SUMMARY
BIBR1532 is a highly specific telomerase inhibitor, however the molecular basis for inhibition is unknown. Here we present the crystal structure of BIBR1532 bound to Tribolium castaneum catalytic subunit of telomerase (tcTERT). BIBR1532 binds to a conserved hydrophobic pocket (FVYL motif) on the outer surface of the thumb domain. The FVYL motif is near TRBD residues that bind the activation domain (CR4/5) of hTER. RNA binding assays show that the human TERT (hTERT) thumb domain binds the P6.1 stem loop of CR4/5 in vitro. hTERT mutations of the FVYL pocket alter wild type CR4/5 binding and cause telomere attrition in cells. Furthermore, the hTERT FVYL mutations V1025F, N1028H and V1090M are implicated in dyskeratosis congenita and aplastic anemia, further supporting the biological and clinical relevance of this novel motif. We propose that CR4/5 contacts with the telomerase thumb domain contribute to telomerase ribonucleoprotein (RNP) assembly and promote enzymatic activity.
INTRODUCTION
Telomerase is a ribonucleoprotein, reverse transcriptase (Gillis et al, 2008) responsible for replicating the ends of chromosomes and maintaining genome integrity. The tcTERT structure consists of four distinct domains (TRBD, fingers, palm and thumb) organized into a ring configuration creating a large interior cavity for RNA template and telomeric DNA binding during telomere elongation (Gillis et al, 2008; Mitchell et al, 2010). The recent EM structure of human telomerase shows that the TERT ring can undergo significant rigid conformational changes allowing for an open and closed configuration possibly required for telomerase ribonucleoprotein (RNP) assembly (Sauerwald et al, 2013). TERT closed-ring configuration is mediated by extensive interactions between the TRBD and the thumb, the two terminal domains of tcTERT (Gillis et al, 2008; Mitchell et al, 2010). The vertebrate TRBD facilitates telomerase ribonucleoprotein assembly via CR4/5 (Bley et al, 2011; Huang et al, 2014) and template boundary element (TBE) (Harkisheimer et al, 2013; Rouda & Skordalakes, 2007) binding.
Strategies to inhibit telomerase generally target either TERT or TER (Harley, 2008; Shay & Keith, 2008; Shay & Wright, 2002), although indirect inhibitors targeting telomerase substrates and regulators are also used to a lesser extent, such as G-guadruplex binders (Hurley et al, 2000; Neidle & Read, 2000). Direct inhibitors of telomerase catalytic activity include both nucleoside as well as non-nucleoside inhibitors (Fletcher et al, 2001; Fletcher et al, 1996; Gomez et al, 1998; Hajek et al, 2005; Liu et al, 2007; Murakami et al, 1999; Strahl & Blackburn, 1994; Yamaguchi et al, 2000; Yamaguchi et al, 2006; Yegorov et al, 1997; Yegorov et al, 1996). BIBR1532 (2-[(E)-3-naphtalen-2-yl-but-2-enoylamino]-benzoic acid) is a non-nucleosidic, non-competitive, small molecule inhibitor of telomerase that is widely and routinely used in studies of telomerase function. BIBR1532 is a highly selective inhibitor of telomerase that has a direct antiproliferative effect on leukemia cells but not in normal hematopoietic stem cells (Damm et al, 2001; El-Daly et al, 2005; Pascolo et al, 2002). Despite extensive use of BIBR1532 in biochemical, kinetic and in vivo studies, its precise mechanism of inhibition of telomerase remains unclear.
The data presented here provides an atomic view of the interaction of TERT with the small molecule inhibitor BIBR1532. It identifies a novel motif (FVYL) on the thumb domain that can be targeted to specifically inhibit telomerase function. It supports the hypothesis that the thumb and TRBD domains of TERT bind CR4/5, contacts that promote telomerase RNP assembly and possibly stabilize the closed configuration of the TERT ring required for full telomerase function.
RESULTS
BIBR1532 binds to a hydrophobic pocket on the outer surface of the thumb domain
Crystals of the tcTERT - BIBR1532 were prepared by soaking the substrate free tcTERT crystals with BIBR1532. tcTERT crystals bound to BIBR1532 diffracted to significantly higher resolution (2.3 Å, Table 1) than the substrate free enzyme (2.7 Å), thus providing the atomic resolution required for detailed analysis of small molecule binding to a protein. The structure was solved by molecular replacement (MR) using the substrate free enzyme PDB ID: 3DU6. Structural comparisons of the substrate free and BIBR1532 bound tcTERT rings showed an overall RMSD of 0.5Å suggesting no major conformational changes to the protein structure upon inhibitor binding.
Table 1.
Data collection and refinement statistics
| tcTERT – BIBR1532 | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å), β (°) | 117.7, 84.9, 123.3, 116.2 |
| Resolution (Å) | 20 - 2.3 (2.42 - 2.3)* |
| Rsym or Rmerge | 12.3 (47.1) |
| I/σI | 4.3 (1.5) |
| Completeness (%) | 99.6 (98.9) |
| Redundancy | 3.0 (2.9) |
| Refinement | |
| Resolution (Å) | 20 - 2.3 |
| No. of reflections | 19815 |
| Rwork/Rfree | 23.5/27.3 |
| No. atoms | |
| Protein | 9,964 |
| Ligand/ion | 50 |
| Water | 430 |
| Mean B value | 48 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 0.1043 |
Values in parentheses are for highest-resolution shell.
