Abstract
Given the wide differences in HIV-1 viral load (VL) setpoint across subjects as opposed to fairly stable VL over time within an infected individual, it is important to identify host and viral characteristics that affect VL setpoint. While recently-infected individuals with multiple phylogenetically-linked HIV-1 founder variants represent a minority of HIV-1 infections, we found in two different cohorts that more diverse HIV-1 populations in early infection were associated with significantly higher VL one year after HIV-1 diagnosis.
Approximately 20-35% of individuals become infected with multiple founder HIV-1 variants1-3. We therefore sought to evaluate whether genetic characteristics of the founder viral populations could influence markers of clinical outcomes. An association between infections with multiple HIV-1 variants and poorer disease outcomes is supported by earlier reports based on heteroduplex mobility assays (HMA)4 and dual HIV infections5-7. More recently, large studies sought to derive HIV-1 sequences through single genome amplification (SGA) from samples collected in acute or early HIV-1 infection to better define the viruses establishing HIV-1 infection, including by enumerating the number of HIV-1 founder variants. The availability of larger, more precise sequence data sets prompted us to test the association between HIV-1 diversity and markers of disease progression using SGA-derived HIV-1 genomic data. We focused on HIV-1 breakthrough infections in the Step and RV144 HIV-1 vaccine efficacy trials (median of six and ten genomes, respectively)8-11, and restricted our analysis to 63 Step trial participants (infected with HIV-1 subtype B) and 100 RV144 trial participants (infected with CRF01_AE) who had VL and CD4+ T cell measurements in the absence of antiretroviral therapy (ART). In both trials, HIV-1 infections were established by a single viral variant in most individuals, with no significant difference in proportions between treatment groups (p ≥ 0.81)10,11. We used two measures of diversity for HIV-1 founder populations: a categorical measure that distinguishes between subjects with a single (homogeneous viral populations) or multiple (heterogeneous viral populations) founder variants, and a continuous measure of env diversity in the envelope gene (env) corresponding to the mean pairwise diversity among sequences from a subject. Regarding the 63 Step study participants, the median diversity was 0.073% (0-0.566%) among the 47 subjects with homogeneous viral populations and 0.593% (0.026-5.98%) for those with heterogeneous populations. For the 100 RV144 participants, median diversity was 0.194% (0.027-0.847%) among the 68 participants with homogeneous founding populations and 0.825% (0.073-4.42%) for those with heterogeneous populations.
Since relevant variables and availability of baseline variables and VL and CD4+ T cell measurements differed between the trials, data were analyzed separately (as previously described8,9,12,13). Linear regression models were used to relate each diversity measure to post-infection endpoints while accounting for baseline subject characteristics. Besides the treatment assignment (vaccine or placebo), statistical models adjusted for multiple co-variates, including sex, HLA genotype (Step only) and a baseline behavioral risk score. Here, we present fully-adjusted results (see Supplementary Tables for unadjusted and partially-adjusted results).
