SUMMARY
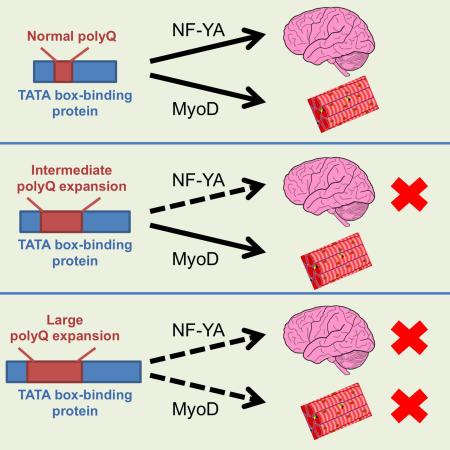
In polyglutamine (polyQ) diseases, large polyQ repeats cause juvenile cases with different symptoms than adult-onset patients, who carry smaller expanded polyQ repeats. The mechanisms behind the differential pathology mediated by different polyQ repeat lengths remain unknown. By studying knock-in mouse models of spinal cerebellar ataxia-17 (SCA17), we found that a large polyQ (105 glutamines) in the TATA box-binding protein (TBP) preferentially causes muscle degeneration and reduces the expression of muscle-specific genes. Direct expression of TBP with different polyQ repeats in mouse muscle revealed that muscle degeneration is mediated only by the large polyQ repeats. Different polyQ repeats differentially alter TBP’s interaction with neuronal and muscle-specific transcription factors. As a result, the large polyQ repeat decreases the association of MyoD with TBP and DNA promoters. Our findings suggest that specific alterations in protein interactions by large polyQ repeats may account for the unique pathology in juvenile polyQ diseases.
Keywords: Polyglutamine, transcription, MyoD, muscle degeneration
INTRODUCTION
Polyglutamine (polyQ) expansion causes at least nine inherited neurodegenerative disorders, including Huntington’s disease (HD), spinocerebellar ataxia (SCA) types 1, 2, 3, 6, 7, and 17, dentatorubral-pallidoluysian atrophy (DRPLA), and spinal bulbar muscular atrophy (SBMA) (Zoghbi and Orr, 2009). Studies of various polyQ disease proteins have shown that expanded polyQ tracts affect the function of the disease proteins, leading to a gain or loss of function (Lim et al., 2008). It is also clear that the function of polyQ proteins can impact disease severity and progression. For example, SCA17, which is caused by polyQ expansion in the TATA box-binding protein TBP, a transcription factor essential for the transcription of a wide range of genes (Vannini and Cramer 2012), is associated with more severe neurological phenotypes than in other polyQ diseases, despite the fact that the repeat in mutant TBP is often shorter than 64Q (van Roon-Mom et al., 2005). In addition, different polyQ diseases display distinct pathology. For example, SBMA is characterized by muscle atrophy (Cortes et al., 2014; Lieberman et al., 2014), which is reported to be moderate or absent in other polyQ diseases.
The selectivity of polyQ toxicity apparently comes from protein context because it determines protein-protein interactions, half-life and stability, subcellular localization, etc. However, the length of the polyQ repeat also seems to modulate the selectivity of polyQ protein toxicity. There is strong evidence that in Huntington’s disease (HD), polyQ repeats larger than 60 glutamines cause juvenile cases, who display different symptoms and have more widespread degeneration in the brain. For example, juvenile HD patients do not display chorea, but they have severe cognitive dysfunction and seizure that is absent in adult HD patients (Vargas et al., 2003; Squitieri et al., 2006). Juvenile HD patient brains also have more nuclear aggregates, whereas adult HD brains have more neuripil aggregates, also suggesting that polyQ lengths mediate different pathogenic pathways (DeFiglia et al., 1997; Gutekunst et al., 1998). Despite the well-known phenomenon of differential pathology and symptoms in early- and adult-onset polyQ diseases, the mechanism underlying this phenomenon has not been investigated rigorously, and understanding it is critical if we are to develop effective therapies for polyQ diseases.
SCA17 is a good candidate for investigating the mechanism behind the cell type-specific pathology in polyQ disease. The CAG repeat in the normal human TBP gene ranges from 30 to 42 units. Expansion of the polyQ tract (>42 glutamines) in TBP induces striking clinical features in SCA17 patients, including ataxia, dystonia, parkinsonism, dementia, and seizures (Bruni et al., 2004; Koide et al., 1999; Nakamura et al., 2001; Rolfs et al., 2003). Marked cerebellar atrophy and Purkinje cell loss are typical in SCA17 patients, with less pronounced neurodegeneration occurring in other brain regions (Bruni et al., 2004; Koide et al., 1999; Nakamura et al., 2001; Rolfs et al., 2003; Toyoshima et al., 2004; Bauer et al., 2004). However, when polyQ repeats exceed 63Q, mutant TBP induces juvenile symptoms, with retarded growth and muscle weakness characterized by impaired laryngeal and sphincter muscle function that results in difficulties in swallowing, talking, and walking, as well as a quick progression of clinical symptoms and early death (Koide et al., 1999; Maltecca et al., 2003). These symptoms are clearly different from those seen in adult-onset SCA17 patients.
We have established SCA17 knock-in mice that express one copy of mutant Tbp with 105Q under the control of Cre recombination. We found that this large polyQ repeat in mutant TBP leads to muscle degeneration, which was verified by selective expression of mutant TBP in muscle. Different polyQ repeat lengths lead to different effects on the interactions of TBP with neuronal and muscle-specific transcription factors. A large (105Q) polyQ repeat decreases the association of TBP with MyoD, a muscle-specific transcription factor (Tapscott, 2005; Guttridge et al., 2000; Di Marco et al., 2005), reducing its stability and association with DNA promoters. Our findings give us new mechanistic insight into the unique pathology caused by large polyQ repeats in polyQ diseases.
RESULTS
Mutant TBP Preferentially Accumulates in the Brain and Muscle in SCA17 Knock-In (KI) Mice
We previously established conditional TBP knock-in mice that express mutant TBP specifically in the brain (Huang et al., 2011). In these KI mice, a stop codon was placed in the front of the translation initiation site for a mutant Tbp gene containing 105 CAGs. This stop codon is flanked by two loxP sites, so it can be removed by Cre to allow expression of mutant TBP. To generate SCA17 KI mice that express mutant TBP ubiquitously at the endogenous level, we crossed floxed SCA17 KI mice with transgenic mice expressing Cre under the control of the EIIa promoter, which drives the expression of Cre in early embryonic cells (Lakso et al., 1996; Holzenberger et al 2000). Floxed male SCA17 KI mice containing the Ella-Cre gene were mated with wild-type female mice. The offspring mice were then genotyped to identify those expressing mutant TBP, which should be derived from the depletion of loxP sites or expression of the mutant Tbp allele at the one-cell stage. F1 male mice expressing the mutant TBP were then mated with wild-type female mice (B6) to generate F2 mice, which carry the mutant Tbp gene without the EIIa-Cre gene. Such knock-in mice express one copy of wild-type Tbp and one copy of mutant Tbp in an inherited manner identical to that seen in SCA17 patients, in whom mutant TBP is ubiquitously expressed (Figure 1A).
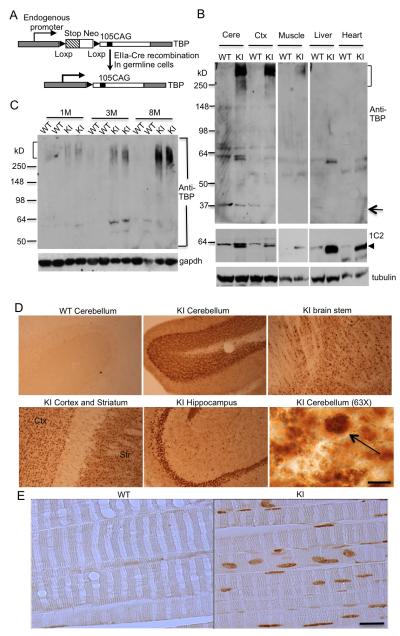
Figure 1. Mutant TBP Preferentially Accumulates in Brain and Muscle in KI Mice.