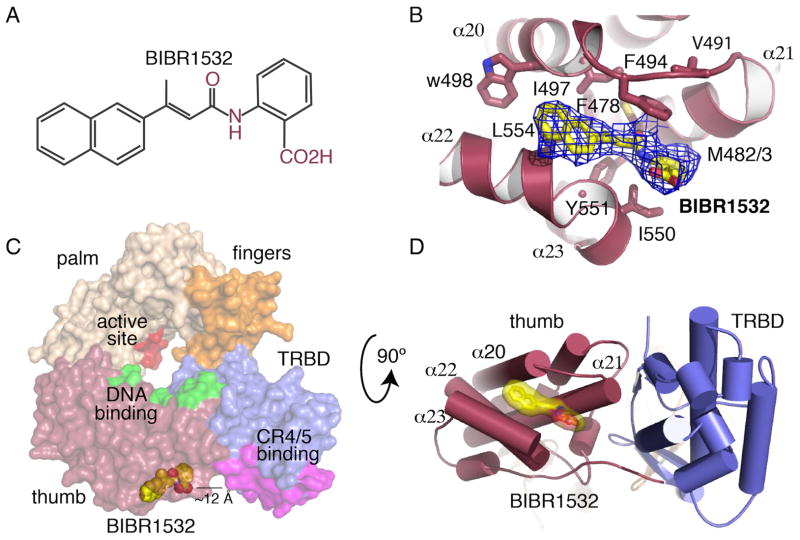
Experimental and simulated annealing (SA) omit maps showed clear, interpretable density for BIBR1532 (Figure 1A) at a shallow but well-defined hydrophobic pocket located on the outer surface of the thumb domain of telomerase and adjacent to the TRBD - thumb interface (Figures 1C and D). This pocket is the result of a small gap, approximately 10 Å wide and 8 Å deep, formed by the organization of the tips and connecting loops of helices α20, α21 and α22, α23 (Figures 1B and D). The gap generated between these two sets of helices arises from an upward rotation of helix α21 with respect to the α22 and α23 plane by approximately 25° (Figure 1D). We refer to this pocket as the FVYL motif/pocket based on the conserved hydrophobic residues that form this motif: F478, V491, Y551 and L554 (Figures 2B–C). The FVYL residues are involved in extensive hydrophobic interactions that stabilize the orientation of the loops and alpha helices surrounding the pocket. F478 is at the center of hydrophobic interactions between helices α20, α21, α22 and α23. V491 pins back the loop connecting α20 and α21 via extensive contacts with L485 and F496. Contacts of V491 with L485 and F496 contribute to the shape of the pocket and keep it in an open configuration. Y551, L445 of α22 interact with L501 of α20 and F478 of α21 respectively also contributing to the positioning of α22 and the shape of the FVYL pocket.
Figure 1. Structure of the tcTERT - BIBR1532 complex.
(A) Molecular structure of BIBR1532. (B) Simulated annealing (SA) omit map (blue mesh) of BBR1532 (yellow stick) bound to the tcTERT thumb domain; residues interacting with the inhibitor are shown in stick. (C) Surface representation of tcTERT showing the active site in red, the thumb DNA binding site in green and the TRBD CR4/5 binding site in magenta. BIBR1532 is shown in yellow spheres (D) Cartoon representation of tcTERT, panel C rotated 90° - BIBR1532 (yellow surface/stick) complex. Panels C and D show BIBR1532 binds in proximity of the TRBD-CR4/5 binding interface.
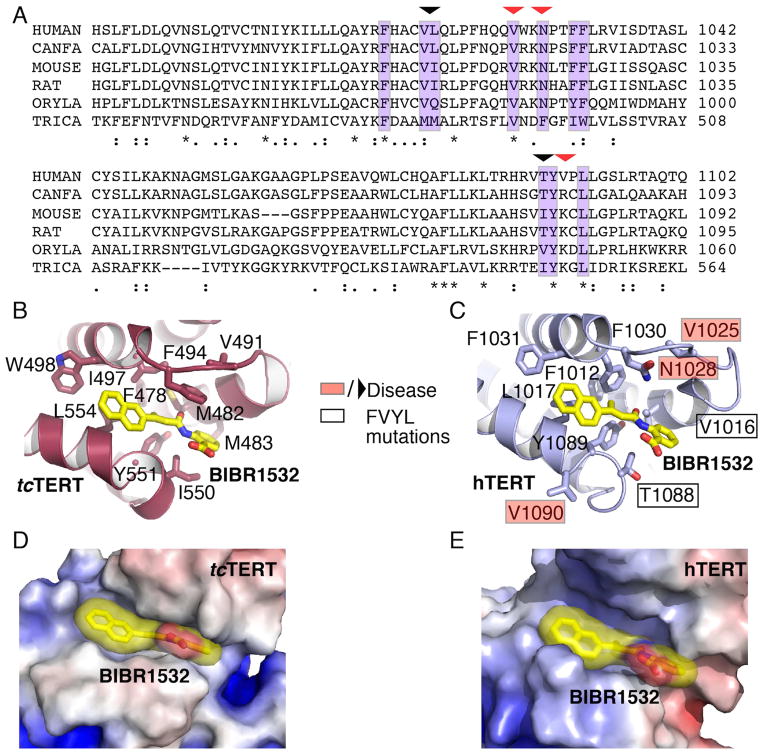
Figure 2. Structural comparison of the tcTERT and hTERT FVYL pockets.
(A) Sequence alignment highlighting (purple color) the residues of tcTERT and hTERT involved in BIBR1532 binding. Residues mutated in this study are shown with an arrow. Naturally occurring missense mutations implicated in human disease are indicated with a red arrow. (B and C) Structural comparison of the tcTERT (crystal structure) and hTERT (PNAS model) FVYL pockets in complex with BIBR1532 (yellow stick); residues involved in inhibitor binding are shown in stick. Black boxes indicate hTERT residues mutated in cell-based and RNA binding assays. Residues associated with human disease are shown in red boxes. (D and E) Electrostatic surface representation of tcTERT and hTERT showing BIBR1532 bound to the FVYL pocket for comparison.
Several conserved and mostly hydrophobic residues line the interior of this pocket, which include M482, M483, F494, I497, W498, I550, Y551 and L554 (Figure 1B). Even though binding of BIBR1532 in this location does not induce significant global conformational changes to the TERT ring (RMSD between the substrate free and BIBR1532 bound structures is 0.5Å), the loop that connects helices α20 and α21 and forms the upper lip of this pocket is moved closer toward the bottom of the pocket by 1Å providing a tighter grip on the small hydrophobic substrate.
The BIBR1532 napthalene group is sandwitched by the hydrophbic side chain of L554, the backbone of G495 and part of F494 (Figures 2C and D). Additional contacts between the napthalene group and the protein are mediated by the side chains of W498 and I497, both of which are located at the back end of the pocket. The methyl moiety of the butynoylamino group points toward the back wall of the pocket and makes van der Walls contacts with the side chains of M482 and Y551. The side chains of F494 and I550 form a hydrophobic pincer that sandwitchs the benzoic group of BIBR1532. Additional contacts with this group involve M483 and the aliphatic part of the side chain of R486. The acid portion of the benzoic moiety of the inhibitor points towards the solvent exposed part of the pocket and does not make contacts with the protein.
Conservation between the human and Tribolium catsaneum BIBR1532, TERT binding pockets
To determine if the FVYL motif is conserved between tcTERT and hTERT, we generated a sequence alignment that consists of tcTERT and vertebrate telomerase sequences including hTERT (Figure 2A). Sequence alignment shows striking similarity in the amino acid composition of the two FVYL pockets. The BIBR binding site residues F478, V491, Y551 and L554 of tcTERT (correspond to F1012, V1025, Y1089 and L1092 of hTERT) are invariant amongst the two telomerases. We also examined the structural conservation of tcTERT and hTERT using the hTERT model published in PNAS (Steczkiewicz et al, 2011). Structural alignment of the tcTERT and hTERT thumb domains shows that the α20, α21, α22 and α23 helices occupy nearly identical positions to create a conserved and solvent exposed hydrophobic pocket (Figures 2B–E). F1012 is located on helix α20, and is at the center of a network of hydrophobic contacts with α21, α22 and α23 (Figure 2C). Y1089 and L1092 make direct hydrophobic contacts with F1012 to stabilize the position of helix α22. On the opposite side of the pocket V1025 holds the loop connecting α20 and α21 in place. Conservation of these key structural elements between tcTERT and hTERT suggests that the organization of the FVYL pocket is conserved across species.