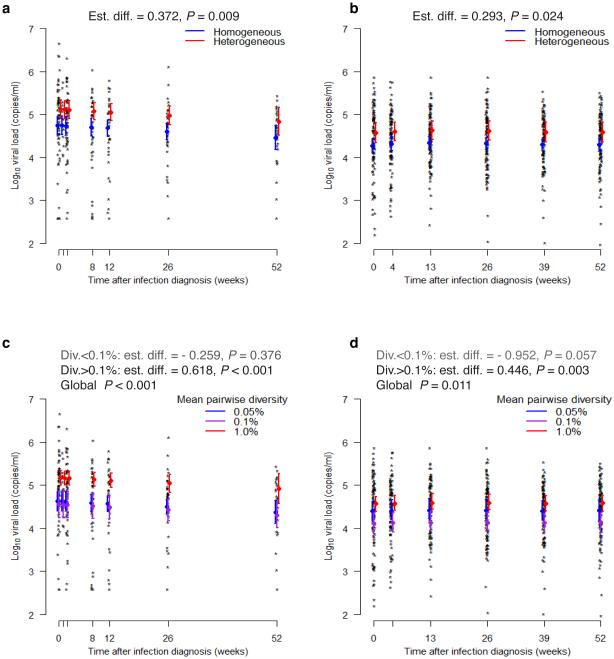
In the Step study, there was no association between sequence diversity and VL at HIV-1 diagnosis, neither when homogeneous vs heterogeneous viral populations were compared (P value = 0.88), nor when env diversity measures were considered (P value = 0.37) (Supplementary Fig. 1 and Supplementary Table 1). When we considered the 276 pre-ART VL measurements obtained during the first year of follow up (median = 5 [1-7] per subject), subjects with heterogeneous founder viral populations showed significantly higher mean VL than subjects with homogeneous founder populations: the estimated difference was 0.37 log10 VL (copies/ml), based on analyses that adjusted for baseline subject characteristics in addition to time since infection diagnosis (P value = 0.01) (Fig. 1). Similar to the categorical data, higher mean VL over the first year was associated with higher env diversity of the founder population (p < 0.001). The relationship was non-linear, such that there was no association between VL and diversity below the cutpoint of 0.1% env diversity (P value = 0.376), and a positive association between VL and env diversity above the cutpoint (p < 0.001). If we limited attention to VL measurements within 3 months of diagnosis, the estimated effects of categorical and continuous diversity measures were larger; while, with all VL measurements out to two years post-diagnosis, the effects were smaller yet still statistically significant for the categorical measure (p < 0.001) or trending for the continuous measure (P value = 0.08; Supplementary Text and Supplementary Table 1).
Figure 1.
Positive relationship between HIV-1 diversity and viral loads over the first year since HIV-1 diagnosis. HIV-1 diversity was measured at the time of HIV-1 diagnosis. Individual log VL values and predicted means and 95% confidence intervals are shown at each post-infection visit. The upper panels correspond to Step (a) and RV144 (b) trial participants with homogeneous or heterogeneous viral populations; the lower panels correspond to low, medium, and high levels of env diversity in Step (c) and RV144 (d) trial participants. Models adjusted for baseline subject characteristics and assumed that the effects of heterogeneity and diversity are constant over time.
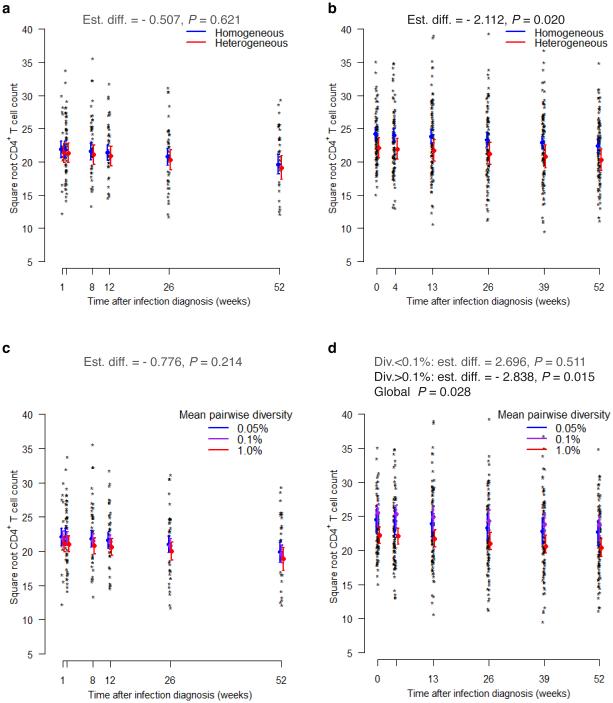
In parallel, we analyzed longitudinal CD4+ T cell counts (216 measurements with a median of 4 [1–6] per subject). There was no significant difference in mean square root CD4+ T cell counts over the first year when analyzing either the categorical (P value = 0.62) or continuous measures of diversity (P value = 0.21) (Fig. 2 and Supplementary Table 1).
Figure 2.
Negative relationship between HIV-1 diversity and CD4+ T cell counts over the first year since HIV-1 diagnosis. HIV-1 diversity was measured at the time of HIV-1 diagnosis. Individual CD4+ T cell count measurements and predicted means and 95% confidence intervals are shown at each post-infection visit. The upper panels correspond to Step (a) and RV144 (b) trial participants with homogeneous or heterogeneous viral populations; the lower panels correspond to low, medium, and high levels of env diversity in Step (c) and RV144 (d) trial participants. Models adjusted for baseline subject characteristics and assumed that the effects of heterogeneity and diversity are constant over time.