(A) The schematic structure of the targeted mouse Tbp gene, which has a stop codon and the neomycin (Neo) resistance gene flanked by two loxP sites to prevent the translation of the mutant Tbp gene. After EIIa Cre recombination, the stop codon and Neo resistance gene are removed, leading to the expression of the mutant Tbp gene with 105 CAGs. (B) Western blot of cerebellum (Cere), cortex (Ctx), muscle, liver, and heart lysates from SCA17 KI (KI) and wild-type (WT) mice at postnatal day 30 (P30). The upper panel shows the blot probed with 1TBP18 antibody, which is against N-terminal TBP. The middle panel shows the blot probed with 1C2 antibody, which is against expanded polyglutamine. The bottom panel shows the same blot probed with γ-tubulin. Arrow indicates endogenous mouse TBP. Arrowhead indicates soluble mutant TBP. (C) Western blot of muscle lysates from KI and WT mice at indicated ages. The blot was probed with 1TBP18 (upper panel) and anti-gapdh (lower panel). Aggregated (bracket) mutant TBP proteins are indicated. (D) 1TBP18 immunostaining showing the selective expression of mutant TBP in the cerebellum, brain stem, hippocampus, cortex, and striatum of 3-month-old KI mouse brain. 1TBP18 at the same concentration did not label the WT cerebellum. Arrow indicates small nuclear aggregates in Purkinje cells at high-power magnification (Scale bar: 10 μm, 63X objective). (E) 1TBP18 immunostaining showing the expression of mutant TBP in 3-month-old KI mouse muscle. WT mouse muscle served as a control. Scale bar: 50 μm.
To confirm that mutant TBP is expressed in the brain and peripheral tissues in SCA17 KI mice, we performed western blotting analysis with two antibodies: anti-TBP (1TBP18), which can recognize aggregated TBP in the stacking gel (Figure 1B upper panel), and 1C2, which reacts more strongly with the soluble form of expanded polyQ proteins (Figure 1B, middle panel). Expression of mutant TBP reduces the level of endogenous TBP (arrow in Figure 1B), as also happened in earlier TBP transgenic and knock-in mouse brains (Friedman et al., 2007; Friedman et al., 2008; Yang et al., 2014). To verify that this indeed occurs in muscle tissue, we immunoprecipitated TBP from WT and KI mouse muscle tissues and found that the presence of mutant TBP also reduced the level of wild-type TBP in the muscle (Figure S1A). RT-PCR revealed that this decrease is associated with a decrease in endogenous mouse TBP transcripts (Figure S1B). Thus, the expression level of TBP is tightly regulated at the transcriptional level because of its critical function in gene transcription.
In SCA17 KI mice, soluble mutant TBP is present in the brain (cerebellum, cortex) and peripheral tissues (muscle, liver, heart). Importantly, aggregated TBP (bracket in Figure 1B), which remains in the stacking gel, is present in the brain tissues and also in muscle, whereas liver and heart tissues, although they express soluble mutant TBP, do not show such aggregated TBP (Figure 1B). We then analyzed the expression of mutant TBP in the muscle tissues of SCA17 KI mice at different ages and found that aggregated TBP accumulates in aged muscle, which is evident by the more abundant aggregated TBP in the stacking gel containing muscle proteins from the older SCA17 KI mice (3 and 8 months) (Figure 1C).
We also performed immunocytochemical analysis to examine the distribution of mutant TBP in the brain and muscle. Mutant TBP is widely expressed in various brain regions, including the cerebellum, brain stem, striatum, and hippocampus, and high magnification reveals that mutant TBP forms small aggregates in the nucleus (Figure 1D). Similarly, mutant TBP also accumulates in the nuclei of skeletal muscle cells and forms small aggregates (Figure 1E). Overall, we see that mutant TBP preferentially accumulates in the nuclei of brain and muscle tissues in SCA17 KI mice.
Mutant TBP in Muscle Causes Degeneration
Gait abnormalities and muscle weakness are seen in some SCA17 patients (Koide et al., 1999; Rolfs et al., 2003). The distribution of mutant TBP in muscle cells led us to examine whether mutant TBP could cause muscle pathology. Haematoxylin and eosin (H&E) staining of muscle tissues revealed normal morphology in the skeletal muscle tissues of wild-type mice, viewed as well-defined muscle cells in which the nuclei are localized peripherally. At 1.5 months of age, SCA17 KI mice showed no distinguishable differences in muscle morphology from wild-type mice (Figure S2A). However, muscle degeneration was seen in SCA17 KI mouse skeletal muscle sections at 3 months of age and became more severe at 7 months; this degeneration was characterized by uneven H&E staining, fragmented muscle morphology, and more importantly, multiple internalized or centralized nuclei, a feature of muscle cells regenerated after degeneration (Figure 2A). Quantitation of muscle cross-sections also verified the degeneration of muscle cells in SCA17 KI mice (Figure 2B). This degeneration appears to be specific to the skeletal muscle in SCA17 KI mice since cardiomyocytes, another type of muscle cell, showed no differences in their morphology between SCA17 KI and wild-type mice at 3 months of age (Figure S2B). Furthermore, we performed electron microscopy studies that revealed typical degeneration changes in the muscle cells of SCA17 mice. These changes include intrafiber Z-band breaks, poorly aligned fibres of the myofibrils, enlarged mitochondria, and swollen spaces between individual muscle cells in cross-sections (Figure 2C). Longitudinal sections also showed degenerated muscle cells in SCA17 mice, as evidenced by disorganized muscle fibers, frequent Z band destruction, and sarcomere disruption, as well as enlarged and swollen mitochondria (Figure 2C).
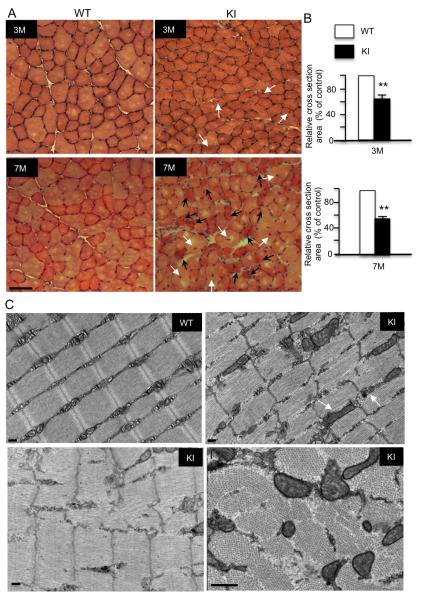
Figure 2. Mutant TBP Induces Muscle pathology.
(A) Haematoxylin and eosin (H&E) cross-section staining of tibialis anterior (TA) muscles from 3-month-old and 7-month-old SCA17 KI mice showing an age-dependent decrease in myofibril size, including severe muscle atrophy (black arrow) and cells containing central nuclei (white arrow), a feature of muscle degeneration. Scale bars: 50 μm. (B) Quantification of cross-sectional area of myofibrils at indicated ages. Values are mean ± SEM of data from 5 mice in each group (**p < 0.01, n = 500). (C) Electron microscopy of WT and SCA17 KI TA muscles. In KI muscle, Z bands are destructed and mitochondria are swollen, abnormally shaped, and enlarged. Scale bars: 0.2 μm.
