Summary
s-adenosylmethionine (SAM) is the sole methyl donor modifying histones, nucleic acids and phospholipids. Its fluctuation impacts hepatic phosphatidylcholine (PC) synthesis or may be linked to variations in DNA or histone methylation. Physiologically, low SAM is associated with lipid accumulation, tissue injury and immune responses in fatty liver disease. However, molecular connections between SAM limitation, methyltransferases and disease-associated phenotypes are unclear. We find that low SAM can activate or attenuate Caenorhabditis elegans immune responses. Immune pathways are stimulated downstream of PC production on a non-pathogenic diet. In contrast, distinct SAM-dependent mechanisms limit survival on pathogenic Pseudomonas aeruginosa. C. elegans undertakes a broad transcriptional response to pathogens and we find that low SAM restricts H3K4me3 at Pseudomonas-responsive promoters, limiting their expression. Furthermore, this response depends on the H3K4 methyltransferase set-16/MLL. Thus, our studies provide molecular links between SAM and innate immune functions and suggest that SAM depletion may limit stress-induced gene expression.
Introduction
Metabolites can regulate signaling and transcriptional pathways by providing small molecules that modulate protein or nucleotide function, coordinating nutrition with physiology. For example, in Caenorhabditis elegans dietary variation in vitamin B12 leads to changes in fertility, lifespan and gene expression allowing adaptation to distinct nutritional sources (Macneil et al., 2013; Watson et al., 2013; Watson et al., 2014). Another metabolite that can affect a broad array of mechanisms is s-adenosylmethionine (SAM). Produced by the 1-carbon cycle (1CC), SAM is the donor for methylation of histones and other proteins, nucleic acids, and is used in phospholipid synthesis (Kaelin and McKnight, 2013; Mato et al., 2008; Vance, 2014) (Figure 1A, see also Table S1 for human orthologs). Reduced 1CC function is strongly associated with metabolic disorders, particularly fatty liver disease, which is characterized by lipid accumulation and immune dysfunction (Lu et al., 2001). SAM deficiency may also underlie liver pathology in alcohol-induced fatty liver disease (ALD) and nutritional limitation of 1CC function accelerates injury in ALD models (Halsted et al., 2002). Mammalian cells have more than 200 genes predicted to encode SAM-dependent methyltransferases (Petrossian and Clarke, 2011). However the mechanisms connecting SAM levels to specific methyltransferases and molecular changes underlying disease phenotypes are unresolved (Kaelin and McKnight, 2013).
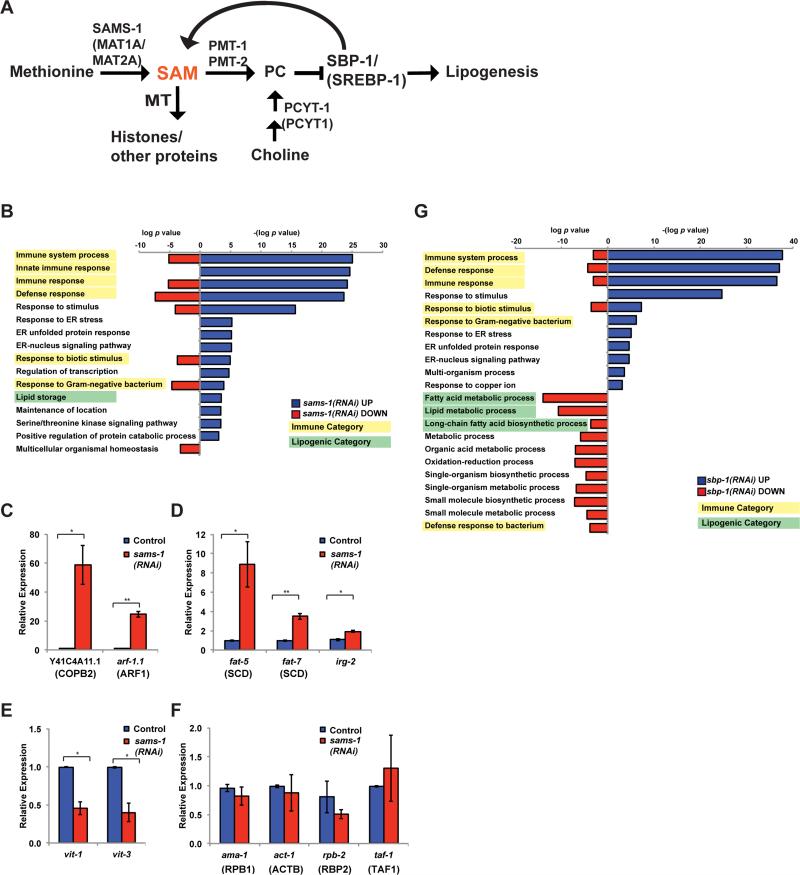
Figure 1. Co-regulation of lipogenic and immune function genes with depletion of SAM.
A. Schematic showing C. elegans pathways producing or utilizing SAM. MT is methyltransferase, SAM is s-adenosylmethionine, PC is phosphatidylcholine. Human names for orthologs are shown in parenthesis, see also Table S1. B. Bar graphs comparing p-values for GO categories of genes regulated more than 2.0 fold after sams-1(RNAi). Downregulated genes are shown in red bars as log p value. To distinguish upregulated genes, -(log p value) was used for blue bars. Immune categories are highlighted in yellow, lipogenic in green. Genes are identified in Table S2, tab: GO Categories. Genes that were upregulated (C-D), down regulated (E) or not changed in microarray studies were validated in qRT-PCR standardized to an exogenous “spike in” mRNA. G. Bar graphs comparing p-values for GO categories of genes regulated more than 2.0 fold after sbp-1(RNAi). Downregulated genes are shown in red bars as log p value. To distinguish upregulated genes, -(log p value) was used for blue bars. Immune categories are highlighted in yellow, lipogenic in green. Genes are identified in Table S3, tab: GO Categories.
We have previously used C. elegans to discern molecular mechanisms linking decreased SAM to lipogenesis and identified a conserved regulatory loop activating the transcription factor SBP-1/SREBP-1 when SAM or the methylated phospholipid phosphatidylcholine (PC) are low (Walker et al., 2011). Here, we find that C. elegans with reduced function of the SAM-synthase sams-1 and concomitant decreases in SAM also have complex immune phenotypes. C. elegans consuming standard E. coli diets have activated immune signatures linked to requirements for SAM in PC production. However, low SAM also impacts histone methylation during acute transcriptional responses, limiting protective gene expression programs upon exposure to the bacterial pathogen Pseudomonas aeruginosa. Thus, phenotypic consequences of SAM deficiency differ as requirements for methylation are changed. Finally, our results suggest that low SAM restricts stress-responsive transcription, potentially linking 1CC dysfunction and tissue injury in metabolic disease.
Results
sams-1 knockdown results in activation of innate immune genes in absence of pathogenic bacteria
In C. elegans, knockdown of the SAM synthase sams-1 decreases SAM and SAH levels around 65% and is associated with increased lipogenesis, decreased fertility and extended lifespan (Hansen, 2005; Walker et al., 2011) (see also Figure S1A). We previously showed that lipogenic gene expression, lipid accumulation and fertility defects in sams-1 animals are linked to synthesis of PC, a methylated phospholipid. Reductions in PC lead to lipid accumulation by activating the transcription factor SBP-1/SREBP-1. To identify other groups of genes that change after sams-1 RNAi and determine if they are also regulated by SBP-1/SREBP-1, we profiled whole genome mRNA expression from sams-1 and sbp-1 RNAi animals (GEO accession: GSE70692). As expected, we found lipogenic genes increased in sams-1(RNAi) animals (Figure 1B, green labels; Table S2). Surprisingly, immune regulators were the most significantly enriched category of upregulated genes (Figure 1B, yellow labels; Table S2). These include components of mitogen activated protein kinase (MAPK) pathways, transcription factors important for immune gene activation as well as infection-response genes, which encode antimicrobial effectors activated upon exposure to pathogens (see Table S2 tab: GO terms) (Engelmann et al., 2011). We validated expression profiling results by quantitative RT-PCR of selected genes normalized to act-1 levels after sams-1(RNAi) (not shown), as well as in sams-1(ok3033) animals (Figure S1B, C). We also normalized qPCR validations to an external “spiked-in” RNA (JNK1a1 alpha, gift of Dr. Roger Davis). These results were similar to the microarray and to the act-1 normalized qPCR for upregulated (Figure 1 C, D), down regulated (Figure 1E) or unchanged genes (Figure 1F) tested. Finally, Lu et al. previously reported changes in some immunity-related genes in mice with a targeted deletion in the SAM synthase MAT1A and fatty liver disease (Lu et al., 2001). Taken together, these results suggest that upregulation of innate immune genes is a major response to methyl donor depletion.
