SUMMARY
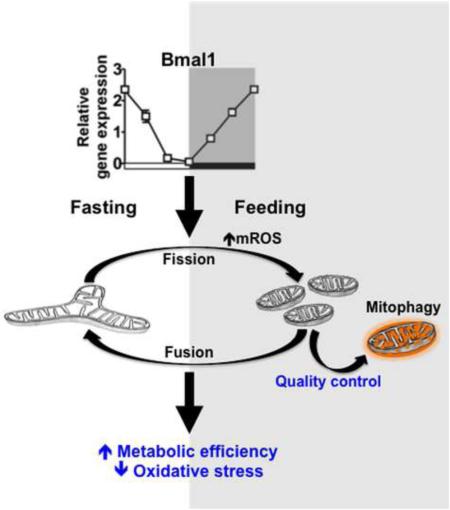
Mitochondria undergo architectural/functional changes in response to metabolic inputs. How this process is regulated in physiological feeding/fasting states remains unclear. Here we show that mitochondrial dynamics (notably fission and mitophagy) and biogenesis are transcriptional targets of the circadian regulator Bmal1 in mouse liver and exhibit a metabolic rhythm in sync with diurnal bioenergetic demands. Bmal1 loss-of-function causes swollen mitochondria incapable of adapting to different nutrient conditions accompanied by diminished respiration and elevated oxidative stress. Consequently, liver-specific Bmal1 knockout (LBmal1KO) mice accumulate oxidative damage and develop hepatic insulin resistance. Restoration of hepatic Bmal1 activities in high fat fed mice improves metabolic outcomes, whereas expression of Fis1, a fission protein that promotes quality control rescues morphological/metabolic defects of LBmal1KO mitochondria. Interestingly, Bmal1 homologue AHA-1 in C. elegans retains the ability to modulate oxidative metabolism and lifespan despite lacking circadian regulation. These results suggest clock genes are evolutionarily conserved energetics regulators.
Keywords: circadian regulation, Mitochondrial dynamics, oxidative stress, Metabolic Diseases
INTRODUCTION
Mitochondria are essential for nutrient metabolism and energy homeostasis. Oxidation of glucose and fatty acids releases NADH and FADH2 that carry high-energy electrons to generate ATP through the electron transport chain (ETC) coupled to the oxidative phosphorylation (OXPHOS) of ADP. An unavoidable side-product of oxidative metabolism is reactive oxygen species (ROS), which can cause detrimental protein and DNA modifications. Accordingly, dysregulated OXPHOS is often associated with obesity and metabolic diseases. Although there is still debate over whether mitochondrial dysfunction is a cause or consequence of the disease state, strong evidence supports its involvement in age-related deteriorations in part through oxidative damage (Balaban et al., 2005; Reznick et al., 2007; Zid et al., 2009). In addition to oxidative metabolism, mitochondrial fusion, fission and selective mitochondrial autophagy (mitophagy), collectively referred to as mitochondrial dynamics, have attracted much attention for their clear roles in mitochondrial quality control and in the development of the aforementioned diseases (Liesa and Shirihai, 2013; Youle and van der Bliek, 2012). The cell biology of mitochondrial dynamics is still under investigation. In cultured cells, fusion promotes an elongated mitochondrial network with increased metabolic efficiency (Liesa and Shirihai, 2013; Molina et al., 2009). Conversely, nutrient overload triggers fission resulting in a fragmented network that favors uncoupled respiration to reduce oxidative stress. These studies demonstrate a new dimension in the regulation of mitochondrial energy metabolism. However, the physiological relevance of these processes and mechanisms controlling them are not completely understood.
Many metabolic genes exhibit rhythmic expression patterns and are under the control of the circadian clock system (Asher and Schibler, 2011; Bass and Takahashi, 2010; Zhang and Kay, 2010). The core clock regulatory unit consists of the Per-Arnt-Sim (PAS) domain-containing transcription factors, aryl hydrocarbon receptor nuclear translocator-like (Arntl or Bmal1) and Circadian Locomotor Output Cycles Kaput (Clock or its homologue Npas2). Bmal1/Clock heterodimers bind to E box elements on promoters of clock output genes through basic helix-loop-helix (bHLH) DNA binding domains. Cryptochrome 1/2 (Cry1/2) and period circadian clock 1/2/3 (Per1/2/3) are induced by Bmal1/Clock to form a negative feedback loop. Additional clock regulators include nuclear receptors Rev-erbα/β and RORα/β/γ as well as several energy-sensing pathways (Bugge et al., 2012; Cho et al., 2012; Gallego and Virshup, 2007). Although the main function of Bmal1/Clock is thought to maintain rhythmic expression of clock output genes, genome-wide binding site mapping of the core clock regulators has provided a clearer picture of the primary action for these circadian regulators (Koike et al., 2012). In mouse liver, Bmal1 protein and mRNA levels are elevated at the dark cycle. The maximal DNA binding of Bmal1/Clock (or Npas2) occurs a few hours later around midday at zeitgeber time 8 (ZT8), while the expression of clock output genes peaks in the light-to-dark cycle transition at ~ZT12. This work sheds new light on a potential role for Bmal1/Clock in metabolic regulation during the dark cycle when nutrient influx and metabolic stress are elevated.
In this study, we provide evidence that the hepatic clock regulates mitochondrial quality control through temporal regulation of mitochondrial dynamics and biogenesis to maintain a healthy mitochondrial population that meets diurnal bioenergetic demands. Disruption of this process induces oxidative damage and metabolic dysregulation. The importance of the identified regulatory mechanism is further supported by a conserved role of Bmal1 homolog aha-1 in C. elegans in regulation of mitochondrial oxidative metabolism.
RESULTS
Mitochondrial dynamics is linked to hepatic nutrient status
Through fusion, fission and mitophagy, mitochondria achieve morphological and functional adaptations to accommodate different metabolic states and energy demands (Liesa and Shirihai, 2013). We found that proteins involved in fission, including fission 1 (mitochondrial outer membrane) homolog (Fis1) and dynamin-1-like protein (Dnm1l or Drp1), and mitophagy, such as BCL2/adenovirus E1B 19kDa interacting protein 3 (Bnip3), were elevated at fed state in mouse liver (Figure S1A). However, the difference between fed and fasted states was dampened when mice were fed a high fat diet (HFD). In addition, there was a substantial increase in the fusion protein mitofusin 1 (Mfn1), suggesting that dysregulated mitochondrial fusion/fission may mediate metabolic dysfunction in diet-induced obesity.