Next, we analyzed data from the RV144 trial. At the time of HIV-1 diagnosis, subjects with heterogeneous founder viral populations showed significantly higher mean VL than subjects with homogeneous populations (0.39 log10 VL, P value = 0.02), and there was a significant positive association between mean VL and increasing env diversity (P value = 0.02) (Supplementary Fig. 1 and Supplementary Table 1). Focusing on the 485 pre-ART VL measurements made within a year of diagnosis (median = 5 [2–6] per subject), we found that subjects with heterogeneous founder viral populations showed significantly higher mean VL over the first year than subjects with homogeneous ones: the estimated difference was 0.29 log10 VL (P value = 0.02) (Fig. 1 and Supplementary Table 1). Similar to the categorical data, higher VL over the first year was associated with increasing env diversity of the founder population (P value = 0.01). The association was non-linear; below the cutpoint of 0.1% env diversity, a borderline-significant negative association between VL and diversity was observed (P value = 0.06), while above the cutpoint a positive association was observed (P value = 0.003). Looking instead at VL measurements either over the first 3 months or the first two years since HIV-1 diagnosis, higher VL was associated with increased heterogeneity (all P values ≤ 0.06; Supplementary Text and Supplementary Table 1).
When RV144 longitudinal CD4+ T cell counts were analyzed, subjects with heterogeneous founder populations showed lower mean square root CD4+ T cell count over the first year of infection compared to subjects with homogeneous ones (P value = 0.02, Fig. 2 and Supplementary Table 1). When env diversity was considered, increasing env diversity was found to be significantly associated with decreasing mean square root CD4+ T cell counts (P value = 0.03). The relationship was non-linear: not significant below the cutpoint of 0.1% env diversity (P value = 0.51) and a negative association above the cutpoint (P value = 0.02).
These results suggest that more heterogeneity in the HIV-1 founder population of recently-infected individuals is associated with higher VL over time, confirming an earlier HMA-based report4. When VL measurements obtained up to two years post-diagnosis were included, the sizes of the effects were smaller, but still significant or trending (P values between < 0.001 and 0.08). Study subjects were vaccine or placebo recipients in the Step and RV144 vaccine trials, yet there was no evidence that the vaccine assignment modified the associations (Q values > 0.3, accounting for multiplicity in interaction tests across endpoints). To address the possibility of post-randomization selection bias, whereby breakthrough vaccine and placebo infected cases are not comparable due to differences in characteristics associated with both HIV-1 infection and post-infection endpoints, we adjusted for covariates potentially predictive of either HIV-1 acquisition or disease progression, such as HLA (for Step) and baseline behavioral risk, and found that including these variables had negligible impact on the results. We also performed sensitivity analyses to show that our sequencing protocol and the lack of precision in the timing of HIV-1 infection dates in both cohorts did not noticeably affect our results (Supplementary Tables 2 and 3; Supplementary Text). The limited depth of sequencing is another potential limitation; however, pyrosequencing data confirmed our estimates of founder variants for 48 of the 63 subjects (selected on the basis of sample availability) in the Step Study (Iyer and Mullins, personal communication).
Some results differed between the two cohorts. In the Step Study, the positive association between VL and multiplicity of founder variants was seen at later time points but not at HIV-1 diagnosis (note that infections were diagnosed earlier in Step compared to RV14414). A similar lack of association between env diversity at a median of six weeks post-infection and contemporary VL was reported previously15. This supports the view that VL setpoint is not a characteristic of the founder virus per se, and highlight that the relationship between the host and the virus is critical in the establishment of the VL setpoint. In addition, we failed to find an association between CD4+ T cell counts and multiplicity of founder variants in the Step Study. To interpret this result, we note that the relationship between CD4+ T cell count and VL was stronger in RV144 (Spearman rank correlation between VL and CD4+ T cell counts residuals = − 0.52) than in the Step Study (− 0.44). It has previously been noted that the link between the two predictors of virulence is not always strong16.