We previously identified a feedback loop stimulating lipogenic SBP-1/SREBP-1-dependent gene expression in sams-1(RNAi) animals (Walker et al, 2011) (see also Figure 1B, G, green highlighting). Innate immune genes may be activated by SREBP proteins in mammalian macrophages (Im et al., 2011), therefore we asked if SBP-1/SREBP-1 was required for immune gene expression after sams-1 RNAi in C. elegans. However these genes were also broadly upregulated after sbp-1 RNAi (Figure 1G). Furthermore, activation of innate immune genes in sams-1(lof) animals occur when sbp-1 is depleted (not shown). Thus, innate immune gene upregulation in sams-1 animals seems unlikely to depend on of feedback loops activating SBP-1/SREBP-1 and may be downstream of distinct SAM and/or PC-dependent pathways.
The PMK-1/p38 MAPK kinase pathway is constitutively active in sams-1 animals
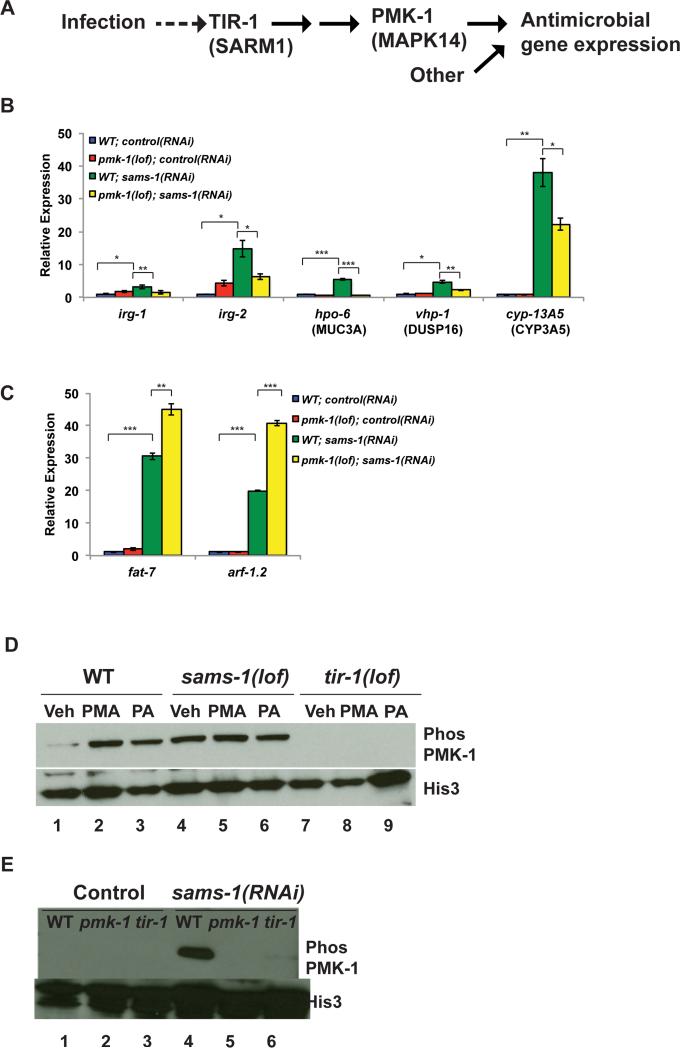
Innate immunity in C. elegans depends on PMK-1, a p38 MAP Kinase important for pathogen responsive expression of many infection response genes (Kim, 2013; Troemel et al., 2006) (Figure 2A). Next, we asked if sams-1-dependent increases in immune gene expression required pmk-1 and found that upregulation was less robust when sams-1 was depleted in pmk-1(lof) mutants (Figure 2B). We also examined expression of irg-1, which is induced independently of pmk-1 upon Pseudomonas exposure (Troemel et al., 2006). Although not identified in our microarray study, irg-1 reproducibly increased after sams-1 RNAi in qRT-PCR experiments, requiring pmk-1 (Figure 2B). Thus, there may be some regulatory distinctions between low SAM and pathogen exposure. Loss of pmk-1 did not limit upregulation of lipogenic genes, or arf-1.1, the gene with the highest-fold increase in sams-1(RNAi) microarrays (Figure 2C), demonstrating that pmk-1 is not generally required for transcriptional changes in sams-1(RNAi) animals.
Figure 2. Constitutive activation of innate immune pathway in after sams-1 depletion.
A. Schematic showing p38/PMK-1 mitogen activated protein kinase signaling during response to bacterial infection in C. elegans (Kim, 2014). B. qRT-PCR comparing innate immune gene expression in sams-1(lof) and pmk-1(lof); sams-1(lof) mutants. C. qRT-PCR comparing expression of a lipogenic (fat-7) or other (arf-1.1) gene highly expressed gene in sams-1(lof) and pmk-1(lof); sams-1(lof) mutants. D. Immunoblot of phospho-PMK-1 in vehicle (Veh), phorbol acid treated (PMA) and Pseudomonas aeruginosa (PA) exposed wild type (WT), sams-1(lof) or tir-1(lof) mutants. Histone 3 shows loading. E. Wild type, pmk-1(lof) or tir-1(lof) animals were exposed to control or sams-1(RNAi) and immunoblotted with antibodies to phosph-PMK-1 or Histone 3. Error bars show standard deviation. Results from Student's T test shown by * <0.05, ** <0.01, *** <0.005.
PMK-1 can be activated by various stresses, however pathogen-responsive phosphorylation depends on a conserved signal transduction pathway downstream of the TIR-1/SARM1 adaptor protein (Couillault et al., 2004; Kim et al., 2002; Troemel et al., 2006) (Figure 2A). To determine if innate immune gene activation was accompanied by PMK-1/p38 MAPK phosphorylation through this pathogen-response pathway, we determined levels of phosphorylated PMK-1/p38 in sams-1(lof) or RNAi animals (Figure 2D, E). As expected, PMK-1/p38 MAPK phosphorylation is induced in response to PMA (a stimulator of p38 MAPKs in C. elegans and mammals (Kawli and Tan, 2008)) (Figure 2D, lanes 1,2) or upon exposure to Pseudomonas (Figure 2D, lanes 1,3) and fails to occur in animals lacking the upstream adaptor tir-1 (Figure 2D, lanes 1-3, 7-9). Strikingly, PMK-1/p38 MAPK was constitutively phosphorylated in sams-1(lof) animals (Figure 2D, lane 4) or after sams-1 RNAi (Figure 2E, lane 4) and phosphorylation was refractory to additional stimulation with PMA or Pseudomonas (Figure 2D, lanes 4-6). Appearance of this band did not occur after pmk-1 was also mutated (Figure 2E, lanes 4,5) and decreased when tir-1 was absent (Figure 2E, lanes 4, 6), suggesting that this PMK-1/p38 MAPK phosphorylation occurs in the context of pathogen-responsive signaling, even in the absence of bacterial stimuli.
