SUMMARY
Functional interactions between gene regulatory factors and chromatin architecture have been difficult to directly assess. Here, we use micrococcal nuclease (MNase) footprinting to probe the functions of two chromatin remodeling complexes. By simultaneously quantifying alterations in small MNase footprints over the binding sites of 30 regulatory factors in mouse embryonic stem cells (ESCs), we provide evidence that esBAF and Mbd3/NuRD modulate the binding of several regulatory proteins. In addition, we find that nucleosome occupancy is reduced at specific loci in favor of subnucleosomes upon depletion of esBAF, including sites of histone H2A.Z localization. Consistent with these data, we demonstrate that esBAF is required for normal H2A.Z localization in ESCs, suggesting esBAF either stabilizes H2A.Z-containing nucleosomes or promotes subnucleosome to nucleosome conversion by facilitating H2A.Z deposition. Therefore, integrative examination of MNase footprints reveals insights into nucleosome dynamics and functional interactions between chromatin structure and key gene regulatory factors.
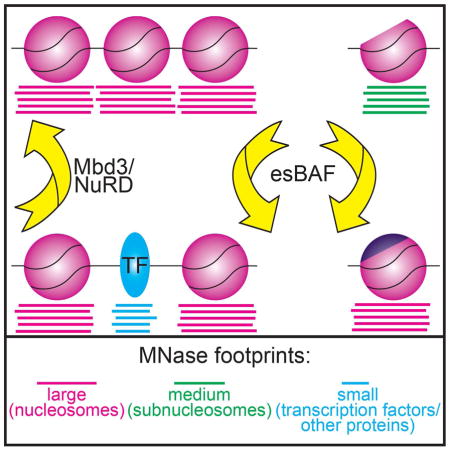
Graphical abstract
INTRODUCTION
In eukaryotes, genomic DNA is packaged with proteins to form chromatin: a repeating array of nucleosomes that each contain 147 bp of DNA wrapped around an octamer of histone proteins composed of a tetramer of H3 and H4 and two H2A and H2B heterodimers (Luger et al., 1997). In some cases, these canonical histone proteins can be replaced with histone variants (such as H2A.Z or H3.3), which contain high sequence similarity to their canonical counterparts, but have somewhat specialized functions in vivo. Regulation of access to factor binding sites through alteration of nucleosome occupancy or positioning is an important mechanism shared among eukaryotes (Almer and Hörz, 1986; Boeger et al., 2003). As a result, most eukaryotic regulatory regions are found within nucleosome-depleted regions (NDRs), which permit binding of regulatory factors and transcription machinery (Mavrich et al., 2008a; Weiner et al., 2010; Yuan et al., 2005).
Nucleosome remodeling factors reposition, deposit, or remove nucleosomes at regulatory regions by altering histone-DNA contacts (Bartholomew, 2014; Racki and Narlikar, 2008). esBAF (Brg1 Associated Factor) is an ESC specific nucleosome remodeling complex that activates transcription of genes and silences transcription near enhancers (Hainer et al., 2015; Ho et al., 2009a; 2009b; 2011), and is necessary for ESC self-renewal and pluripotency (Fazzio et al., 2008; Ho et al., 2009a; Kidder et al., 2009; Schaniel et al., 2009). The Mbd3/NuRD (Nucleosome Remodeling and Deacetylase) complex creates repressive chromatin structure and is required for normal ESC differentiation (Denslow and Wade, 2007; Kaji et al., 2006; 2007; Yildirim et al., 2011). Interestingly, esBAF and NuRD antagonistically regulate many overlapping gene targets, resulting in moderate levels of expression (Yildirim et al., 2011).
While nucleosome positioning and occupancy have been examined in multiple systems (Carone et al., 2014; Li et al., 2012; Mavrich et al., 2008b; Schones et al., 2008; Valouev et al., 2011), very little is known about regulation of subnucleosomes – histone-DNA structures that lack some components of the histone octamer. Hexasomes (one H3/H4 tetramer and one H2A/H2B dimer) and half-nucleosomes (either an H3/H4 tetramer or half an H3/H4 tetramer and one H2A/H2B dimer) have been observed in vivo (Rhee et al., 2014). However, the conditions under which subnucleosomes form, the mechanisms underlying their assembly, and the roles of nucleosome remodeling factors in regulating interchange of subnucleosome and nucleosome structures are unknown.
