Abstract
Escherichia coli O157:H7 is a notorious foodborne pathogen due to its low infectious dose and the disease symptoms it causes, which include bloody diarrhea and severe abdominal cramps. In some cases, the disease progresses to hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS), due to the expression of one or more Shiga toxins (Stx). Isoforms of Stx, including Stx2a, are encoded within temperate prophages. In the presence of certain antibiotics, phage induction occurs, which also increases the expression of toxin genes. Additionally, increased Stx2 accumulation has been reported when O157:H7 was cocultured with phage-susceptible nonpathogenic E. coli. This study characterized an E. coli O157:H7 strain, designated PA2, that belongs to the hypervirulent clade 8 cluster. Stx2a levels after ciprofloxacin induction were lower for PA2 than for the prototypical outbreak strains Sakai and EDL933. However, during coculture with the nonpathogenic strain E. coli C600, PA2 produced Stx2a levels that were 2- to 12-fold higher than those observed during coculture with EDL933 and Sakai, respectively. Germfree mice cocolonized by PA2 and C600 showed greater kidney damage, increased Stx2a accumulation in feces, and more visible signs of disease than mice given PA2 or C600 alone. These data suggest one mechanism by which microorganisms associated with the colonic microbiota could enhance the virulence of E. coli O157:H7, particularly a subset of clade 8 strains.
INTRODUCTION
Escherichia coli can be either nonpathogenic, as exemplified by the bacteria associated with the normal human gut microflora, or pathogenic, classified as intestinal or extraintestinal (1). The intestinal pathogenic E. coli group is further subdivided into six pathotypes, among which enterohemorrhagic E. coli (EHEC) is a causative agent of bloody diarrhea, hemorrhagic colitis (HC), and hemolytic uremic syndrome (HUS). Shiga toxins (Stx) are one of the defining virulence factors of E. coli O157:H7 (2, 3). The stx genes are carried within lambdoid prophages downstream of the promoter for late gene expression. The toxin is released after phage-induced lysis of the bacterial cell (4). Shiga toxins are approximately 70-kDa proteins, consisting of an enzymatically active A subunit and a pentameric B subunit. The A subunit is an N-glycosidase and inactivates the 60S ribosomal subunit. This leads to inhibition of protein synthesis in the eukaryotic cell (5). There are two antigenically distinct Stx proteins, designated Stx1 and Stx2 (6). Despite having an amino acid identity of 57%, mouse and primate studies have shown that Stx2 causes more severe disease outcomes than Stx1 (7, 8). Several allelic variants of Stx2 have been described, based on the amino acid differences in the A and B subunits. The most common forms of Stx2 expressed by E. coli O157:H7 human isolates are Stx2a and Stx2c (9, 10).
Since its first report in 1983 (11), the serotype O157:H7 rapidly became a notorious foodborne pathogen due to its severe disease outcomes and an infectious dose of less than 100 CFU (12). Cattle are the main reservoir, and transmission to humans often occurs through contaminated food and water, or sometimes person-to-person contact (13). The best-studied O157:H7 strains are Sakai and EDL933, which were isolated from early outbreaks and were the first to be fully sequenced (14, 15). Sakai was implicated in an outbreak associated with radish sprouts and caused illness in over 9,000 children in Sakai city, Japan (16). EDL933, on the other hand, was linked to consumption of tainted hamburgers in Michigan (11).
Octamer-based genome scanning (OBGS) (17), lineage-specific polymorphism assay (LSPA) (18), and single nucleotide polymorphism (SNP)-based methods (19) have successfully classified E. coli O157:H7 strains into informative subgroups. OBGS and LSPA were used to define three lineages. Lineage I is associated with strains expressing higher levels of Stx2 (20) that are common human isolates in the United States (21), while lineage II strains are more likely to be of bovine origin and express lower Stx2 levels than the other two lineages (17, 20). The third lineage described is designated I/II (22). A SNP-based method was also used to subtype E. coli O157:H7 strains into nine clades (19). The strains clustering within clade 8 demonstrate increased virulence potential (19), as patients presenting HUS symptoms were more likely to be infected with strains from this group. A larger study confirmed this observation, noting that clade 6 and clade 8 strains were more frequently associated with HUS than were strains from other clades (23).
Stx2 expression requires induction of a stx2-bearing prophage (4), many of which adsorb to the essential outer membrane protein BamA (24), and is enhanced by DNA-damaging agents, including antibiotics, UV light, and H2O2 (25–27). Increased phage and Stx2 accumulation also occurs when E. coli O157:H7 is cocultured with phage-sensitive E. coli strains (28). The model proposed by Gamage et al. suggested that a lytic infection of these strains by phage that are spontaneously released by E. coli O157:H7 increases the number of phage particles, as well as Stx2 levels. This model was developed using a single strain of E. coli O157:H7, although stx2-bearing phage are known to be highly polymorphic (29–31). We hypothesized here that strain-specific differences between these phage would impact the level of toxin amplification observed.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used in the study are listed in Table 1. The E. coli O157:H7 strains with “PA” designations were human isolates obtained from the Pennsylvania Department of Health and were characterized previously (9). The bacteria were routinely propagated in Luria-Bertani (LB) broth at 37°C, while culture stocks were maintained at −80°C in 10% glycerol. For coincubation assays, modified LB broth supplemented with calcium chloride (CaCl2) at a final concentration of 10 mM was used.
TABLE 1.
Strains used in the study
| Strain | Relevant characteristics | Reference |
|---|---|---|
| E. coli O157:H7 | ||
| Sakai | stx1 stx2a; clade 1, lineage I | 14 |
| EDL933 | stx1 stx2a; clade 3, lineage I | 15 |
| PA2 | stx2a; clade 8, lineage I/II | 9 |
| PA3 | stx2a stx2c; clade 8, lineage I/II | 9 |
| PA8 | stx2a; clade 8, lineage I/II | 9 |
| PA9 | stx2a; clade 8, lineage I/II | 9 |
| PA13 | stx2a stx2c; clade 8, lineage I/II | 9 |
| PA19 | stx2a; clade 8, lineage I/II | 9 |
| PA24 | stx1; clade 8, lineage I/II | 9 |
| PA25 | stx2a; clade 8, lineage I/II | 9 |
| PA28 | stx2a stx2c; clade 8, lineage I/II | 9 |
| Other E. coli | ||
| C600 | K-12 derivative | 43 |
| JM109 | Host for plaque assays | 44 |
Induction and supernatant collection.