Innate immune activation is downstream of PC in sams-1 animals
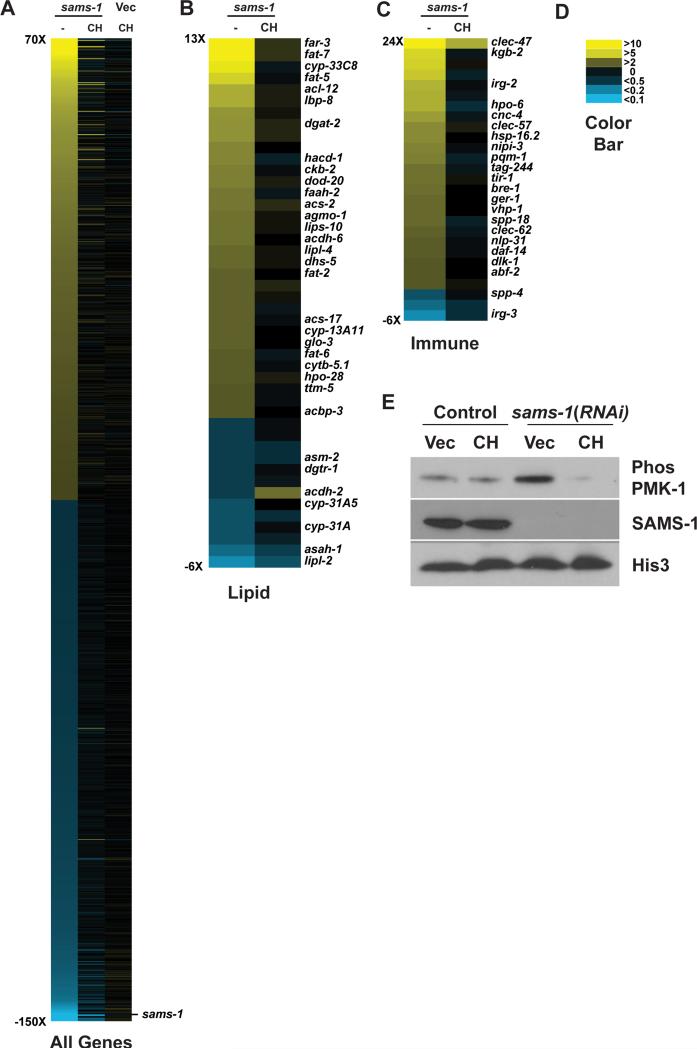
SAM has not been previously identified as an effector of PMK-1/p38 MAPK signaling. Therefore, we asked which methylation-dependent processes (Figure 1A) might link SAM to innate immune activation. To distinguish between PC-dependent or –independent effects, we provided dietary choline, which enables methylation independent PC production (Vance, 2014), returns PC levels to wild type levels (Figure S1D) and robustly rescues lipogenic phenotypes after sams-1(RNAi) (Walker et al., 2011). C. elegans lack a betaine hydoxymenthltransferase ortholog, making it unlikely that choline act to regenerate SAM in nematodes (Wasmuth et al., 2008). First, we profiled genome wide mRNA expression in sams-1(RNAi) and sams-1(RNAi) animals treated with choline (GEO accession: GSE70693). Choline had few significant effects on gene expression in control animals (Figure 3A, Vec/CH; Table S3). Surprisingly, most gene expression changes after sams-1(RNAi) were returned to wild type levels by choline (Figure 3A). As in our previous array study (Figure 1B) we found that many lipogenic and innate immune genes were upregulated (Figure 3 B,C; Figure S1B, C). Next, to determine if blocking PC synthesis downstream of SAM was necessary and sufficient to affect innate immune gene expression, we performed RNAi of the rate-limiting enzyme for PC production, pcyt-1/PCYT1. We found that pcyt-1 RNAi was sufficient to activate lipogenic genes (as in (Walker, et al. 2011)), innate immune genes (Figure S1 E-J) and also induced PMK-1 phosphorylation (not shown). There was little significant change in the expression of lipogenic or immune genes in sams-1(lof); pcyt-1(RNAi) animals, as would be expected if these sams-1 and pcyt-1 act in the same pathway. We noted that choline treatment caused increased expression of innate immune genes in wild type animals exposed to pcyt-1 RNAi. Importantly, choline was no longer sufficient to rescue gene expression increases in sams-1(lof); pcyt-1(RNAi) animals, demonstrating that PC biosynthetic enzymes are required for choline rescue of sams-1 immune phenotypes. Finally, we found that PMK-1/p38 MAPK phosphorylation returned to control levels in sams-1(RNAi) animals supplemented with choline (Figure 3E). Thus, mechanisms activating innate immunity in low SAM appear directly linked to PC production.
Figure 3. Restoration of PC synthesis through dietary choline rescues innate immune phenotypes in sams-1(RNAi) animals.
A. Heat map of genome wide expression changes in sams-1(RNAi), sams-1(RNAi) rescued by choline, or vector-only control (Vec) animals supplemented with choline. Heat map showing changes in expression level and choline rescue of lipogenic (B) and immune function (C) genes. Color values are shown in D. E. Control (Vec) or sams-1(RNAi) animals which were grown on normal media or media supplemented with dietary choline (CH) were immunoblotted with antibodies recognizing phospho-PMK-1, SAMS-1 or Histone 3.
sams-1(lof) animals die rapidly when exposed to Pseudomonas
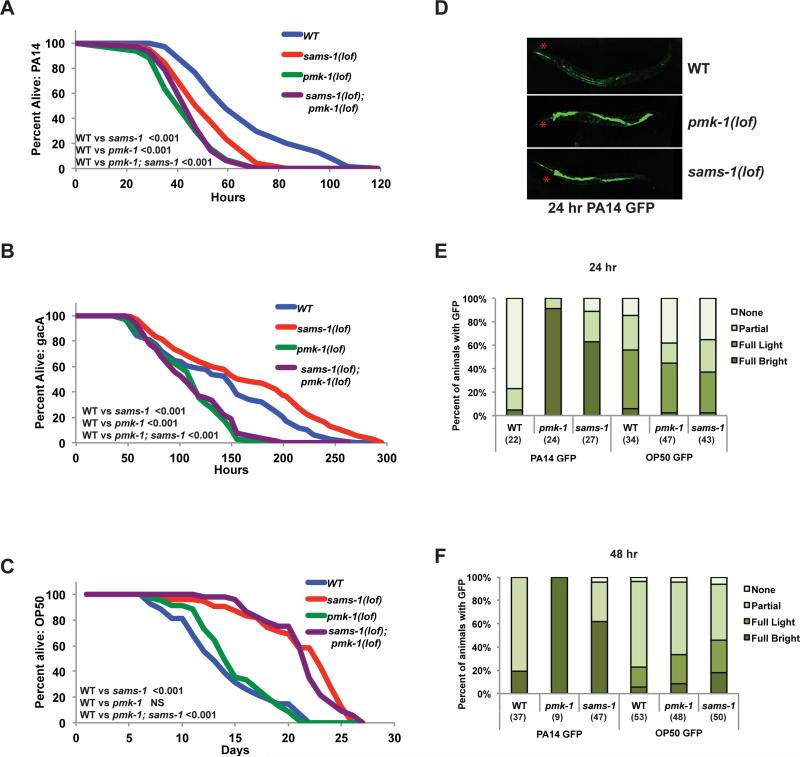
The presence of phosphorylated of PMK-1 and increases in immune genes suggest that sams-1 animals could resist pathogenic challenge. To test this, we exposed sams-1(lof) animals to virulent Pseudomonas (PA14). Surprisingly, sams-1(lof) animals died faster than control animals on PA14 (Figure 4A; Table S4). Survival of sams-1(lof); pmk-1(lof) animals was identical to pmk-1(lof) alone. Pseudomonas represents a distinct nutritional source from E. coli, therefore we asked if the bacterial food source or the strain virulence affected survival by exposure to the attenuated strain Pseudomonas gacA (Tan et al., 1999). In contrast to poor survival on PA14, we found that sams-1(lof) animals lived slightly longer than wild type on Pseudomonas gacA and that pmk-1 was important for this effect (Figure 4B, Table S4). This suggests that poor survival on PA14 is linked to pathogenicity rather nutritional content and that the constitutive phosphorylation of PMK-1 could play a role in sams-1(lof) animals in moderate or weakly pathogenic contexts. sams-1(RNAi) animals live longer than wild type animals when fed E. coli; while this effect appears linked to dietary restriction, the mechanisms remain unclear (Hansen, et al. 2005). Links between innate immunity and lifespan extension (Kurz and Tan, 2004) prompted us to ask if pmk-1 was also needed for lifespan extension of sams-1(lof) on E. coli. However, pmk-1 was dispensable for lifespan extension (Figure 4C), demonstrating that the control of lifespan in sams-1 animals differ from effects on innate immunity.