Mutants of the telomerase FVYL pocket lead to telomere attrition
To determine the role of the FVYL motif in telomerase function, we engineered a series of single hTERT mutants designed to perturb the natural substrate binding properties of the FVYL pocket. Some of these mutations were modeled after the naturally occurring mutations N1028H and V1090M associated with human disease. We also identified small solvent exposed hTERT residues in the FVYL pocket that could be converted into bulky aromatic residues without disrupting the overall fold of the protein. These mutations include V1016F, N1028W, T1088F, and T1088W (Figures 2A–C). Modeling of the larger side chains into the hTERT structure indicates that these mutant residues can adopt conformations that obstruct or enhance substrate binding but do not destabilize the hTERT structure.
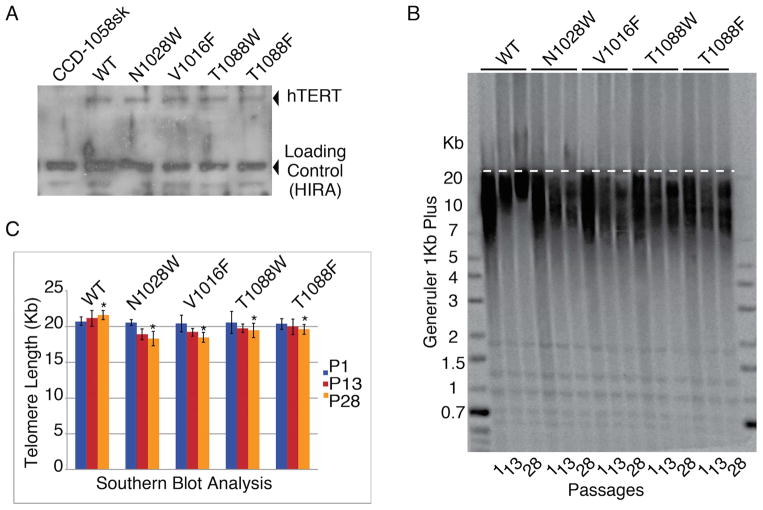
We transfected human fibroblasts (CCD-1058Sk, ATCC) with either wild type (WT) hTERT, mutant hTERT, or empty vector (mock treated). All cellines retained normal morphology, however fibroblasts transfected with WT or mutant hTERT continued dividing even after the mock treated cells senesced. This suggests the mutant proteins retained some level of proliferative activity. Western blot analysis showed that the proteins were expressed at similar levels demonstrating that the mutant proteins were correctly folded and therefore not degraded (Figure 3A).
Figure 3. Southern blots analysis of CCD-1058Sk infected cells with wild type and FVYL, mutant hTERTs.
(A) Western blot showing expression levels of WT and mutant hTERT proteins in CCD-1058Sk cells. (B) Southern blot analysis of FVYL pocket WT and mutant telomerases showing clear loss of telomere length with time (passages 1, 13, and 28). (C) Bar graph analysis of the telomere length for each celline at passages 1, 13 and 28 (TeloTool (MATLAB)) (Gohring et al, 2014). The standard deviation (error) associated with measurements and the statistical significance of the data are shown; *P<0.01 (two-tailed paired Student’s t-test).
Southern blot analysis of DNA isolated from fibroblasts transfected with WT hTERT shows robust telomere lengthening at passages 18 and 23 (Figures 3B and C). In contrast, telomeric DNA isolated from fibroblasts transformed with hTERT mutants (V1016F, N1028W, T10180F and T10180W) showed gradual attrition (Figures 3B and C).
Occlusion of the FVYL pocket of telomerase results in chromosomal abnormalities associated with telomere shortening in vivo
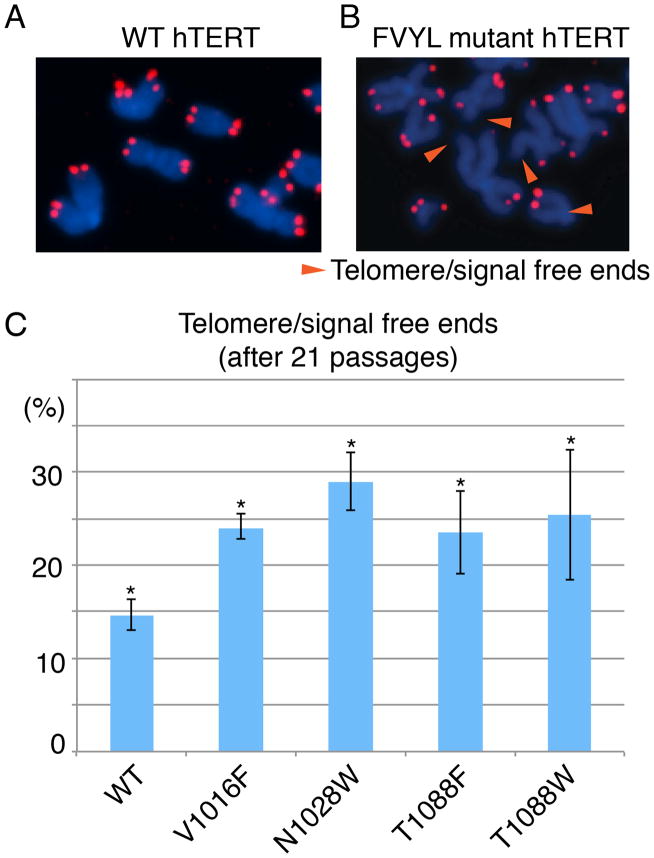
To further establish the role of the FVYL motif in telomerase function and telomere maintenance, we asked if the telomerase mutations V1016F, N1028W, and T1080F lead to chromosomal abnormalities associated with chromosome free ends and fusions. We assayed for defective telomere phenotypes observable by fluorescence in situ hybridization (FISH) analysis, in human fibroblasts (CCD-1058Sk) immortalized with WT and mutant (V1016F, N1028W, T10180F and T10180W) human telomerases. We prepared metaphase spreads and counted the frequency of telomere fusions, fragile telomeres, and telomere/signal free ends (TFE). We observed a significantly increased proportion of TFE (Figures 4A–C), and a slight increase in fragile telomeres, in the mutant cell lines. Cells expressing WT telomerase showed 11% of chromosomes with TFE, compared with 24% in V1016F, 28.0% in N28W, 23.5% in T1088F and 25% in T1088W (Figure 4C).