E. coli O157:H7 strains were grown overnight and diluted to an optical density at 600 nm (OD600) of 0.05 using LB broth. For phage induction, ciprofloxacin was added to the LB broth to a final concentration of 45 ng/ml. The growth curve for both the induced and uninduced cultures was profiled for 8 h, with samples collected every 2 h. The OD600 was measured using a DU730 spectrophotometer (Beckman Coulter, Pasadena, CA), while phage and Stx2 levels were measured for each collected sample as described below.
Stx2 quantification using receptor ELISA.
For Stx2 quantification, samples were treated with 6 mg/ml of polymyxin B (PMB) and incubated at 37°C for 5 min to lyse cells and release intracellular Stx2. The PMB-treated samples were centrifuged at 4,000 × g for 10 min, and the collected supernatant was either used immediately or stored at −80°C. Receptor enzyme-linked immunosorbent assay (RELISA) uses ceramide trihexoside (CTH), a Gb3 analogue, as a capture for Stx (32). The 96-well polystyrene microtiter strip plates (Thermo Scientific, Waltham, MA) were coated with 2.5 μg of CTH (Matreya Biosciences, Pleasant Gap, PA). The plates were incubated at 4°C overnight with blocking buffer composed of 4% bovine serum albumin (Sigma-Aldrich, St. Louis MO) in 0.01 M phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBST). Diluted or undiluted samples were dispensed in triplicate and placed on a shaking incubator at room temperature (RT) for 1 h. Monoclonal mouse anti-Stx2 (Santa Cruz Biotechnology, Santa Cruz, CA), which recognizes the A subunit, was added to a final concentration of 0.1 μg/ml and incubated at room temperature for 1 h. After 5 washes with PBST, goat anti-mouse IgG peroxidase conjugate was added to a final concentration of 0.1 μg/ml and incubated at RT for 1 h. Subsequently, 100 μl of 1-Step Ultra TMB (3,3′,5,5′ tetramethylbenzidine) (Thermo Fisher, Waltham, MA) was added to each well, and the plates were incubated for 10 min at room temperature. Finally, 100 μl of stop solution (2 M H2SO4) was added, and the OD450 was measured. Wells containing supernatant from E. coli O157:H7 strain PA24 (9), which produces only Stx1, served as a negative control. With each run, a 2-fold serially diluted Stx2a of known concentration was used to construct a standard curve. This standard was made by filter sterilizing supernatant from 30 ml of ciprofloxacin-induced E. coli O157:H7 strain PA11, followed by quantification using recombinant Stx2a (BEI Resources, Manassas, VA) to make a standard curve. Aliquots of the supernatant (300 μl) were individually frozen at −80°C, and typical concentrations were 20 μg Stx2a/ml. Each aliquot was thawed only once. Total protein was measured using the Bradford reagent (Amresco, Solon, OH).
Plaque assay.
A double-overlay agar method for measuring plaques was adapted from Islam et al. (33). Phage from induced and uninduced cultures were harvested as described in that publication. The supernatant was centrifuged at 4,000 × g for 10 min, and serial dilutions were made in SM buffer (0.1 M NaCl, 50 mM Tris-Cl, 8 mM MgSO4, and 0.01% gelatin). The indicator strain JM109 (200 μl) was added to 100 μl of phage and mixed in modified LB agar-soft agar (0.75% agar). The soft-agar mixture was overlaid on modified LB agar and incubated at 42°C to allow overexpression of the phage receptor bamA gene (24). Plaques were counted after 16 h of incubation.
Coincubation experiments.
The coincubation assays were developed based on experiments by Gamage et al. (28). Briefly, overnight cultures of E. coli O157:H7 and C600 were diluted in LB broth to an OD600 of 0.03. For control experiments, O157:H7 or C600 was present as a monoculture, while coculture setups contained both O157:H7 and C600 diluted in LB broth to a final OD600 of 0.03 each. The 6-well plates (BD Biosciences Inc., Franklin Lakes, NJ) containing modified LB agar served as a base for the modified LB broth, on which 1 ml of monoculture or coculture setup was overlaid and incubated at 37°C. The E. coli O157:H7 strains PA2, Sakai, and EDL933 were either incubated as monocultures or cocultured with C600. Samples for enumerating cell density and Stx2 levels were harvested every 3 h after the 6-h time point. Viable-cell counts were measured by plating onto Sorbitol MacConkey agar (SMaC), a differential medium where O157:H7 appears as white colonies and C600 appears as red colonies. Stx2 levels were quantified using the RELISA described above. To test Stx2 accumulation when other clade 8 strains were cocultured with C600, E. coli O157:H7 strains from the collection previously characterized by Hartzell et al. (9) were chosen. PA2, PA8, PA9, PA19, and PA25, which carry stx2a only, and PA3, PA13, and PA28, which carry both the stx2a and stx2c alleles, were tested. The monoculture and coincubation samples were harvested after 16 h of incubation, and the cell counts from three biological repeats were enumerated from 108 dilutions. For the clade 8 coincubation, the cell counts were reported as the percentage of C600 in the total E. coli population.
Mucus assays.
Three Angus cows and one Jersey-Holstein mix were slaughtered at the Penn State Meat Laboratory or Rising Spring Mills Meat Co. (Rising Spring Mills, PA), and 3 feet from the rectum and distal colon were sequestered to collect mucus. The mucus was autoclaved, and approximately 2.5 ml was added to each well in a 6-well polystyrene plate (BD Biosciences Inc., Franklin Lakes, NJ). The inoculum was prepared by resuspending overnight cultures in PBS to wash off residual medium from the overnight culture. Dilution was done in PBS to a final OD600 of 0.03. The monocultures and cocultures of PA2, EDL933, and C600 were assembled as described above. A negative control with mucus in PBS was also included. Growth was profiled on SMaC every 3 h until 16 h. After 16 h, the mucus was collected in Eppendorf tubes and vortexed at maximum speed for 1 min to homogenize the mucus. Subsequently, centrifugation was done for 15 min at 4,000 × g to collect the supernatant. ELISA was done using the Premier EHEC ELISA kit (Meridian Biosciences Inc., Cincinnati, OH), which detects both Stx1 and Stx2.
