patients with missing information: 81 with recurrence; 39 without recurrence
Patients who received intravesical chemotherapy but no BCG
Individual SNPs and Cumulative Effects of Unfavorable Genotypes in Cancer Recurrence after TUR Treatment Only
Among the 114 SNPs examined, 7 were significantly associated with bladder cancer recurrence in TUR-only patients (Table 2). The most significant SNP was rs3746162 in GPX4 (HR 5.43, 95 % CI 2.19–13.46, P = 0.0003), and after adjustment for multiple comparisons, this SNP remained statistically significant with false discovery rate set at 5 % (Q = 0.05), indicating that this finding was robust. In stratified analysis by tumor grade, we found rs3746162 was significantly associated with recurrence in patients with low grade but not high grade tumors (HR 6.97, 95 % CI 2.69–18.05, P = 6.25 × 10−5 and HR 1.32, 95 % CI 0.52–3.35, P = 0.564, respectively; Supplementary Table S2). In contrast, rs1006771 of GSTT2 and rs17614751 of GSTA4 were significantly associated with recurrence in high-grade but not low-grade patients.
TABLE 2.
Selected SNPs and the cumulative effect of unfavorable genotypes on recurrence in NMIBC patients receiving treatment of TUR alone
| SNP | Gene | Genotype | MOI* | Recurrence WW/WV/VV |
No recurrence WW/WV/VV |
HR (95 % CI)b | P | Q |
|---|---|---|---|---|---|---|---|---|
| rs3746162 | GPX4 | G/A | rec | 59/22/7 | 26/19/0 | 5.43 (2.19–13.46) | 0.0003 | 0.013 |
| rs451774 | GPX5 | A/G | rec | 41/36/11 | 21/24/0 | 2.45 (1.24–4.84) | 0.010 | 0.237 |
| rs1006771 | GSTT2 | T/G | rec | 38/38/12 | 17/26/2 | 2.24 (1.17–4.27) | 0.015 | 0.237 |
| rs2144698 | GSTA2 | G/T | dom | 53/26/9 | 16/19/10 | 0.61 (0.40–0.95) | 0.029 | 0.248 |
| rs367836 | GSTA4 | A/C | dom | 19/50/19 | 15/22/8 | 1.81 (1.06–3.08) | 0.029 | 0.248 |
| rs2180314 | GSTA2 | G/C | add | 44/32/12 | 14/15/16 | 0.72 (0.54–0.97) | 0.031 | 0.248 |
| rs17614751 | GSTA4 | G/A | dom | 80/8/0 | 35/10/0 | 0.45 (0.21–0.96) | 0.038 | 0.248 |
| Group (number of unfavorable genotypesa) | Recurrence | No recurrence | HR (95 % CI)b | P | MST (mo) |
|---|---|---|---|---|---|
| Low-risk group (0–2) | 30 | 29 | 1 (reference) | 21.7 | |
| Medium-risk group (3) | 7 | 24 | 2.44 (1.34–4.44) | 0.003 | 5.3 |
| High-risk group (4–6) | 8 | 38 | 2.53 (1.51–4.25) | 0.0004 | 5.9 |
| P trend | 0.0003 |
SNPs that continued to have a significant effect after correcting for multiple comparisons by Q value with a false discovery rate of ≤ 10 % are in boldface
rec recessive, dom dominant, add additive, WW homozygous wild-type genotype, WV heterozygous variant genotype, VV homozygous variant genotype, HR hazard ratio, CI confidence interval, MST median survival time, MOI model of inheritance
The model with the smallest P value
Unfavorable genotypes: GPX4 rs3746162 (vv); GPX5 rs451774 (vv); GSTT2 rs1006771 (vv); GSTA2 rs2144698 (ww); GSTA4 rs367836 (wv+vv); GSTA2 rs2180314 (vv); GSTA4 rs17614751 (ww)
Adjusted by age, sex, smoking status, cancer stage, and cancer grade
We assessed the cumulative effect of unfavorable genotypes identified from the individual SNP analysis on bladder cancer recurrence (GPX4:rs3746162 (AA); GPX5:rs451774 (GG); GSTT2:rs1006771 (GG); GSTA2:rs2144698 (GG); GSTA4: rs367836 (AC+CC); GSTA2:rs2180314 (CC); GSTA4:rs17614751 (GG)). We observed that increased risk of cancer recurrence was associated with increased numbers of these genotypes. Table 2 shows results for recurrence risk and recurrence-free survival in patients in the TUR group. Compared with the low-risk group (patients with 0–2 unfavorable genotypes), the medium-risk group (patients with three unfavorable genotypes) and high-risk group (patients with 4–6 unfavorable genotypes) had HRs for recurrence of 2.44 (95 % CI 1.34–4.44, P = 0.003) and 2.53 (95 % CI 1.51–4.25, P = 0.0004), respectively (Ptrend = 0.0003). Kaplan–Meier recurrence-free survival curves (Supplementary Fig. S1) showed that the high-risk group and the medium-risk group had recurrence-free median survival times (MSTs) of 5.9 months and 5.3 months, respectively, which were much shorter than that of the low-risk group (MST = 21.7 months, Plog-rank = 0.003).