SCA17 is known to have a significant impact on the viability of Purkinje cells in the cerebella of patients (Koide et al., 1999; Nakamura et al., 2001; Rolfs et al., 2003; Bauer et al., 2004; Bruni et al., 2004; Toyoshima et al., 2004). The striking pathology in SCA17 KI muscle cells raises the important issue of whether this degeneration results from the degeneration of the central nervous system, which controls muscle viability and activity. It is possible that mutant TBP might affect the neuromuscular junction (NMJ), which then results in reduced muscle activity and muscle atrophy. We therefore examined the ultrastructure of the NMJ using electron microscopy (EM). EM examination revealed SCA17 mice had normal NMJ morphology, which displays a clear synaptic junction with normal abundance of synaptic vesicles compared with the NMJ of wild-type mouse (Figure S3). α-Bungarotoxin labeling of the neuromuscular junction revealed no difference between WT and SCA17 KI mice (Figure S4A). Also, immunostaining of spinal cord neurons with antibodies to NeuN and ChAT revealed no differences between SCA17 KI and WT mice (Figure S4B, C). All these findings suggest that mutant TBP is unlikely to cause neuromuscular dysfunction or spinal cord neuronal loss that leads to muscle atrophy.
To provide more rigorous evidence for the primary effect of mutant TBP in muscle cells, we crossed floxed SCA17 KI mice with transgenic mice that express Cre under the control of the muscle creatine kinase (MCK) promoter, which drives the expression of Cre selectively in skeletal muscle cells (Bruning et al., 1998). The expression of mutant TBP in muscles in the crossed (muscle-KI) mice was verified by immunocytochemistry, which showed that mutant TBP in muscle-KI mice is restricted to skeletal muscle cells and is less abundant than in SCA17 KI muscle, likely due to the fact that mutant TBP is not expressed in non-muscle cells in muscle-KI mice (Figure 3A). By performing immunostaining of the brain cortex tissues of wild-type, SCA17 KI, and muscle-KI mice, we verified that mutant TBP in muscle-KI mice is absent in the brain, whereas SCA17 KI mouse brain displayed intensive nuclear TBP staining (Figure 3B). High-magnification micrographs revealed that mutant TBP also formed nuclear aggregates in muscle-KI skeletal myocytes in the same manner as in the muscle cells of SCA17 KI mice (Figure 3C). Importantly, we also found degeneration of skeletal muscle cells in the muscle-KI mice, as characterized by multiple centralized nuclei and atrophic muscle cells with reduced size (Figure 3D-E). Thus, by examining both SCA17 KI and muscle-KI mouse models, we find compelling evidence that mutant TBP expression in skeletal muscle cells can cause muscle atrophy, pointing to a primary effect of mutant TBP in peripheral tissues.
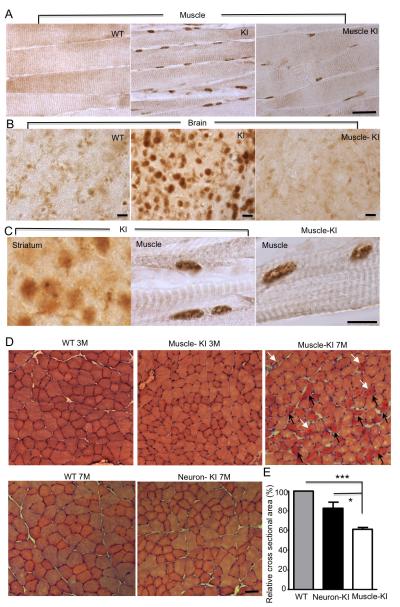
Figure 3. Expression of Mutant TBP in Muscle Also Leads to Muscle Degeneration.
(A) 1TBP18 staining showing the expression of mutant TBP in 3-month-old SCA17 KI (KI) and muscle-KI mouse muscles. (B) Mutant TBP was detected in the SCA17 KI brain, but not WT or muscle-KI brain. (C) High-power magnification (630x) photographs showing small nuclear aggregates in SCA17 KI mouse striatum and muscle, as well as muscle-KI mouse muscle. (D) H&E cross-sectional staining of TA muscles from WT, muscle-KI, and neuron-KI (nestin-KI) mice at indicated ages. Atrophic muscle cells (black arrow) and cells containing centralized nuclei (white arrow) are indicated. (E) Quantification of cross-sectional areas of myofibrils in WT, neuron-KI, and muscle-KI skeletal muscle tissues (n = 500, *p < 0.01; ***p < 0.001). Scale bars: (A): 100 μm; (B): 20 μm; (C): 20 μm; (D): 50 μm. Data are represented as mean ±SEM.
Mutant TBP Weakens Muscle and Causes Movement Abnormalities and Early Death
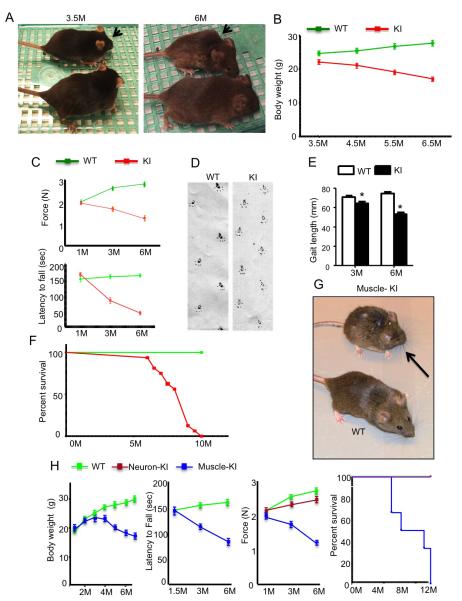
Given the muscle atrophy in SCA17 KI mice, we wondered whether this peripheral pathology might contribute to the progression of disease and more severe symptoms. Indeed, SCA17 KI mice showed age-dependent symptoms, including hunchback appearance (Figure 4A) and reduced body weight (Figure 4B). Because of the muscle atrophy in these mice, we tested grip strength, which reflects the muscle strength of mouse legs. We found a significant reduction of grip strength in SCA17 KI mice (Figure 4C). As a result, rotarod performance, which can be affected by reduced muscle strength, was poor in SCA17 KI mice (Figure 4C). Also, we detected an abnormal gait in SCA17 KI mice, which reflects both ataxia and muscle atrophy (Figure 4D-E). Furthermore, the life span of SCA17 KI mice was shorter than wild-type mice (Figure 4F), indicating that muscle atrophy makes an important contribution to severe disease symptoms.
Figure 4. Mutant TBP Causes Phenotypes Related to Muscle Degeneration.
(A) SCA17 KI mouse at 6 months of age (arrow) was smaller and poorly groomed relative to WT littermate. Such phenotypes were not seen in SCA17 KI mouse at 3.5 months. (B) Changes in body weight of KI mice compared with WT mice (n = 16 each group, sex matched). (C) Grip strength (upper panel) and non-accelerating rotarod performance (low panel) of KI and WT mice at 1, 3, and 6 months of age. (D) Representative walking footprint patterns of 6-month-old WT and KI mice. (E) Quantification of stride length test showing that KI mice display shorter strides compared with the evenly spaced footprints of WT mice (n = 10). (F) Survival plot showing a reduced lifespan of SCA17 KI mice relative to WT mice (n = 16 each group). (G) The muscle-KI mouse at 7 months of age (arrow) was smaller and poorly groomed relative to WT littermate. (H) From left to right, body weight, non-accelerating rotarod performance, grip strength, and survival plot showing reduced body weight, movement deficit, decreased grip strength, and shortened life span of muscle-KI mice compared with age-matched WT or Neuron-KI (nestin-KI) mice (n = 16 each group, sex matched). Data are represented as mean ±SEM.