Figure 4. Fluorescence in situ hybridization (FISH) data of CCD-1058Sk infected cells with wild type and FVYL, mutant hTERTs.
FISH data at passage 21 showing chromosomes of CCD-1058Sk cells transformed with (A) WT hTERT and (B) FVYL pocket mutant hTERT; chromosomes with free telomere ends are indicated with an orange arrow; (C) Percentage of telomere/signal free ends in WT and FVYL mutant hTERT transfected CCD-1058Sk cells. The standard deviation (error) associated with an average of 1,500 chromosome measurements for each cell line carrying the WT or each of the FVYL pocket mutant hTERTs and the statistical significance of the data are shown; *P<0.05 (two-tailed paired Student’s t-test).
TERT mutants that occlude the FVYL pocket exhibit reduced telomerase activity
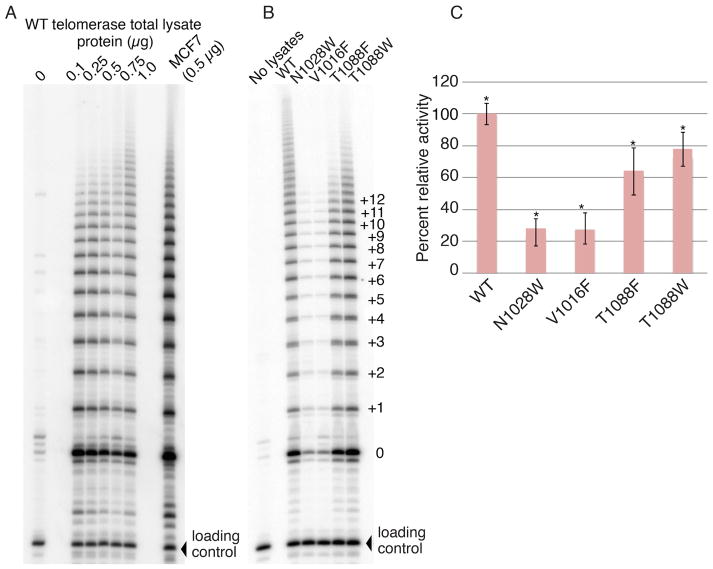
Next, we sought to establish whether the FVYL pocket residues directly influence the catalytic activity of telomerase. To this end we carried out telomeric repeat amplification protocol (TRAP) assays using cell lysates of fibroblasts expressing either WT or one of the FVYL mutant (V1016F, N1028W, T10180F and T10180W) hTERTs proteins. Consistent with the southern and FISH data, the V1016F and N1028W mutations showed significantly reduced telomerase activity (>70%) compared to the WT enzyme (Figures 5A–C). The T1088F and T1088W mutants showed 60–70% of WT TRAP activity even though these mutants do not support robust wild type telomere extension in fibroblast cells (Figures 5A–C).
Figure 5. Telomerase activity assays of CCD-1058Sk infected cells with wild type and FVYL, mutant hTERTs.
(A) TRAP activity of WT telomerase in 0, 0.25, 0.5, 0.75 and 1 μg total lysate protein concentration. (B) TRAP gel showing the activity of WT telomerase alongside the mutants N1028W, V1016F, T1088F, and T1088W in 0.5 μg of total lysate protein (C) Bar graph showing the relative activity, in terms of band intensity, of the wild type and FVYL mutant telomerases. The standard deviation (error) associated with 3 measurements and the statistical significance of the data are shown; **P<0.005 (two-tailed paired Student’s t-test).
The BIBR1532 binding pocket of TERT binds hTER
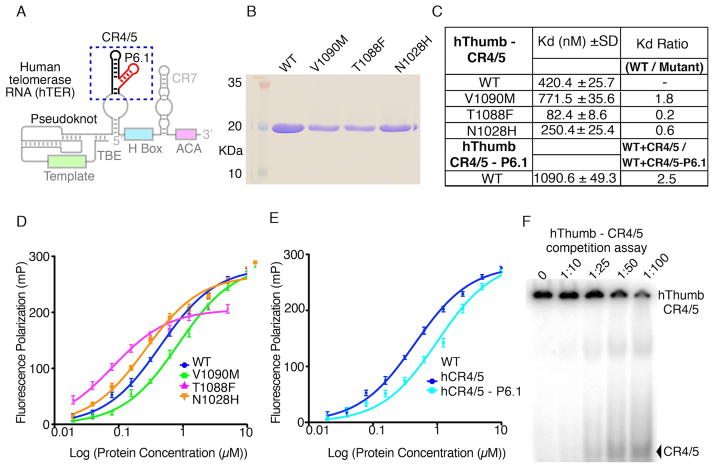
The structural data presented here shows that the FVYL motif is located in proximity (~12 Å) of the CR4/5 binding interface of TRBD (Figure 1C). The proximity of the BRIBR1532 binding pocket of TERT to the TRBD – CR4/5 binding interface suggested that the thumb domain may also be making contacts with the activation domain of CR4/5 and in particular the P6.1 stem loop (Figure 6A). To test this hypothesis, we expressed and purified the WT and mutant (T1088F, N1028H and V1090M) hThumb proteins to homogeneity (Figure 6B). We tested these proteins for CR4/5 binding using fluorescence polarization (FP) assays in the presence of 30-fold excess of cold tRNA competitor and competition assays (Figures D and E). The WT hThumb domain has a Kd of ~0.4μM for the CR4/5, while the mutant hThumb proteins displayed an overall 2–5 fold change in CR4/5 binding (Figure 6C). To determine if P6.1 is contributing to hThumb binding we performed FP assays of WT hThumb with CR4/5 lacking the P6.1 stem loop. The WT hThumb domain showed a 2.5 fold loss of binding affinity for CR4/5 minus P6.1 suggesting that P6.1 plays an important role in TERT-TER association.
Figure 6. Human thumb domain - RNA binding data.
(A) Schematic of the hTER RNA showing conserved motifs in color. The CR4/5 and P6.1 motifs are highlighted with a blue dashed box. The template, pseudoknot and other conserved regions are also shown faded. (B) SDS PAGE gel of the WT and mutant hThumb proteins used in this study. (C) Table of WT and mutant hThumb-TBE binding constants calculated using Prism5 (GraphPad Software). (D) FP data of the CR4/5 with WT, N1028H, T1088F and V1090M hThumb proteins. (E) FP data of the CR4/5 minus P6.1 stem loop with WT hThumb. (F) SDS page gel of the competition assay of hThumb – CR4/5 complex with cold CR4/5.
DISCUSSION
Treatment of cancer cells with BIBR1532 leads to progressive telomere shortening, cell proliferation arrest after several weeks of drug treatment and senescence. Understanding the mechanism of telomerase inhibition by small molecule inhibitors such as BIBR1532 on an atomic level will assist in our effort to identify and develop useful telomerase inhibitors as potential therapeutics for cancer.