Here we take an integrative approach to survey the functions of two chromatin remodeling complexes with key roles in ESC pluripotency, utilizing MNase footprinting to reveal nucleosome footprints (135–165 bp), subnucleosome footprints (100–130 bp), and footprints of regulatory factors (≤80 bp), as previously described (Carone et al., 2014; Henikoff et al., 2011; Kent et al., 2011). Using this method, we analyzed the chromatin structure of ESCs depleted of important factors to determine their roles in controlling nucleosome and subnucleosome architecture, as well as regulatory factor occupancy. We provide evidence that esBAF and Mbd3/NuRD modulate the binding of several regulatory factors and we specifically demonstrate that esBAF is required for Klf4 occupancy in ESCs. Furthermore, we find in the absence of esBAF, the abundance of subnucleosomes is increased at the expense of nucleosomes at specific loci, most notably at sites of H2A.Z localization. Consistent with these results, we find that H2A.Z occupancy is strongly decreased in the absence of esBAF. These data suggest esBAF promotes nucleosome occupancy by stabilizing H2A.Z-containing nucleosomes (to prevent conversion of nucleosomes into subnucleosomes) or promoting H2A.Z deposition, potentially by facilitating the functions of H2A.Z deposition factors.
These findings reveal that, by quantifying changes in the abundance of MNase footprints, one can quickly and easily uncover interactions between chromatin remodeling proteins and gene regulatory factors, which can subsequently be validated by standard approaches. Furthermore, changes in subnucleosome size footprints relative to nucleosome footprints provide insights into the regulation of intra-nucleosome architecture, which have been elusive. Therefore, MNase footprinting is a powerful tool for the study of chromatin dynamics in living cells.
RESULTS AND DISCUSSION
Alterations in footprinting of multiple regulatory proteins upon loss of esBAF or Mbd3/NuRD function
Previously, we used deep sequencing of DNA footprints protected from MNase digestion to map nucleosomes in ESCs depleted of esBAF and Mbd3/NuRD (Hainer et al., 2015). To test the roles of these complexes in regulation of transcription factor and chromatin regulatory factor binding, we focused on the information provided by the small reads (≤80 bp) obtained from these MNase-seq experiments. Proof-of-concept analyses have been performed in yeast and mammalian cells, showing that peaks of small MNase footprints correspond to binding sites for factors determined by independent methods, such as ChIP-seq (Carone et al., 2014; Henikoff et al., 2011; Kent et al., 2011). Therefore, we plotted the average abundance of small read footprints (≤80 bp) from EGFP, Mbd3, and Smarca4 KD ESCs that mapped to 32 distinct genomic regions: the experimentally determined binding sites of 30 key components of the ESC gene regulatory network (including Brg1 itself, as a positive control), annotated transcription start sites (TSSs), and a random selection of nucleosome-bound regions as a negative control (Figure S1 and Supplemental Data S1A).
We performed several analyses to assess data quality. First, we quantified changes in small read footprints directly over the factor binding sites and used available ChIP-seq data for Mbd3 and Brg1 to distinguish changes in factor occupancy directly due to loss of Mbd3/NuRD or esBAF function (Figure 1A). For both esBAF and Mbd3/NuRD, we observed minimal alterations in footprinting at factor binding sites at which Brg1 and Mbd3 were not highly enriched (Figure 1A). As a positive control, small read footprints were dramatically changed at the empirically determined binding sites of Brg1 upon Smarca4 KD. Finally, as a negative control, we observed no changes in small reads over a random sampling of nucleosomes, demonstrating alterations in esBAF and Mbd3/NuRD are confined to specific regions of the genome.
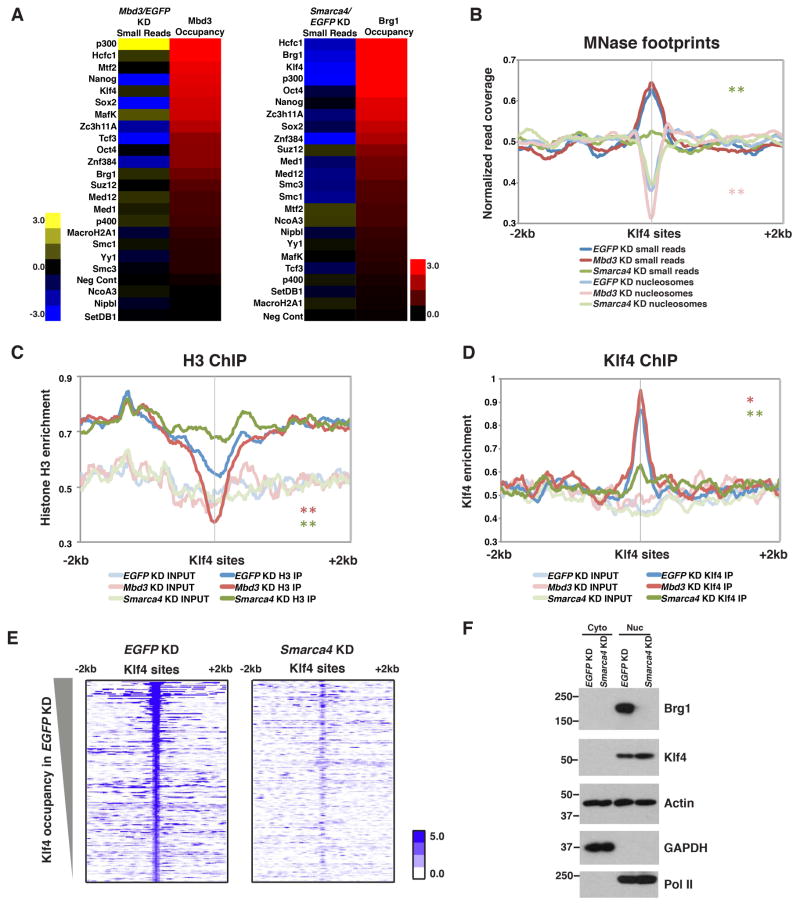
Figure 1. Klf4 binding is esBAF-dependent.