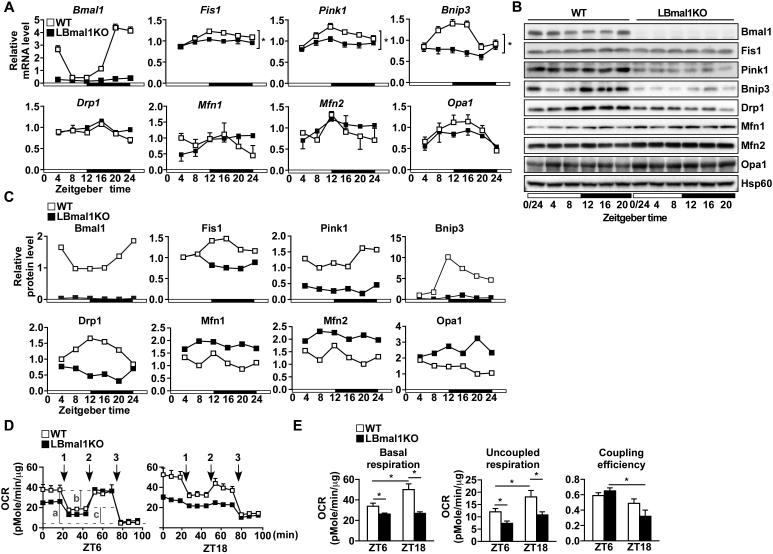
Many metabolic pathways in mouse liver exhibit a pre-programmed, diurnal rhythm that coincides with the feeding/fasting cycle. To assess whether mitochondrial dynamics is subjected to circadian regulation, we generated mice with liver-specific deletion of the Bmal1 gene, the master clock regulator, using albumin-cre (LBmal1KO: Bmal1f/f × albumin-cre versus control: Bmal1f/f). mRNA levels of many mitochondrial dynamics genes exhibited a diurnal pattern in wild type (WT) livers peaking at the beginning of the dark or feeding cycle (Figure 1A). With the exception of Drp1, the rhythmic expression of fission/mitophagy genes [Fis1, PTEN induced putative kinase 1 (Pink1) and Bnip3] was Bmal1-dependent, while mRNA levels of fusion genes [Mfn1, Mfn2 and optic atrophy 1 (Opa1)] were not significantly different between WT and LBmal1KO mice. Fission/mitophagy proteins showed a similar diurnal pattern in WT liver (Figures 1B-1C). Their levels failed to cycle and were substantially reduced in LBmal1KO liver. In contrast, fusion proteins were increased in LBmal1KO mice, indicating that hepatic mitochondrial dynamics is controlled by both Bmal1-dependent (notably fission and mitophagy) and independent mechanisms and is subjective to post-transcriptional regulation. The impact of this diurnal regulation on hepatic bioenergetics was assessed in freshly isolated hepatocytes at light and dark cycles, corresponding to the physiological fasted and fed states, respectively. In WT hepatocytes, the basal oxygen consumption rate (OCR) was higher at ZT18 than at ZT6, which was accompanied by an increase in uncoupled respiration but not coupling efficiency (Figures 1D-1E). LBmal1KO hepatocytes not only lost diurnal variation of OXPHOS activity but also showed a significant reduction in basal and uncoupled respiration as well as coupling efficiency, notably at ZT18 compared to WT cells. In addition, mitochondrial respiration driven by pyruvate/malate (Complex I), succinate/rotenone (Complex II), or ascorbate/TMPD (Complex IV) was also diminished in LBmal1KO mitochondria (Figure S1B-S1C). These results suggest that Bmal1-controlled rhythmic mitochondrial metabolism is important in maintaining energy homeostasis and implicate a role for Bmal1 in HFD-induced dysregulation of mitochondrial dynamics (Figure S1A), as HFD has been shown to reduce Bmal1 activity (Eckel-Mahan et al., 2013; Kohsaka et al., 2007).
Figure 1. Rhythmic expression of mitochondrial dynamics genes in the liver is regulated by Bmal1.
(A) Diurnal mRNA expression of genes involved in mitochondrial dynamics in WT and LBmal1KO livers determined by real-time PCR. (B) Western blot analyses of mitochondrial dynamics proteins throughout the day. Liver samples were collected every 4 hours for 24 hours. Pooled samples (n=3-4/time point/genotype, see also Table S4) were used for each time point. The white and black bar represents light cycle and dark cycle, respectively. Zeitgeber time ZT0: lights on; ZT12: lights off. (C) Western blot signals in (B) were quantified and normalized to the loading control (Hsp60). (D) Bioenergetics assays in hepatocytes from control and LBmal1KO mice isolated at ZT6 and ZT18. Numbers 1-3 refer to the time course of adding oligomycin, FCCP, and antimycin A/rotenone. a, b and c in ZT6 WT hepatocytes indicate the basal oxygen consumption rate (OCR), ATP turnover and uncoupled respiration (proton leak), respectively. (E) The basal OCR, uncoupled respiration, and coupling efficiency (ATP turnover/basal OCR) calculated based on data in (D). Data presented as mean ± SEM. *p<0.05.
Hepatic Bmal1 regulates diurnal mitochondrial remodeling
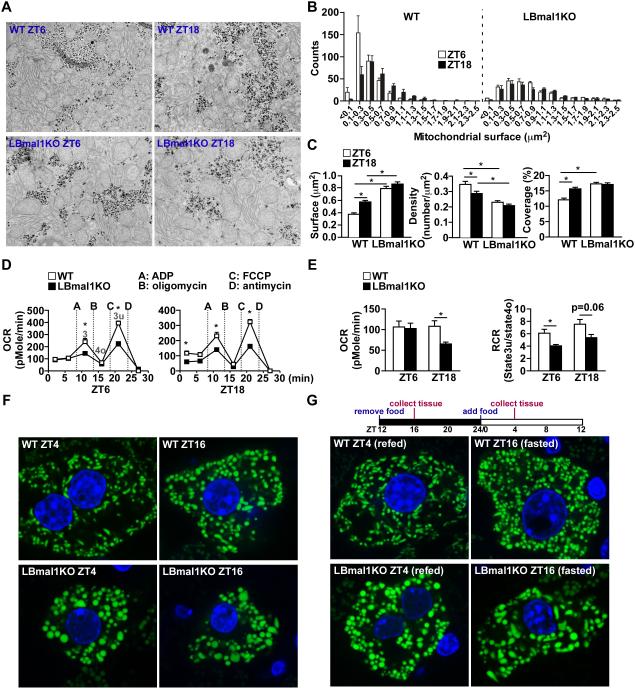
Electron microscopy (EM) was employed to study the Bmal1-mediated regulatory mechanism of mitochondrial dynamics at cellular and organelle levels. In WT livers, the average surface area and the overall coverage (percentage) of mitochondria increased at the fed state (ZT18) in comparison to the fasted state (ZT6, Figures 2A-C). LBmal1KO mitochondria exhibited an enlarged or swollen phenotype that remained unchanged at both time points. In addition, mitochondria isolated from LBmal1KO livers showed a reduction in OCR in all stages (basal, state 3, and state 3u), compared to those from WT livers (Figures 2D-2E). This defect was more pronounced at ZT18. Respiratory control ratio (RCR, state 3u/state 4o) further indicated reduced mitochondrial function in LBmal1KO livers (Figure 2E).
Figure 2. Diurnal mitochondrial remodeling is abolished in LBmal1KO liver.
(A) Representative electron microscopy images of liver sections from WT and LBmal1KO mice (n=3/time point/genotype) at ZT6 and ZT18. (B) Mitochondrial size distribution calculated from EM images in 1000 μm2 surface area/liver. (C) The mitochondrial surface (average), density, and coverage calculated from EM images at ZT6 and ZT18. (D) Coupling assay in mitochondria isolated from WT and LBmal1KO livers at ZT6 and ZT18. A-D refer to the time course of adding ADP, oligomycin, FCCP, and antimycin A. Stages 3, 4o, and 3u respiration are indicated. (E) The basal oxygen consumption rate (OCR, left panel) and respiratory control ratio (RCR=3u/4o, right panel) determined based on data in (D). (F) Images of liver tissue sections showing GFP-tagged mitochondria in WT and LBmal1KO mice fed ad libitum and sacrificed at ZT4 or ZT16. Green: Cox8a-GFP. Blue: nucleus stained with DAPI. (G) Images of mitochondrial network from fasted/refed mice. Food was removed at ZT12. Mice were sacrificed at ZT16 (fasted) or refed at ZT0 and sacrificed at ZT4 (see top panel for the experimental design). Data presented as mean ± SEM. *p<0.05.