If muscle degeneration does contribute critically to the severe symptoms of SCA17 KI mice, we should also see similar movement abnormalities in muscle-KI mice. Indeed, muscle-KI mice at the age of 7 months had a hunchback appearance similar to SCA17 KI mice (Figure 4G). They also had an age-dependent reduction in body weight, poor rotarod performance, and reduced grip strength; moreover, they had a shorter life span, as they died within one year (Figure 4H). Comparing our previously established SCA17 mice that express mutant TBP in the brain via nestin-Cre (nestin-KI) and the SCA17 KI mice established in this current study, both SCA17 KI and muscle-KI mice show more severe phenotypes and earlier onset (3-4 months) than nestin-KI mice, which experience later (12 months) onset and milder phenotypes including body weight loss and poor rotarod performance as characterized in our previous studies (Huang et al., 2011). In addition, both SCA17 KI and muscle-KI mice experience early death (Table S1). Thus, the expression of TBP in skeletal muscle cells can cause progressive phenotypes and leads to the early death of mice.
The late-onset muscle degeneration and symptoms in SCA17 KI mice suggest that mutant TBP may only affect adult muscle cells. To validate this idea, we also crossed floxed SCA17 KI mice to transgenic mice expressing Cre in muscle progenitor or satellite cells, which are able to differentiate to adult muscle cells, under the control of the Pax7 promoter. The crossed mice expressed mutant TBP selectively in muscle satellite cells that are located between the basal lamina and sarcolemma of muscle fiber (Figure S5A). These mice, however, did not show any abnormal growth and impaired movement compared with wild type mice (Figure S5B, C).
Mutant TBP Abnormally Interacts with MyoD and Reduces Its Level
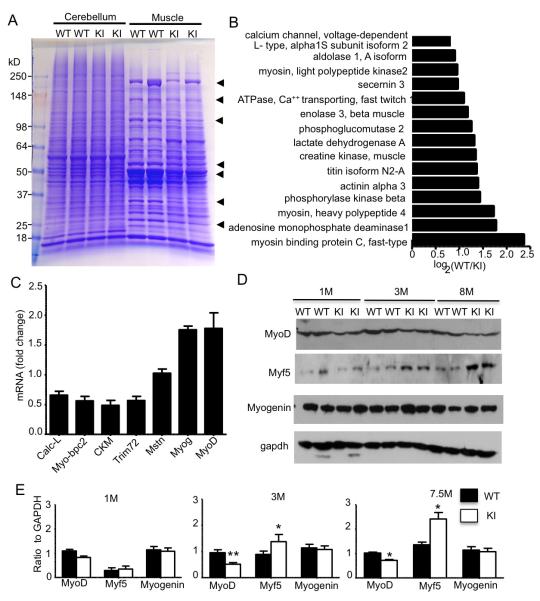
To investigate the mechanism by which mutant TBP causes muscle atrophy, we first used proteomics for protein profiling. Muscle tissues from SCA17 KI and wild-type mice at 6 months of age were isolated for analysis. Coomassie blue staining of the mouse hindlimb skeletal muscle tissue samples from WT and SCA17 KI mice revealed a number of proteins decreased specifically in KI muscles; however, there were no obvious differences between SCA17 KI and WT samples in cerebellum tissues (Figure 5A). Mass spectrometry, which uncovered 2493 proteins and 12,310 peptides, revealed a decrease in the number of muscle-specific proteins, such as myosin light polypeptide kinases-2, titin isoform N2-A, and muscle creatine kinase, in SCA17 KI muscle (Figure 5B). Because TBP is a transcription factor, the decreased expression of these muscle-specific proteins likely occurs at the transcriptional level. We therefore also performed microarrays for gene expression profiling in the cerebellum and skeletal muscle of SCA17 KI and WT mice. We analyzed a total of 35,240 transcripts in SCA17 KI muscle and cerebellum and found more genes with altered expression in muscle than in the cerebellum (Table-S2, S3). Also, more muscle-specific genes were upregulated (12) or downregulated (18) than neuronal-specific genes (2 upregulated, 5 downregulated) (Table-S4). Quantitative PCR analysis verified there was a decrease in the mRNA levels for muscle-specific genes, such as Calc-L, Myo-bpc2, CKM, and Trim72, in SCA17 KI mouse muscles (Figure 5C).
Figure 5. Mutant TBP Downregulates Muscle-Specific Protein Expression by Reducing MyoD Protein Level.
(A) Coomassie blue staining showing the protein composition of cerebellum and muscle tissue from WT and KI mice at 6 months of age. Arrows indicate proteins that show decreased levels in SCA17 KI muscles compared with age-matched WT muscles. (B) Mass spectrometry analysis identified several muscle-specific genes that are downregulated in SCA17 KI muscle. (C) Relative mRNA expression levels were determined by quantitative real-time PCR for calcium channel, voltage-dependent L type, alpha 1S subunit (Calc-L), myosin-binding protein C, fast type (Myo-bpc2), creatine kinase, muscle (CKM), tripartite motif-containing 72 (Trim72), myostatin (Mstn), myogenin (Myog), and MyoD in the TA muscle from SCA17 KI mice compared with WT mice at 3 months of age. (D) Western blot analysis of MyoD, Myf5, and myogenin expression in TA muscles of 1-, 3-, and 8-month-old KI and WT mice. (E) The densitometric ratios of indicated protein to GAPDH. (*p < 0.05, **p < 0.01). Data are represented as mean ±SEM.
Muscle-specific gene expression is regulated by a few transcription factors (MyoD, Myf5, myogenin, and MRF4), which make up the myogenic bHLH transcription factor family and are specifically expressed in skeletal muscle (Tapscott et al., 1998). We found that, in SCA17 KI mice, transcripts of myogenin and MyoD were increased (Figure 5C), suggesting that these muscle-specific transcription factors are upregulated due to the decreased levels of muscle-specific gene products. We were able to obtain antibodies to detect MyoD, Myf5, and myogenin in mouse tissues, so we focused on the expression of these transcription factors in the skeletal muscle tissues of SCA17 KI mice. Western blotting revealed that only MyoD, but not myogenin, was decreased in SCA17 KI muscles (Figure 5D-E). The expression of Myf5 appeared to increase in old SCA17 mouse muscle, perhaps because of a compensatory up-regulation mechanism by the decreased levels of MyoD and muscle specific proteins. These results suggest it is the decreased protein level of MyoD that results in the downregulation of a number of muscle-specific genes and subsequent muscle degeneration.
PolyQ Length-Dependent Muscle Degeneration and Interaction of TBP with MyoD
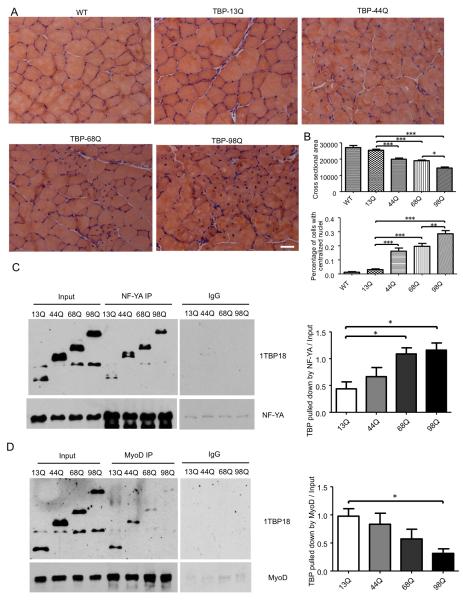
The striking retarded growth and muscle atrophy are not seen in SCA17 patients who express 43-55 polyQ TBP repeats. Given that large repeats (63Q and 66Q) in TBP cause juvenile SCA17 cases and also lead to retarded growth and muscle weakness phenotypes (Koide et al., 1999; Maltecca et al., 2003), we assume that a large polyQ repeat in TBP can cause the unique muscle atrophy phenotype. To test this idea, we generated AAV vectors that express TBP containing 13Q, 44Q, 68Q, or 98Q and injected them into the tibialis anterior (TA) muscle of wild-type mice. One month after injection, muscle histology revealed mutant TBP with the larger polyQ repeat elicited more severe muscle degeneration, which is shown by the greater reduction in the cross-sectional area of muscle fibers and increase in the percentage of muscle cells with centralized nuclei (Figure 6A, B). Thus, the extent of muscle degeneration depends on the TBP polyQ repeat length.