(A) Log2 fold change of small read (left: Mbd3/EGFP KD, right: Smarca4/EGFP KD) spanning 200 bp directly over binding peaks sorted by either Mbd3 (left) or Brg1 (right) occupancy. (B) Aggregation plot of normalized small reads (≤80 bp) and nucleosome size reads (135–165 bp) of MNase-seq upon EGFP, Mbd3, or Smarca4 KD over Klf4 binding sites. Klf4 binding sites were called from GEO:GSE11431 (Chen et al., 2008). Asterisks (*, **) indicate p-values (<0.05, <0.001) reflecting statistical significance of changes in MNase footprints over the relative to EGFP KD, colored as indicated in key. (C) Histone H3 ChIP-seq in EGFP, Mbd3, or Smarca4 KD ESCs shown as normalized occupancy aggregated over Klf4 binding sites. (D) Klf4 ChIP-seq in EGFP, Mbd3, or Smarca4 KD ESCs shown as normalized Klf4 occupancy aggregated over Klf4 binding sites. (E) Heatmaps of Klf4 occupancy over Klf4 binding sites in EGFP (left) and Smarca4 (right) KD. Occupancy is indicated as log2(normalized reads). (F) Equal levels of nuclear Klf4 in EGFP and Smarca4 KD ESCs confirmed by Western blotting. GAPDH and Pol II are specificity controls for cytoplasmic and nuclear fractions, respectively.
Smarca4 KD resulted in a substantial reduction in small read accumulation at several sites (Figure 1A and Supplemental Data S1A), consistent with the function of esBAF to create open chromatin structure to facilitate binding of regulatory factors and the general transcription machinery (Ho et al., 2009a; 2009b; 2011; Novershtern and Hanna, 2011; Yildirim et al., 2011). While KD of Mbd3 resulted in more subtle changes in MNase footprinting at most sites relative to Smarca4 KD, we observed a strong increase in peaks of small reads at p300 binding sites in Mbd3 KD cells, consistent with the antagonistic roles of Mbd3/NuRD and p300 in enhancer function (Pasini et al., 2010; Reynolds et al., 2011). Therefore, alterations in small read profiles from MNase-seq data suggest that both esBAF and Mbd3/NuRD are important regulators of transcription factor and chromatin regulatory factor binding.
esBAF is required for Klf4 binding
Although alterations in small read profiles at transcription factor binding sites imply altered binding of transcription factors themselves, these changes could alternatively result from altered binding of cofactors that co-occupy the same binding sites, or loss of esBAF or Mbd3/NuRD footprints when these factors are knocked down. To distinguish between these possibilities, we tested one functional interaction by an independent method. One of the factors that appeared most strongly affected by Smarca4 KD was Klf4 – small MNase footprints over Klf4 binding sites were strongly reduced upon Smarca4 KD, while Mbd3 KD had a very modest increase (Figure 1A–B). Klf4 plays a critical role in maintenance of the ESC gene regulatory network (Kim et al., 2012; Schuh et al., 1986; Takahashi and Yamanaka, 2006), and Klf4 binding sites are highly bound by Brg1, consistent with the possibility that esBAF promotes Klf4 binding (Figure 1A).
When we examined the nucleosome size (135–165 bp) MNase footprints over Klf4 binding sites, we observed a small increase in nucleosome footprints upon Smarca4 KD (Figure 1B), suggesting that esBAF may promote Klf4 binding in part by clearing its binding sites of nucleosomes. To test this prediction, we performed ChIP-seq for histone H3 and Klf4 in EGFP, Mbd3, and Smarca4 KD ESCs. Consistent with our MNase footprinting data, we observed increased histone H3 occupancy over Klf4 binding sites upon Smarca4 KD and decreased histone H3 occupancy upon Mbd3 KD (Figure 1C). These data are consistent with changes we observed in nucleosome size footprints at Klf4 binding sites, and demonstrate that esBAF is required to maintain open chromatin structure over these regions. Furthermore, ChIP-seq of Klf4 showed a dramatic reduction in Klf4 occupancy over Klf4 binding sites in Smarca4 KD cells and a slight increase in Mbd3 KD cells (Figure 1D–E), demonstrating that alterations in small read abundance over Klf4 binding sites upon Smarca4 and Mbd3 KD, directly reflect alterations in Klf4 binding. Depletion of Brg1 does not result in reduced levels or altered intracellular localization of Klf4, ruling out these potential indirect effects on Klf4 binding (Figure 1F).