Individual SNPs and Cumulative Effects of Unfavorable Genotypes in Cancer Recurrence after TUR and BCG Treatment
In the 191 patients who received intravesical BCG therapy, 15 SNPs showed significant effects on recurrence (Table 3). The most significant SNP was rs7265992 in GSS (HR 3.43, 95 % CI 1.56–7.56, P = 0.002). Four SNPs remained significance after adjusting for multiple comparisons. These four SNPs were all located in the GSS gene, and two of them (rs7260770 and rs4911455) were strongly linked with r2 = 1.0. When stratified by tumor grade, rs6060124 of GSS was significantly associated with recurrence in patients with both low- and high-grade tumors (HR 4.11, 95 % CI 1.25–13.58, P = 0.02 and HR 2.79, 95 % CI 1.28–6.06, P = 0.01, respectively), whereas two SNPs in GPX5 and GSTM3 and eight variants in GSS, GSR, GSTT2, GSTM3, and GSTM4 were associated with recurrence in low-grade and high-grade patients, respectively (Supplementary Table S3).
TABLE 3.
Selected SNPs and the cumulative effect of unfavorable genotypes on recurrence in NMIBC patients receiving BCG after TUR
| SNP | Gene | Genotype | MOI* | Recurrence WW/WV/VV |
No recurrence WW/WV/VV |
HR (95 % CI)b | P | Q |
|---|---|---|---|---|---|---|---|---|
| rs7265992 | GSS | G/A | rec | 81/21/8 | 60/20/1 | 3.43 (1.56–7.56) | 0.002 | 0.050 |
| rs6060124 | GSS | C/A | rec | 61/36/12 | 44/34/3 | 2.80 (1.44–5.47) | 0.003 | 0.050 |
| rs7260770c | GSS | G/A | rec | 56/41/13 | 41/35/5 | 2.56 (1.34–4.90) | 0.005 | 0.050 |
| rs4911455c | GSS | A/C | rec | 56/40/13 | 40/35/5 | 2.54 (1.32–4.87) | 0.005 | 0.050 |
| rs377514 | GPX5 | T/C | dom | 93/15/2 | 56/24/1 | 0.50 (0.29–0.87) | 0.013 | 0.104 |
| rs6088662 | GSS | T/G | add | 77/30/3 | 44/31/6 | 0.64 (0.45–0.93) | 0.017 | 0.115 |
| rs8190996 | GSR | C/T | rec | 31/59/20 | 17/41/23 | 0.56 (0.34–0.92) | 0.022 | 0.121 |
| rs4970737 | GSTM3 | C/G | rec | 54/50/6 | 41/32/8 | 0.39 (0.16–0.92) | 0.031 | 0.121 |
| rs4970774 | GSTM3 | A/C | rec | 26/68/16 | 20/47/14 | 0.55 (0.31–0.96) | 0.036 | 0.121 |
| rs4970776 | GSTM3 | T/A | rec | 34/62/14 | 29/40/12 | 0.53 (0.29–0.97) | 0.038 | 0.121 |
| rs10133290 | GPX2 | A/C | rec | 64/36/10 | 49/28/4 | 2.08 (1.04–4.17) | 0.039 | 0.121 |
| rs11101992 | GSTM3 | A/C | add | 65/37/8 | 56/21/4 | 1.39 (1.02–1.91) | 0.040 | 0.121 |
| rs1571858 | GSTM3 | G/A | rec | 50/49/11 | 36/36/9 | 0.51 (0.26–0.98) | 0.043 | 0.121 |
| rs560018 | GSTM4 | A/G | rec | 52/42/16 | 32/36/11 | 1.76 (1.01–3.08) | 0.046 | 0.121 |