Our structural data shows that BIBR1532 binds to a well-defined hydrophobic pocket on the outer surface of the thumb domain. The key residues in structural organization of the FVYL motif are conserved across species and the pocket is solvent accessible for substrate binding (Figures 1D and 2D and E). Although this report is the first structural and biochemical characterization of the FVYL pocket, mutations to FVYL residues have previously been implicated in human disease. The hTERT FVYL mutations V1025F, N1028H, and V1090M (Figures 2A and C) are associated with bone marrow syndrome failure and severe aplastic anemia (Garcia et al, 2007; Yamaguchi et al, 2005). V1025 is a conserved residue of the FVYL motif and plays a critical role in stabilizing the open conformation of the BIBR1532-binding pocket. N1028, forms part of the loop that connects α20 and α21 and the aliphatic part of its side chain makes contacts with the butynoylamino group of BIBR1532. V1090 forms part of a solvent accessible loop that connects helices α22 and α23 and is not involved in direct contact with the rest of the protein. A previous study demonstrated that the V1090M mutation leads to a dramatic loss of telomerase activity (Yamaguchi et al., 2005), which is consistent with our data.
Our cell-based assays clearly show that the FYVL pocket mutations V1016F, N1028W, T1088F, and T1088W lead to telomere shortening (Figures 3B and C), telomere free ends (Figures 4A–C), and loss of telomerase activity (Figures 5B and C). V1016 occupies a rigid position on helix α20 and occupies part of the back wall of the FYVL pocket (Figure 2C). The large hydrophobic side chain of the V1016F mutation most likely occludes the FVYL pocket thus affecting substrate binding and telomerase activity. N1028 forms part of helix α21 and is located at the center of the entry of the FYVL pocket, and therefore the N1028W mutation also disrupts substrate binding and enzyme activity (Figure 2C). In contrast, T1088 is part of the loop that links helices α22 and α23 and therefore this coil is likely more flexible compared to the helical residues V1016 and N1028 (Figure 2C). Thus, the aromatic residues introduced by the T1088F and T1088W mutations may move aside to accommodate substrate binding, resulting in limited loss of activity of these mutants.
Notably, although FVYL mutations reduce hTERT activity and prevent telomere extension, the FVYL mutant hTERT still allowed fibrobalsts to bypass their normal senescence point. This data is in agreement with previous studies on BIBR1532, which show that it takes approximately 120 days for BIBR1532 treated leukemia cells to enter senescence (Damm et al, 2001; El-Daly et al, 2005; Pascolo et al, 2002). The lag time to reaching senescence suggests that telomerase retains some pro-proliferative activity and cancer cells only senesce after extensive telomere attrition. FVYL mutations show similar effects to BIBR1532 treatment on both cell proliferation and hTERT activity, further supporting the role of the FVYL pocket in BIBR1532 binding.
Core functions of the thumb domain of telomerase involve single stranded nucleic acid binding (Hossain et al, 2002; Mitchell et al, 2010) elongation complex formation and polymerase activity (Banik et al, 2002; Hossain et al, 2002; Huard et al, 2003; Mitchell et al, 2010; Peng et al, 2001), characteristics shared among reverse transcriptases and DNA polymerases. In the case of telomerase, DNA binding is mediated by the conserved thumb helix and loop motifs, both of which face the interior cavity of the TERT ring (Gillis et al, 2008; Mitchell et al, 2010). Because of the critical role that the thumb domain plays in DNA binding, we considered the possibility that BIBR1532 binding disrupts the DNA binding pocket. However, the FVYL pocket is located at least 25 Å away from the thumb helix and loop motifs (Figure 1C). Moreover, inhibitor binding to the FYVL pocket does not induce conformational changes that would affect DNA binding by this domain (RMSD 0.5).
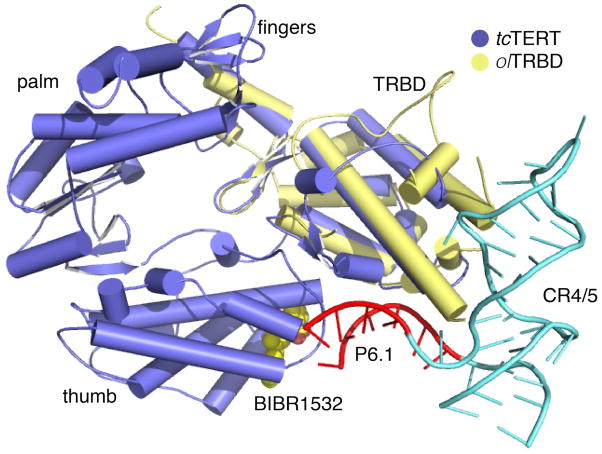
Interestingly, the FVYL pocket is located adjacent to the thumb - TRBD binding interface (Gillis et al, 2008; Mitchell et al, 2010) and in proximity to the CR4/5 RNA-binding surface of TRBD (Bley et al, 2011; Huang et al, 2014) (Figure 1C), which led us to investigate binding between FVYL and the CR4/5. The results of our FP binding assays show that mutations to the hTERT FVYL motif exert a strong influence on binding to the CR4/5 element of hTER. For example, the T1088F hThumb mutant binds the CR4/5 with 5-fold higher affinity than WT hThumb, suggesting that a large aromatic residue in the FVYL motif is positioned to make favorable interactions with substrate RNA. Similarly, the N1028H mutant binds the RNA with approximately 2 fold higher affinity also suggesting favorable interactions between the RNA substrate and the larger pyrimidine-like side chain of histidine. In contrast the V1090M mutant has a reduced affinity for the RNA substrate suggesting interference with the association of hTERT with CR4/5. It is interesting that the above mutations (N1028H, T1088F and V1090M) have opposite effects in RNA binding and yet they all reduce telomerase activity (Yamaguchi et al, 2005). Telomerase activity is tightly regulated so that proper length of telomeres is maintained. Telomere length regulation is directly dependent on the proper assembly of the telomerase RNP complex. Subtle changes in the residue composition of TERT or TER can affect proper telomerase RNP assembly, telomerase activity leading to cell immortalization or senescence. Using an overlay of tcTERT and the structure of the TRDB-CR4/5 complex we identified the stem loop, P6.1 of CR4/5 as a potential candidate for hThumb binding. The RNA binding assays presented here show that WT hThumb binds CR4/5 with 2-fold higher affinity than the CR4/5-P6.1 (Figures 6C–E). An overlay of the tcTERT crystal structure with the olTRBD-CR4/5 (RMSD between tcTRBD and olTRBD is 2.6 Å, which is within the range observed for telomerase TRBDs across species; e.g. 2.3 Å for olTRBD or trTRBD and Tetrahymena thermophila TRBD) places the P6.1 loop region within coordinating distance of the FVYL pocket (Figure 7). The data presented here and the physical proximity of the FVYL pocket to the CR4/5 binding surface of TRBD suggest that P6.1 is the natural substrate of the FVYL pocket.