We conclude that esBAF functions directly to promote Klf4 occupancy by maintaining open chromatin structure over Klf4 binding sites. These findings confirm that changes in small read profiles from MNase-seq experiments can uncover alterations in factor occupancies when mapped over experimentally determined peaks from ChIP-seq datasets. Future studies following up additional functional interactions predicted by these data should provide additional insights into the ESC gene regulatory network.
esBAF and Mbd3/NuRD regulate factor binding by modulating nucleosome occupancy over factor binding sites
We previously found that esBAF activates expression of many genes by reducing promoter-proximal nucleosome occupancy and facilitating binding of RNA Polymerase II (RNAPII), whereas Mbd3/NuRD acts oppositely (Hainer et al., 2015; Yildirim et al., 2011). To test whether these complexes modulate binding of regulatory proteins by similar mechanisms, we examined the effect of Smarca4 or Mbd3 KD on the abundance of nucleosome footprints at regulatory factor binding sites (many of which are far from promoters). Since nucleosome occupancy often inhibits binding of regulatory proteins, we plotted the changes in small read footprints versus nucleosome footprints in Mbd3 (Figure 2A) or Smarca4 (Figure 2B) KD cells, relative to control, for all 30 sets of binding sites.
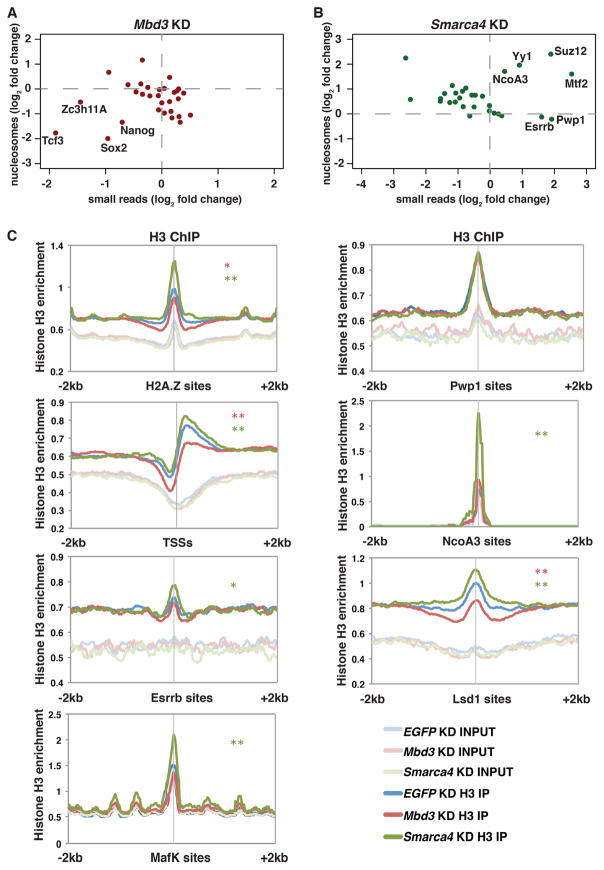
Figure 2. esBAF and Mbd3/NuRD complexes regulate nucleosome occupancy over factor binding sites.
(A,B) Scatterplot of log2 fold change values of nucleosome size reads versus small reads in Mbd3 (A) or Smarca4 (B) KD ESCs relative to EGFP KD. (C) Aggregation plots of histone H3 ChIP-seq over a subset of factor binding sites of EGFP, Mbd3, and Smarca4 KD ESCs. Asterisks (*, **) indicate p-values as described in Figure 1.
Similar to promoters, Mbd3 KD resulted in decreased, and Smarca4 KD resulted in increased average abundance of nucleosome footprints at the binding sites of most factors (Figure 2A–B; compare points above and below horizontal lines). Also consistent with the requirement of most regulatory proteins for a nucleosome-free binding site, changes in the abundance of small read footprints anti-correlated with changes in nucleosome size footprints (Figure 2A–B). These data indicate that esBAF promotes factor binding by creating open chromatin structure and Mbd3/NuRD inhibits factor binding by creating a closed chromatin environment, consistent with the known biological functions of these factors (Ho et al., 2011; Novershtern and Hanna, 2011; Reynolds et al., 2011; Yildirim et al., 2011).