| rs15864 | GSTM3 | G/C | rec | 49/53/8 | 40/32/9 | 0.47 (0.22–0.99) | 0.048 | 0.121 |
| Group (number of unfavorable genotypesa) | Recurrence | No recurrence | HR (95 % CI)b | P | MST (mo) |
|---|---|---|---|---|---|
| Low-risk group (1–4) | 30 | 20 | 1 (reference) | ||
| Medium-risk group (5–6) | 39 | 70 | 2.38 (1.39–4.07) | 0.002 | 8.6 |
| High-risk group (7–10) | 9 | 29 | 6.20 (3.15–12.18) | 1.22 × 10−07 | 7.1 |
| P trend | 1.37 × 10−07 |
SNPs that continued to have a significant effect after correcting for multiple comparisons by Q value with a false discovery rate of ≤ 10 % are in boldface
rec recessive, dom dominant, add additive, WW homozygous wild-type genotype, WV heterozygous variant genotype, VV homozygous variant genotype, HR hazard ratio, CI confidence interval, MST median survival time, MOI model of inheritance
The model with the smallest P value
Unfavorable genotypes: GSS rs7265992 (vv); GSS rs6060124 (vv); GSS rs7260770 (vv); GSS rs4911455 (vv); GPX5 rs377514 (ww); GSS rs6088662 (ww); GSR rs8190996 (ww + wv); GSTM3 rs4970737 (ww + wv); GSTM3 rs4970774 (ww + wv); GSTM3 rs4970776 (ww + wv); GPX2 rs10133290 (vv); GSTM3 rs11101992 (vv); GSTM3 rs1571858 (ww + wv); GSTM4 rs560018 (vv); GSTM3 rs15864 (ww + wv)
Adjusted by age, sex, smoking status, cancer stage, and cancer grade
rs4911455 is in strong linkage with rs7260770, r2 = 0.99
Table 3 also shows the cumulative effects of unfavorable genotypes on recurrence in the BCG group (GSS:rs7265992 (AA); GSS:rs6060124 (AA); GSS:rs7260770 (AA); GSS:rs4911455 (CC); GPX5:rs377514 (TT); GSS:rs6088662 (TT); GSR:rs8190996 (CC+CT); GSTM3:rs4970737 (CC+CG); GSTM3:rs4970774 (AA+AC); GSTM3:rs4970776 (TT+TA); GPX2:rs10133290 (CC); GSTM3:rs11101992 (CC); GSTM3:rs1571858 (GG+GA); GSTM4:rs560018 (GG); GSTM3:rs15864 (GG+GC)). Compared with the low-risk group (patients with 1–4 unfavorable genotypes), the medium-risk group (patients with five or six unfavorable genotypes), and high-risk group (patients with 7–10 unfavorable genotypes) had HRs of recurrence of 2.38 (95 % CI 1.39–4.07, P = 0.002) and 6.20 (95 % CI 3.15–12.18, P = 1.22 × 10−07), respectively (Ptrend = 1.37 × 10−07). Kaplan–Meier recurrence-free survival curves (Fig. 1) indicated the high-risk group had a recurrence-free MST of 7.1 months, whereas the medium-risk group had MST of 8.6 months and the low-risk group had MST of greater than 100 months (more than half of the patients were recurrence-free throughout the study period; Plog-rank = 0.0005).
FIG. 1.