Figure 7. Model of TERT, CR4/5 binding.
Overlay of the crystal structures of tcTERT (PDB ID: 3DU6) and olTRBD-CR4/5 (PDB ID: 4026). tcTERT is shown in blue, the olTRBD in yellow and CR4/5 in cyan. The P6.1 stem loop of CR4/5 interacting with the FVYL pocket is shown in red.
Contacts between the FVYL pocket and CR4/5 most likely promote telomerase RNP assembly and enzymatic activity. Another possibility is that CR4/5 binding between FVYL and the CR4/5 stabilizes the closed TERT ring configuration also contributing to the enzymatic activity of telomerase. The EM structure of human telomerase shows that the TERT ring can adopt open and closed conformations (Sauerwald et al, 2013). It is possible that contacts between the CR4/5 and the Thumb and TRBD domains stabilize the closed configuration of the TERT ring thus promoting function. Binding of BIB1532 to the FVYL motif disrupts CR4/5 thumb binding and prevents proper telomerase RNP assembly.
METHODS
Protein crystallization and data collection
The tcTERT protein was expressed and purified as described previously (Gillis et al, 2008; Mitchell et al, 2010). The purified tcTERT was concentrated to 40 mg/ml and dialyzed in 10 mM Tris-HCl (pH 7.5), 0.1 M KCl, and 1 mM TCEP prior to crystallographic studies. tcTERT was co-crystallized with single-stranded DNA consisting of 3 telomeric repeats (TCAGG)3 at 18° C by sitting drop, vapor diffusion. The crystallization buffer contained 1.3 M NaNO3 and 0.1 M Tris-HCl (pH 8.5). The fully formed crystals were stabilized in a solution containing 1.3 M NaNO3, 0.1 M Tris-HCl (pH 8.5) and 30% glycerol overnight. To these crystals was then added trace amounts of BIBR1532 powder and the two were incubated for 10 hours prior to harvesting. The inhibitor soaked tcTERT crystals were flash frozen in liquid N2 and data was collected at the NSLS X25 beam line and processed using MOSFLM as implemented in Elves (Holton & Alber, 2004). The model was refined using REFMAC5 (Murshudov et al, 1997). Figures were prepared in PyMOL (http://www.pymol.org).
Cell culture
Human cell culture studies were carried out in CCD-1058sk cells, grown in Dulbucco’s modified eagle medium (DMEM; Gibco) and 10% fetal bovine serum (FBS; Gibco). WT and mutant hTERT genes in the pLU-EF1a-iBLAST vector, which confers blasticidin S resistance, were delivered by infection with lentiviral vectors. Infected cell lines were grown in the presence of 5 μg/mL blasticidin S to ensure that they continued plasmid expression. Cells were trypsinized, counted, and 1×106 cells re-plated 3 times per week.
Western blot analysis
We isolated approximately 3×106 CCD-1058Sk cells of the WT and each mutant telomerase by centrifugation. We lysed the cells in 300 μL of pre-chilled RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% Sodium Deoxycholate, 1% Triton X-100). Lysates were centrifuged for 15 minutes at 20,000 × g and the supernatant saved. We quantified the lysate protein concentration using a Bradford assay (Bradford, 1976), and 25 μg of each sample were run on a polyacrylamide gel. We then blotted the protein onto a polyvinylidene fluoride (PVDF) membrane (Perkin Elmer Health Sciences) by electrophoretic blot at 200 mA for 3 hours at 4° C in 20% Methanol, 125 mM Tris-HCl, 1.25 M glycine and 0.5% SDS. We blocked the membrane with TBST (100 mM Tris-HCl, pH 7.5, 2M NaCl, 0.5% Tween-20) and 5% bovine serum albumin (BSA), washed TBST, and then incubated overnight with monoclonal anti-FLAG antibody M2 produced in mouse (Sigma) in TBST at 4° C. We further washed the membrane with TBST, incubated with TERT Antibody (H-231): sc-7212 (Santa Cruz Biotechnology) for 2 hours, and washed again with TBST. The chemiluminescent HRP reaction was activated using SuperSignal West Pico Substrate (Thermo Scientific), and the image developed with CL-exposure film (Thermo Scientific).
Southern blot analysis of hTERT transfected fibroblasts (CCD-1058Sk)
Genomic DNA was extracted from the WT and FVYL mutant hTERT transfected fibroblasts using a QIAamp DNA-mini Kit (Qiagen). 10 μg of Genomic DNA from each sample was digested overnight at 37° C using 20 U of MboI (NEB) and 20 U of AluI (Invitrogen). Digested DNA was precipitated in 10 mM NaOAc, 1 mM MgCl2, and 75% ethanol overnight at −20° C, then pelleted and resuspended in DNase free water. 3 μg of each sample were run on a 0.5% agarose gel in 1X Tris acetate EDTA (TAE) buffer for 4.5 hours at 120 V. The gel was sequentially washed with 0.25 M HCl, denaturing buffer (0.5 M NaOH, 1.5 M NaCl), and neutralization buffer (0.5 M Tris-HCl, 3 M NaCl, pH 7.5), then blotted onto hybridization transfer membrane (Genescreen Plus™, Perkin Elmer Health Sciences). DNA was cross-linked to the membrane using a UV Stratalinker 1800 (Stratagene), and hybridized with 0.2 nM of 32P labeled DNA probe (TTAGGG)4 overnight at 42° C in 25 mL of church buffer (7% SDS, 0.25M Na2PO4 pH 7.2, 1 mM EDTA, 1% w/v BSA). The membrane was washed with 20 mM Na2PO4 pH 7.2, 1% w/v SDS and 1 mM EDTA, exposed to a phosphor imager and the image developed on a typhoon scanner (GE Healthcare). The telomere length was calculated using the software TeloTool (MATLAB) (Gohring et al, 2014)
Fluorescence in situ hybridization (FISH)
We treated fibroblast cells transfected with WT and mutant telomerase with 100 μg/ml of colcemid for 7 hrs. The cells were grown to 70% confluence after 21 passages on a 10 cm plate. Cells were then trypsinized to detach them from the plate, pelleted and treated in a hypertonic environment (75 mM KCl for 30 mins at 37° C) to rupture them. We fixed the cells in 10 ml of 3:1 methanol:acetic acid solution and stored at 4° C. Metaphase spreads were fixed in 4% formaldehyde (Sigma), treated with 1 mg/ml pepsin in 10 mM glycine, pH 2.0 at 37° C (Sigma) and fixed again with 4% formaldehyde. The slides were then dehydrated with ethanol, air dried, and hybridized with 20 μL of 200 nM telomeric-Cy5 peptide nucleic acid (PNA) probe (TelC-Cy5 - PNA biosciences) (LiCor) according to the manufacturer’s instructions. Slides were counterstained with DAPI and imaged using a Nikon E600 upright microscope.