Although nucleosome footprints negatively correlate with small read footprints overall, there are exceptions at several locations (compare Supplemental Data S1A with Supplemental Data S1B), suggesting that nucleosome occupancy does not inhibit the binding of some factors (Figure 2A–B). At many of these sites, there are clear peaks of nucleosome size footprints centered on factor binding sites (Supplemental Data S1B), consistent with this model. Importantly, the presence of nucleosomes over several of these sites is predicted by their functions. PRC2 binds and methylates histone H3K27 within nucleosomes (Margueron and Reinberg, 2011; Simon and Kingston, 2009), consistent with the co-localization of its Suz12, Ezh2, and Mtf2 subunits (Zhang et al., 2011) with nucleosome footprints. Similarly, Pwp1 (a WD40-repeat containing protein) occupies regions marked with H4K20me3 and regulates H4K20 methylation at some sites (Shen et al., 2015) and Ring1b (an E3 ubiquitin ligase) monoubiquitinates H2AK119 (Wang et al., 2004). Furthermore, Lsd1 is a histone H3 lysine demethylase (Shi et al., 2004). Finally, Esrrb has been shown to bind within regions occupied by nucleosomes (Teif et al., 2012), and NcoA3 interacts with Esrrb, co-occupying some locations on chromatin (Percharde et al., 2012).
To validate these findings, we analyzed our histone H3 ChIP-seq data in control, Mbd3, and Smarca4 KD cells over a subset of factor binding sites (Figure 2C). As positive controls, we found that sites of H2A.Z incorporation have peaks of histone H3, whereas TSSs are depleted of histone H3 immediately upstream of the TSS and have strong peaks of occupancy over the +1 nucleosome location (Figure 2C). Consistent with our MNase footprinting data, we observed a strong peak of histone H3 occupancy over factor binding sites found to have nucleosome size MNase footprints, demonstrating that these sites are indeed occupied by nucleosomes (Figure 2C).
For one factor examined, the role of nucleosome architecture in regulating factor binding has not been addressed. MafK is a leucine zipper transcription factor that, to our knowledge, has not been shown to bind nucleosomes. Here, we found a peak of nucleosome size reads over MafK sites and our histone H3 ChIP-seq data support these findings, confirming that MafK binds DNA within nucleosome occupied regions. Together, these data confirm that while most regulatory factors bind to nucleosome-depleted regions of the genome, some do not. In addition, these data suggest that differential affinities of factors for nucleosome bound DNA must be taken into account in studies examining their biochemical functions and roles within gene regulatory networks.
esBAF regulates nucleosome-subnucleosome interconversion at specific sites
To globally address if and how Mbd3/NuRD and esBAF regulate the composition of nucleosomes in ESCs, we compared nucleosome footprints (135–165 bp, Supplemental Data S1B) to intermediate size footprints (100–130 bp, Supplemental Data S1C) over the same factor sites. The intermediate size fragments could result from either large non-histone protein complexes or nonstandard nucleosomes (i.e. hexasomes or half-nucleosomes). Consistent with the latter possibility, the profiles of nucleosome and intermediate size footprints (hereafter, subnucleosomes) were strongly positively correlated in both Mbd3 and Smarca4 KD cells (Figure 3A,B).
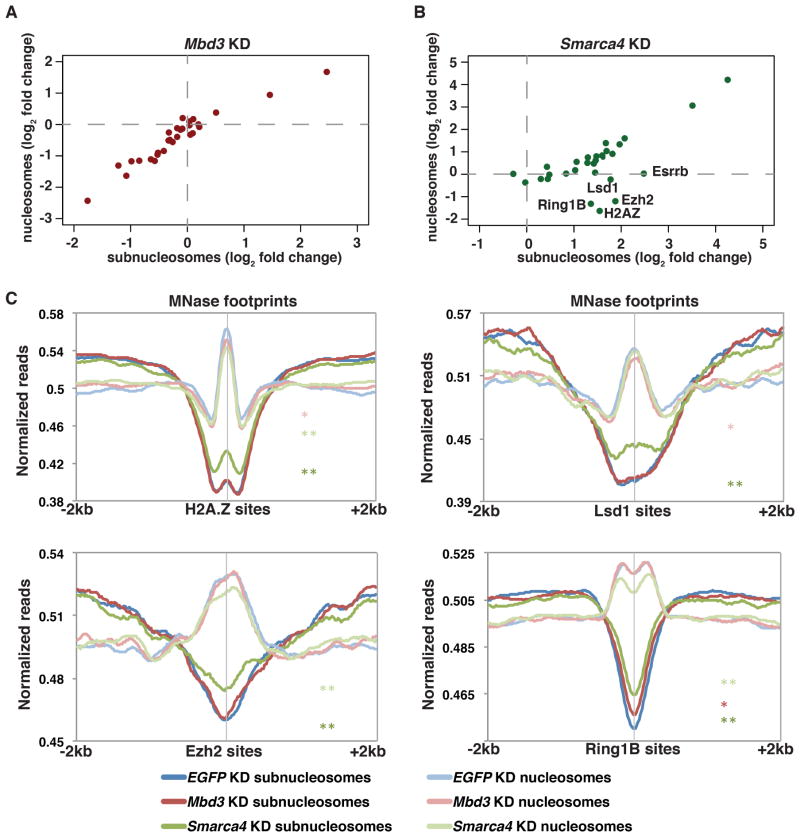
Figure 3. esBAF promotes nucleosome formation at the expense of subnucleosomes.