Animal experiments.
Male and female germfree Swiss-Webster mice 3 to 5 weeks of age were raised in the University of Michigan germfree colony. They were housed in soft-sided bubble isolators and fed autoclaved water and laboratory chow ad libitum. The mice were infected orally with ∼106 CFU of LB medium-cultured bacteria. Following inoculation, the mice were placed in sterile microisolator cages within the germfree isolators. The microisolator cages were then aseptically removed from the isolators and kept in a laminar flow hood for the duration of the experiment. Throughout the experiment, the mice received sterile food, water, and bedding to maintain germfree conditions, except for the infecting E. coli strains. All animal experiments were conducted with the approval of the University of Michigan Animal Care and Use Committee.
The mice were given one of three inocula: C600 alone, PA2 alone, or C600 followed by PA2 1 week later. They were weighed prior to C600 inoculation, prior to PA2 inoculation, and just prior to euthanasia 6 days after PA2 inoculation. The mice were evaluated daily for evidence of illness (dehydration, ruffled coat, or reluctance to move) and were euthanized 6 days after PA2 inoculation or when they became moribund. Prior to euthanasia, evidence of illness was recorded, and at necropsy, samples were collected for bacterial culture, Stx2 ELISA, and histologic examination. For bacterial culture, samples of the cecal contents were weighed, serially diluted in sterile LB broth, and cultured on SMaC plates. For quantification of Stx2, the cecal content was stored at −20°C until evaluation with a Premier EHEC ELISA kit. For histologic examination, samples of the right and left kidneys were bisected and emersion fixed in 10% neutral buffered formalin. The colon was removed and fixed in its entirety. Tissues were paraffin embedded, and 5-μm sections were stained with hematoxylin and eosin and scored for histologic lesions.
Histologic scoring.
Acute renal tubular necrosis was scored as previously described (34). Briefly, scores were as follows: 0, no necrosis; 1, occasional dilated tubules with cellular debris; 2, necrotic tubules in most histologic fields; 3, necrotic tubules in all fields with minimal normal cortex present. Colon necrosis and neutrophilic infiltration were scored as a weighted average. For this, each 200× microscopic field was scored separately for the presence of epithelial necrosis or neutrophils in the lamina propria. Necrosis was scored as follows: 0, no necrotic cells; 1, scattered necrotic epithelial cells; 2, many necrotic cells in the surface epithelium and most glands and scattered necrotic cells in the lumen; 3, severe necrosis of most of the surface epithelium extending into many glands with rafts of necrotic epithelial cells in the lumen. Neutrophils were scored as follows: 0, none; 1, scattered neutrophils in the lamina propria; 2, occasional clusters or prominent infiltration of neutrophils between glands; 3, sheets of neutrophils infiltrating much of the superficial lamina propria. Each field was scored separately, and a weighted average was calculated by dividing the number of fields with each score by the score and adding the result.
Data analysis.
Microsoft Excel was used to calculate the mean, standard deviation, and standard error, while GraphPad Prism 8 software was used to calculate P values.
RESULTS
Ciprofloxacin induces less Stx2a production in PA2 than in EDL933 and Sakai.
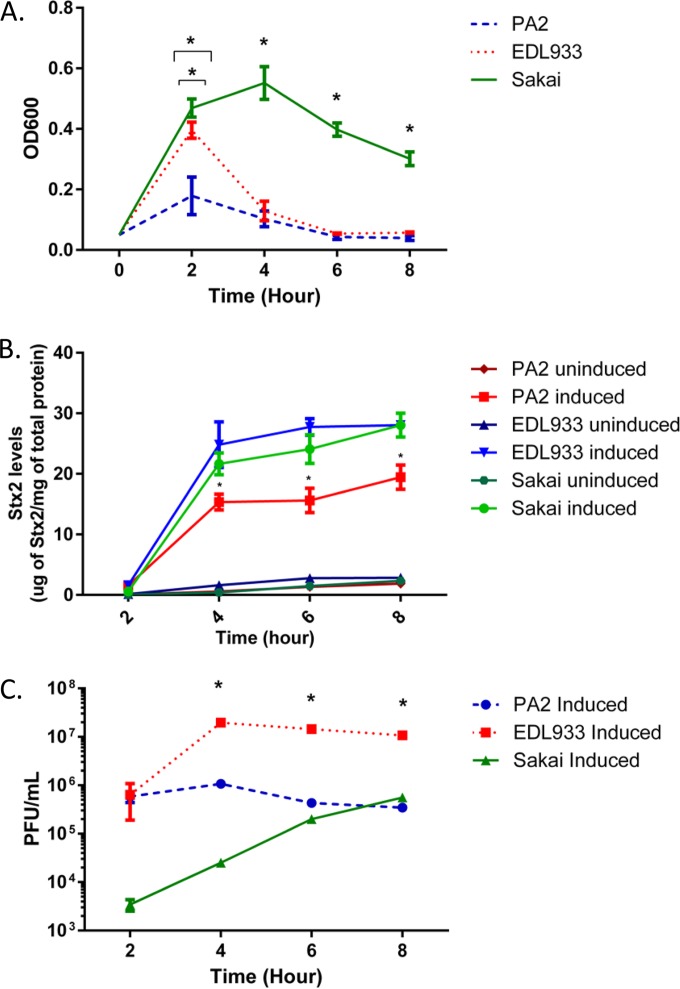
We began by comparing the kinetics of induction, Stx2a, and phage production between E. coli O157:H7 strains EDL933, Sakai, and PA2. The last is a clade 8 strain previously characterized in our laboratory (9) for which we have the full genome sequence (GenBank accession no. NZ_AOEL00000000). Initial sequence analysis indicated the stx2a-bearing phage from PA2 is distinct from those found in the EDL933 and Sakai genomes (unpublished data). Strains PA2, EDL933, and Sakai did not show significant differences in their growth in LB medium in the absence of antibiotics (data not shown). However, the growth profiles were different when the lytic cycle was induced by growth in subinhibitory concentrations of ciprofloxacin (Fig. 1A). Both PA2 and EDL933 reached their peak OD600s at hour 2, although this maximum was approximately 2-fold lower for PA2 than for EDL933. Sakai, on the other hand, reached its highest OD600 at hour 4, followed by a steady decline in turbidity. An increase in toxin production was observed over the time course, and as expected, the induced samples produced significantly higher (P < 0.0001) Stx2a levels than their uninduced counterparts (Fig. 1B). Among the induced samples, Sakai produced significantly less (P < 0.005) Stx2a than EDL933 only at the hour 4 time point. PA2 consistently produced less Stx2a than both EDL933 and Sakai from hour 4 until hour 8 (P < 0.005). EDL933 produced significantly more (P < 0.0001) plaques than PA2 and Sakai at hours 4 through 8 (Fig. 1C).