To distinguish between the contribution of circadian regulation and nutrient availability to the observed changes in mitochondrial shapes and activities, liver samples from mice under constant fasting were analyzed. Western blotting demonstrated that the rhythmic mitochondrial fission/mitophagy protein expression were largely unaffected by fasting (Figure S2A). Morphological alteration of mitochondria between day and night in WT hepatocytes and swollen mitochondria in the LBmal1KO liver were evident under EM (Figures S2B). While the temporal and genotypic differences in respiration in isolated liver cells were diminished by constant fasting, the basal OCR of LBmal1KO hepatocytes at ZT18 remained lower than that of WT cells (Figure S2C). These findings indicate that whereas Bmal1 controls pre-programmed mitochondrial remodeling, mitochondrial respiration is further modulated by the nutrient/energy substrate status. To further validate the rhythmic architectural alteration of mitochondria is independent of nutrient intake, in vivo hepatic mitochondrial network was visualized with Cox8a-GFP protein (adCox8a-GFP) through adenovirus-mediated gene delivery. In WT livers, the mitochondrial network showed a day/night shift in morphology under ad libitum feeding (Figure 2F). This rhythmic pattern was not disturbed by altered nutrient availability, in which food was removed at ZT12 and replenished at the following ZT0 (Figure 2G). LBmal1KO mitochondria remained enlarged regardless of the time and feeding condition. Collectively, these results suggest that the circadian clock controls daily mitochondrial remodeling in anticipation of the upcoming feeding/fasting cycles. Dysregulation of this regulatory mechanism in LBmal1KO mice leads to dysfunctional mitochondria, likely due to inability to adapt to the fluctuating metabolic flux.
Bmal1 confers mitochondrial metabolic flexibility
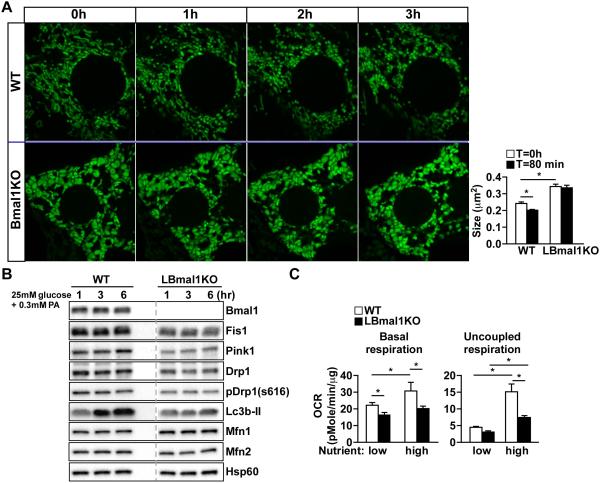
We next sought to establish an ex vivo model to study the impact of rhythmic remodeling on mitochondrial oxidative metabolism in response to changes in nutrient influx and to understand the underlying cause of the swollen phenotype of LBmal1KO mitochondria. We tagged mitochondria with adCox8a-GFP ex vivo in primary hepatocytes from WT and LBmalKO mice isolated at ZT12, when the expression of Bmal1 targets is high. LBmal1KO hepatocytes retained the swollen mitochondrial morphology (similar results were obtained with an anti-Cox1 antibody, data not shown) and had reduced levels of fission/mitophagy proteins (Fis1, Pink1 and Drp1/phospho-Drp1 s616) as well as an increase in Mfn1 but not Mfn2 (Figures 3A-3B), compared to WT hepatocytes. Time-lapse imaging showed that when cultured in a high nutrient condition to mimic the feeding state (25 mM glucose/0.3 mM palmitic acid), WT hepatocytes exhibited a progressive increase in fragmented mitochondrial morphology (Figure 3A), followed by a substantial increase in Lc3b-II protein expression indicative of mitochondrial autophagy (Figure 3B). This observation was absent in LBmal1KO hepatocytes, suggesting a defective quality control. Cellular bioenergetics studies demonstrated that in WT hepatocytes, increased metabolic flux led to a higher basal OCR primarily due to increased uncoupled respiration (Figure 3C), a phenomenon not seen in LBmal1KO cells. These results appear to recapitulate the difference in respiration between WT and Bmal1KO hepatocytes isolated at ZT6 and ZT18 (Figure 1E).
Figure 3. Bmal1-controlled mitochondrial dynamics regulates metabolic flexibility.
(A) Representative time-lapse confocal images of the mitochondrial network in WT or LBmal1KO primary hepatocytes. Cells were cultured in low nutrient (5.5 mM glucose) for 1 hr and switched to high nutrient (25 mM glucose/0.3 mM palmitic acid). Cox8a-GFP adenovirus was used to tag mitochondria. Right panel: The average mitochondrial size (n=10). (B) Western blotting of mitochondrial dynamics proteins in WT and LBmal1KO primary hepatocytes cultured in high nutrient for the indicated times. Samples were run on the same gel with lanes omitted for clarity (indicated with the dotted line). pDrp1(s616): phosphor-Drp1 at ser616 indicative of increased Drp1 activity. (C) The basal oxygen consumption rate (OCR) and uncoupled respiration of WT and LBmal1KO primary hepatocytes cultured in low or high nutrient medium for 4 hours. Data presented as mean ± SEM. *p<0.05.
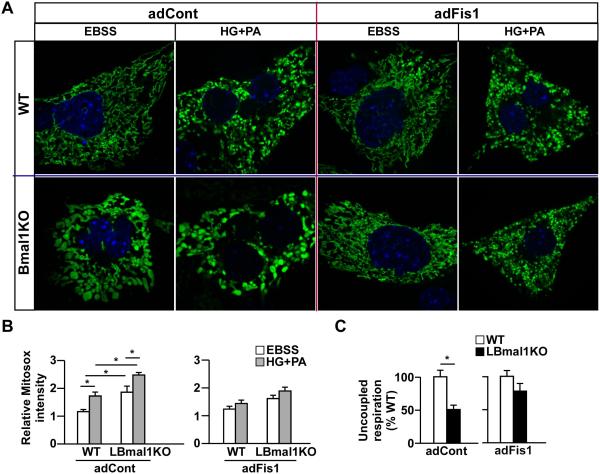
As mentioned earlier, mitochondrial fission promotes uncoupled respiration as a means to reduce oxidative stress, which when elevated, triggers mitochondrial quality control mechanisms to remove damaged mitochondrial components (Liesa and Shirihai, 2013; Molina et al., 2009). Defective quality control can lead to mitochondrial swelling (Bueno et al., 2015; Poole et al., 2008). To examine whether restoring mitochondrial dynamics could rescue the defects associated with LBmal1KO, we expressed Fis1 protein, which was down-regulated in the LBmal1KO liver, using adeno-Fis1 (adFis1) in primary hepatocytes. Hepatocytes were cultured for 3 hours in Earle’s balanced salt solution (EBSS), a minimal nutrient condition used to force mitochondrial fusion and set the basal oxidative stress level (Liesa and Shirihai, 2013; Molina et al., 2009). As expected, WT hepatocytes formed an elongated network under EBSS (Figure 4A), which became fragmented when switching to medium containing high glucose/palmitic acid (25 mM glucose/0.3 mM palmitic acid, HG+PA). MitoSOX Red staining indicated that HG+PA increased the mitochondrial ROS level, compared to that of EBSS (Figures 4B and S3A). LBmal1KO mitochondria were incapable of remodeling even under the extreme low-nutrient medium and showed higher levels of basal and HG+PA-induced mitochondrial ROS. Interestingly, adFis1 normalized LBmal1KO mitochondrial morphology, eliminating the swollen phenotype and enabling adaptive morphological changes in EBSS or HG+PA (Figure 4A). adFis1 also lowered mitochondrial ROS and restored the uncoupled respiration in Bmal1KO hepatocytes (Figures 4B-4C). Collectively, these findings suggest that dysregulated mitochondrial dynamics is causative to metabolic inflexibility and elevated oxidative stress in LBmal1KO hepatocytes.