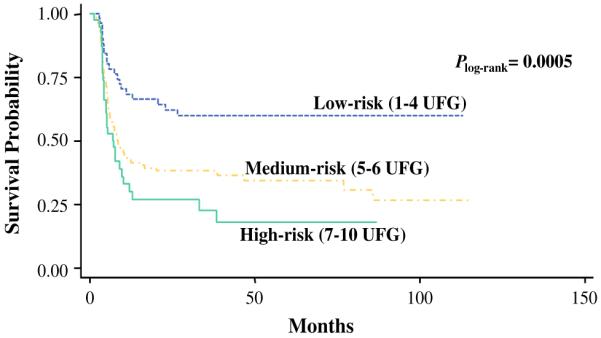
Kaplan–Meier estimate of NMIBC recurrence-free survival by level of risk according to the number of unfavorable genotypes (UFG) in the TUR and BCG treated group
Survival Tree Analysis
To explore the potential high-order gene–gene interactions between the GSH pathway SNPs, we performed a survival tree analysis using the significant SNPs identified in the individual SNP analysis. In the TUR group, no interaction was found between the seven significant SNPs. In the BCG group, the tree structure resulted in four terminal nodes with a range of low- to high-risk subgroups (Fig. 2; Supplementary Table S4). The initial split was rs377514 in GPX5 followed by splits at rs8190996 in GSR and rs6088662 in GSS. Using the low-risk group (node 1) as a reference, the HR was 2.36 (95 % CI 0.26–21.33, P = 0.445) for the medium-risk group (node 2), and 7.97 (95 % CI 1.10–57.80, P = 0.040) for the high-risk group (nodes 3 and 4; Ptrend = 0.003).
FIG. 2.
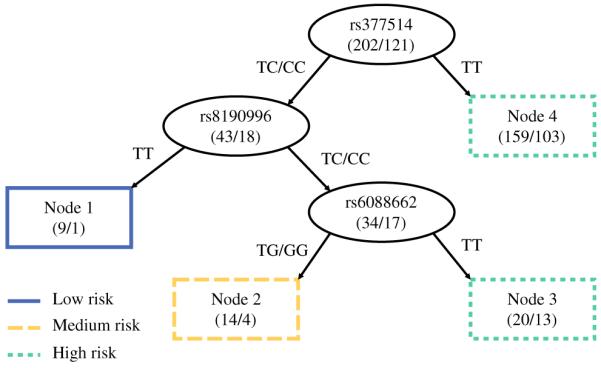
Survival tree analysis of NMIBC recurrence after TUR and BCG therapy. Four nodes were generated which could be divided into three groups of low, medium, and high risk
DISCUSSION
In this report, we have identified several genetic variants in GSH pathway with possible predictive value in NMIBC patients treated with TUR and BCG treatment. We systematically evaluated the effects of 114 SNPs in 21 GSH pathway genes on NMIBC recurrence. One SNP (rs3746162) in GPX4 was significantly associated with bladder cancer recurrence after TUR treatment. Four SNPs (rs7265992, rs6060124, rs7260770, and rs4911455) in GSS were significantly associated with bladder cancer recurrence after TUR and BCG treatment. To our knowledge, this is the first study to systemically evaluate the association between GSH pathway variants and NMIBC outcome.