FIG 1.
Growth, Stx2a accumulation, and phage production by ciprofloxacin-induced and uninduced strains. (A and B) Overnight cultures of E. coli O157:H7 strains PA2, EDL933, and Sakai were inoculated into LB medium supplemented to 45 ng/ml ciprofloxacin, and phage induction was visualized by the optical density. (A) At hour 2, PA2 and EDL933 showed significantly lower OD600 values (P < 0.0001 and P = 0.005, respectively) than Sakai. At hour 4, Sakai showed a significantly higher OD600 (P < 0.0001) than EDL933 and PA2. Asterisks highlight data points where statistical significance was observed. (B) Stx2a accumulation in the same experiment. The Stx2a levels were normalized to total protein. The error bars represent standard errors of the mean (SEM) from three biological repeats, and the asterisks indicate that PA2 produced less Stx2a than both EDL933 and Sakai between hour 4 and hour 8 (P < 0.005). (C) Overnight cultures of E. coli O157:H7 strains PA2, EDL933, and Sakai were inoculated into fresh media supplemented to 45 ng/ml of ciprofloxacin, and plaque assays were performed at the indicated time points. The error bars represent SEM from three biological repeats, and the asterisks indicate significantly more (P < 0.0001) plaques were produced by EDL933 than by PA2 and Sakai at hours 4 through 8.
Coincubation with E. coli strain C600 amplifies Stx2a in PA2 and EDL933 in vitro.
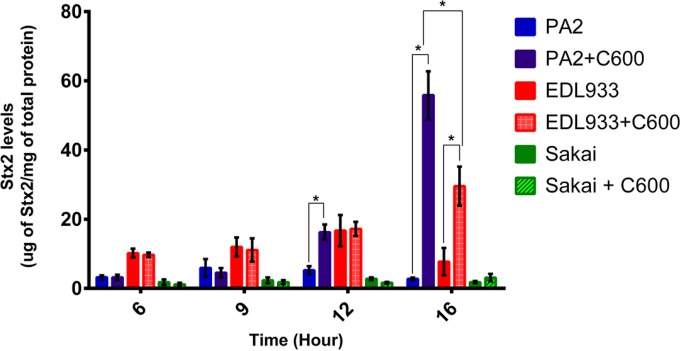
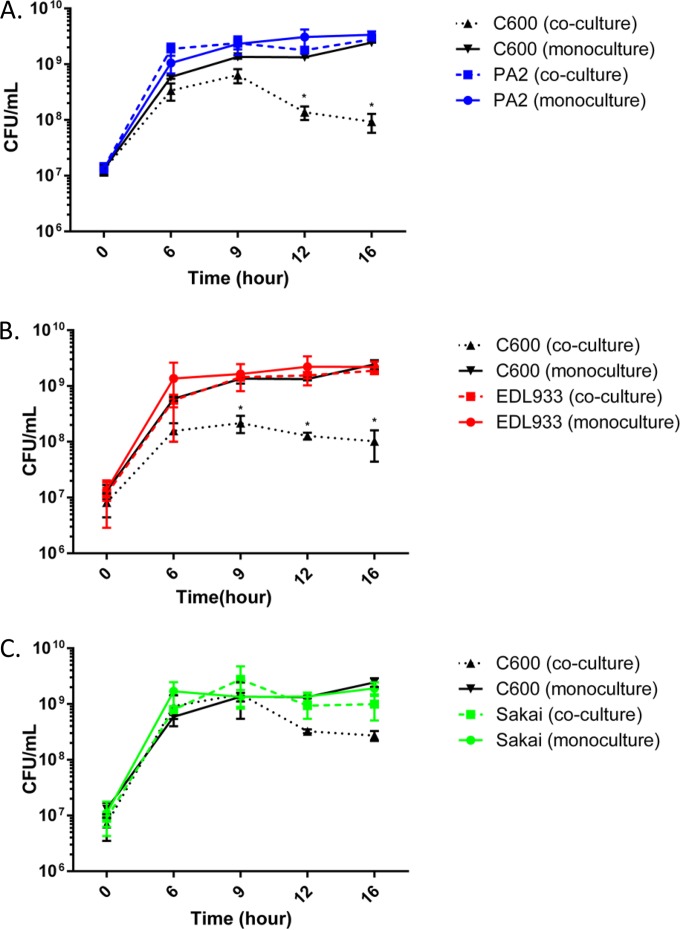
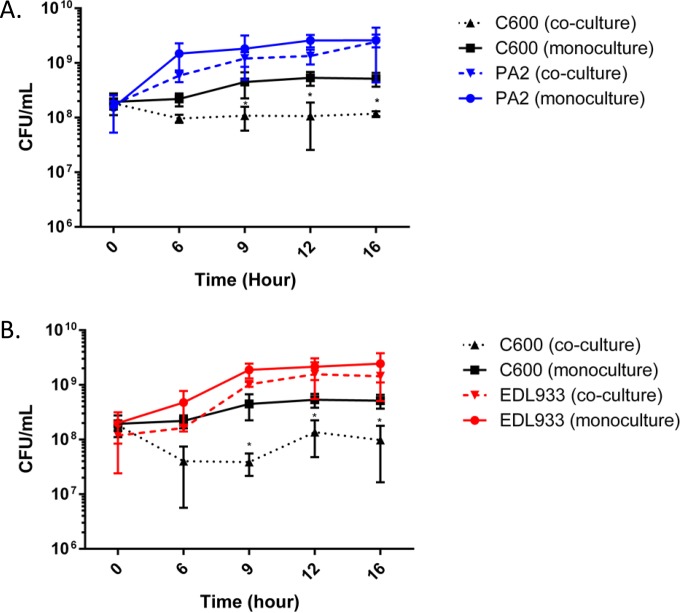
We investigated the strain-specific differences in Stx2a levels when the three O157:H7 strains were individually cocultured with C600. There was no difference in Stx2a levels between the monoculture and coculture samples of all three strains between hours 6 and 9; however, from hour 12 onward, a significant increase in Stx2a levels was noted for the PA2 coculture setup (Fig. 2). At hour 16, both PA2 and EDL933 had significantly higher Stx2a levels in the coculture samples than O157:H7 monoculture controls. On the other hand, Stx2a levels in the Sakai monoculture and the coculture with C600 were statistically indistinguishable. A significantly higher (P < 0.05) amplification was reported for PA2 cocultures than for monoculture controls, with the difference being 20.3 (±1.2)-fold compared to EDL933 and Sakai, which showed 2.7 (±0.4)- and 1.6 (±0.4)-fold differences, respectively. Cell counts of PA2, EDL933, Sakai, and C600 were enumerated in monoculture and coculture to profile the population dynamics during the course of coincubation (Fig. 3). When PA2 and C600 were cocultured, a sharp reduction in C600 cell counts was observed between hours 9 and 16 (Fig. 3A). Similarly, the C600 cell counts in the EDL933-plus-C600 coculture were consistently lower than when C600 was present alone (Fig. 3B). In contrast, coculture with Sakai did not significantly reduce the colony counts of C600 at any time (Fig. 3C).