Figure 4. Fis1 overexpression normalizes mitochondrial morphology and superoxide production in LBmal1KO primary hepatocytes.
(A) Representative confocal images of the mitochondrial network in WT or LBmal1KO primary hepatocyte that were first infected with adCox8a-GFP, followed by adFis1 or control virus (empty vector, adCont). Cells were cultured either in EBSS or 25 mM glucose/0.3 mM palmitic acid (HG+PA). Blue: DAPI staining of nucleus. (B) Superoxide production assessed by MitoSOX Red normalized to MitoTracker green fluorescence in WT and LBmal1KO primary hepatocytes (representative images shown in Figure S3). Hepatocytes were transduced with adFis1 or adCont and cultured in EBSS or HG+PA. Results were quantified from 12 cells per group. (C) Uncoupled respiration measured by the Seahorse bioenergetics analyzer in WT and LBmal1KO hepatocytes transduced with adCont or adFis1 under HG+PA. Data presented as mean ± SEM. *p<0.05.
Mitochondrial function is a primary transcriptional target of the hepatic clock
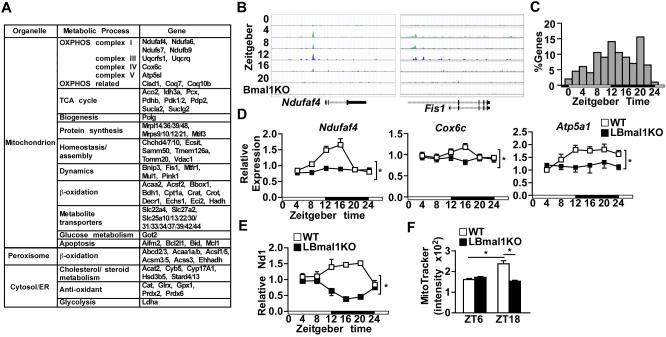
The circadian clock regulates temporal expression of many metabolic pathways through direct and indirect mechanisms. We analyzed the published chromatin immunoprecipitation sequencing (ChIP-seq) data of Bmal1, Clock and Cry1 in mouse liver (Koike et al., 2012) to determine whether the circadian clock controls transcription of mitochondrial dynamics genes. Similar results were obtained from another Bmal1 ChIP-seq study (Figure S4B and Table S1) (Rey et al., 2011). Gene ontology analysis revealed that mitochondrion was the top cellular component targeted by the hepatic clock (Figures S4A). Fis1, Bnip3, Pink1, and mitochondrial fission regulator 1 (Mtfr1, involved in fission) were among the 211 genes identified in the mitochondrial pathway (Figure 5A and Table S1). Others included genes involved in oxidative metabolism [ETC, complex assembly, and TCA cycle], mitochondrial biogenesis and protein synthesis, and fatty acid β-oxidation. Drp-1, Mfn1/2, and Opa1, on the other hand, did not appear to be direct targets of the hepatic clock, consistent with the circadian gene expression results (Figure 1A).
Figure 5. The oxidative metabolism pathway is a primary transcriptional target of the hepatic clock.
(A) Partial list of the 211 genes involved in mitochondrial function that are bound by Bmal1, Clock, and Cry1 (see also Table S1). (B) ChIP-seq signal of representative Bmal1 target genes in WT and Bmal1 knockout (Bmal1KO) liver. Original data were derived from published sources (Koike et al., 2012). (C) The frequency distribution of peak mRNA expression throughout the day of the 211 genes involved in mitochondrial function. (D) Diurnal expression of mitochondrial oxidative metabolism genes in control and LBmal1KO mice. Liver samples were collected every 4 hr for 24 hr (n=3-4/time point/genotype). The white and black bar represents light cycle and dark cycle, respectively. (E-F) Mitochondrial biogenesis in liver samples isolated at different time points determined by the relative Nd1 DNA content (E) or by flow cytometry analysis of MitoTracker Green staining. Data presented as mean ± SEM. *p<0.05.
To validate the transcriptional regulation of these additional clock targets in oxidative metabolism, we used transient deletion or over-expression strategies to manipulate the activity of Bmal1. Acute hepatic Bmal1 deletion achieved by tail vein injection of Cre adenovirus in Bmal1 f/f mice (adCre-LBmal1KO) resulted in down-regulation of genes encoding OXPHOS, TCA cycle, and β-oxidation (Figure S4C), whereas Bmal1 over-expression in the liver using Bmal1 adenovirus (adBmal1) up-regulated most of these genes (Figure S4D). Acute Bmal1 gene deletion and over-expression were associated with decreased and increased hepatic mitochondrial biogenesis, respectively, as determined by the relative DNA content of mitochondrial NADH dehydrogenase 1 (mt-Nd1) (Figures S4C-S4D). In Hepa 1-6 cells (a mouse hepatoma cell line), Bmal1 knockdown (shBmal1) lowered respiration (Figures S4E). Knockdown of negative regulators of Bmal1, including Cry1, Cry2, Per1, and Per2 had an opposite effect (Figure S4F). By contrast, over-expression of Bmal1 (adBmal1) or Cry1 (adCry1) respectively enhanced or decreased OCR (Figure S4G). These data demonstrate that the hepatic clock controls oxidative metabolism, in addition to mitochondrial dynamics.
The published ChIP-seq data also demonstrated the specificity of Bmal1 binding on regulatory regions of predicted target genes, such as NADH dehydrogenase (ubiquinone) 1 alpha subcomplex assembly factor 4 (Ndufaf4) and Fis1 [Figure 5B, data derived from (Koike et al., 2012)]. Based on the Circa database (Pizarro et al., 2013), the majority of the 211 genes in the liver peaked at ~ZT12 (Figure 5C). Similar to Fis1, Bnip3, and Pink1, the expression of OXPHOS genes [(e.g., Ndufaf4, cytochrome c oxidase subunit VIc (Cox6c), and Atp5a1], also showed diurnal expression with the highest expression at the dark cycle (Figure 5D). The cell-autonomous cyclical expression could be demonstrated in dexamethasone-synchronized Hepa 1-6 cells, which was lost in shBmal1 cells (Figure S4H). Interestingly, mitochondrial biogenesis (determined by Nd1 DNA content and flow cytometry of MitoTracker Green staining) also showed a Bmal1-dependent diurnal pattern similar to that of dynamics and OXPHOS genes (Figures 5E-5F). These results indicate that the circadian clock controls energy metabolism through coupled regulation of mitochondrial biogenesis and dynamics in preparation for increased nutrient intake at the fed state. Since reduced complex I activity promotes mitochondrial ROS production (Verkaart et al., 2007), the increased oxidative stress in LBmal1KO liver cells (Figure 4B) is likely a combined effect of dysregulated mitochondrial dynamics and oxidative metabolism.