Telomerase repeat amplification protocol (TRAP) assay
Total cell lysates were prepared using CHAPS lysis buffer (10 mM Tris-HCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM benzamidine, 1 mM phenylmethylsulfonylfloride (PMSF), 5 mM β–mercaptoethanol, 0.5% CHAPS, 10% Glycerol). WT telomerase lysates containing 0 (no lysate), 0.25, 0.5, 0.75 and 1 μg total protein were incubated in TRAP extension buffer ((20 mM Tris-HCl pH 8.3, 63 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 0.005% TWEEN-20, 0.1 μg BSA, 32U RNasin, 1 μM TS primer (5′-AATCCGTCGAGCAGAGTT-3′), 0.1 mM dNTPs, 0.1 mM dithiothreitol (DTT), 0.1 mM PMSF, 0.1mM benzamidine)) for 1 hour at 37° C, then 10 nmol of a 32P labeled loading control (single-stranded, yeast telomeric DNA, 26 bases long) was added to each sample. DNA was isolated by phenylchloroform extraction, and precipitated in 10 mM NaOAc, 1 mM MgCl2, and 75% ethanol overnight at −20° C, and pelleted and resuspended in DNase free water. The DNA samples were resuspended and PCR amplified in 50 μL of TRAP PCR buffer ((1X Coral Loading buffer, 0.2 mM dNTPs, 2 μM CX primer (′5-CCCTTACCCTTACCCTTACCCTAA-3′) containing 1.25 U Taq DNA Polymerase (Qiagen)). We then added 10 μL of formamide loading buffer (95% formamide, 4.5% dH2O, 0.25% bromophenol Blue, and 0.25% xylene cyanol) to each sample and incubated at 95° C for 10 minutes. Samples were run on a 10% polyacrylamide (19:1), 7 M urea gel for 2 hours at 70W. The gel was fixed with 20% ethanol and 5% acetic acid, vacuum dried, and imaged as described above. We subsequently used the above procedure to perform a TRAP assay for the WT and the mutant telomerases at 0.5 μg of total lysate protein. Telomerase activity and standard deviation were calculated from three measurements each. ImageJ was used to determine band intensities on the gel and the three measurements for each band were averaged and standard deviation was calculated for each repeat. The band intensity averages and standard deviations were then normalized to the intensity of the loading control. Finally, the wild type, mean average was set to 100% activity and the mutant averages were calculated as a percentage of it.
Human thumb domain protein expression and purification
The human thumb (hThumb - CTE) domain, consisting of residues 943 to 1132, was cloned into a modified vector expressing a hexahistidine tag (his-tag) followed by a TEV cleavable small ubiquitin-like modifier (SUMO) fusion protein at its N terminus. The protein was overexpressed in E. coli ScarabXpress-1 (T7Lac) cells (Scarab Genomics) at 16° C overnight using 1mM IPTG (isopropyl-β-D-thiogalactopyranoside; Gold Biotechnology). Cells were harvested by centrifugation and resuspened in a buffer containing 95%: 1 M KCl, 25 mM Tric-HCl, 5% glycerol, 0.1 mM benzamidine, 0.1 M phenylmethylsulfonyl fluoride (PMSF), pH 7.5 and 5%: 0.5 M KCl, 25 mM Tric-HCl, 300 mM imidazole, 5% glycerol, 0.1 mM benzamidine, 0.1M phenylmethylsulfonyl fluoride (PMSF), pH 7.5 prior to lysis via sonication. The hThumb protein was then purified over a Ni-Nitrilotriacetic acid resin (Ni.NTA; MCLab) column. The his-SUMO-tag was cleaved by Tobacco Etch Virus endopeptidase (TEV) overnight at 4° C. We further purified the protein over a porous HS resin (Applied Biosystems) column using a gradient of 25 mM Tris.HCl, 0.5 – 1.5 M KCl, 5% glycerol, 1mM dithiothreitol (DTT), pH 7.5. The mutant constructs V1090M, N1028H, and T1088F were overexpressed and purified using the same procedure as the wild type protein.
Fluorescence polarization (FP) assays
We performed FP, hThumb – CR4/5 (CCCGCCTGGAGGCCGCGGTCCGCCGCGAAGAGTTGGGCTCTGTCAGCCGCGGG) and CR4/5 minus P6.1 binding reactions in 15 μl samples using an Envision Xcite Multilabel Plate Reader (Perkin Elmer). The binding reactions were carried out in a buffer containing 20 mM Tris.HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM EDTA, 2 mM DTT, 1 mg/mL BSA, 5% v/v glycerol, and 75 nM cold tRNA competitor (Life technologies - Ambion yeast tRNA). The RNA probes were purchased with a 5′ 6-FAM label from IDT and Dharmacon. The final probe concentration used was 2.5 nM, while the hThumb protein concentration ranged from 0 to 5 μM. The reactions were incubated at room temperature for five minutes and pipetted in triplicate into a black 384 well optiplate (PerkinElmer). The reactions were excited with 480 nm light and the emissions were measured at 535 nm light. The milipolarization values were calculated by the Envision operating software (PerkinElmer). The data was fit and the binding constants were determined with a one-site binding, nonlinear regression model using PRISM 5.0 (GraphPad Software, San Diego California USA, www.graphpad.com).
RNA competition assays
The CR4/5 RNA substrate was purchased from Dharmacon and labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P] ATP then purified through a G25 column. Increasing concentrations of the WT hThumb protein was incubated with 1 nM of the 32P labeled CR4/5 on ice for 30 minutes and in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM dithiothreitol (DTT), 1 mM MgCl2, 5% glycerol, 1 mM EDTA, 30 nM cold tRNA (Life Technologies; Ambion yeast tRNA) competitor, and 0.01% Triton X-100 for 30 min. For the competition assay, increasing concentrations of unlabeled CR4/5 was added to the reactions containing 250 nM of hThumb protein. hThumb concentrations were calculated using a standard Bradford assay. The reactions were then loaded onto 6% DNA retardation PAGE gels and run at 100V for 1 hr. The gels were fixed in 20% methanol–10% acetic acid and then vacuum-dried and exposed using a storage phosphor screen (GE Healthcare).
HIGHLIGHTS.