Figure 4. Reduced resistance to Pseudomonas aeruginosa in sams-1(lof) animals.
Representative Kaplan-Meir plot comparing survival of (WT), sams-1(lof), pmk-1(lof) and pmk-1(lof); sams-1(lof) mutants exposed to pathogenic Pseudomonas aeruginosa, PA14 (A), the attenuated Pseudomonas strain gacA (B) or E. coli OP50 (C). NS is not significant. Additional statistics are available in Table S4. All strains in panels A, B, and C were raised on cdc-25(RNAi) to prevent egg laying. D. Fluorescent micrograph showing Pseudomonas load (PA14 GFP) after 24 hour exposure in intestines of wild type, pmk-1(lof) and sams-1(lof) mutants. Red asterisks show pharynx position. Representative experiments showing quantitation of PA14 GFP and OP50 GFP after 24 (E) or 48 (F) hours exposure. Number of animals is shown in parentheses. “Partial” refers to light GFP in a section of the intestine, “full light” to light GFP along the length of the intestine and “full bright” to strong GFP in the entire intestinal tract.
Sensitivity to pathogens can arise from mechanisms including failure of the PMK-1/p38 MAP kinase pathway or clearance of intestinal bacteria via effects on defecation (Kim, 2013). The constitutive activation of PMK-1 (Figure 2D, E) in sams-1 animals suggests that signaling to PMK-1 is intact, but insufficient for full protection. We also examined bacterial load in sams-1(lof) animals and found more Pseudomonas (PA14 GFP) in the gut lumen after 24 or 48 hours with additional intestinal distension, another hallmark of Pseudomonas sensitivity (Irazoqui et al., 2010) (Figure 4D-F). General mechanisms regulating bacterial processing were not grossly affected, as E. coli (OP50 GFP) levels were similar in sams-1(lof) animals and wild type (Figure 4E, F, not shown). Thus, mechanisms limiting survival of sams-1(lof) animals on Pseudomonas appear distinct from previously described pathogen responses.
Failure of transcriptional response to Pseudomonas in sams-1(lof) animals
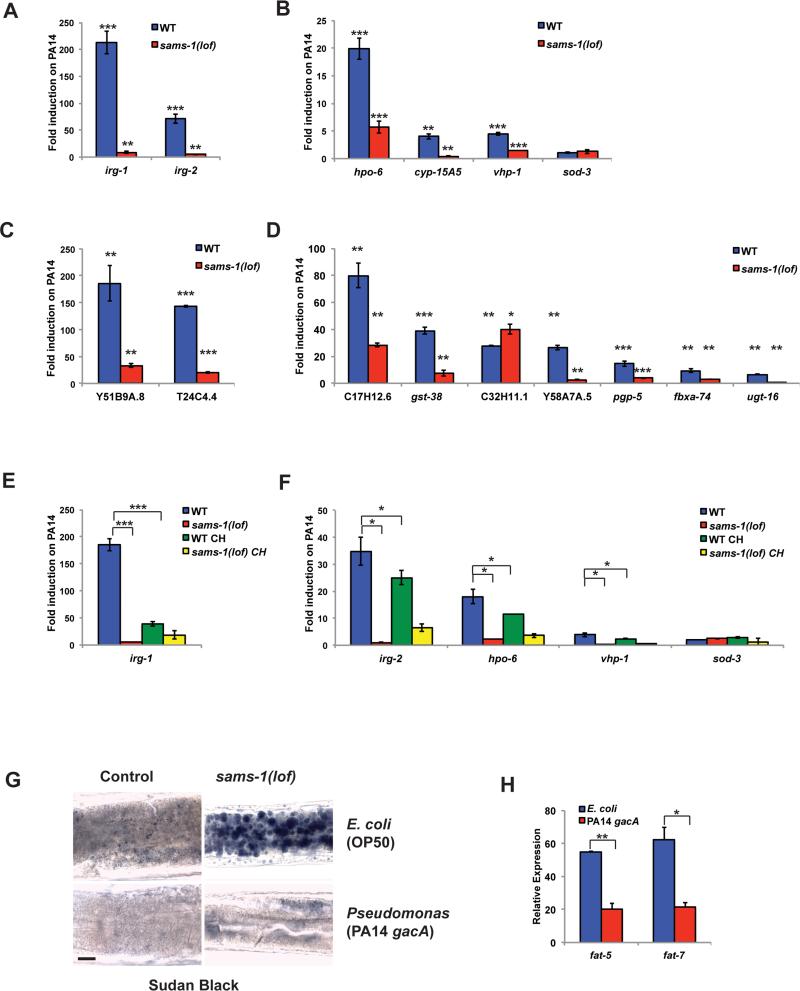
Despite activation of the protective PMK-1/p38 MAPK pathway, sams-1(lof) animals die rapidly on Pseudomonas. This paradox suggests SAM-depleted animals are deficient in other aspects of the pathogen response. To understand what these impairments might be, we examined the transcriptional dynamics for the innate immune genes that increase in expression upon contact with Pseudomonas, referred to as infection response genes. Upon exposure to virulent Pseudomonas, wild type C. elegans rapidly remodel gene expression, with some transcripts rising 200 fold within 4-8 hours (Troemel et al., 2006) (see also Figure 5A-D). Although infection response genes are upregulated in sams-1 animals fed E. coli, the increase is moderate (2-5 fold, with a few as high as 10 fold) (Figure 1B; Figure S1C). Remarkably, induction of these genes did not occur in sams-1(lof) animals exposed to virulent Pseudomonas (Figure 5A, B). This effect did not extend to genes specific to other stresses, as the oxidative stress response gene, sod-3, remained unchanged. To determine if failure to induce these genes was linked to higher basal expression on E. coli, we examined Pseudomonas-responsive genes from Troemel, et al. (2006) that were expressed normally in sams-1 animals fed E. coli. We found these infection response genes also failed to increase in sams-1(lof) animals exposed to Pseudomonas (Figure 5C, D). Thus, the transcriptional response to Pseudomonas is blunted when SAM is depleted and suggests that a breakdown in stress-inducible gene expression underlies pathogen sensitivity in sams-1(lof) animals.
Figure 5. sams-1(lof) animals lack a transcriptional response to pathogens.
qRT-PCR comparing induction of infection response genes by Pseudomonas in wild type (WT) or sams-1(lof) mutants after 6 hours of exposure to PA14 compared to the value on E. coli (OP50). Infection response genes were selected from innate immune genes with moderate induction on E. coli (OP50) after sams-1(RNAi) (A, B) or were selected from Troemel et al. (2006) (C, D). E, F. Induction of infection response genes in sams-1(lof) and sams-1(lof) choline (CH) rescued animals were compared by qRT-PCR. Lipid droplet accumulation shown by Sudan Black staining in anterior intestine (G) and lipogenic gene expression shown by qRT-PCR (H) comparing C. elegans maintained on E. coli and those raised on E. coli and transferred at L4 to Pseudomonas gacA for 48 hours. Error bars show standard deviation. Results from Student's T test shown by * <0.05, ** <0.01, *** <0.005.