Since different studies have shown considerable effects of viral genotype on VL setpoint, between 6%17 and 59%18, a better characterization of viral factors potentially impacting VL setpoint is critical to our understanding of HIV-1 pathogenesis. Although the size of the HIV-1 genetic effect estimated in our analyses can be considered modest (0.29-0.37 log10), our findings were replicated in two independent cohorts with different distributions of subject ethnicity, route of HIV-1 transmission and infecting HIV-1 subtype. Besides, a difference of 0.3 log10 in VL setpoint is clinically relevant regarding both disease progression19 and HIV-1 transmission (a decrease in VL of 0.74 log10 (95% CI 0.60 to 0.97) was estimated to reduce by 50% the risk of heterosexual transmission20). Further studies are needed to define host specificities that predict an individual’s propensity to acquire a multi-variant HIV-1 infection. While our study cannot be used to determine whether certain individuals were predisposed to acquire multiple HIV-1 variants, the fact that individuals replicating multiple HIV-1 variants presented higher VL illustrates the consequences of the initial steps of HIV-1 infection for clinical disease progression, and suggests that limiting HIV-1 founder heterogeneity could be a goal for prophylactic interventions.
Online Methods
Ethics Statement
The Step Study (HVTN502) and RV144 vaccine trials were registered at ClinicalTrials.gov and assigned the registration numbers NCT00095576, and NCT00223080, respectively. The RV144 protocol was approved by the ethics committees of the Ministry of Public Health, the Royal Thai Army, Mahidol University, and the Human Subjects Research Review Board of the U.S. Army Medical Research and Materiel Command. The Step protocol was approved by the ethics committees of each trial site (Sydney, Australia; Rio de Janeiro, Brazil; Sao Paulo, Brazil; Montreal, Canada; Toronto, Canada; Vancouver, Canada; Santo Domingo, Dominican Republic; Port-au-Prince, Haiti; Kingston, Jamaica; Iquitos, Peru; Lima, Peru; San Juan, Puerto Rico; United States: Atlanta, GA; Birmingham, AL; Boston, MA; Chicago, IL; Denver, CO; Houston, TX; Los Angeles, CA; Miami, FL; New York, NY; Newark, NJ; Philadelphia, PA; Rochester, NY; Saint Louis, MI; San Francisco, CA; Seattle, WA). Written informed consent was obtained from all volunteers.
HIV-1 genetic data
Breakthrough infections were sequenced via endpoint dilution PCR from plasma samples collected at HIV-1 diagnosis (except for one Step Study subject sampled 28 days after diagnosis, and 6 RV144 subjects sampled on average 26 days later), with five to ten near full-length genome sequences per individual10,11. Breakthrough HIV-1 sequences were obtained from 68 Step trial participants and 121 RV144 trial participants. Exclusion criteria for our study were: lack of availability of VL and CD4+ T cell measurements, lack of sequence data, being on ART, not being infected by the prevalent HIV-1 subtype, being female in Step. Thus, the cohorts included 63 male participants from the Step Study infected with HIV-1 subtype B (one female, one non-subtype B infection, one subject on ART at diagnosis, and two subjects with only one sequence available were excluded), and 100 RV144 participants infected with CRF01_AE (114 subjects were enrolled in the post-infection follow-up study, three subjects who lacked HIV-1 sequence data, and 11 who were infected with non-CRF01_AE viruses were excluded).
For each intra-host dataset, inspection of sequence alignments, phylogenetic tree topologies and sequence diversity measures were used to categorize infections as established by a single founder, referred to as homogeneous viral population, or multiple founder variants (heterogeneous viral population).
The primary variables used to measure the multiplicity of HIV-1 founder variants were an indicator of homogeneous or heterogeneous infection and the mean pairwise nucleotide diversity in env (mean percent distance between all pairs of sequences for a subject, calculated using the general time reversible model of nucleotide substitution in HyPhy). Mean pairwise diversity was analyzed on the log10 scale, with values of 0 set to 0.0065, the midpoint between the lowest positive value of 0.013 and 0. Piecewise linear splines for log mean pairwise sequence diversity (hereafter “env diversity") were considered as candidate predictors. Where the data did not support the inclusion of piecewise-linear terms, models included only the linear terms.