Cancer therapeutics targeting telomerase
Structure of TERT in complex with the allosteric inhibitor BIBR1532
BIBR1532 binds the thumb domain of TERT
BIBR1532 inhibits telomerase function by disrupting TERT-RNA binding
Acknowledgments
The research was funded by the NIGMS (5 R01 GM088332-03) and The Wistar Cancer Center Support Grant (P30 CA10815).
Footnotes
Author Contribution: E.S., C.B., C.R. H.H. and M.H. designed the experiments; C.B., M.H. and M.S. performed the cell based assays; C.R. carried out the RNA binding assays; M.H. solved the crystal structure with guidance from E.S.; H.H. expressed and purified the hThumb domain; All authors contributed to data analysis and wrote the manuscript. The authors declare no conflict of interest.
Note: The coordinates and structure factors of the tcTERT - BIBR1532 complex have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) databank and the PDB ID number is 5CQG.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banik SS, Guo C, Smith AC, Margolis SS, Richardson DA, Tirado CA, Counter CM. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Molecular and cellular biology. 2002;22:6234–6246. doi: 10.1128/MCB.22.17.6234-6246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley CJ, Qi X, Rand DP, Borges CR, Nelson RW, Chen JJ. RNA-protein binding interface in the telomerase ribonucleoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, Priepke H, Niestroj C, Daiber C, Enenkel B, Guilliard B, Lauritsch I, Muller E, Pascolo E, Sauter G, Pantic M, Martens UM, Wenz C, Lingner J, Kraut N, Rettig WJ, Schnapp A. A highly selective telomerase inhibitor limiting human cancer cell proliferation. The EMBO journal. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daly H, Kull M, Zimmermann S, Pantic M, Waller CF, Martens UM. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood. 2005;105:1742–1749. doi: 10.1182/blood-2003-12-4322. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, Cathers BE, Ravikumar KS, Mamiya BM, Kerwin SM. Inhibition of human telomerase by 7-deaza-2′-deoxyguanosine nucleoside triphosphate analogs: potent inhibition by 6-thio-7-deaza-2′-deoxyguanosine 5′-triphosphate. Bioorganic chemistry. 2001;29:36–55. doi: 10.1006/bioo.2000.1194. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, Salazar M, Chen SF. Human telomerase inhibition by 7-deaza-2′-deoxypurine nucleoside triphosphates. Biochemistry. 1996;35:15611–15617. doi: 10.1021/bi961228v. [DOI] [PubMed] [Google Scholar]
- Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic acids research. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Gohring J, Fulcher N, Jacak J, Riha K. TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic acids research. 2014;42:e21. doi: 10.1093/nar/gkt1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez DE, Tejera AM, Olivero OA. Irreversible telomere shortening by 3′-azido-2′,3′-dideoxythymidine (AZT) treatment. Biochemical and biophysical research communications. 1998;246:107–110. doi: 10.1006/bbrc.1998.8555. [DOI] [PubMed] [Google Scholar]
- Hajek M, Matulova N, Votruba I, Holy A, Tloust’ova E. Inhibition of human telomerase by diphosphates of acyclic nucleoside phosphonates. Biochemical pharmacology. 2005;70:894–900. doi: 10.1016/j.bcp.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A Motif in the Vertebrate Telomerase N-terminal Linker of TERT Contributes to RNA Binding and Telomerase Activity and Processivity. Structure. 2013;21:1–9. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci U S A. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Singh S, Lue NF. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative “thumb” domain. The Journal of biological chemistry. 2002;277:36174–36180. doi: 10.1074/jbc.M201976200. [DOI] [PubMed] [Google Scholar]
- Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ, Lei M. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21:507–512. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard S, Moriarty TJ, Autexier C. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic acids research. 2003;31:4059–4070. doi: 10.1093/nar/gkg437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- Liu X, Takahashi H, Harada Y, Ogawara T, Ogimura Y, Mizushina Y, Saneyoshi M, Yamaguchi T. 3′-Azido-2′,3′-dideoxynucleoside 5′-triphosphates inhibit telomerase activity in vitro, and the corresponding nucleosides cause telomere shortening in human HL60 cells. Nucleic acids research. 2007;35:7140–7149. doi: 10.1093/nar/gkm859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- Murakami J, Nagai N, Shigemasa K, Ohama K. Inhibition of telomerase activity and cell proliferation by a reverse transcriptase inhibitor in gynaecological cancer cell lines. Eur J Cancer. 1999;35:1027–1034. doi: 10.1016/s0959-8049(99)00037-4. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, Garin-Chesa P, Rettig WJ, Damm K, Schnapp A. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J Biol Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- Peng Y, Mian IS, Lue NF. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Molecular cell. 2001;7:1201–1211. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–460. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Keith WN. Targeting telomerase for cancer therapeutics. Br J Cancer. 2008;98:677–683. doi: 10.1038/sj.bjc.6604209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Telomerase: a target for cancer therapeutics. Cancer Cell. 2002;2:257–265. doi: 10.1016/s1535-6108(02)00159-9. [DOI] [PubMed] [Google Scholar]
- Steczkiewicz K, Zimmermann MT, Kurcinski M, Lewis BA, Dobbs D, Kloczkowski A, Jernigan RL, Kolinski A, Ginalski K. Human telomerase model shows the role of the TEN domain in advancing the double helix for the next polymerization step. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9443–9448. doi: 10.1073/pnas.1015399108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl C, Blackburn EH. The effects of nucleoside analogs on telomerase and telomeres in Tetrahymena. Nucleic acids research. 1994;22:893–900. doi: 10.1093/nar/22.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Kawarai M, Takeshita Y, Ishikawa F, Saneyoshi M. Inhibition of human telomerase by nucleotide analogues bearing a hydrophobic group. Nucleic acids symposium series. 2000:175–176. doi: 10.1093/nass/44.1.175. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Liu X, Ogawara T, Inomata M, Saneyoshi M. Telomerase inhibition by 3′-azido-2′, 3′-dideoxynucleoside 5′-triphosphates and telomere shortening in human cultured cells by the corresponding nucleosides. Nucleic acids symposium series (2004) 2006:271–272. doi: 10.1093/nass/nrl135. [DOI] [PubMed] [Google Scholar]
- Yegorov YE, Chernov DN, Akimov SS, Akhmalisheva AK, Smirnova YB, Shinkarev DB, Semenova IV, Yegorova IN, Zelenin AV. Blockade of telomerase function by nucleoside analogs. Biochemistry (Mosc) 1997;62:1296–1305. [PubMed] [Google Scholar]
- Yegorov YE, Chernov DN, Akimov SS, Bolsheva NL, Krayevsky AA, Zelenin AV. Reverse transcriptase inhibitors suppress telomerase function and induce senescence-like processes in cultured mouse fibroblasts. FEBS letters. 1996;389:115–118. doi: 10.1016/0014-5793(96)00533-9. [DOI] [PubMed] [Google Scholar]