Figure 6. PolyQ Repeat Length-Dependent Muscle Degeneration and Interactions with Different Transcription Factors.
(A) Two-month-old wild-type (WT) mice were injected with AAV viruses encoding TBP with different polyQ lengths (13Q, 44Q, 68Q, and 98Q) into the tibialis anterior (TA) muscle. One month after injection, TA muscle was collected and stained using H&E. Scale bar: 40 μm. (B) Cross-sectional area of muscle cells and percentage of muscle cells with centralized nuclei from mice injected with different AAV viruses (n = 10 images per group, one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001). (C, D) HEK293 cells were co-transfected with NF-YA (C) or MyoD (D) and TBP with different polyQ lengths (13Q, 44Q, 68Q, and 98Q). A rabbit polyclonal antibody was used to pull down NF-YA or MyoD, and 1TBP18 antibody was used to detect TBP in the pull-down lysate. The results were quantified by normalizing the amount of TBP pulled down to input (n = 3, one-way ANOVA, *p < 0.05). Data are represented as mean ± SEM.
We know that the interaction between TBP and MyoD is required for the transcriptional activity of MyoD (Heller et al., 1998). Thus, it would be important to know whether a large polyQ repeat can reduce the association of TBP with MyoD to a greater extent, such that the large polyQ repeat selectively causes the muscle atrophy phenotype in SCA17. We therefore examined the association of MyoD with TBP containing different lengths of the polyQ repeat in transfected HEK293 cells via immunoprecipitation. We reported previously that polyQ expansion increases the association of mutant TBP with the transcription factor nuclear factor-YA (NF-YA) and affects its transcriptional activity on the expression of chaperone proteins in neuronal cells (Huang et al., 2011). We compared the interactions of mutant TBP with MyoD and NF-YA under the same immunoprecipitation conditions. The comparison revealed that the larger polyQ repeat indeed caused a greater reduction in the association of TBP with MyoD, although it increased the binding of TBP to NF-YA to a great extent (Figure 6C, D). Thus, different polyQ repeats may confer different conformations of TBP, leading to the differential association of mutant TBP with other transcription factors.
Large PolyQ Repeats in TBP Affect MyoD Levels
TBP reportedly interacts with MyoD to stabilize its association with DNA promoter (Heller et al., 1998). We hypothesized that the large polyQ repeat may decrease the binding of TBP to MyoD, thereby reducing its association with the DNA promoter and promoting the degradation of MyoD. To test this hypothesis, we first examined whether the half-life of MyoD is shortened in the presence of TBP-105Q because of an accelerating degradation. Cycloheximide-chase experiments on cultured C2C12 cells, a muscle cell line, revealed that MyoD was degraded faster in the presence of mutant TBP (Figure S6A, B). Inhibiting the proteasome with MG132 could increase the level of MyoD in TBP-105Q-transfected cells (Figure S6C), suggesting that MyoD is cleared out by the ubiquitin-proteasome-system. Unlike other interacting proteins that can be sequestered into nuclear TBP inclusions, MyoD appears to be diffuse even in the presence of TBP aggregates in transfected cells, indicating that soluble mutant TBP interacts with MyoD (Figure S6D). Because the association of TBP with MyoD stabilizes MyoD (Heller et al., 1998) and the large polyQ repeat reduces this association, the decreased association of mutant TBP with MyoD may promote the degradation of MyoD and reduce its half-life.
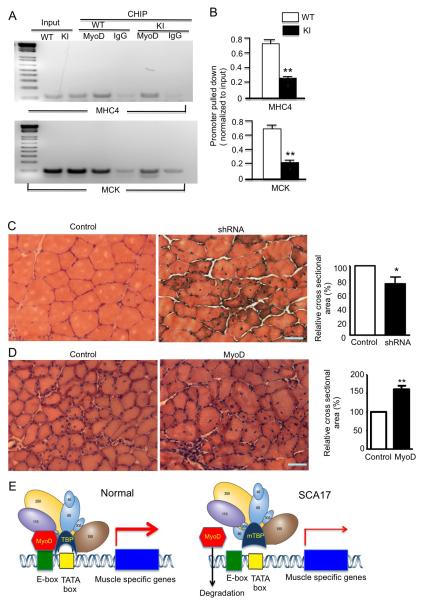
We suspected the decreased interaction between mutant TBP and MyoD might affect the association of MyoD with DNA promoter to reduce its transcriptional activity. To test this idea, we performed a chromatin immunoprecipitation (ChiP) assay that has been widely used to measure the DNA-protein interaction. Compared with normal TBP, mutant TBP indeed reduced the association of MyoD with the DNA promoter for the expression of the muscle-specific proteins MHC4 and MCK (Figure 7A). Quantification of ChiP assays validated the decreased association of MyoD with the promoter in the presence of mutant TBP (Figure 7B).
Figure 7. Mutant TBP with 105Q Causes MyoD Dysfunction.
(A) In vivo ChIP assay examining MyoD occupancy of the DNA promoters for mouse myosin heavy chain 4 (MHC4) and mouse muscle creatine kinase (MCK) in SCA17 KI and WT TA muscle tissues. Cross-linked chromatin materials were precipitated by anti-MyoD antibody, and then subjected to polymerase chain reaction with primers for the promoter regions of mouse MHC4 or MCK that bind MyoD. (B) Quantification of PCR results from ChIP assays evaluating MyoD association with DNA promoters (mean ± SEM, n = 3). ChIP values relative to WT for the MHC4 and MCK promoters in TA muscles from WT and SCA17 KI mice at 3 months of age are presented (n = 5 **p < 0.01). (C) H&E cross-section staining of tibialis anterior (TA) muscles from 3-month-old WT mice, which had been injected with lentiviral (Lv) Lv-GFP or Lv-MyoD-shRNA for 45 days. MyoD-shRNA-injected TA muscle showing a decrease in myofibril size in the viral-injected area. Quantification of cross-sectional area of myofibrils that were injected with Lv-GFP (control) or Lv-MyoD-shRNA. Values are mean ± SEM of data from 5 mice in each group. Values are relative to control cross-sectional area (* p < 0.05, n = 150). (D) H&E cross-sectional staining of tibialis anterior (TA) muscles from 3-month-old WT mice 1.5 months after injection of AAV1-GFP or AAV1-MyoD, which is under the control of human actin 1 (ACT1) promoter. Scale bars: 40 μm. AAV1-MyoD-injected TA muscle showing an increase in myofibril size in the viral-injected area compared to the control side. Quantification of cross-sectional area of myofibrils (**p < 0.01, n = 500). Values are relative to the cross-sectional area of control muscle injected with AAV1-GFP. (E) Summary cartoon of the effect of the large polyQ repeat (105Q) on the association of mutant TBP with MyoD. MyoD normally binds the E-box of DNA promoters and links with other transcriptional factors to trigger the expression of muscle-specific genes. When mutant TBP carries a large polyQ repeat, its decreased binding to MyoD diminishes the association of MyoD with DNA promoter and transcriptional complex, resulting in the reduced expression of muscle genes and associated muscle atrophy.