Liver Bmal1 modulates oxidative stress and maintains metabolic homeostasis
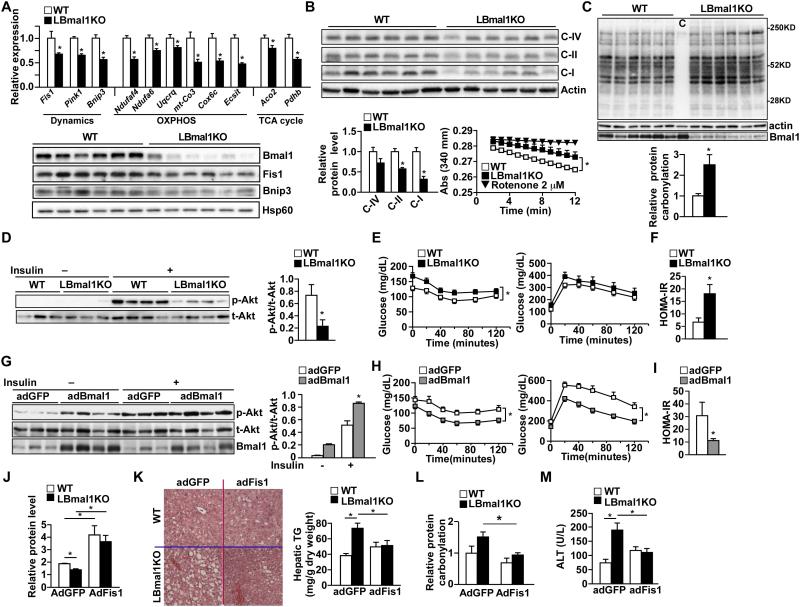
Results described thus far implicate a critical role for Bmal1-controlled mitochondrial function in energy metabolism. To determine the effect of Bmal1 loss-of-function on metabolic homeostasis, we challenged WT and LBmal1KO mice with HFD. LBmal1KO mice gained more weight after 16 weeks of high fat feeding, compared to control animals (Figure S5A). There was no difference in food intake or physical activity (Table S2). LBmal1KO mice also had elevated levels of circulating triglycerides (TG), cholesterol, and free fatty acids (Figures S5B). As expected, Bmal1 target genes in mitochondrial dynamics and oxidative metabolism were down-regulated in the liver of LBmal1KO mice and the differences in expression was greater when tissues were collected at ZT12 (Figure 6A), compared to ZT6 (Figure S5C). LBmal1KO livers showed lowered OXPHOS proteins, notably complex I, which was accompanied by a reduction in the complex I activity (NADH-coenzyme Q reductase, Figure 6B) and exacerbated fatty liver conditions (Figure S5D-S5E).
Figure 6. Hepatic Bmal1 regulates oxidative stress, lipid homeostasis, and insulin response.
(A) Expression of Bmal1 targets involved in mitochondrial function in liver from WT and LBmal1KO mice on high fat diet (HFD) for 5 months. Samples were collected at ZT12 (n=6/genotype). Upper panel: mRNA expression determined by real-time PCR. Lower panel: Western blot analyses. (B) OXPHOS protein levels in livers from HFD fed WT and LBmal1KO mice (n=6/genotype). Upper panel: Western blot analysis. C-I, II, and IV: ETC complex I (Ndufa9), II (Sdha), and IV (Cox4). Lower left panel: quantification of protein levels normalized to actin. Lower right panel: Complex I activity using mitochondria isolated from livers of WT and LBmal1KO mice. Rotenone is used as a negative control. (C) Oxidative damage assessed by levels of protein carbonylation in liver lysate from WT and LBmal1KO mice (n=6) fed a HFD for 5 months using Western blotting. c: negative control liver lysate from a WT mouse omitting DNPH substrates. Quantification normalized to actin is shown at the lower panel. (D) Western blot analyses of hepatic insulin signaling. Akt phosphorylation was examined in livers collected prior to and 5 min after insulin injection (n=3-4). The level of insulin-stimulated phospho-Akt (p-Akt) was quantified and normalized to that of the total Akt (t-Akt, right panel). (E) ITT (left) and GTT (right) in high fat fed WT and LBmal1KO mice (n=7-8). (F) The HOMA-IR assessment. (G) Western blot analyses of liver insulin signaling in high fat fed control (adGFP) and hepatic Bmal1 over-expression (adBmal1) mice (n=3-4) (H-I) ITT, GTT, and HOMA-IR in adGFP and adBmal1 mice (n=5-6). (J)-(M) Effect of hepatic Fis1 overexpression in WT and LBmal1KO mice. Mice (HFD fed for 5 months; n=6/genotype) were given adGFP (control) or adFis1 for 1 week. (J) Quantification of hepatic Fis1 protein levels. (K) Left panel: liver histology with H & E staining; Right panel: hepatic triglyceride (TG) content. (L) Relative protein carbonylation in liver samples (n=4, see also Figure S5G). (M) Serum alanine aminotransferase (ALT) activity. Data presented as mean ± SEM. *p<0.05.
Protein carbonylation analyses of liver lysate demonstrated an increase in oxidative damage in LBmal1KO livers (Figure 6C). Increased oxidative stress is linked to insulin resistance (Houstis et al., 2006). We found a significant reduction in insulin-stimulated Akt phosphorylation in LBmal1KO livers, compared to WT livers (Figure 6D). LBmal1KO mice also had higher fasting glucose (WT: 128.8 ± 5.1 mg/dL; LBmal1KO: 167.6 ± 11.6 mg/dL, p<0.05) and insulin levels (WT: 0.69 ± 0.22 ng/mL; LBmal1KO: 1.58 ± 0.29 ng/mL, p<0.05). Despite hepatic insulin resistance, LBmal1KO mice performed moderately worse than control animals in insulin tolerance test (ITT) and there was no difference in glucose tolerance test (GTT, Figure 6E). Of note, the glucose phenotype could be confounded by reduced gluconeogenic gene expression reported in chow fed LBmal1KO mice (Lamia et al., 2008; Rudic et al., 2004), which we have also observed (data not shown). In fact, defective mitochondrial function may also contribute to impaired gluconeogenesis, as the conversion of pyruvate to oxaloacetate is mediated by mitochondrial phosphoenolpyruvate carboxylase and glutamate oxaloacetate transaminase (Got) (Mendez-Lucas et al., 2013). Got2 was identified as a Bmal1 target (Figure 5A) and its expression was dysregulated in the LBmal1KO liver (data not shown). Nevertheless, the homeostasis model assessment-estimated insulin resistance (HOMA-IR) result suggested that LBmal1KO mice were more insulin resistant compared to WT mice (Figure 6F). Gain-of-function studies using adBmal1 were performed in mice that had already received HFD for 4 months. Within 10 days, adBmal1 lowered fasting glucose concentrations (adGFP: 194.0 ± 20.7 mg/dL; adBmal1: 143.3 ± 6.8 mg/dL, p<0.05), reduced hepatic fat content (Figure S5F), and increased both basal and insulin-stimulated Akt phosphorylation in the liver (Figure 6G). The improvement in GTT and ITT (Figure 6H), together with the lower fasting insulin concentration (adGFP: 3.93 ± 1.06 ng/mL; adBmal1: 1.57 ± 0.27 ng/mL, p<0.05) and HOMA-IR (Figure 6I) demonstrated increased insulin sensitivity in adBmal1 mice. Lastly, liver Fis1 over-expression (an ~3-fold increase at the protein level) using adFis1 normalized hepatic TG content, reduced oxidative damage, and improved overall liver function (based on serum alanine transaminase, ALT) in HFD fed LBmal1KO mice (Figures 6J-6M). These loss/gain-of-function and rescue studies suggest that Bmal1-controlled mitochondrial function modulates oxidative stress and maintains hepatic and systemic metabolic homeostasis.