(A,B) Scatterplot of log2 fold change values of nucleosome size reads versus medium size reads in Mbd3 (A) or Smarca4 (B) KD ESCs relative to control ESCs. (C) Aggregation plots of relative nucleosome (135–165 bp) and subnucleosome (100–130 bp) occupancy of MNase-seq data upon EGFP, Mbd3, or Smarca4 KD over various factor binding sites. Asterisks (*, **) indicate p-values as described in Figure 1.
Interestingly, although subnucleosomes and nucleosomes were strongly correlated at all regions examined upon Mbd3 KD (Figure 3A), KD of Smarca4 resulted in alterations to subnucleosome footprints that were uncoupled from alterations in nucleosome footprints at some sites (Figure 3B). At four locations where Smarca4 KD results in either decreased (Ezh2, Ring1B, and H2A.Z sites) or had no effect on (Lsd1 sites) nucleosome occupancy, subnucleosome footprints are increased (Figure 3C). Importantly, histone H3 occupancy measured by ChIP-seq (which likely cannot discriminate between nucleosomes and subnucleosomes) is increased over Lsd1, Ezh2, Ring1B, and H2A.Z binding sites upon Smarca4 KD (Figure 2C, S3), confirming that subnucleosome footprints at these sites reflect the presence of histones. Taken together, these data show that Smarca4 KD results in higher subnucleosome occupancy and reduced nucleosome occupancy at a subset of genomic locations, suggesting esBAF is necessary for either maturation of subnucleosomes to nucleosomes or prevention of nucleosome disassembly at these sites.
Brg1 is required for normal H2A.Z localization
While the mechanisms underlying subnucleosome-nucleosome interconversion are unknown, prior reports suggest that hexasomes are composed of an H3/H4 tetramer and a single H2A/H2B dimer (Rhee et al., 2014; Weintraub et al.,1975). These findings suggest that regulation of H2A/H2B (or H2A variant/H2B) dimer incorporation could be responsible for subnucleosome maturation. Due to our observation that subnucleosomes were enriched at H2A.Z sites upon Smarca4 KD, we hypothesized that one role of esBAF in subnucleosome regulation could be through regulation of H2A.Z occupancy or incorporation at specific locations throughout the genome.
In mammals, H2A.Z is incorporated into nucleosomes by p400 and SRCAP via exchange of H2A/H2B dimers for H2A.Z/H2B dimers (Cai et al., 2005; Park et al., 2010; Ruhl et al., 2006; Wong et al., 2007) and these nucleosomes are enriched near specific genomic features, including enhancers and promoters (Mavrich et al., 2008b; Zilberman et al., 2008). H2A.Z nucleosomes play key roles in gene regulation, although the effect of H2A.Z incorporation on nucleosome stability and dynamics, and their specific effects on transcription by RNAPII are controversial (Abbott et al., 2001; Jin and Felsenfeld, 2007; Park et al., 2004; Suto et al., 2000; Thambirajah et al., 2006; Thakar et al., 2010; Zhang et al., 2005).
We considered the possibility that esBAF regulates H2A.Z occupancy at enhancers and promoters either directly or indirectly. To test this possibility, we performed histone H2A.Z ChIP-seq in EGFP, Mbd3, and Smarca4 KD cells. In EGFP KD cells, H2A.Z localization was similar to that observed previously in ESCs, confirming the specificity of our datasets (Figure 4A and S4A). We found that Smarca4 KD led to decreased H2A.Z occupancy when one examines either all H2A.Z binding sites or TSSs in particular (Figure 4A–B). Although Smarca4 KD also resulted in decreased H2A.Z occupancy at Lsd1 binding sites (Figure S4B), it had no effect at Pwp1 sites (Figure 4C) and modestly increased H2A.Z occupancy at MafK sites (Figure S4B), demonstrating that esBAF is required for H2A.Z occupancy at some, but not all, of its locations throughout the genome. Although we found no evidence that depletion of Smarca4 alters p400 occupancy (Supplemental Data S1A), Smarca4 KD also resulted in increased subnucleosome footprinting over p400 binding sites (Supplemental Data S1C), consistent with the role of esBAF in regulation of H2A.Z localization.