Moderate increases of innate immune genes after sams-1(RNAi) on E. coli appears linked to both SAM and PC, as dietary choline returns gene expression to wild type levels (Figure 3C; Figure S1C). We next asked if induction defects were related directly PC production or were instead due to deficiencies in other methylation-dependent processes. We differentiated SAM/PC and SAM PC-independent pathways using several criteria. First, we examined dynamics of Pseudomonas-induced gene expression when C. elegans were raised on choline as SAM-independent source of PC. We found that choline supplementation did not rescue the failure of infection response gene induction on Pseudomonas in sams-1(lof) animals (Figure 5E, F). Second, Pseudomonas, unlike E. coli, synthesizes PC (Malek et al., 2012). This PC appears sufficient to rescue lipid droplet formation and lipogenic gene expression in sams-1 animals exposed to attenuated Pseudomonas strains for 48 hours (Figure 5G, H), suggesting that C. elegans fed Pseudomonas are not PC restricted. Thus, defects in infection response gene induction seem unlikely to be linked to PC production and rather is likely to depend on other SAM-dependent methyltransferases.
Alterations in H3K4 methylation in sams-1 animals
Although most gene expression changes in sams-1(RNAi) animals in basal conditions on E. coli are linked to changes in PC (Figure 3A), we reasoned that rapid transcriptional remodeling after contact with Pseudomonas might reveal requirements for SAM in histone methylation, as modifications are reconfigured along with gene expression. We focused on activating methylation marks such as H3K4me3 that are associated with high level gene expression in a variety of species, including C. elegans (Shilatifard, 2012). H3K4me3 has been shown to vary as SAM levels change in cultured cells (Shyh-Chang et al., 2013), as well as in mouse liver (Kraus et al., 2014). To determine if H3K4 methylation was an important determinant in pathogen-induced gene expression, we examined several parameters. First, we used immunostaining to compare levels of H3K4me3 wild type and sams-1(RNAi) animals and found broad decreases in the global H3K4me3 levels in intestinal cells (Figure 6A, B; Figure S2A). Specificity of H3K4me3 can be seen by staining on autosomes, but not transcriptionally inactive X chromosome in pachytene nuclei (Bean et al., 2004) (Figure S2B). This decrease was unaffected by dietary choline (Figure 6 A, B; Figure S2A), providing further evidence that decreases in H3K4 methylation are independent of PC-based pathways and could be directly affected by SAM levels.
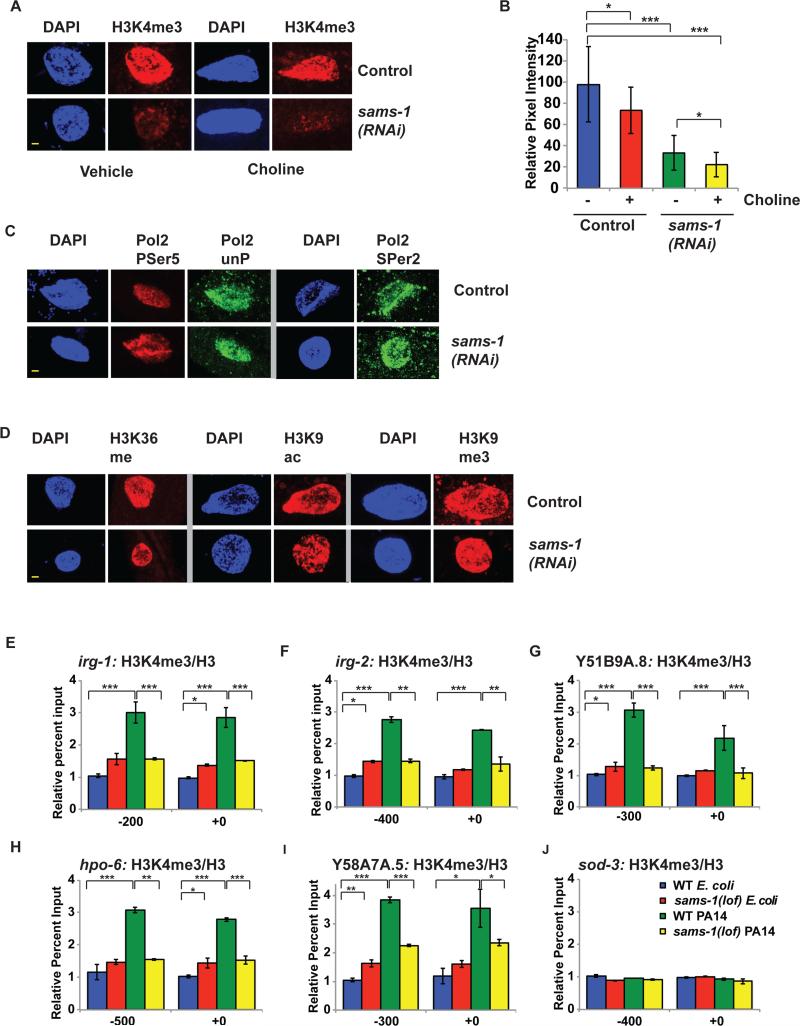
Figure 6. Infection response genes do not accumulate activating histone methylation marks in sams-1(lof) mutants exposed to Pseudomonas.
A. H3K4me3 is diminished in nuclei of intestinal cells after sams-1(RNAi) and also in choline treated sams-1(RNAi). Yellow bar shows 2 microns. B. Quantitation of immuoflourescence showing an average of pixel intensity over area for 8-12 nuclei per sample. C. Immunostaining comparing markers of active phosphorylated RNA Polymerase II (Pol II PSer 5, PSer 2) with total Pol II (unP) See Figure S2 for quantitation. D. Other histone modifications associated with active transcription (H3K36me and H3K9ac) or with heterochromatin (H3K9me3) within intestinal nuclei in control or sams-1(RNAi) animals. See Figure S2 for quantitation. E-J. Chromatin immunoprecipitation comparing levels of H3K4me3 on infection response or control genes grown on E. coli (OP50) or Pseudomonas (PA14) in wild-type (WT) or sams-1(lof) mutants. Input levels were normalized to the WT E. coli value on the upstream primer pair. Numerical representation of primer location is based on translational start site. Legend in J refers to all panels. Error bars show standard deviation. Results from Student's T test shown by * <0.05, ** <0.01, *** <0.005.
Although changes in H3K4 methylation are not predicted to affect transcription globally in basal conditions (Weiner et al., 2012), we confirmed that reduction in SAM does not have global effects on transcription by examining nuclear levels of transcribing RNA Pol II (Figure 6C; Figure S2C-F). Neither promoter-associated Pol II phosphorylated on Ser5 of the C-terminal domain (CTD) or Pol II CTD phospho-Ser2 (enriched in open reading frames) (Hsin and Manley, 2012) differed significantly between sams-1(RNAi) and controls. Furthermore, nuclear levels of H3K36 methylation, which like Pol II Ser2, maps to open reading frames of actively transcribed genes (Shilatifard, 2006), or H3K9 acetylation, which is specific to promoter regions (Shilatifard, 2006) appeared equivalent in control and sams-1(RNAi) animals (Figure 6 D; Figure S2 G-H). We also found no changes in a repressive methylation mark, H3K9me3 (Figure 6D; Figure S2I). Thus, it appears that most basal transcriptional functions in E. coli-fed sams-1(RNAi) intestinal nuclei occur normally despite decreases in SAM levels and nuclear staining with antibodies to H3K4me3.
We next sought to determine if the dynamic responses of infection response genes upon Pseudomonas exposure were accompanied by changes in H3K4me3. Using chromatin immunoprecipitations with antibodies specific to H3K4me3 (see Figure S2I: pcaf-1 5’ and 3’, (Xiao et al., 2011)), we found that wild type animals increased H3K4me3 levels at infection response genes upon exposure to virulent Pseudomonas (Figure 6E-I), consistent with robust transcriptional activation of these genes (Figure 5A-D). Importantly, increases in H3K4me3 did not occur in sams-1(lof) animals exposed to Pseudomonas, correlating strongly with the failure to induce these genes.