Regulation of oxidative metabolism is an evolutionarily conserved Bmal1 function
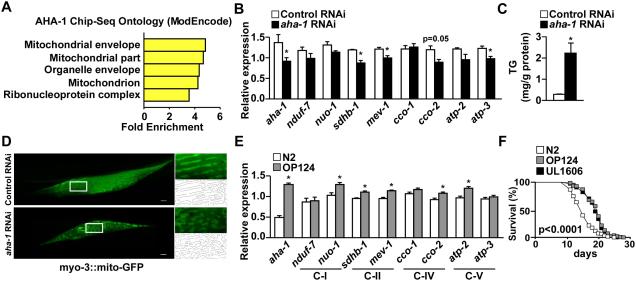
We next used C. elegans as a model system to assess whether regulation of mitochondrial respiration is a primary metabolic function of Bmal1. These worms lack a well-defined clock regulatory network (Hasegawa et al., 2005; van der Linden et al., 2010): they do not have Cry-like molecules but contain a Bmal1 homologue, aha-1 (Figures S6A-S6B), which has no circadian function (Banerjee et al., 2005). In fact, while many C. elegans genes can be entrained by light or temperature (van der Linden et al., 2010), there was only a small overlap between light entrained, temperature entrained and AHA-1 bound genes (Figure S6C). Similar to its mammalian counterpart, gene ontology analysis of AHA-1 Chip-seq at the L4 stage from the ModEncode project (Gerstein et al., 2010) demonstrated an enrichment of genes involved in mitochondrial functions (Figure 7A and Table S3). aha-1 RNAi reduced the expression of OXPHOS genes and increased worm TG content (Figures 7B-7C). Furthermore, aha-1 knockdown caused disorganization of mitochondrial networks in the muscle of mitochondrial reporter worms (Figure 7D). By contrast, mitochondrial oxidative metabolism genes were up-regulated by AHA-1 over-expression (strain OP124, Figure 7E). Consistent with a short lifespan of whole body Bmal1 knockout mice (Kondratov et al., 2006), aha-1 gene deletion caused early death at L1 stage (Huang et al., 2004). Therefore, we determined survival in two strains expressing AHA-1 either through stable integration (OP124) or with an extra-chromosome array (UL1606). AHA-1 over-expression prolonged lifespan to the same extent in both OP124 and UL1606 worms (Figure 7F). These data indicate that regulation of mitochondrial respiration is a conserved metabolic function of Bmal1-like molecules that is critical for maintaining healthspan and is separable from their role as a circadian regulator.
Figure 7. C. elegans Bmal1 homologue AHA-1 modulates oxidative metabolism and lifespan.
(A) Gene ontology analysis of AHA-1 ChIP-seq data. (B) Expression analyses of potential AHA-1 target genes in OXPHOS based on the ChIP-seq analysis (see also Table S3) determined by real-time PCR in control (control RNAi) and aha-1 knockdown (aha-1 RNAi) worms. (C) Assessment of triglyceride (TG) content. (D) Representative fluorescence microscopy images showing muscle mitochondrial organization in mitochondrial reporter worms fed control or aha-1 RNAi. (E) OXPHOS gene expression in control (N2 strain) and aha-1 gain-of-function worms (strain OP124). C-I, II, IV and V: Mitochondrial Complex-I, II, IV and V. (F) Lifespan of control and two separate strains of aha-1 gain-of-function worms (OP124 and UL1606). Data presented as mean ± SEM. *p<0.05.
DISCUSSION
In this study, we identify mitochondrial dynamics and oxidative metabolism as primary transcriptional targets of the circadian clock. Diurnal Bmal1 activity enables synchronized regulation of mitochondrial biogenesis and architectural remodeling in the liver to accommodate the nutrient influx at the fasting-to-feeding transition and maintain post-prandial mitochondrial quality control. Disruption of this regulatory mechanism leads to enlarged, dysfunctional mitochondria that are less responsive to metabolic input and have elevated ROS level. As a result, hepatic Bmal1 gene deletion increases oxidative stress and causes fatty liver and insulin resistance. The function in regulation of mitochondrial oxidative metabolism is well conserved in a C. elegans Bmal1-like protein, AHA-1, which also modulates longevity. Of note, a metabolic cycle (occurring in 4-5 hours) has been described in yeast under a constant low-glucose culture condition (Tu et al., 2005), in which a reductive, nonrespiratory phase is followed by an oxidative, respiratory phase with increased oxygen consumption. This rhythmic respiration is facilitated by temporal expression of mitochondrial OXPHOS genes, suggesting the compartmentalization of mitochondrial oxidative metabolism in time is evolutionarily advantageous to optimize metabolic output (Tu and McKnight, 2007).
Recent studies have demonstrated that architectural/organizational changes of mitochondria serve specific metabolic purposes such that fusion increases metabolic efficiency, whereas fission promotes uncoupled respiration (Liesa and Shirihai, 2013; Youle and van der Bliek, 2012). Our study suggests that mitochondrial dynamics is coupled to the daily fasting/feeding cycle in part through Bmal1 dependent gene/protein regulation. Mitochondrial morphology and network exhibit cyclical changes in mouse liver based on EM and Cox8a-GFP images. Several genes involved in mitochondrial biogenesis and fission are transcriptional targets of Bmal1 and are up-regulated at ZT12. It is likely that oxidative stress builds up through fuel burning during feeding. The pre-programmed induction of mitochondrial fission and mitophagy helps dissipate oxidative stress and eliminate damaged mitochondrial components. Mitochondrial biogenesis is also increased at the transition from the light to the dark cycle to increase the oxidative capacity and/or to replenish mitochondria following mitophagy. Therefore, the diurnal mitochondrial dynamics constitutes a daily “refreshing” cycle to maintain functional mitochondria. The amplitude of diurnal expression for Bmal1 targets seems to be greater at the protein level than that of mRNA, suggesting the involvement of post-transcriptional regulation. Consistent with this notion, it has been reported that the circadian clock regulates ribosomal biogenesis (Jouffe et al., 2013). Our ChIP-seq analysis also identifies several genes in mitochondrial protein synthesis that are bound by the core molecular clock (Figure 5A). Furthermore, a recent study demonstrates that cytosolic Bmal1 interacts with the translational machinery to stimulate translation at the active cycle (Lipton et al., 2015). These observations indicate a preponderant role for Bmal1-regulated protein synthesis following target mRNA expression in mediating circadian physiology.
While the rhythmic mitochondrial remodeling in vivo is dependent on diurnal Bmal1 activity and independent of substrate availability, the mitochondrial network could be manipulated by altering nutrient conditions in cultured hepatocytes. As reported previously for β-cells (Liesa and Shirihai, 2013; Molina et al., 2009), the mitochondrial network of WT hepatocytes could be “reset” to a fused or elongated morphology in EBSS, followed by a fragmented morphology when switching to HG+PA. LBmal1KO mitochondria lack these adaptations and retain the swollen morphology. Interestingly, defective quality control, such as in Pink1 mutations (Bueno et al., 2015; Poole et al., 2008), has been shown to cause mitochondrial swelling. We then tested whether over-expression of Fis1 to promote fission/mitophagy could rescue LBmal1KO phenotypes. Fis1 expression not only eliminates the swollen shape but also restores the functional and morphological plasticity of LBmal1KO hepatocytes. This manipulation also dampens elevated levels of basal and high nutrient-induced ROS production in LBmal1KO mitochondria. Furthermore, Fis1 over-expression reduces hepatic lipid accumulation and normalized liver function in LBmal1KO mice. Of note, adFis1 does not appear to interfere with the fusion/fission process in WT hepatocytes under EBSS/HG+PA conditions, possibly because of the moderate increase in Fis1 protein (~3-fold) over the endogenous level. It is currently unclear what causes the swollen/enlarged mitochondrial phenotype in LBmal1KO mice. The facts that several fission/mitophagy genes are Bmal1 transcriptional targets and Fis1 over-expression restores the function of LBmal1KO mitochondria suggest that defective fission/mitophagy may be causative to the abnormal morphology. In addition, sustained oxidative stress has been shown to inhibit mitophagy (Seillier et al., 2015). adFis1 reduces mitochondrial ROS levels and oxidative damage in liver cells. As such, Fis1 over-expression may eliminate the block in mitophagy and quality control in LBmal1KO hepatocytes both by normalizing Fis1 expression and lowering ROS levels, thereby resuming the dynamic fusion-fission process and metabolic flexibility of mitochondria. The molecular details of the Fis1 rescuable morphological phenotype of LBmal1KO mitochondria require further investigation.