The enzyme glutathione synthetase, encoded by the GSS gene, is involved in GSH synthesis. It is known that overexpression of GSS failed to increase intracellular GSH, but GSS deficiency could result in decreased GSH levels and cause dramatic metabolic consequences.10 Genetic variations in GSS may affect GSS production and/or function, leading to a deficiency in GSH. In our study, two of the significant SNPs in GSS that were related to poor outcomes for NMIBC patients after BCG therapy were located in the intron region. Their functional significance is not yet understood. One of them (rs7265992) has already been found to be associated with the survival of small-cell lung cancer patients after platinum-based chemotherapy.15
GPX is an enzyme family whose function is to reduce lipid hydroperoxides to their corresponding alcohols and to reduce free hydrogen peroxide to water. This reducing function helps protect cells from oxidative damage.16 The eight isozymes in this family are encoded by various genes (GPX1-8).17 SNP caused by transversion of cytosine to thymine at codon 198 of GPX1 (resulting in an amino acid substitution of proline to leucine) was associated with an increased risk of bladder and lung cancers but prolonged recurrence-free survival in NMIBC.18,19 According to Chiong et al., the GPX1 CT genotype was associated with decreased recurrence time after intravesical BCG therapy.20 The same group had previously found that the combination of BCG and antioxidants caused a reduction in reactive oxygen species and increased cytotoxicity in bladder cancer cells, suggesting that a nonimmunological effect of BCG treatment also may contribute to the antitumor response.20,21
GPX4 is a selenium-containing enzyme. Selenium can be incorporated at amino acid 21 to form selenocysteine, which is in the catalytic center of GPX4. Selenium incorporation requires a specific RNA structure in the 3′ untranslated region (3′UTR) of the GPX4 mRNA. Therefore, the SNPs in the GPX4 3′UTR may affect the function of GPX4. Bermano et al. reported that the SNP rs713041, located in GPX4 3′UTR, was associated with the risk of colorectal cancer. They also showed that different genotypes of rs713041 led to different mRNA expression and enzyme activities.22 In our study, the most significant SNP for the TUR group (rs3746162) also was located in the 3′UTR of GPX4. This may provide the functional basis for the association of this SNP, although its biological significance remains to be determined in future experiments. Interestingly, the association of rs3746162 was highly significant in patients with low-grade but not high-grade tumors (Q < 0.05), whereas the association of rs1006771 of GSTT2 was significant in high-grade but not low-grade patients, indicating the predictive value of these SNPs based on treatment and tumor characteristic.
There are several significant SNPs in GST genes related to cancer recurrence in the TUR or BCG groups. GSTs play a major role in phase II detoxification by catalyzing the conjugation of GSH with a number of different compounds, many of which are carcinogenic.11 Human cytosolic GSTs are highly polymorphic and can be divided into six classes: alpha (GSTA), mu (GSTM), omega (GSTO), pi (GSTP), theta (GSTT), and zeta (GSTZ).10 SNPs in GST genes have been widely studied and are correlated with an increased risk of various cancers. We found that two SNPs in GSTA2 (rs2144698 and rs2180314) were significantly correlated with NMIBC recurrence after TUR treatment. In a previous study, Ning et al. did not find any association with prostate cancer after evaluating various GSTA2 genetic polymorphisms.23 Andonova et al. genotyped two SNPs in GSTA2 (rs2180314 and rs6577), and they did not find any association with breast cancer risk.24 However, the same gene, and even the same SNP, may have different influences on different cancer types. More studies are needed to understand how GSTA2 may impact cancer risk and prognosis. GSTM3 SNPs have been more widely studied in various cancer types. Schnakenberg et al. reported that homozygous wild-type genotypes of a GSTM3 SNP (rs36120609) in intron 6 were significantly protective against bladder cancer.25 However, the same SNP in GSTM3 was reported to have no impact on bladder cancer risk in a study by Golka et al.26 In a study by Yu et al. investigating breast cancer risk in a Han Chinese population, the GG genotype in GSTM3:rs4970737 was a protective factor.27 We also found that the GG genotype in GSTM3:rs4970737 could protect NMIBC cancer patients from early recurrence after TUR and BCG therapy.
We also evaluated the cumulative effects of GSH pathway SNPs on the risk of bladder cancer recurrence. We observed a dose-dependent correlation between the number of unfavorable genotypes and recurrence risk. Altogether, these results support the hypothesis that NMIBC recurrence after definitive treatment is a polygenic process.
Besides recurrence, we evaluated the association of the GSH SNPs with NMIBC progression, which is defined as recurring tumor progressing in tumor grade or stage or leading to cystectomy. Overall, there were a total of 105 cases showing progression and 307 cases without progression. Our findings showed a few SNPs with significant association, but none of the associations were significant after consideration of multiple testing. This might be due to the small sample size or suggests the unique role of the GSH pathway genes in NMIBC recurrence.