To test whether MyoD levels are indeed critical for the muscle atrophy seen in SCA17 mice, we used lentiviral or AAV1 vectors to suppress MyoD via siRNA or overexpress MyoD in skeletal muscle tissues by focal injection of these viral vectors into the skeletal muscle of mouse hind legs. We first confirmed that the MyoD viruses could alter the expression of MyoD in the muscle cell line C2C12 (Figure S7A, B). These vectors coexpressed with EGFP or ECFP so the injected muscle area could be defined (Figure S7C). The expression of lentiviral siRNA in wild-type mouse muscle for 1.5 months led to a reduction in muscle cross-sectional areas, a phenotype similar to muscle degeneration (Figure 7C). An even more important experiment was testing whether overexpression of MyoD could alleviate muscle degeneration in SCA17 KI mice. Thus, mouse skeletal muscles were injected with AAV1-MyoD for 1.5 months, and we found that, compared with AAV1-GFP (control) injection, AAV1-MyoD significantly increased the muscle cross-sectional areas or reduced muscle degeneration in SCA17 KI mice at 3 months of age (Figure 7D). Since MyoD binds the E-box sequence (CANNTG) in promoters of downstream muscle target genes to drive the transcription of these muscle-related genes in collaboration with other transcription factors, we propose that the large polyQ repeat decreases the association of TBP with MyoD, thereby weakening the association of MyoD with DNA, resulting in the reduction of muscle-specific proteins and causing muscle degeneration, a peripheral phenotype that is specifically mediated by large polyQ repeats in SCA17 (Figure 7E).
DISCUSSION
One mystery of polyglutamine (polyQ) neurodegenerative diseases is how large polyQ repeats in these diseases cause pathology and symptoms in juvenile patients that are different from those in adult-onset patients, who express mutated proteins with intermediate polyQ expansion, often shorter than 55 glutamines. A typical example is Huntington’s disease (HD), which is caused by polyQ expansion in the ubiquitously expressed protein, huntingtin. Most HD patients carry 42-55 CAG repeats in the HD gene and show symptoms in the middle of life. About 10 percent of HD cases carry larger polyQ repeats (>65) and develop symptoms younger than the age of 20 years. Although both the early- and adult-onset forms of HD are characterized by dementia, movement disorder, and personality changes, juvenile HD patients do not display chorea as adult-onset patients. Instead, juvenile HD patients are often rigid and stiff and exhibit severe cognitive dysfunction and seizure that is not seen in adult HD patients (Vargas et al., 2003; Squitieri et al., 2006). In the brains of juvenile HD patients, there are more nuclear aggregates and fewer neuropil aggregates than in adult HD brains (DeFiglia et al., 1997; Gutekunst et al., 1998), and also more widespread degeneration, suggesting that polyQ repeat lengths mediate different pathogenic pathways.
Like HD and other polyQ diseases, most SCA17 patients carry 43-48 CAGs in the TBP gene with reduced penetrance or late age of onset of symptoms. Although these symptoms are characterized as ataxia, dementia, and involuntary movements, larger repeats (>63 CAG) apparently cause mental deterioration, gait abnormalities, muscle weakness, and retarded growth at age 13 or younger (Koide et al., 1999; Maltecca et al., 2003). The clinical symptoms in juvenile SCA17 patients are similar to those in SAC17 KI mice, also suggesting that glutamine repeat length can determine the nature of the pathology in polyQ diseases. It has been reported that a large CAG repeat can produce polyalanine and polyserine peptides via RAN translation or repeat associated non-ATG translation (Cleary & Ranum, 2013). However, our conditional SCA17 KI mice do not show any symptoms and muscle phenotypes when expanded polyQ repeat expression is prevented by the stop codon, supporting the idea that the muscle degeneration is caused by mutant TBP with expanded polyQ repeats.
The rarity of SCA17 patients has allowed for only limited documentation of histological examinations from very few postmortem SCA17 patient brains, and the peripheral pathology of SCA17 patients has gone unreported. By establishing SCA17 KI mice expressing a large polyQ repeat (105Q), we uncovered strong evidence for muscle degeneration that is apparently caused by the expression of mutant TBP in muscle cells and its adverse effect on MyoD. First, we found no evidence for degeneration of the neuromuscular junction in those SCA17 KI mice that begin to show muscle degeneration at 3 months of age. Second, the selective expression of mutant TBP in muscles in muscle-KI mice also caused muscle degeneration and clinical phenotypes. Third, direct injection of viral vector expressing mutant TBP into the mouse muscle tissue led to muscle degeneration. Finally, muscle degeneration in SCA17 mice could be rescued by overexpressing MyoD, a muscle tissue-specific transcription factor.
Several different transgenic SCA17 rodent models expressing polyQ repeats (64Q, 71Q, and 109Q) have been established and show age-dependent ataxia phenotypes, with no reports of muscle atrophy (Chang et al., 2011; Kelp et al., 2013; Portal et al., 2013). However, these rodent models express mutant TBP under the control of the neuronal prion promoter (Portal et al., 2013; Kelp A et al., 2013) or the Purkinje-specific protein (Pcp2/L7) gene promoter (Chang et al., 2011), suggesting that the muscle degeneration seen in SCA17 requires the expression of mutant TBP in muscle cells. In support of this idea, our previously established SCA17 mouse model, which expresses mutant TBP-105Q in neuronal cells, does not develop the severe muscle atrophy and symptoms of SCA17 KI mice (Huang et al., 2011). The muscle phenotype seen in our SCA17 KI mice apparently depends on the expression of TBP with a larger polyQ repeat (105Q). First, there have been no reports of muscle atrophy or related symptoms in adult-onset SCA17 patients who carry polyQ repeats shorter than 55 glutamines. Second, by expressing TBP containing different polyQ repeat lengths in mouse skeletal muscle, we found that only the large repeat induces severe muscle degeneration. We suspected the large polyQ repeat could cause a unique protein conformational change that confers a specific gain or loss function to TBP. To test this idea, we compared the interactions of mutant TBP containing different polyQ repeats with the neuronal transcription factor NF-YA and the muscle transcription factor MyoD. The comparison shows that the large polyQ repeat can increase the interaction of mutant TBP with NF-YA, but reduces the interaction of TBP with MyoD. This finding provides a biochemical basis to explain why the large polyQ repeat in TBP can induce unique pathological changes in the peripheral tissues that may hardly be seen in adult patients, who carry mutant TBP with shorter expanded polyQ repeats.
Our studies identified MyoD as an important target in SCA17 muscle atrophy. MyoD is believed to be activated during muscle cell differentiation and regeneration (Hawke and Garry, 2001). Yet despite its critical role in muscle differentiation, genetic depletion of the mouse MyoD gene does not lead to obvious muscle atrophy in mice (Rudnicki et al., 1992). Muscle genesis and differentiation during early embryonic stages are controlled by different transcription factors; the basic helix-loop-helix myogenic regulatory factors MyoD, Myf5, myogenin, and MRF4 play critical roles in skeletal muscle development. After muscle cell fibers have formed, MyoD in satellite cells is thought to be activated only during the regeneration of muscle cells or in response to muscle injury (Hawke and Garry, 2001). In addition, MyoD is present in myotubes, and its level in myotubes is critical for maintaining muscle size and regeneration (Dedkov et al., 2003; Ishido et al., 2004; Legerlotz and Smith 2008; Tanaka et al., 2008; Andrews et al., 2010). Myogenesis is known to require switching of the core promoter recognition complex, TFIID, and MyoD targets specific TBP-associated factor (TAF3) and TBP-related factor (TRF3) to activate myogenin transcription (Deato and Tjian, 2007; Deato et al., 2008). Although little is known about the function of MyoD and its relationship with TBP in adult muscle cells, our studies suggest that mutant TBP affects muscle only in adult mice. This is because our SCA17 mice, like juvenile SCA17 patients with 63Q or 66Q in TBP (Koide et al., 1999; Maltecca et al., 2003), are born normally without muscle weakness, with muscle degeneration occurring several months later. Mutant TBP accumulates in muscle nuclei and becomes misfolded with age, consistent with its late-onset effect on MyoD function in adult muscles.