aha-1 is the closest C. elegans homologue of the mammalian Bmal1 and is found to be a key transcriptional regulator of mitochondrial oxidative metabolism in the current study. Worms with aha-1 knockdown show a disorganized mitochondrial network similar to that seen with drp-1 knockdown (homologue of the mammalian fission gene Drp1) (Labrousse et al., 1999), mirroring the role of its mammalian counterpart in the control of mitochondrial dynamics. However, unlike Bmal1, the binding partner (if there is any) has not been identified. In line with the notion that C. elegans lacks a robust circadian clock system (van der Linden et al., 2010), aha-1 is also not involved in circadian behavior regulation (Banerjee et al., 2005). These results may indicate that the circadian role of Bmal1 is an extension to its primordial function in regulating mitochondrial activity. Alternatively, Bmal1 may serve as an important regulator of mitochondrial respiration independent of circadian regulation. In fact, acute deletion or over-expression of Bmal1 in mouse liver or in Hepa 1-6 cells is sufficient to modulate oxidative metabolism. Both aha-1 deletion worms (Huang et al., 2004) and whole body Bmal1 (and Clock) knockout mice have shortened lifespan (Dubrovsky et al., 2010; Kondratov et al., 2006). AHA-1 over-expression increases OXPHOS gene expression and extends lifespan in C. elegans. It is possible that oxidative damages associated with dysfunctional mitochondria due to loss of Bmal1/Clock activity contribute to the aging phenotypes in mice. The effect of Bmal1 or AHA-1 on the expression of certain OXPHOS genes is modest. Nevertheless, moderate alterations in several genes in rate-limiting steps of the common pathway can result in major differences in physiological outcomes. The translation stimulating activity of Bmal1 discussed earlier may further amplify the gene effects. Collectively, our study suggests that synchronization of mitochondrial biogenesis and dynamics with nutrient status by Bmal1 is key to sustain energy homeostasis and healthspan. Therefore, identification of small molecules or physiological pathways that increase Bmal1 activity may provide new therapeutic strategies to treat metabolic diseases
EXPERIMENTAL PROCEDURES
Animal studies
Mice of the C57BL/6J background (the Jackson Laboratory) were used in this study. Detailed information for each experimental cohort and metabolic studies are described in Table S4 and Supplemental Experimental Procedures. Liver-specific Bmal1 knockout mice were generated by crossing albumin-cre transgene to Bmal1f/f mice. Animals were on a chow diet or high fat diet (60% fat, F3282, BioServ) and housed in a barrier facility with 12-hour light and dark cycles. ZT0: lights on at 6 am; ZT12: lights off at 6 pm. For circadian/diurnal studies, mice between 8-12 weeks old were sacrificed every 4 hours for 24 hours (Liu et al., 2013). Animals under constant fasting were fasted 24 hours prior to the first time point of tissue collection. For metabolic studies, mice were fed a high fat diet for 4-6 months starting at 8-12 weeks of age. Glucose and insulin tolerance tests were performed on overnight fasted animals. Mice were given 1.5 g/kg body weight glucose (GTT) or 1 U/kg insulin (ITT). Protein carbonylation to assess oxidative damage was conducted using the OxyBlot protein oxidation detection kit according to manufacturer’s instruction (Millipore). Briefly, protein carbonyl groups were detected by incubation with 2,4-dinitrophenylhydrazine (DNPH) to form DNP-derivatized protein products, followed by Western blotting with an anti-DNP antibody. For Bmal1, Cre, GFP (serving as a control) and Fis1 adenovirus approaches, 109 pfu/mouse was injected through the tail vein. Subsequent metabolic characterizations were carried out within 7-14 days post injection. All animal studies were approved by the Harvard Medical Area Standing Committee on Animals.
In vitro/ex vivo studies
Isolation of hepatocytes/liver mitochondria, generation of Hepa 1-6 stable lines, and Seahorse Bioenergetics analyses are described in Supplemental Experimental Procedures. For cellular bioenergetics, after measurements of basal OCR (a in Figure 1D) the sequential addition of oligomycin, FCCP, and antimycin A/rotenone determined ATP turnover (b in Figure 1D), proton leak (c in Figure 1D), and maximal respiration. The coupling efficiency was determined by the ratio of ATP turnover to basal mitochondrial respiration. For mitochondria coupling assays, addition of ADP, oligomycin, FCCP, and antimycin A determined stages 3, 4o, and 3u respiration (respiratory control ratio=3u/4o), respectively.
Electron and confocal microscopy
Analyses of mitochondrial surface, coverage, and density with electron microscopy are described in Supplemental Experimental Procedures. For spinning disk confocal fluorescent microscopy, mitochondria were visualized using adenovirus carrying Cox8a-GFP, which contains mitochondrial matrix localization sequences derived from Cox8a (Perales-Clemente et al., 2011). Expression of adCox8a-GFP in WT and LBmal1KO livers (chow fed, male mice) were achieved through tail vein injection. Animals were sacrificed 72 hours after virus injection. One group of animals was fed ad libitum and sacrificed at ZT4 or ZT16. A second group of animals was fasted at ZT12 and either sacrificed at ZT16 or refed at the following ZT0 and sacrificed at ZT4. Cryosections (5 micron) of liver samples counter-stained with DAPI were used for confocal microscopy. For the primary hepatocytes experiments, cells were plated in William E media with 2 mM glutamine, 5% FBS. Experiments were started 12 hours after plating. For adenovirus experiments, cells were transduced 4 hours after plating and media was refreshed 8 hours later. For exposure to different nutrient conditions, hepatocytes were cultured in EBSS (Earle’s balanced salt solution with 5.5 mM glucose) or HG+PA (DMEM with 25 mM glucose and 0.3 mM palmitic acid) for the indicated time course.
Assessment of ROS production
WT and LBmal1KO primary hepatocytes were treated with adFis1 or control virus (empty vector). Cells were cultured in EBSS or HG+PA for 3 hours. Cells were co-stained with 100 nM Mitotracker Green FM (Invitrogen M7514) and 5 μM MitoSOX Red (Invitrogen M36008) in EBSS for 15 min before imaging. MitoSOX fluorescence was quantified with ImageJ and normalized to Mitotracker green fluorescence (see Figure S3 for representative images). Results were the average of 12 cells per group.
Complex I activity
Complex I (NADH oxidase/Co-enzyme Q reductase) was measured using the MitoCheck Complex I Activity Assay kit (Cayman chemical 700930). The rate of NADH oxidation, proportional to the activity of complex I, was determined by a decrease in absorbance at 340 nm over 15 min in the presence of ubiquinone and potassium cyanide to inhibit complex IV and prevent oxidation of ubiquinone. Rotenone (2 μM), which inhibits complex I activity, was used as a negative control. Bovine heart mitochondria from the kit were used as a positive assay control (data not shown).