We also examined H3K4me3 levels on control genes with similar expression levels on E. coli and Pseudomonas. The oxidative stress response gene sod-3, the control gene pcaf-1 and non-intestinally expressed gene odr-10 showed equivalent H3K4me3 methylation in both control and PA14 samples (Figure 6J; Figure S2J, K), showing that H3K4me3 hypermethylation upon Pseudomonas exposure was linked to dynamic increases in expression of those genes. In apparent contrast to significant decreases in global H3K4me3 seen in immunofluorescence of sams-1(RNAi) intestinal nuclei, H3K4me3 was not decreased in basal conditions on infection response genes, pcaf-1, sod-3 or odr-10. Therefore we compared H3K4me3 levels at the start site for genes expressed at high levels in the intestine as well as other tissues (ama-1, rpb-2, act-1, arf-1.1 and taf-1), an intestinal specific gene (ges-1) and a gene not expressed in hermaphrodites (her-1) with control intragenic primers (Figure S2 I). We found that H3K4me3 was significantly enriched at the control gene start sites in both wild type and sams-1(lof) for the intestinally expressed genes, suggesting that the broad decrease in global H3K4me3 could reflect other genes or differences in sensitivity between immunofluorescence and chromatin immunoprecipitation. Taken together, our assays suggest that deficits in SAM have the greatest effects on promoters with dynamic changes expression and in H3K4me3.
set-16/MLL is critical for transcriptional response to pathogenic stress
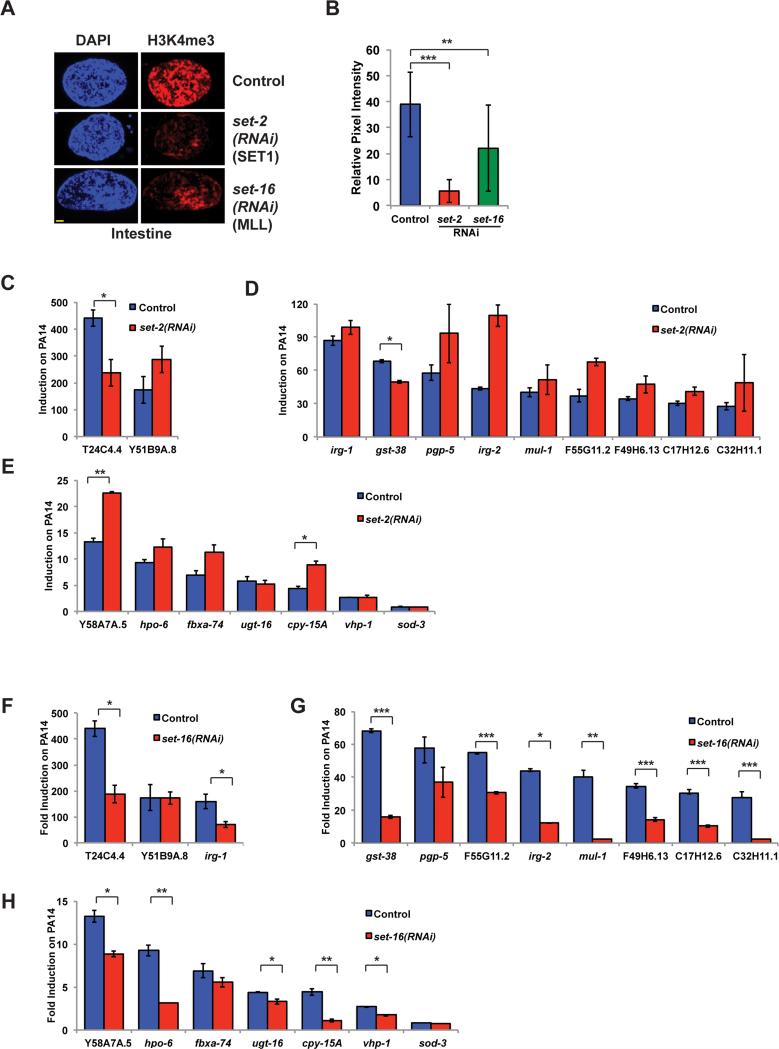
H3K4 modification may occur through the action of several methyltransferases which function in the COMPASS methyltransferase complex (Shilatifard, 2012). C. elegans contains orthologs of catalytic COMPASS complex components such as set-2/SET1 and set-16/MLL (Wenzel et al., 2011) which function in germline-dependent transgenerational regulation of lifespan and also in development (Greer et al., 2010; Greer et al., 2011; Li and Kelly, 2011; Xiao et al., 2011). We next asked which H3K4 methyltransferases were most important for global histone methylation in the intestine and for induction of infection response genes. set-2/SET1 is reported to function as the major H3K4 methyltransferase in the distal germline as well as during development (Li and Kelly, 2011; Xiao et al., 2011). Consistent with these observations, H3K4me3 is significantly reduced in set-2(RNAi) intestinal nuclei (Figure 7A, B), although the characteristic autosome-specific staining pattern can be seen in pachytene nuclei (Figure S3A). Strikingly, infection response gene induction after set-2/SET1(RNAi) occurs normally, suggesting set-2/SET1 does not play an essential role in the transcriptional response to Pseudomonas (Figure 7C-E). In contrast, set-16/MLL knockdown showed a more moderate decreases in global nuclear levels of H3K4me3 in intestinal nuclei (Figure 7A, B), but was necessary for complete induction of majority of infection response genes (Figure 7F-H). As with sams-1, we found that set-16 was necessary for H3K4me3 at infection response gene promoters (Figure S3 B-E) and that methylation was similar between control and set-16(RNAi) samples on control genes (Figure S3 F). Consistent with this observation, candidate lists from an RNAi screen for genes important for irg-1::GFP induction on Pseudomonas include T12D8.1 (identified as set-16) (Estes et al., 2010). This suggests genes requiring set-16/MLL-dependent methylation may be preferentially affected by SAM depletion during the transcriptional response to pathogens.
Figure 7. set-16/MLL is important for expression of infection response genes upon Pseudomonas exposure.
A. Immunostaining of intestinal nuclei with antibodies to H3K4me3 after RNAi of set-2 or set-16. Yellow bar shows 2 microns. B. Quantitation of immuoflourescence showing an average of pixel intensity over area for 8-12 nuclei per sample. Requirement for set-2/SET1 (B-D) or set-16/MLL (E-G) for induction of infection response genes upon a 6 hour exposure to PA14 compared to E. coli (HT115). Error bars show standard deviation. Results from Student's T test shown by * <0.05, ** <0.01, *** <0.005.
Discussion
The 1-carbon cycle links dietary levels of folate and B12 to nucleotide production, protection from oxidative stress and synthesis of the methyl donor SAM (Lu and Mato, 2008). Diverse cellular pathways are impacted by methylation, including epigenetic modification of histones or DNA (Kaelin and McKnight, 2013), phospholipid production (Vance, 2014), RNA modification (Ha and Kim, 2014) and the reach of non-histone protein methylation is only partly described (Moore et al., 2013). Many studies addressing physiological roles of methylation have focused on inactivating specific methyltransferases complexes, however, reductions in SAM due to dietary deficiencies, genetic polymorphisms or environmental factors may not impact all methyltransferases equivalently, therefore it has been difficult to discern how changes in SAM levels interface with molecular mechanisms.
We have made important new connections between SAM levels and phenotypes associated with metabolic disease by determining which methylation-dependent pathways are most sensitive to SAM depletion on different diets and stress conditions. In C. elegans living on E. coli, sams-1(RNAi) or loss of function results in accumulation of lipid droplets (Walker et al., 2011), lifespan extension (Hansen, 2005) and upregulation of innate immune signatures. The upregulation of lipogenesis or PMK-1-dependent innate immunity appears linked to decreased PC, as both phenotypes are rescued by dietary choline. However, lipogenesis and innate immune activation appear regulated by distinct mechanisms downstream of PC, since innate immune genes do not appear to be activated by the SBP-1/SREBP-1 transcription factors driving lipogenesis. PC is both an integral membrane component and a substrate for phospholipases involved in signaling (Vance 2014). Low PC could non-specifically alter basic membrane properties and induce PMK-1 similarly to mechanisms sensing epidermal wounding (Zugasti et al., 2014) or the action of pore forming toxin (Huffman et al., 2004). However, as in the case of low cholesterol activation of SREBP in mammals (Osborne and Espenshade, 2009), levels of membrane lipids may also act as part of highly specific molecular mechanisms to change protein activity. Stimulation of innate immunity has been suggested as the “second hit” in the development of fatty liver disease after lipid accumulation (Jin et al., 2013), therefore, finding that both lipogenic and specific innate immune gene expression programs can be affected by changes in SAM/PC levels is striking and suggests that these distinct phenotypes could be regulated by similar metabolic dysfunctions.