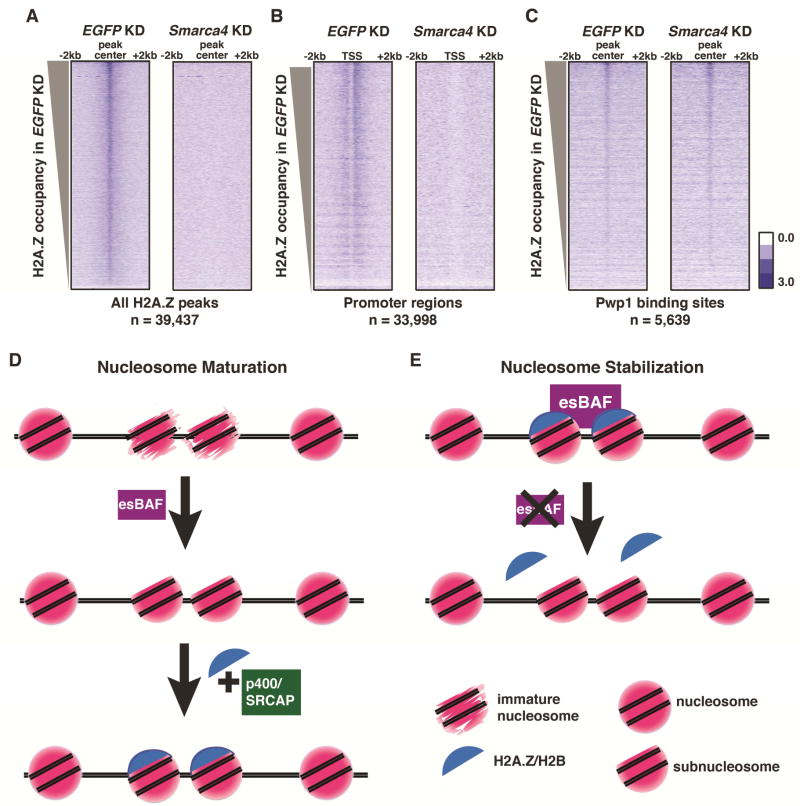
Figure 4. Brg1 is required for H2A.Z occupancy at a subset of locations.
(A–C) Heatmaps of H2A.Z occupancy over indicated regions in EGFP (left) and Smarca4 (right) KD. Binding sites were taken from the following datasets: H2A.Z (GEO:GSE34483), TSSs (mm9), and Pwp1 (GEO:GSE59389). Occupancy is indicated as log2(normalized reads). (D) Maturation model of subnucleosome-nucleosome transition by esBAF. esBAF is required for converting subnucleosomes into a form suitable for H2A.Z/H2B incorporation by p400 or SRCAP, potentially by organizing wrapping of DNA around the H3/H4 tetramer, promoting accessibility of H3/H4 binding interfaces, or another mechanism. (E) Stabilization model of subnucleosome regulation by esBAF. esBAF prevents disassembly of nucleosomes into subnucleosomes (such as hexasomes or half-nucleosomes) by preventing loss of H2A.Z/H2B or H2A/H2B dimers from the histone octamer. Maturation and stabilization of subnucleosomes could be occurring simultaneously and esBAF could regulate H2A.Z occupancy based on interactions with additional regulatory factors.
Taken together, these data suggest that esBAF is required for H2A.Z occupancy at some locations. Upon Smarca4 KD, H2A.Z is strongly depleted, and subnucleosomes partially replace nucleosomes at several regions where H2A.Z is normally enriched. Whether esBAF promotes H2A.Z incorporation by facilitating the functions of SWR1-family complexes, either directly or indirectly, or is required for the stability of H2A.Z-containing nucleosomes, remains unresolved (Figure 4D–E).
Conclusions
Chromatin remodeling enzymes have been examined for their roles in regulation of nucleosome architecture in many cell types. However, their effects on intra-nucleosome architecture, as well as their roles in regulation of DNA-binding proteins, are not fully understood. Here we showed that use of a single footprinting method, MNase-seq, combined with available factor occupancy data, uncovers dynamic regulation of factor binding and subnucleosome structures that can be confirmed by traditional approaches. We focused on two antagonistically functioning chromatin regulators, esBAF and Mbd3/NuRD, to gain a deeper understanding of their roles in modulating ESC chromatin architecture. However, this method should be broadly applicable as a screen for functional interactions between chromatin regulators and the gene regulatory network in any organism/cell type where transcription factor binding data are available.
EXPERIMENTAL PROCEDURES
Cell culture
E14 mouse ESCs were cultured as previously described (Chen et al., 2013). RNAi-mediated KD was performed with endoribonuclease III digested siRNAs (esiRNAs) as previously described (Fazzio et al., 2008) using Lipofectamine 2000 (Invitrogen). KDs were performed for 48 hours.
RT-qPCR
RNA was isolated using TRIzol reagent (Invitrogen) and used in a cDNA synthesis reaction with random hexamers (Promega). cDNA was used as a template in quantitative PCR reactions using a FAST SYBR mix (KAPA Biosystems) on an Eppendorf Realplex with Mbd3, Smarca4, or GAPDH specific primers (Hainer et al., 2015).
Western blotting
Whole cell lysates were extracted using WE16th buffer (25 mM Tris [pH 7.5], 125 mM NaCl, 2.5 mM EDTA, 0.05% SDS, 0.5% NP-40, 10% w/v glycerol). Nuclear and cytoplasmic fractions were isolated using the NE-PER extraction kit (Thermo Scientific) following the manufacturers instructions. 30 μg of lysate were separated by SDS-PAGE, transferred to nitrocellulose (Life Sciences), and assayed by immunoblotting. The antibodies used to detect proteins were: anti-Brg1 (1:1000, Bethyl A300-813A), anti-Mbd3 (1:1000, Bethyl A302-529A), anti-Klf4 (1:1000, Millipore AB4138), anti-Pol II (1:1000, Santa Cruz sc-899), anti-GAPDH (1:5000, Cell Signaling 2118), and anti-actin (1:50,000, Sigma A1978).