Gene expression and mitochondrial DNA quantification
Relative expression was determined by SYBR Green based real-time PCR using 36B4 as an internal standard. A relative standard curve was used to calculate the expression level. Primers used for gene expression studies are listed in Table S5. Relative mitochondrial DNA content were determined by real-time PCR using primers specific to mitochondrial gene Nd1 and normalized to 36B4 DNA. For flow cytometry, freshly isolated hepatocytes were incubated with glucose free DMEM media containing 10 mM pyruvate with 100 nM MitoTracker green FM (Invitrogen) for 30 minutes.
Antibodies and Western blotting
Primary antibodies were obtained from the following sources: phospho-Akt (9271), total Akt (9272), Lc3b (8899, with higher affinity to Lc3b-II), and Phospho-DRP1 (Ser616) (4494) from Cell Signaling Technology; Bnip3 (ab109362), Mfn1 (ab126575), Mfn2 (ab50838), Opa1 (ab42364), and Fis1 (ab71498) from Abcam; Bmal1 (sc-8550), Drp1 (sc-32898), and Pink1 (sc32584) from Santa Cruz Biotechnology; total OXPHOS primary antibody cocktail (458099) from Invitrogen. Hsp60 (Abcam 45134), β-tubulin (Cell Signaling 2128), or β-actin (Cell Signaling 4967) was included as loading control and for normalization. Western blot signal was detected using a Bio-Rad imaging system and quantified by ImageJ.
C. elegans studies
N2 Bristol was the wild type control strain. Two aha-1 over-expression strains, OP124 [unc-119(ed3); wgIs124(aha-1::TY1::EGFP::3xFLAG(92C12) + unc-119(+))] and UL1606 [unc-119(ed3); leEx1606(aha-1::GFP + unc-119(+))], as well as the mitochondrial reporter strain SJ4103 [zcIs14(myo-3p::GFP(mit))] were provided by the caenorhabditis genetics center (CGC). Worms were maintained at 20°C on nematode growth media with OP50-1 bacteria. RNAi, imaging and lifespan experiments (Mair et al., 2011) are described in Supplemental Experimental Procedures. TG was determined from chloroform:methanol extracts using enzymatic assays. For gene expression, RNA was isolated from worms at the L4/young adult stage and the relative expression was determined by real-time PCR using pmp-3 for normalization.
Published ChIP-seq data analysis
Published liver ChIP-seq data were downloaded from gene expression omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). AHA-1 Chip-seq data at L4 stage was obtained from the ModEncode project. Data analyses are described in Supplemental Experimental Procedures.
Statistical analyses
Statistical analyses comparing two parameters (between treatments or genotypes) in the cell-based work were conducted using two-tailed Student’s t test. Two-parameter analyses for samples from in vivo studies (where Gaussian distribution could not be ascertained) were determined using Mann-Whitney test. Statistics for multiparameter analyses were determined by one-way analysis of variance (ANOVA) followed by Bonferroni post-hoc tests. Two-way ANOVA was used to determine statistical significance for GTTs and ITTs. For lifespan in worms, the log-rank Mantel-Cox test was used. Values are presented as the mean ± S.E.M. For in vitro assays, the mean and S.E.M. were determined from 5 biological replicates for one representative experiment. Experiments were repeated at least three times. P<0.05 was considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Robert V. Farese and Tobias Walther for help in imaging studies and Dr. Marek Wagner for assistance in stable cell line generation. We are grateful to Dr. Gokhan Hotamisligil and his lab members for technical supports: Dr. Ana Paula Arruda for help with the Seahorse bioenergetics assays and Dr. Karen Inouye for metabolic cages studies. N.H.K. is supported by National Institutes of Health Interdisciplinary Training in Genes and Environment grant T32ES016645; R.K.A. supported by Herchel Smith Graduate Fellowship; N.K. supported by an American Heart Association predoctoral fellowship; and K.B. supported by NIH fellowship F32AG044944. W.B.M is supported by Ellison Medical Foundation and NIG grant R01AG044346. This work was funded through American Diabetes Association grant 1-14-BS-122 (C-.H.L) and NIH grant R01DK075046 (C-.H.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 Figures, 5 Tables and Supplemental Experimental Procedures.
AUTHOR CONTRIBUTIONS
D.J. and S.L. designed and performed most experiments together. S.L. analyzed published ChIP-seq and expression data. K.B. and W.B.M. conducted worm experiments and interpreted the results. N.H.K., R.K.A., U.U., A.H, X.K., and X.L. contributed to expression data and hepatic lipid content analyses. N.H.K. generated the Cox8-mCherry mitochondrial reporter construct. N.K. conducted time-lapse confocal microscopy and interpreted the results. M.R.G. prepared reagents. D.J., S.L., and C-.H.L. performed data analyses and wrote the manuscript.
REFERENCE
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell. Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, Oxidants, and Aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Kwok A, Lin SY, Slack FJ. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev. Cell. 2005;8:287–295. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, Corey C, Herazo-Maya JD, Sembrat J, Lee JS, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J. Clin. Invest. 2015;125:521–538. doi: 10.1172/JCI74942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging (Albany NY) 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Saigusa T, Tamai Y. Caenorhabditis elegans opens up new insights into circadian clock mechanisms. Chronobiol. Int. 2005;22:1–19. doi: 10.1081/cbi-200038149. [DOI] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- Huang X, Powell-Coffman JA, Jin Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development. 2004;131:819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell. Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell. Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, Sahin M. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell. 2015;161:1138–1151. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Lucas A, Duarte JA, Sunny NE, Satapati S, He T, Fu X, Bermudez J, Burgess SC, Perales JC. PEPCK-M expression in mouse liver potentiates, not replaces, PEPCK-C mediated gluconeogenesis. J. Hepatol. 2013;59:105–113. doi: 10.1016/j.jhep.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina AJ, Wikstrom JD, Stiles L, Las G, Mohamed H, Elorza A, Walzer G, Twig G, Katz S, Corkey BE, et al. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes. 2009;58:2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales-Clemente E, Fernandez-Silva P, Acin-Perez R, Perez-Martos A, Enriquez JA. Allotopic expression of mitochondrial-encoded genes in mammals: achieved goal, undemonstrated mechanism or impossible task? Nucleic Acids Res. 2011;39:225–234. doi: 10.1093/nar/gkq769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu ZX, Dong J, Mustard KJ, Hawley SA, et al. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell. Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillier M, Pouyet L, N'Guessan P, Nollet M, Capo F, Guillaumond F, Peyta L, Dumas JF, Varrault A, Bertrand G, et al. Defects in mitophagy promote redox-driven metabolic syndrome in the absence of TP53INP1. EMBO Mol. Med. 2015;7:802–818. doi: 10.15252/emmm.201404318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. The yeast metabolic cycle: insights into the life of a eukaryotic cell. Cold Spring Harb. Symp. Quant. Biol. 2007;72:339–343. doi: 10.1101/sqb.2007.72.019. [DOI] [PubMed] [Google Scholar]
- van der Linden AM, Beverly M, Kadener S, Rodriguez J, Wasserman S, Rosbash M, Sengupta P. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000503. doi: 10.1371/journal.pbio.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkaart S, Koopman WJ, van Emst-de Vries SE, Nijtmans LG, van den Heuvel LW, Smeitink JA, Willems PH. Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim. Biophys. Acta. 2007;1772:373–381. doi: 10.1016/j.bbadis.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.