FIG 2.
Stx2a levels after coincubation with C600. E. coli O157:H7 strains were incubated statically at 37°C, either alone or with C600. Samples were harvested and assayed for Stx2a. The error bars indicate SEM from three biological repeats, and the asterisks indicate significant (P < 0.05) differences between samples.
FIG 3.
Population dynamics in cocultured samples. E. coli O157:H7 strains were statically incubated at 37°C, either alone or with C600. Samples were harvested every 3 h from 6 h onward and plated onto sorbitol MacConkey agar. On this medium, C600 grows as red colonies and O157:H7 as white colonies. The error bars represent SEM from three biological repeats. The C600 coincubation samples marked with asterisks are significantly lower than the C600 monoculture control (P < 0.01).
We also developed an ex vivo model to investigate whether toxin amplification seen during PA2-plus-C600 coculture was repeatable in environments other than laboratory media. In autoclaved bovine rectal mucus, there was a reproducible increase in Shiga toxin production in coculture compared to the monoculture controls. The increases in total toxin production comparing coculture to monoculture experiments were 4.3 (±1.2)-fold (1 standard deviation) for PA2, and 2.2 (±0.5)-fold for EDL933. As seen in laboratory media, growth suppression of C600 was observed when the strain was cocultured with either E. coli O157:H7 strain (Fig. 4A and B).
FIG 4.
Stx2 levels and percentages of C600 after coincubation of clade 8 strains with C600. Clade 8 strains carrying genes for stx2 (PA2, PA8, PA9, PA19, and PA25) or stx2 and stx2c (PA3, PA13, and PA28) were cocultured with C600 for 16 h. (A) Stx2 levels quantified from the cocultured samples demonstrated that PA2 and PA8 significantly (P < 0.01) amplified Stx2a in comparison to monoculture controls. (B) The cell count for C600 relative to that for O157:H7 showed that during coculture, PA8 reduced the number of C600 colonies to the same extent as PA2. The error bars indicate SEM.
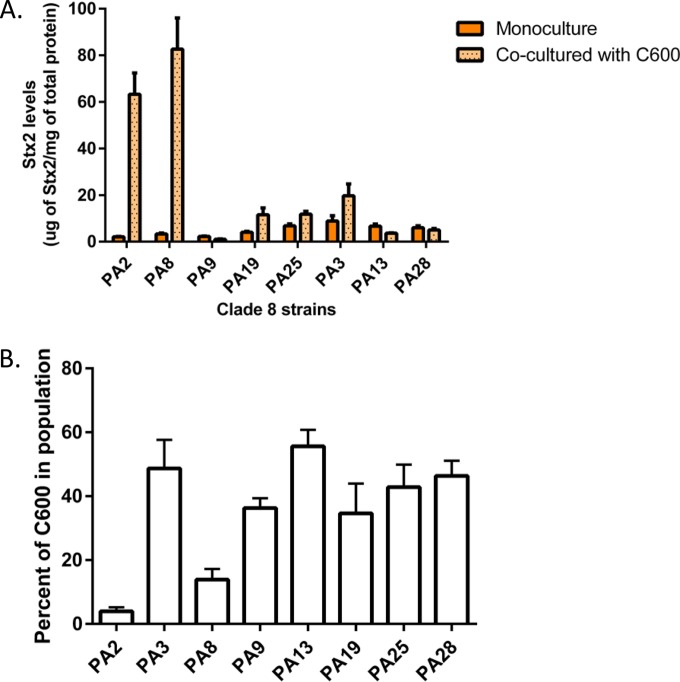
Toxin amplification occurs with a limited number of clade 8 strains.
Since cocultures of PA2 and C600 showed a drastic increase in Stx2a levels compared to PA2 alone, we next hypothesized that other clade 8 strains would also amplify Stx2a. We chose eight clade 8 strains from our previously characterized collection (9). PA2 carries only the stx2a gene; therefore, we selected four other clade 8 strains that also carry stx2a only, namely, PA8, PA9, PA19, and PA25. Three other strains, PA3, PA13, and PA28, carrying both stx2a and stx2c, were also chosen. Stx2 quantification demonstrated that PA8 cocultured with C600 also increased Stx2 levels by 24.1 (±2.5)-fold compared to monoculture controls (Fig. 5A). Other strains, including PA3, PA19, and PA25, showed increased Stx2 levels by 2.3 (±0.1)-, 2.7 (±1.1)-, and 1.9 (±0.5)-fold, although the numbers were not statistically significant compared to the E. coli O157:H7 monoculture controls. Cell counts were quantified for E. coli O157:H7 and C600 at the end of the coculture experiments. Both PA2 and PA8 outgrew E. coli C600, reducing the nonpathogenic strain from the initial 50% of the total bacterial population to 4.0% (±0.1.3%) and 14.0% (±3.3%), respectively (Fig. 5B). Therefore, only a minority of clade 8 strains show the same toxin amplification phenotype observed with cocultured PA2 and C600.