There are a few limitations in our study. First, our sample size, although adequately powered for overall analysis, may be limited in subgroup analyses. Nevertheless, several top significant associations with biological plausibility were found in patients stratified by treatment, even after adjustment for multiple testing to minimize false discoveries. Second, the SNPs genotyped in this study are mostly tagging SNPs and most likely not the true causal variant. Therefore, functional characterizations are needed to ascertain the underlying biological mechanisms for the significant associations. Due to the significant challenges of obtaining a comparable clinical cohort with similar study design and treatment regimens with follow-up protocol, we were unable to conduct a replication analysis at this time. Further independent or external validation will be necessary to verify our findings.
CONCLUSIONS
We have conducted the first study to evaluate systemically the association between genetic variation in GSH pathway and NMIBC recurrence. We also determined the effect of GSH pathway SNPs on clinical outcome in various treatment and tumor grade subgroups. Once validated, these findings may provide urologists additional genetic information for clinical assessment and treatment decisions. Nevertheless, the underlying mechanisms through which these genes or SNPs affect the clinical behavior of bladder cancers require further study. Future investigations in other populations and detailed functional characterization are needed to establish GSH pathway variants as predictive or prognostic markers for NMIBC.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH Grants U01 CA127615, R01 CA74880, P50 CA91846, R01 CA131335, and MD Anderson’s Cancer Center Support Grant CA016672.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1245/s10434-015-4431-5) contains supplementary material, which is available to authorized users.
CONFLICT OF INTEREST All authors declare no conflict of interest.
REFERENCES
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.van Rhijn BW, Burger M, Lotan Y, et al. Recurrence and progression of disease in non-muscle-invasive bladder cancer: from epidemiology to treatment strategy. Eur Urol. 2009;56(3):430–42. doi: 10.1016/j.eururo.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Heney NM, Nocks BN, Daly JJ, Blitzer PH, Parkhurst EC. Prognostic factors in carcinoma of the ureter. J Urol. 1981;125(5):632–636. doi: 10.1016/s0022-5347(17)55143-5. [DOI] [PubMed] [Google Scholar]
- 4.Klan R, Loy V, Huland H. Residual tumor discovered in routine second transurethral resection in patients with stage T1 transitional cell carcinoma of the bladder. J Urol. 1991;146(2):316–8. doi: 10.1016/s0022-5347(17)37779-0. [DOI] [PubMed] [Google Scholar]
- 5.Bohle A, Brandau S. Immune mechanisms in bacillus Calmette-Guerin immunotherapy for supercial bladder cancer. J Urol. 2003;170(3):964–9. doi: 10.1097/01.ju.0000073852.24341.4a. [DOI] [PubMed] [Google Scholar]
- 6.Davis JW, Sheth SI, Doviak MJ, Schellhammer PF. Superficial bladder carcinoma treated with bacillus Calmette-Guerin: progression-free and disease specific survival with minimum 10-year followup. J Urol. 2002;167:494–500. doi: 10.1016/S0022-5347(01)69072-4. 2 Pt 1. discussion 501. [DOI] [PubMed] [Google Scholar]
- 7.Lamm DL, Blumenstein BA, Crawford ED, et al. A randomized trial of intravesical doxorubicin and immunotherapy with bacille Calmette-Guerin for transitional-cell carcinoma of the bladder. N Engl J Med. 1991;325(17):1205–9. doi: 10.1056/NEJM199110243251703. [DOI] [PubMed] [Google Scholar]
- 8.Catalona WJ, Ratliff TL. Bacillus Calmette-Guerin and superficial bladder cancer. Clinical experience and mechanism of action. Surg Ann. 1990;22:363–78. [PubMed] [Google Scholar]