Clinical data
Pre-ART VL measurements from the first year of follow up were obtained at weeks 0, 1, 2, 8, 12, 26, 52, 78 and 104 in the Step trial, and at months <1, 1, 3, 6, 9, 12, 18, 24 in RV144. Longitudinal CD4+ T cell counts were measured at the same post-infection visits (except for the diagnostic visit in Step).
Statistical Methods
The VL endpoints were log10 VL at HIV-1 diagnosis (corresponded to the time of HIV sequencing), and longitudinal VL; both were censored at ART initiation. Longitudinal pre-ART CD4+ T cell counts were analyzed on the square root scale. Linear regression models were used to relate each predictor (homogeneous or heterogeneous infection; env diversity) to each post-infection endpoint. Wald tests were used to test for statistical interactions between each predictor and the vaccine or placebo assignment. Q values were calculated to account for multiplicity in interaction tests across endpoints; we considered Q < 0.20 to be significant, implying that up to 20% of the “significant” interaction results are expected to be false positives. (See results in Supplementary Text).
For the Step Study analyses of longitudinal VL and CD4+ T cell counts, weighted generalized estimating equations (GEE) models with exchangeable working correlation were used. GEE models account for the correlation between longitudinal measurements on the same subject. Observations were weighted with respect to the inverse probability of having an observed pre-ART VL and CD4+ T cell count measurement, which was factored as the product of: 1) the probability of not dropping out; 2) the probability of not initiating ART given not dropping out; and 3) the probability of not missing the visit given not dropping out or initiating ART. Each probability was modeled using a separate logistic GEE regression model with independence working correlation. Weeks post-infection, year of diagnosis, geographic region, age, and an indicator of all 3 vaccinations received were used to predict dropout; weeks post-infection, geographic region, Ad5 seropositivity, an indicator of all 3 vaccinations received, and previous VL were used to predict ART initiation; and geographic region and previous VL and CD4+ T cell count were used to predict missing visits. The following covariates were included as specified: treatment assignment, Ad5 seropositivity, self-reported circumcision status, HSV-2 serostatus, age under or over 30 years, Australian or North American residence, HLA group and baseline behavioral risk score. HLA group was defined as “protective" (B27, B57, B*58:01), “unfavorable" (B*35:02, *35:03, *35:04, B53, or homozygous in at least one locus), or “neutral" (all others) (a “protective” allele assigns a subject in the “protective” HLA group irrespective of the other alleles). The baseline risk score is defined as the number of risk behaviors reported at baseline among the following: more than 2 male partners, any drug use over the last 6 months, unprotected anal sex with a male partner, unprotected vaginal sex with a female partner, evidence of a sexually transmitted disease, or exchanging sex for money, all over the 6 months prior to enrollment. Two individuals from North America without circumcision status were considered circumcised. Where specified, models also adjusted for linear terms of days post-diagnosis.
For RV144 analyses of longitudinal VL and CD4+ T cell counts, multiple imputation was used to fill in missing values and GEE was used for modeling with AR1 working correlation structure. The following covariates were included where specified: 6 centered polynomial terms of days since infection diagnosis, calendar period of infection diagnosis [2003-2005, 2006, 2007, 2008-2009], age, gender, and baseline behavioral risk [low, medium, high]. The VL model was not adjusted for time-dependent CD4+ T cell count, nor were CD4+ T cell counts adjusted for VL; this was a departure from previous analyses of the VL and CD4+ data13. Centered polynomial terms of days since infection diagnosis were included: one polynomial (linear) term was used for months 0-3, 3 polynomial terms in analyses of months 0-12, and 4 polynomial terms in analyses of months 0-24. The baseline risk score depended on the following risk factors: number of partners, unprotected sex with regular or casual sex partner, with sex worker, with same sex partner or with injecting drug user, injection drug use, sharing needles, symptoms of a sexually transmitted disease. No HLA adjustment was done in the RV144 cohort because there is limited information about which HLA alleles are protective, unfavorable, or neutral in the Thai CRF01_AE infected population. Our comparison of the genotypes from 74 placebo recipients who became infected in the RV144 trial to the genotypes of 450 placebo recipients21 who remained uninfected did not identify any HLA alleles with frequencies greater than 5% that associated with HIV-1 acquisition (Fisher’s exact test p ≥ 0.05, q > 0.20).