Exposure of sams-1 animals to Pseudomonas, on the other hand, elicits a distinct set of metabolic and stress related phenotypes. In surprising contrast to innate immune activation and lifespan extension when fed E. coli, sams-1(lof) animals die rapidly when exposed to virulent Pseudomonas and do not implement critical transcriptional responses to bacterial stress. Despite global decreases in H3K4me3, we do not find evidence that basal transcriptional processes are altered. Rather, the biological effects of decreased SAM emerge when new transcription to combat Pseudomonas becomes essential. H3K4me3 accumulation accompanies infection response gene induction in wild type animals, but is absent in sams-1(lof) animals, suggesting transcriptional consequences of H3K4me3 deficiency are evident under conditions that promote large scale changes in gene expression.
It is also striking that the H3K4 methyltransferase set-16/MLL, which has a less dramatic effect on total methylation than set-2/SET1, has a more critical role inducing infection response genes in response to Pseudomonas. Interestingly, MLL has been proposed to bookmark genes that have the highest dynamic range of gene expression as cells exit mitosis, while SET1 may be important for maintaining H3K4 methylation (Blobel et al., 2009). Several studies have shown global H3K4 methylation likely to be affected when SAM is limiting (Kraus et al., 2014; Shyh-Chang et al., 2013), however, we find that SAM depletion specifically mirrors set-16/MLL-dependent effects on stress induced gene expression.
Extensive studies have highlighted the importance of chromatin-regulatory complexes affecting histone methylation (Shilatifard, 2012; Weiner et al., 2012), or the need SAM to support PC synthesis in the liver (Vance, 2014). However mechanistic links between SAM levels and phenotypes associated with metabolic disease have been difficult to discern. Our studies in the C. elegans pathogen response suggest that sams-1 can preferentially affect set-16-dependent H3K4 methylation. Interestingly, Twobin et al. have found that a different SAM synthase, sams-3, is important for repressive H3K9 methylation (Towbin et al., 2012), suggesting methyltransferases might access distinct pools of SAM or require interactions with specific synthases. Several important studies have focused on effects of altering histone methylation in C. elegans through inactivating H3K4 methylation complexes and found transgenerational germline-dependent consequences on lifespan (Greer et al., 2010; Greer et al., 2011). Our studies demonstrate that SAM depletion significantly impacts physiology by limiting acute transcriptional responses to stress. Finally, as depletion of SAM affects multiple methylation-dependent pathways, understanding physiological impacts of methyl donor limitation requires integration of phospholipid and chromatin mediated effects on cellular function.
Experimental Procedures
C. elegans strains and RNAi constructs
N2 (wild type), pmk-1(ku25), tir-1(tm3036), OP50, and OP50 GFP were obtained from the Caenorhabditis Genetics Center; PA14, PA14 gacA and PA14 GFP were gifts from the Ausubel lab. Normal growth media (NGM) was used unless otherwise noted, choline was added to 30 mM where described. HA1975 sams-1(ok3033) is described in Walker, et al (2011). Control RNAi is L4440, X-5P21 was used for sams-1(RNAi); set-2 and set-16 RNAi were performed with Orfeome clone C26E6.9 (Rual et al., 2004) and III-6D12 from the Ahringer RNAi library (Simmer et al., 2003), respectively.
Gene expression analysis
L4/young adult C. elegans were lysed in 0.5% SDS, 5% β-ME, 10 mM EDTA, 10 mM Tris-HCl ph7.4, 0.5 mg/ml Proteinase K, then RNA was purified with Tri-Reagent (Sigma). RNA for microarrays was purified by RNAeasy columns (Qiagen). For control, sams-1 and sbp-1(RNAi) comparisons (Figure 1B, G), Affymetrix Gene 1.1 C. elegans exon tiling arrays were used. Affymetrix C. elegans arrays (GeneChip C. elegans Genome Arrays) were used for comparison of control, sams-1(RNAi), control choline, and sams-1(RNAi) choline samples (Figure 3A-C). cDNA synthesis and array hybridization were carried out by the UMASS Microarray core facility. For whole genome analysis, significant genes were defined as changing >2.0 fold with a p-value <0.05. GoRilla (http://cbl-gorilla.cs.technion.ac.il) (Eden et al., 2009) and Revigo (http://revigo.irb.hr) were used for GO term analysis. cDNA for quantitative RT-PCR was prepared with the Invitrogen Transcriptor kit. cDNA was standardized to act-1, except for Figure 1C-F where a “spike in” control was used. Briefly, 20 picograms of in vitro translated (Ambion Megascript) human JNK1alpha1 RNA was added to C. elegans samples before cDNA synthesis and PCR was quantitated against this absolute standard. Primers for qPCR available upon request. Representative experiments from at least three repetitions are shown.
Immunoblottting
C. elegans at the L4/Young adult transition were sonicated in RIPA (50 mM Tris, pH 8.0, 150 mM sodium chloride, 1% NP40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) containing 1 mM DTT, Complete Protease Inhibitors (Roche), Phosphostop (Roche) and ALLN (Calbiochem). Equal amounts of protein were separated, transferred to nitrocellulose and probed with antibodies to phospho p38 (Cell Signaling), sams-1 (MAT1A 4D11, Novus Biologicals), Histone 3 (Cell Signaling). Immune complexes were visualized with ImmobilonTM Luminol Reagent (Millipore).
Lipid analysis
For lipid analysis by thin-layer chromatography (TLC) and gas chromatography-mass spectrometry (GC-MS), mid-L4 larvae were harvested and snap-frozen in liquid N2, stored at −80°C before lipids extracted, separated, and quant ified as described (Hou et al., 2014).
Pseudomonas treatment
Pseudomonas strains were grown 15 hours in liquid culture, spread to edges of slow kill agar in 60mM tissue culture plates, and prepared as in (Powell and Ausubel, 2008). For sterilization, gravid hermaphrodites were placed on cdc-25(RNAi) for 6 hours before bleaching to cdc-25(RNAi) plates (Shapira and Tan, 2008). More than 100 L4 larvae were added to replicate plates of PA14 or PA14 gacA and were incubated at 25 degrees. OASIS was used to generate Kaplan-Meyer plots (Yang et al., 2011). In parallel with pathogenicity assays, more than 30 C. elegans young adults sterilized by cdc-25(RNAi) were plated onto NGM/OP50 plates and incubated at 20°C. For analysi s of PA14 GFP or OP50 GFP, animals were removed at 24 and 48 hours and imaged on a Leica TCS SPE II. For images of whole worms, composite images were pasted to grey (DIC) or black (GFP) backgrounds. For analysis Pseudomonas-induced effects on gene expression, protein levels or chromatin modification, C. elegans were grown on E. coli until L4/young adult, then re-plated on to E. coli or PA14 and incubated for 6 hours at 25°C.
Immunofluorescence
For H3K4me3, dissected intestines were incubated in 2% paraformaldehyde, freeze cracked then treated with −20°C ethanol before washing in P BS, 1% Tween-20, and 0.1% BSA (Li and Kelly, 2011). For H3K9me3, H3K9ac or H3K36, −20°C Methanol was used and washes or blocking were performed in PBS, 1% Triton-X100 and 0.1% BSA. For RNA Pol II, slides were freeze cracked, fixed in −20°methanol, then 3. 7% formaldehyde and incubated in PBS/Triton blocking solution (Walker et al., 2007). Images were taken on a Leica SPE II at identical gain settings within experimental sets. Quantitation was derived for pixel intensity over nuclear area for at least 10 nuclei. Levels corrections in Adobe Photoshop were performed uniformly across experimental sets.