- 9.Oddens JR, van der Meijden AP, Sylvester R. One immediate postoperative instillation of chemotherapy in low risk Ta, T1 bladder cancer patients. Is it always safe? Eur Urol. 2004;46(3):336–8. doi: 10.1016/j.eururo.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43(2):143–81. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 11.Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem Funct. 2004;22(6):343–52. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 12.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134(3):489–92. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 13.Chen M, Hildebrandt MA, Clague J, et al. Genetic variations in the sonic hedgehog pathway affect clinical outcomes in non-muscle-invasive bladder cancer. Cancer Prev Res (Phila) 2010;3(10):1235–45. doi: 10.1158/1940-6207.CAPR-10-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Singer B. Recursive partitioning in the health sciences. Springer; New York: 1999. [Google Scholar]
- 15.Sun Z, Chen J, Aakre J, et al. Genetic variation in glutathione metabolism and DNA repair genes predicts survival of small-cell lung cancer patients. Ann Oncol. 2010;21(10):2011–6. doi: 10.1093/annonc/mdq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paz-y-Mino C, Munoz MJ, Lopez Cortes A, et al. Frequency of polymorphisms pro198leu in GPX-1 gene and ile58thr in MnSOD gene in the altitude Ecuadorian population with bladder cancer. Oncol Res. 2010;18(8):395–400. doi: 10.3727/096504010x12644422320780. [DOI] [PubMed] [Google Scholar]
- 17.Toppo S, Vanin S, Bosello V, Tosatto SC. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid Redox Signal. 2008;10(9):1501–14. doi: 10.1089/ars.2008.2057. [DOI] [PubMed] [Google Scholar]
- 18.Zhao H, Liang D, Grossman HB, Wu X. Glutathione peroxidase 1 gene polymorphism and risk of recurrence in patients with superficial bladder cancer. Urology. 2005;66(4):769–74. doi: 10.1016/j.urology.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 19.Ichimura Y, Habuchi T, Tsuchiya N, et al. Increased risk of bladder cancer associated with a glutathione peroxidase 1 codon 198 variant. J Urol. 2004;172(2):728–32. doi: 10.1097/01.ju.0000130942.40597.9d. [DOI] [PubMed] [Google Scholar]
- 20.Chiong E, Kesavan A, Mahendran R, et al. NRAMP1 and hGPX1 gene polymorphism and response to bacillus Calmette-Guerin therapy for bladder cancer. Eur Urol. 2011;59(3):430–7. doi: 10.1016/j.eururo.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 21.Pook SH, Esuvaranathan K, Mahendran R. N-acetylcysteine augments the cellular redox changes and cytotoxic activity of internalized mycobacterium bovis in human bladder cancer cells. J Urol. 2002;168(2):780–5. [PubMed] [Google Scholar]
- 22.Bermano G, Pagmantidis V, Holloway N, et al. Evidence that a polymorphism within the 3′UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. Genes Nutr. 2007;2(2):225–32. doi: 10.1007/s12263-007-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning B, Wang C, Morel F, et al. Human glutathione S-transferase A2 polymorphisms: variant expression, distribution in prostate cancer cases/controls and a novel form. Pharmacogenetics. 2004;14(1):35–44. doi: 10.1097/00008571-200401000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Andonova IE, Justenhoven C, Winter S, et al. No evidence for glutathione S-transferases GSTA2, GSTM2, GSTO1, GSTO2, and GSTZ1 in breast cancer risk. Breast Cancer Res Treat. 2010;121(2):497–502. doi: 10.1007/s10549-009-0589-5. [DOI] [PubMed] [Google Scholar]
- 25.Schnakenberg E, Breuer R, Werdin R, Dreikorn K, Schloot W. Susceptibility genes: GSTM1 and GSTM3 as genetic risk factors in bladder cancer. Cytogenet Cell Genet. 2000;91(1–4):234–8. doi: 10.1159/000056851. [DOI] [PubMed] [Google Scholar]
- 26.Golka K, Schmidt T, Seidel T, et al. The influence of polymorphisms of glutathione S-transferases M1 and M3 on the development of human urothelial cancer. J Toxicol Environ Health A. 2008;71(13 14):881–6. doi: 10.1080/15287390801988087. [DOI] [PubMed] [Google Scholar]
- 27.Yu KD, Fan L, Di GH, et al. Genetic variants in GSTM3 gene within GSTM4-GSTM2-GSTM1-GSTM5-GSTM3 cluster influence breast cancer susceptibility depending on GSTM1. Breast Cancer Res Treat. 2010;121(2):485–96. doi: 10.1007/s10549-009-0585-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