TBP levels are tightly regulated, since floxed SCA17 KI mice, which express one copy of the normal TBP gene, or are equivalent to heterozygous TBP knockout mice, show the same levels of normal TBP as wild-type TBP mice, which carry two copies of the normal TBP gene (Friedman et al., 2007; Friedman et al., 2008; Yang et al., 2014). Similarly, in SCA17 KI mouse brain and muscle, expressing one copy of mutant TBP can downregulate the level of endogenous normal TBP (Figure 1 and Figure S1). This fact suggests that, while mutant TBP preserves some important normal functions during early development, it can cause a gain of toxic function and also reduces the level of normal TBP to induce a loss of function of normal TBP. TBP is known to interact with MyoD to stabilize its association with DNA promoter (Heller et al., 1998), and the stability of MyoD is critical for muscle cell function. For example, TNFalpha destabilizes MyoD and induces apoptosis of muscle cells (Degens, 2010). In adult muscle cells, when polyQ expansion in TBP reduces its association with MyoD and when normal TBP is also decreased because of the expression of mutant TBP, more MyoD is likely to dissociate from a protein-DNA complex, and then be subjected to degradation by the ubiquitin-proteasome system. The delicate regulation of MyoD by TBP may explain the specific role of TBP in muscle cells and the adverse effect of mutant TBP on gene transcription in muscle cells. Because of its low level in myotubes, the function of MyoD in adult muscle cells may be particularly vulnerable to mutant TBP, especially when the function of TBP is affected by a large glutamine repeat.
There are differing extents of muscle atrophy in various polyQ diseases. SBMA is characterized by adult-onset muscle weakness and lower motor neuron degeneration, and transgenic mouse models of SBMA also show that muscle atrophy can be caused by the expression of mutant protein in muscle (Cortes et al., 2014; Lieberman et al., 2014). HD mouse models also show muscle atrophy at the late disease stages, which may be caused by defective energy metabolism and oxidation associated with the re-expression of the HDAC4-DACH2-myogenin axis (Mielcarek et al., 2015). The muscle atrophy in our SCA17 KI mice indicates that a large polyQ expansion in TBP selectively alters its specific function to affect MyoD in muscle cells. Because this dysfunction is dependent on the length of the polyQ repeat in TBP, our findings suggest that polyQ repeat lengths can differentially alter the function of the polyQ proteins, offering an explanation for the differential pathology and symptoms in adult-onset and early-onset polyQ diseases, which are caused by different polyQ repeat lengths. Also, characterization of animal models expressing large polyQ repeats should not be limited to the brain and needs to consider the possible peripheral pathology. Moreover, our findings also suggest that the treatment of juvenile polyQ diseases involving large polyQ repeats should take into account the widespread and severe pathology that may occur in peripheral tissues.
EXPERIMENTAL PROCEDURES
Mice
All mice were bred and maintained in the animal facility at Emory University under specific pathogen-free conditions in accordance with institutional guidelines of the Institutional Animal Care and Use Committee at Emory University. The TBP105Q floxed mice and TBPKI-NestinCre mice were generated as described before (Huang et al., 2011). To generate germline knock-in mice, heterozygous TBP105Q floxed mice were crossed with EIIa-Cre transgenic mice (The Jackson Laboratory, B6.FVB-Tg (EIIa-Cre) C5379Lmgd/J); this line carries a Cre transgene under the control of the adenovirus EIIa promoter that targets expression of Cre recombinase to the early mouse embryo. Cre-mediated recombination occurs in a wide range of tissues, including the germ cells that transmit the genetic alteration to progeny. To generate muscle TBP knock-in mice, heterozygous TBP105Q floxed mice were crossed with CKmm-Cre transgenic mice (The Jackson Laboratory, B6.FVB (129S4)—Tg(Ckmm-cre) 5Khn/J). These transgenic mice have the Cre recombinase gene driven by the muscle creatine kinase (MCK or Ckm) promoter. Cre activity is observed in skeletal and cardiac muscle. When bred with mice containing a loxP-flanked mutant TBP sequence, Cre-mediated recombination will result in skeletal and cardiac muscle turn-on of the mutant TBP gene. TBP105Q floxed mice were also crossed with Pax7-Cre mice (The Jackson Laboratory, B6;129-Pax7tm1(cre)Mrc/J) to generate mice that express TBP105Q in muscle satellite cells. Primers used for genotyping are listed here. For mutant TBP: forward: 5′-CCA CAG CCT ATT CAG AAC ACC-3′; reverse: 5′-AGA AGC TGG TGT GGC AGG AGT GAT-3′. For Cre: forward 5′-GCG GTC TGG CAG TAA AAA CTA TC-3′; reverse: 5′-TGT TTC ACT ATC CAG GTT ACG G-3′.
Collection of Muscles and Morphometric Measurements
Mice were sacrificed, and tibialis anterior (TA) muscles were removed, embedded in OCT mounting medium, and frozen in 2-methybutane cooled in liquid nitrogen. Serial cross-sections were collected onto gelatin-subbed slides at 400- to 500-μm intervals along the entire length of the muscle and analyzed histologically by H&E staining. Serial 15-μm sections were collected along the entire length of the muscle and stained with H&E. Analyses and photography were performed using a Zeiss Axioplan microscope equipped with a video camera and AxioVision Rel. 4.8 software. To quantitatively analyze cross-sectional area, we used imaging software ImageJ (NIH). The cross-sectional area is determined in the muscle belly, and anatomical landmarks of each muscle were used to find the same region in different samples.
Statistical Analysis
Results generated from 3 or more independent experiments are expressed as the mean±SD and were analyzed for statistical significance using GraphPad Prism 5 software. Statistical significances were calculated based on t-tests, and a P-value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by NIH grants (AG19206 and NS041449 to XJL, NS095279 and NS0405016 to SHL). This research project was supported in part by the Viral Vector Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. We thank Heju Zhang at the Transgenic Mouse and Gene Targeting core at Emory for generating the TBP floxed mouse line, Duc M. Duong and Nicholas T. Seyfried at ENNCF Proteomics Core at Emory University for mass spectrometry analysis, Yi Hong at Robert P. Apkarian Integrated Electron Microscopy Core for electron microscopy analysis, Lin Mei at Georgia Health Sciences University for providing C2C12 cells and the human ACT1 promoter, Grace K. Pavlath for advice, Benjamine J. Redpath and Naureen Mith for technical assistance, and Cheryl Strauss for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information includes Expanded Experimental Procedures, 5 figures, and 4 tables.
Author Contributions
S.H., S.Y., S.L. and X.J.L. designed research. S.H. and S.Y. performed experiments. S.H., S.Y., S.L. and X.J.L. analyzed the data and wrote the manuscript. J.G. and Sen Yan assisted with generating reagents. M.A.G. assisted with mouse breeding.