Sudan Black staining
Second day adult C. elegans were fixed in 1% paraformaldehyde before three freeze thaw cycles. Samples were washed in 25%, 50% and 75% Ethanol before staining overnight in a 1:1 dilution of Sudan Black in 75% ethanol. After rehydration, imaging was performed by brightfield microscopy on a Leica SPE II.
Chromatin Immunoprecipitations
L4/young adult C. elegans were collected and frozen at −80°C. Frozen pellet s were ground and resuspended in 5 volumes of hypotonic lysis buffer (250mM Sucrose, 10mM KCl, 1.5mM MgCl2, 1mM EGTA, 1mM DTT, plus complete protease inhibitors), dounced with pestle A, then nuclei were separated by centrifugation. Crosslinking reactions, performed in 1% formaldehyde, were stopped by 100mM glycine before sonication. 10 micrograms of lysate was pre-cleared with Protein A/G Dynabeads (Invitrogen), then incubated with antibodies to H3K4me3 (Cell Signaling) or Histone 3 (Cell Signaling). Immune complexes were precipitated with Protein A/G (Invitrogen). For quantitative PCR, a standard curve for each primer pair was used to determine the amount of DNA/Ct value, percent of input was determined and used for comparison between samples.
Supplementary Material
Highlights.
Immmunity markers increase in SAM deficient C. elegans on a standard diet.
Immune activation is not protective, as Pseudomonas rapidly kills sams-1 animals.
sams-1 animals fail to mount a transcriptional response to pathogens.
sams-1 and set-16/MLL are critical for H3K4me3 in response to Pseudomonas.
Acknowledgements
We would like to acknowledge Dr. Yvonne Edwards for bioinformatics and Xun Shi for technical assistance with lipid analysis. We are grateful to Drs. Tom Fazzio, Oliver Rando (UMASS) and Francesca Palladino (Ecole Normale Superieure de Lyon, Lyon, France) for advice on chromatin IP protocols. We also thank Dr. Jonathan Ewbank for discussing our microarray data and to the lab of Dr. Frederick Ausubel for Pseudomonas strains. We thank Dr. Marian Walhout for invaluable comments on the manuscript, as well as Drs. Thomas Fazzio and Dr. Brendan Kiernan for critical reading. Some C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC), funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the NIH through R01DK084352 to A.K.W and R01DK74114 to J.W., and by CIHR MOP-93713, NSERC RGPIN 386398-13 and a Canada Research Chair to S.T..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Conceptualization: A.K.W.; Methodology: S.T, J.L.W and A.K.W.; Formal Analysis: A.K.W; Investigation: W.D., L.J.S., N.B., N.S.H. J.L.W. S.T. and A.K.W.; Resources, W.D., N.S.H.; Writing – Original Draft: A.K.W.; Writing - Review and Editing, W.D., L.J.S., J.L.W., S.T., and A.K.W.; Funding acquisition, J.L.W., S.T. and A.K.W.
The authors declare no competing financial interests.
References
- Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou J-F, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nature immunology. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, Waterston RH, Hillier LW, Ewbank JJ. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PloS one. 2011;6:e19055. doi: 10.1371/journal.pone.0019055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Halsted CH, Villanueva JA, Devlin AM, Niemela O, Parkkila S, Garrow TA, Wallock LM, Shigenaga MK, Melnyk S, James SJ. Folate deficiency disturbs hepatic methionine metabolism and promotes liver injury in the ethanol-fed micropig. Proc Natl Acad Sci U S A. 2002;99:10072–10077. doi: 10.1073/pnas.112336399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New Genes tied to Endocrine, Metabolic and Dietary Regulation of Lifespan from a Caenorhabditis elegans RNAi Screen. PLoS Genetics. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou NS, Gutschmidt A, Choi DY, Pather K, Shi X, Watts JL, Hoppe T, Taubert S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci U S A. 2014;111:E2271–2280. doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SS, Yousef L, Blaschitz C, Liu JZ, Edwards RA, Young SG, Raffatellu M, Osborne TF. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011;13:540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS pathogens. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Henao-Mejia J, Flavell RA. Innate immune receptors: key regulators of metabolic disease progression. Cell metabolism. 2013;17:873–882. doi: 10.1016/j.cmet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawli T, Tan MW. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nature immunology. 2008;9:1415–1424. doi: 10.1038/ni.1672. [DOI] [PubMed] [Google Scholar]
- Kim DH. Bacteria and the aging and longevity of Caenorhabditis elegans. Annu Rev Genet. 2013;47:233–246. doi: 10.1146/annurev-genet-111212-133352. [DOI] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT, Pirinen E, Pulinilkunnil TC, Gong F, Wang Y, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz CL, Tan MW. Regulation of aging and innate immunity in C. elegans. Aging cell. 2004;3:185–193. doi: 10.1111/j.1474-9728.2004.00108.x. [DOI] [PubMed] [Google Scholar]
- Li T, Kelly WG. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS genetics. 2011;7:e1001349. doi: 10.1371/journal.pgen.1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, Avila MA, Kanel G, Mato JM. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Mato JM. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. Journal of Gastroenterology & Hepatology 23 Suppl. 2008;1:S73–77. doi: 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneil LT, Watson E, Arda HE, Zhu LJ, Walhout AJM. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153:240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek AA, Wargo MJ, Hogan DA. Absence of membrane phosphatidylcholine does not affect virulence and stress tolerance phenotypes in the opportunistic pathogen Pseudomonas aeruginosa. PloS one. 2012;7:e30829. doi: 10.1371/journal.pone.0030829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato JM, Martínez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annual review of nutrition. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- Moore KE, Carlson SM, Camp ND, Cheung P, James RG, Chua KF, Wolf-Yadlin A, Gozani O. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell. 2013;50:444–456. doi: 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Molecular & cellular proteomics : MCP. 2011;10:M110 000976. doi: 10.1074/mcp.M110.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods in molecular biology (Clifton, NJ) 2008;415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Tan M-W. Genetic analysis of Caenorhabditis elegans innate immunity. Methods in molecular biology (Clifton, NJ) 2008;415:429–442. doi: 10.1007/978-1-59745-570-1_25. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. Plos Biology. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance DE. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochimica et biophysica acta. 2014;1838:1477–1487. doi: 10.1016/j.bbamem.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Walker AK, Boag PR, Blackwell TK. Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Developmental Biology. 2007;304:382–393. doi: 10.1016/j.ydbio.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ, et al. A conserved SREBP-1 /phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J, Schmid R, Hedley A, Blaxter M. On the extent and origins of genic novelty in the phylum Nematoda. PLoS neglected tropical diseases. 2008;2:e258. doi: 10.1371/journal.pntd.0000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, Macneil LT, Arda HE, Zhu LJ, Walhout AJM. Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell. 2013;153:253–266. doi: 10.1016/j.cell.2013.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, Macneil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJM. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell. 2014;156:759–770. doi: 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Chen HV, Liu CL, Rahat A, Klien A, Soares L, Gudipati M, Pfeffner J, Regev A, Buratowski S, et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS biology. 2012;10:e1001369. doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D, Palladino F, Jedrusik-Bode M. Epigenetics in C. elegans: facts and challenges. Genesis (New York, NY : 2000) 2011;49:647–661. doi: 10.1002/dvg.20762. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Bedet C, Robert VJP, Simonet T, Dunkelbarger S, Rakotomalala C, Soete G, Korswagen HC, Strome S, Palladino F. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8305–8310. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PloS one. 2011;6:e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugasti O, Bose N, Squiban B, Belougne J, Kurz CL, Schroeder FC, Pujol N, Ewbank JJ. Activation of a G protein-coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nature immunology. 2014;15:833–838. doi: 10.1038/ni.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.