FIG 5.
Cell counts in the ex vivo mucus model. Shown is the time course of PA2 and C600 (A) and EDL933 with C600 (B) cell counts when grown as cocultures or monocultures. The cultures were grown for 16 h total, and colony counts were measured by plating onto sorbitol MacConkey agar. On this medium, O157:H7 colonies are white, while C600 colonies are red. The asterisks indicate time points where C600 counts under coculture conditions were lower (P < 0.01) than E. coli O157:H7 counts. The error bars indicate SEM.
Increased lethality and Stx2a levels in germfree mice cocultured with C600.
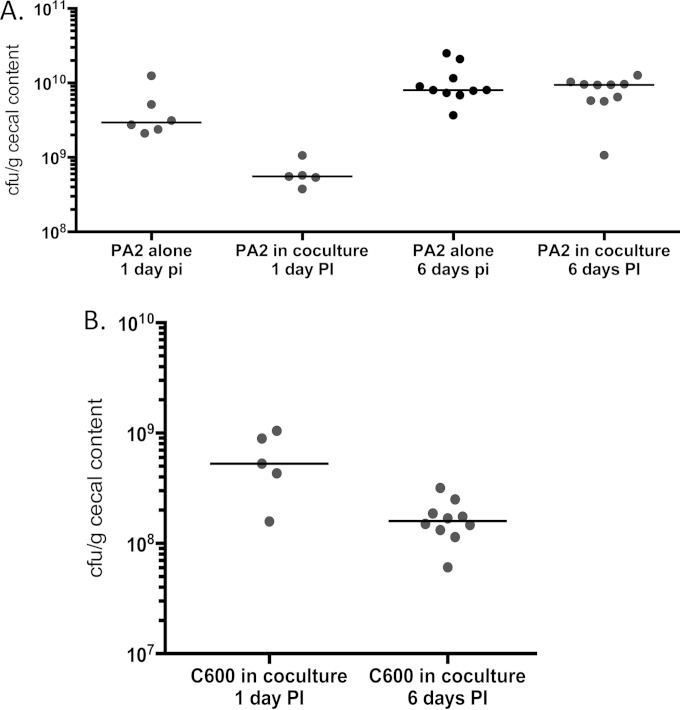
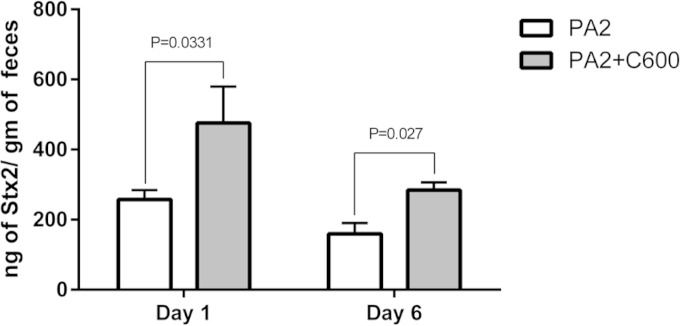
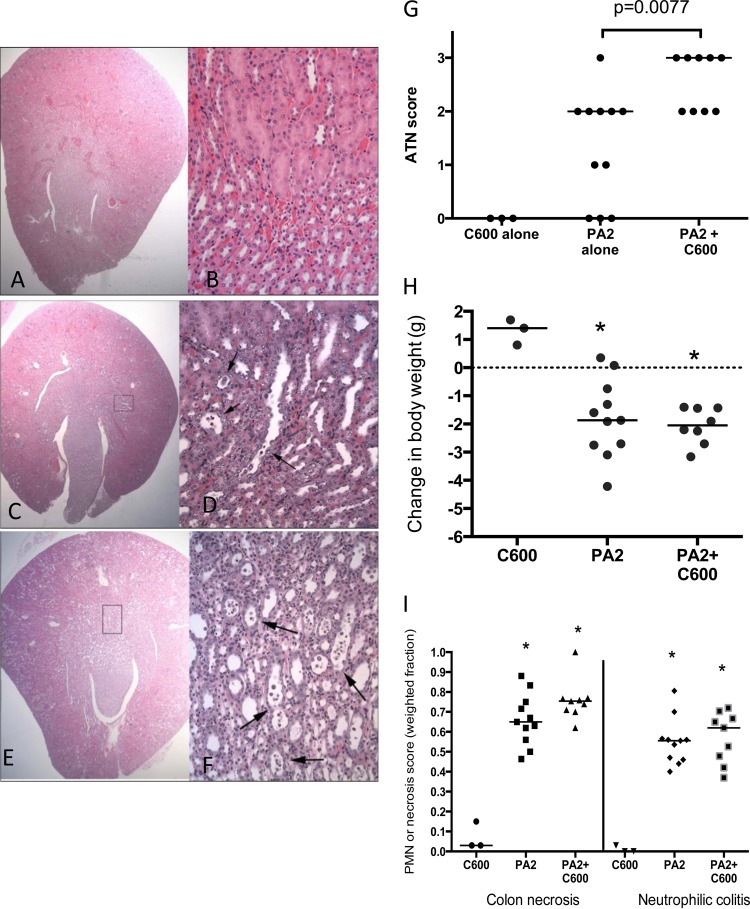
To investigate whether toxin amplification could be observed in an in vivo system, we used a germfree mouse model (34). Germfree mice precolonized with C600 were infected with PA2 and euthanized after 6 days, while mice inoculated with PA2 and C600 alone served as controls. When infected alone, PA2 (Fig. 6A) and C600 (data not shown) colonized mice to approximately 109 and 1010 CFU/g of cecal contents, respectively, after 6 days, although in the same period, coculture counts of C600 were reduced to approximately 2 × 108 CFU/g (Fig. 6B). Mice precolonized with C600 6 days prior to PA2 infection appeared sicker and showed signs of becoming moribund (Table 2). The PA2-inoculated mice, on the other hand, appeared healthy at the time of being euthanized. Consistent with the observations in vitro, the Stx2a levels recovered from the feces of PA2-plus-C600-infected mice were significantly higher than those from mice infected with PA2 alone on both day 1 (P < 0.05) and day 6 (P < 0.005) (Fig. 7). Moreover, the histopathological data demonstrated more extensive acute tubular necrosis (ATN) in the kidneys of mice infected with PA2 plus C600 than in those infected with PA2 alone (Fig. 8A to G). Infection with E. coli O157:H7 caused weight loss, colon epithelial cell necrosis, and neutrophilic colitis compared to mice infected with C600 (P < 0.05), but scores were statistically indistinguishable between mice given PA2 alone and those coinoculated with PA2 and C600 (Fig. 8H and I).