Supplementary Material
Acknowledgements
We would like to thank the volunteers in the Step Study and RV144 trial for their contribution to this research.
Footnotes
Author Contributions
H.J. and M.R. designed and performed experiments, analyzed data and wrote the manuscript. S.T., A.D., J.H., L.C., S.G.S., S.B., M.J.M., R.J.O., R.P., S.R.-N., S.N., P.P., J.K., M.L.R., N.L.M., J.H.K., P.B.G. oversaw the vaccine trials and clinical aspects. J.T.H., R.T., N.F., M.J.M., M.L.R., N.L.M., J.I.M., J.H.K., P.B.G. edited the manuscript.
Competing Financial Interests
Sequencing and analysis were performed under grants from the National Institute of Allergy and Infectious Diseases: US Public Health Service grant AI41505; Interagency Agreement Y1-AI-2642-12 with the US Army Medical Research and Material Command; NIH grant 2R37AI05465-10 to P.B.G. This work was also supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the US Department of Defense. M.R., S.T., R.T., and M.L.R. are employees of the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. The opinions expressed herein are those of the authors and should not be construed as official or representing the views of the US Department of Defense or the Department of the Army. This does not alter our adherence to policies on sharing data and materials.
References
- 1.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahams MR, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, et al. High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sagar M, et al. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J Virol. 2003;77:12921–12926. doi: 10.1128/JVI.77.23.12921-12926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb GS, et al. Lancet. 2004;363:619–622. doi: 10.1016/S0140-6736(04)15596-7. [DOI] [PubMed] [Google Scholar]
- 6.Jost S, et al. HIV Super-Infection: Rapid Replacement of AE Subtype by B Subtype; 9th Conference on Retroviruses and Opportunistic Infections; Seattle, Washington. 2002. Vol. Abstract: 757-W. [Google Scholar]
- 7.Altfeld M, et al. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature. 2002;420:434–439. doi: 10.1038/nature01200. [DOI] [PubMed] [Google Scholar]
- 8.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009 doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 10.Rolland M, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolland M, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012 doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janes H, et al. MRKAd5 HIV-1 Gag/Pol/Nef Vaccine-Induced T-Cell Responses Inadequately Predict Distance of Breakthrough HIV-1 Sequences to the Vaccine or Viral Load. PLoS ONE. 2012;7:e43396. doi: 10.1371/journal.pone.0043396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rerks-Ngarm S, et al. Extended evaluation of the virologic, immunologic, and clinical course of volunteers who acquired HIV-1 infection in a phase III vaccine trial of ALVAC-HIV and AIDSVAX B/E. J Infect Dis. 2013;207:1195–1205. doi: 10.1093/infdis/jis478. [DOI] [PubMed] [Google Scholar]
- 14.Edlefsen PT, Gilbert PB, Rolland M. Sieve analysis in HIV-1 vaccine efficacy trials. Curr Opin HIV AIDS. 2013;8:432–436. doi: 10.1097/COH.0b013e328362db2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rieder P, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin Infect Dis. 2011;53:1271–1279. doi: 10.1093/cid/cir725. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez B, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. Jama. 2006;296:1498–1506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 17.Hodcroft E, et al. The contribution of viral genotype to plasma viral set-point in HIV infection. PLoS Pathog. 2014;10:e1004112. doi: 10.1371/journal.ppat.1004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alizon S, et al. Phylogenetic approach reveals that virus genotype largely determines HIV set-point viral load. PLoS Pathog. 2010;6:e1001123. doi: 10.1371/journal.ppat.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellors JW, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 20.Lingappa JR, et al. Estimating the impact of plasma HIV-1 RNA reductions on heterosexual HIV-1 transmission risk. PLoS One. 2010;5:e12598. doi: 10.1371/journal.pone.0012598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prentice HA, et al. HLA class I, KIR, and genome-wide SNP diversity in the RV144 Thai phase 3 HIV vaccine clinical trial. Immunogenetics. 2014;66:299–310. doi: 10.1007/s00251-014-0765-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.