MNase-seq analysis
We re-analyzed MNase-seq footprinting data for ESCs depleted of the indicated factors that were previously published (Hainer et al., 2015). See Supplemental Experimental Procedures for details.
ChIP-seq
ChIP experiments and single-end libraries of ChIP-enriched DNA were prepared as previously described (Chen et al., 2013). See Supplemental Experimental Procedures for details.
Supplementary Material
Acknowledgments
We thank P. Chen, L. Ee, and K. McCannell for insightful discussions and critical reading of this manuscript. We thank H-H. Ng from the Genome Institute of Singapore for the Klf4 antibody. This work was supported by the T32CA130807 training grant and a Leukemia and Lymphoma postdoctoral fellowship to S.J.H. and NIH HD072122 to T.G.F. T.G.F. was a Pew Scholar in the Biomedical Sciences and is a Leukemia and Lymphoma Society Scholar. The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript. All deep sequencing was performed at the UMass Medical School Core facility on a HiSeq2000 supported by 1S10RR027052-01. New genomic datasets have been deposited within GEO (accession number GSE68400).
Footnotes
AUTHOR CONTRIBUTIONS
S.J.H. and T.G.F. designed all experiments. S.J.H. carried out all experiments and performed analyses of genomic data. S.J.H. and T.G.F. wrote the manuscript.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. The Journal of Biological Chemistry. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. The EMBO Journal. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Molecular Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. The Journal of Biological Chemistry. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. High-Resolution Mapping of Chromatin Packaging in Mouse Embryonic Stem Cells and Sperm. Developmental Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Hung JH, Hickman TL, Coles AH, Carey JF, Weng Z, Chu F, Fazzio TG. Hdac6 regulates Tip60-p400 function in stem cells. eLife. 2013;2:e01557. doi: 10.7554/eLife.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi Screen of Chromatin Proteins Identifies Tip60-p400 as a Regulator of Embryonic Stem Cell Identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Gu W, Carone BR, Landry BD, Rando OJ, Mello CC, Fazzio TG. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015;29:362–378. doi: 10.1101/gad.253534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. PNAS. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. PNAS. 2009a;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. PNAS. 2009b;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nature Cell Biology. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nature Cell Biology. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Research. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim MO, Kim SH, Cho YY, Nadas J, Jeong CH, Yao K, Kim DJ, Yu DH, Keum YS, Lee KY, et al. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nature Structural & Molecular Biology. 2012;19:283–290. doi: 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Research. 2008a;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008b;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Hanna JH. esBAF safeguards Stat3 binding to maintain pluripotency. Nature Cell Biology. 2011;13:886–888. doi: 10.1038/ncb2311. [DOI] [PubMed] [Google Scholar]
- Park JH, Sun X-J, Roeder RG. The SANT domain of p400 ATPase represses acetyltransferase activity and coactivator function of TIP60 in basal p21 gene expression. Mol Cell Biol. 2010;30:2750–2761. doi: 10.1128/MCB.00804-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. The Journal of Biological Chemistry. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Research. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lavial F, Ng J-H, Kumar V, Tomaz RA, Martin N, Yeo J-C, Gil J, Prabhakar S, Ng H-H, et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–2298. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Current Opinion in Genetics & Development. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. The EMBO Journal. 2011;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014;159:1377–1388. doi: 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh R, Aicher W, Gaul U, Côté S, Preiss A, Maier D, Seifert E, Nauber U, Schröder C, Kemler R, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- Shen J, Jia W, Yu Y, Chen J, Cao X, Du Y, Zhang X, Zhu S, Chen W, Xi J, et al. Pwp1 is required for the differentiation potential of mouse embryonic stem cells through regulating Stat3 signaling. Stem Cells. 2015;33:661–673. doi: 10.1002/stem.1876. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thakar A, Gupta P, McAllister WT, Zlatanova J. Histone variant H2A.Z inhibits transcription in reconstituted nucleosomes. Biochemistry. 2010;49:4018–4026. doi: 10.1021/bi1001618. [DOI] [PubMed] [Google Scholar]
- Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausió J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. The Journal of Biological Chemistry. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger MIW, Mallm JP, Marth C, Höfer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nature Structural & Molecular Biology. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Research. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Palter K, Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975;6:85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Wong MM, Cox LK, Chrivia JC. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. The Journal of Biological Chemistry. 2007;282:26132–26139. doi: 10.1074/jbc.M703418200. [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Research. 2011;39:e35–e44. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD Complex Regulates Expression of 5-Hydroxymethylcytosine Marked Genes in Embryonic Stem Cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-Scale Identification of Nucleosome Positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.