FIG 6.
Colonization of germfree mice. Mice were orally inoculated with approximately 106 CFU/ml of PA2 in the monoculture control. In the coculture experiment, germfree mice were precolonized with C600, followed by inoculation with PA2 7 days later. Fecal cell counts for PA2 (A) and C600 (B) are reported for day 1 and day 6 postinoculation (pi or PI) with strain PA2. The medians are indicated by the horizontal lines. The detection limit for plate counts was approximately 100 to 1,000 CFU/ml.
TABLE 2.
Qualitative observation of disease in germfree mice 6 days postinfection with E. coli O157:H7 PA2 with and without E. coli C600
| E. coli inoculum | No. healthy | No. sick | No. moribund/dead |
|---|---|---|---|
| PA2 only | 11a | 0 | 0 |
| PA2 + C600 | 2 | 5 | 3 |
The PA2-plus-C600-infected mice had a higher morbidity and mortality rate than mice infected with PA2 alone (P < 0.05 by chi-square test).
FIG 7.
Stx2a levels in feces of germfree mice. Stx2a levels were quantified, using ELISA, from the feces of PA2- and the PA2-plus-C600-infected mice on day 1 and day 6 postinoculation. The error bars represent SEM from different mice.
FIG 8.
Coculture with PA2 and C600 causes increased kidney damage in germfree mice. (A to F) Hematoxylin- and eosin-stained sections. (A, C, and E) Cross sections of right kidney (original magnification, ×200). (B, D, and F) Corticomedulary junction (original magnification, ×2,000). (A and B) Mouse colonized with C600. No lesions are present. (C and D) Mouse colonized with PA2. The cross section of the right kidney reveals occasional dilated tubules. Higher magnification of the boxed area demonstrates mild tubular epithelial necrosis and regeneration (arrows). (E and F) Mouse colonized with C600 followed by PA2. The cross section of the right kidney demonstrates widespread tubular dilation. Higher magnification of the boxed area demonstrates severe, widespread tubular epithelial necrosis and regeneration (arrows). (G) Acute tubular necrosis scores. Hematoxylin- and eosin-stained renal sections from mice infected with either C600 or PA2 and PA2 plus C600. The mice were scored as described in Materials and Methods. (H and I) Body weight change (H) and necrosis (I) scores measured 6 days postinoculation with PA2 and/or C600. The asterisks indicate that measurements from mice infected with PA2 or PA2 plus C600 were statistically different (P < 0.05) than those from mice infected with C600 alone.
DISCUSSION
Previous studies described how Stx2a production increases when O157:H7 is cocultured with phage-susceptible E. coli (28, 35). Our work augmented the understanding of this phenotype by demonstrating that the level of amplification depends upon the strain of E. coli O157:H7 used. Moreover, we identified clade 8 O157:H7 strains that produce low Stx2a levels compared to EDL933 and Sakai (Fig. 2 and 5A) but show a drastic increase in Stx2a accumulation when cocultured with the nonpathogenic E. coli strain C600 (Fig. 3 and 5A).
We used the prototypical strains Sakai and EDL933 to characterize the phenotypes of PA2 and other clade 8 E. coli O157:H7 strains. The first two were the first O157:H7 strains to be fully sequenced (14, 15). Sakai is a lineage I, clade 1 strain carrying both stx1 and stx2a and was responsible for an outbreak that affected over 9,000 children in Sakai city, Japan. EDL933 is a clade 3, lineage I strain that also carries stx1 and stx2. It was implicated in the multistate hamburger outbreak in 1982 that caused at least 47 illnesses (11). Studies on Stx2 expression showed that human clinical isolates belonging to lineage I or I/II produce higher Stx2 levels than cattle-derived lineage II isolates (17, 20). Further classifying the strains based on SNP typing identified a subset called clade 8, which is associated with higher incidence of HUS than other clades (19, 23). An example of a large outbreak caused by a clade 8 strain is the multistate spinach outbreak in 2006, with 205 cases and 3 casualties reported (36). It resulted in hospitalizations in over 60% of cases and a 13% HUS rate (19). The isolates PA2 and PA8 belong to a clade 8, lineage I/II strain and carry only stx2a.
According to the model proposed by Gamage et al., spontaneously induced phage infect susceptible strains of E. coli, leading to lysis and enhanced Stx2a production (28). We initially hypothesized that E. coli O157:H7 strains that were high Stx2a producers in the presence of ciprofloxacin (Fig. 2) would also be the highest producers when cocultured with C600. However, we were surprised to see that the lowest-Stx2a-producing strain, PA2, showed the highest toxin levels in coculture experiments (Fig. 3). Bacterial cell count data showed reduced numbers of the susceptible host when cocultured with PA2, further supporting the assumption that phage-mediated lysis of C600 was occurring (Fig. 4A). Moreover, we found that addition of supernatants from ciprofloxacin-induced PA2 cultures to E. coli C600 resulted in Stx2a production (data not shown). This leads to the conclusion that the fold amplification observed after coculture with C600 is not dependent on the basal Stx2a levels expressed by O157:H7. Toxin amplification could also be recapitulated when E. coli O157:H7 strains were cocultured with C600 in an ex vivo model using sterile bovine rectal mucus (data not shown). The fold amplification was less than that observed in laboratory media but provided further support for pursuing in vivo studies. We suggest that the amplification differences observed may be driven by polymorphisms between the stx2a-converting phage. It was noted previously that genes within the early regulation and replication regions differ between these phage in EDL933 and Sakai (29). The sequencing of the phage genomes from PA2 and PA8 (unpublished data) revealed that the stx2a-converting phage differ only by the position of an IS629 element, and both are related to the stx2a-converting phage VT2_phi272 (31). VT2_phi272 is more closely related to a phage from E. coli O104:H4 than it is to the one from EDL933 and Sakai (31, 37).