REFERENCES
- Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Laccone F, Rolfs A, Wullner U, Bosch S, Peters H, Liebscher S, Scheible M, Epplen JT, Weber BH, et al. Trinucleotide repeat expansion in SCA17/TBP in white patients with Huntington’s disease-like phenotype. J Med Genet. 2004;41:230–232. doi: 10.1136/jmg.2003.015602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni AC, Takahashi-Fujigasaki J, Maltecca F, Foncin JF, Servadio A, Casari G, D’Adamo P, Maletta R, Curcio SA, De Michele G, et al. Behavioral disorder, dementia, ataxia, and rigidity in a large family with TATA box-binding protein mutation. Arch Neurol. 2004;61:1314–1320. doi: 10.1001/archneur.61.8.1314. [DOI] [PubMed] [Google Scholar]
- Bruning JC, Michael MD, Winnay JN, Hayashi T, Horsch D, Accili D, Goodyear LJ, Kahn CR. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- Chang YC, Lin CY, Hsu CM, Lin HC, Chen YH, Lee-Chen GJ, Su MT, Ro LS, Chen CM, Hsieh-Li HM. Neuroprotective effects of granulocyte-colony stimulating factor in a novel transgenic mouse model of SCA17. Journal of neurochemistry. 2011;118:288–303. doi: 10.1111/j.1471-4159.2011.07304.x. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22(R1):R45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, Ling SC, Guo LT, Hung G, Tsunemi T, Ly L, Tokunaga S, Lopez E, Sopher BL, Bennett CF, et al. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron. 2014;82:295–307. doi: 10.1016/j.neuron.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Molecular cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes & development. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedkov EI, Kostrominova TY, Borisov AB, Carlson BM. MyoD and myogenin protein expression in skeletal muscles of senile rats. Cell Tissue Res. 2003;311:401–416. doi: 10.1007/s00441-002-0686-9. [DOI] [PubMed] [Google Scholar]
- Degens H. The role of systemic inflammation in age-related muscle weakness and wasting. Scandinavian journal of medicine & science in sports. 2010;20:28–38. doi: 10.1111/j.1600-0838.2009.01018.x. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Di Marco S, Mazroui R, Dallaire P, Chittur S, Tenenbaum SA, Radzioch D, Marette A, Gallouzi IE. NF-kappa B-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol Cell Biol. 2005;25:6533–6545. doi: 10.1128/MCB.25.15.6533-6545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MJ, Shah AG, Fang ZH, Ward EG, Warren ST, Li S, Li XJ. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat Neurosci. 2007;10:1519–1528. doi: 10.1038/nn2011. [DOI] [PubMed] [Google Scholar]
- Friedman MJ, Wang CE, Li XJ, Li S. Polyglutamine expansion reduces the association of TATA-binding protein with DNA and induces DNA binding-independent neurotoxicity. J Biol Chem. 2008;283:8283–8290. doi: 10.1074/jbc.M709674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Ferrante RJ, Li XJ, Hersch SM. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. Journal of applied physiology (Bethesda, Md : 1985) 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- Heller H, Bengal E. TFIID (TBP) stabilizes the binding of MyoD to its DNA site at the promoter and MyoD facilitates the association of TFIIB with the preinitiation complex. Nucleic acids research. 1998;26:2112–2119. doi: 10.1093/nar/26.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Lenzner C, Leneuve P, Zaoui R, Hamard G, Vaulont S, Le Bouc Y. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic acids research. 2000;28(21):e92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ling JJ, Yang S, Li XJ, Li S. Neuronal expression of TATA box-binding protein containing expanded polyglutamine in knock-in mice reduces chaperone protein response by impairing the function of nuclear factor-Y transcription factor. Brain. 2011;134:1943–1958. doi: 10.1093/brain/awr146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido M, Kami K, Masuhara M. In vivo expression patterns of MyoD, p21, and Rb proteins in myonuclei and satellite cells of denervated rat skeletal muscle. Am J Physiol Cell Physiol. 2004;287:C484–493. doi: 10.1152/ajpcell.00080.2004. [DOI] [PubMed] [Google Scholar]
- Kelp A, Koeppen AH, Petrasch-Parwez E, Calaminus C, Bauer C, Portal E, Yu-Taeger L, Pichler B, Bauer P, Riess O, et al. A novel transgenic rat model for spinocerebellar ataxia type 17 recapitulates neuropathological changes and supplies in vivo imaging biomarkers. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:9068–9081. doi: 10.1523/JNEUROSCI.5622-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide R, Kobayashi S, Shimohata T, Ikeuchi T, Maruyama M, Saito M, Yamada M, Takahashi H, Tsuji S. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum Mol Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerlotz K, Smith HK. Role of MyoD in denervated, disused, and exercised muscle. Muscle Nerve. 2008;38:1087–1100. doi: 10.1002/mus.21087. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Yu Z, Murray S, Peralta R, Low A, Guo S, Yu XX, Cortes CJ, Bennett CF, Monia BP, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell reports. 2014;7:774–784. doi: 10.1016/j.celrep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltecca F, Filla A, Castaldo I, Coppola G, Fragassi NA, Carella M, Bruni A, Cocozza S, Casari G, Servadio A, et al. Intergenerational instability and marked anticipation in SCA-17. Neurology. 2003;61:1441–1443. doi: 10.1212/01.wnl.0000094123.09098.a0. [DOI] [PubMed] [Google Scholar]
- Mielcarek M, Toczek M, Smeets CJ, Franklin SA, Bondulich MK, Jolinon N, Muller T, Ahmed M, Dick JR, Piotrowska I, et al. HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy. PLoS genetics. 2015;11:e1005021. doi: 10.1371/journal.pgen.1005021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Jeong SY, Uchihara T, Anno M, Nagashima K, Nagashima T, Ikeda S, Tsuji S, Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Portal E, Riess O, Nguyen HP. Automated home cage assessment shows behavioral changes in a transgenic mouse model of spinocerebellar ataxia type 17. Behavioural brain research. 2013;250:157–165. doi: 10.1016/j.bbr.2013.04.042. [DOI] [PubMed] [Google Scholar]
- Rolfs A, Koeppen AH, Bauer I, Bauer P, Buhlmann S, Topka H, Schols L, Riess O. Clinical features and neuropathology of autosomal dominant spinocerebellar ataxia (SCA17) Ann Neurol. 2003;54:367–375. doi: 10.1002/ana.10676. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Squitieri F, Ciarmiello A, Di Donato S, Frati L. The search for cerebral biomarkers of Huntington’s disease: a review of genetic models of age at onset prediction. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2006;13:408–415. doi: 10.1111/j.1468-1331.2006.01264.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Yamaguchi A, Fujikawa T, Sakuma K, Morita I, Ishii K. Expression of mRNA for specific fibroblast growth factors associates with that of the myogenic markers MyoD and proliferating cell nuclear antigen in regenerating and overloaded rat plantaris muscle. Acta Physiol (Oxf) 2008;194:149–159. doi: 10.1111/j.1748-1716.2008.01866.x. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Toyoshima Y, Yamada M, Onodera O, Shimohata M, Inenaga C, Fujita N, Morita M, Tsuji S, Takahashi H. SCA17 homozygote showing Huntington’s disease-like phenotype. Ann Neurol. 2004;55:281–286. doi: 10.1002/ana.10824. [DOI] [PubMed] [Google Scholar]
- Vargas AP, Carod-Artal FJ, Bomfim D, Vazquez-Cabrera C, Dantas-Barbosa C. Unusual early-onset Huntingtons disease. Journal of child neurology. 2003;18:429–432. doi: 10.1177/08830738030180061301. [DOI] [PubMed] [Google Scholar]
- Yang S, Huang S, Gaertig MA, Li XJ, Li S. Age-dependent decrease in chaperone activity impairs MANF expression, leading to Purkinje cell degeneration in inducible SCA17 mice. Neuron. 2014;81:349–365. doi: 10.1016/j.neuron.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J Biol Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.