Gamage et al. (35) also tested whether the toxin amplification phenotype could be recapitulated in streptomycin-treated mice and obtained encouraging although inconclusive results. The study showed that 8 of 36 stool samples from mice precolonized with a phage-susceptible commensal E. coli strain prior to inoculation with E. coli O157:H7 had Shiga toxin levels above the limit of detection. Mice given the E. coli O157:H7 strain alone were more commonly toxin positive (17 of 36 stool samples). However, toxin quantifications were similar between the two groups. As pathogen colonization was approximately 50-fold less in cocultured mice, this suggested that the commensal may enhance toxin production (35). In our study, we used the established germfree mouse model (34) to limit the inherent variability when a complex microbiota is present. Our results clearly demonstrate that E. coli C600 adversely affects disease outcome in this model. As hypothesized, the Stx2a levels observed in vivo when PA2 was cocolonized with C600 was significantly higher than with PA2 alone (Fig. 7). In parallel with elevated Stx2a, cocolonized mice showed more severe signs of disease and were more likely to die than the mice inoculated with PA2 alone (Table 2). In addition, the histopathological data showed more extensive renal tubular necrosis in the PA2-plus-C600 mice than in the mice colonized by PA2 alone.
Given the dramatic in vitro and in vivo data related to strain PA2; the known link between Stx2a and development of severe outcomes, such as HUS (2, 3); and reports that strains of clade 8 are particularly virulent (19, 23), we tested whether the Stx2a amplification could be generalized to other clade 8 strains. We included strains that produce Stx2a only and strains that produce both Stx2a and Stx2c, as these are the two toxin genotypes most correlated with HUS development (23). We observed that not all clade 8 strains show increased Stx2a production during coculture (Fig. 5A), suggesting either that the mechanism described here is not essential for severe-disease development or that C600 is not an appropriate host for all Stx2-converting phage. As mentioned above, the phage from PA2 and PA8 are highly related; however, we do not yet have complete genome sequences for the phage from the other strains to assess whether the lack of amplification is due to genetic polymorphisms.
Taken together, these findings create a compelling argument to reconsider the appropriateness of assessing the virulence potential of O157:H7 strains solely by quantifying Stx2a production by pure cultures (20). There are 107 to 109 CFU/g E. coli bacteria in the human intestine (38) that vary both in susceptibility to phage infection (28) and in other mechanisms by which commensal E. coli enhances Shiga toxin production in vitro (39). A further exploration of the interactions between E. coli O157:H7 and the plethora of bacterial species present in the gut is needed to appreciate how the gut microbiome affects the virulence of this foodborne pathogen. It was also shown previously that fecal Bacteroides thetaiotaomicron alters Stx2 expression (40–42), although two studies (40, 42) report decreases while one (41) report shows an increase. It is noteworthy, though, that the first two studies quantified Stx2 after ciprofloxacin induction while the third study did not induce the phage during coculture. Regardless, these studies and the current report highlight the complex interactions affecting Stx2 production when Shiga toxin 2-producing E. coli interacts with other gut microorganisms.
ACKNOWLEDGMENT
This work was funded by USDA-NIFA grant 2010-65201-20619 to E.G.D.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Boerlin P, Mcewen SA, Wilson JB, Johnson RP, Gyles CL, Ewen SAMC, Boerlin-petzold F. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol 37:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ethelberg S, Olsen KEP, Scheutz F, Jensen C, Schiellerup P, Engberg J, Petersen AM, Olesen B, Gerner-Smidt P, Mølbak K. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg Infect Dis 10:842–847. doi: 10.3201/eid1005.030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner PL, Neely MN, Zhang X, Acheson WK, Waldor MK, Friedman DI, Acheson DWK. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J Bacteriol 183:2081–2085. doi: 10.1128/JB.183.6.2081-2085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandvig K, Garred O, Prydz K, Kozlov JV, Hansen SJ, van Deurs B. 1992. Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature 358:510–512. doi: 10.1038/358510a0. [DOI] [PubMed] [Google Scholar]
- 6.Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O'Brien AD. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun 53:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, Samuel JE. 1993. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect Immun 61:3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegler RL, Obrig TG, Pysher TJ, Tesh VL, Denkers ND, Taylor FB. 2003. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr Nephrol 18:92–96. [DOI] [PubMed] [Google Scholar]
- 9.Hartzell A, Chen C, Lewis C, Liu K, Reynolds S, Dudley EG. 2011. Escherichia coli O157:H7 of genotype lineage-specific polymorphism assay 211111 and clade 8 are common clinical isolates within Pennsylvania. Foodborne Pathog Dis 8:763–768. doi: 10.1089/fpd.2010.0762. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich AW, Bielaszewska M, Zhang W-L, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis 185:74–84. doi: 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- 11.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen M. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 12.Tilden J Jr, Young W, McNamara AM, Custer C, Boesel B, Lambert-Fair MA, Majkowski J, Vugia D, Werner SB, Hollingsworth J, Morris JG Jr. 1996. A new route of transmission for Escherichia coli: infection from dry fermented salami. Am J Public Health 86:1142–1145. doi: 10.2105/AJPH.86.8_Pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caprioli A, Morabito S, Brugère H, Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res 36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 15.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 16.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol 150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Nietfeldt J, Benson AK. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci U S A 96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Kovar J, Kim J, Nietfeldt J, Smith DR, Moxley RA, Olson ME, Fey D, Benson AK, Fey PD. 2004. Identification of common subpopulations of non-sorbitol-fermenting, β-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl Environ Microbiol 70:6846–6854. doi: 10.1128/AEM.70.11.6846-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci U S A 105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Laing CR, Zhang Z, Hallewell J, You C, Ziebell K, Johnson RP, Kropinski AM, Thomas JE, Karmali M, Gannon VPJ. 2010. Lineage and host source are both correlated with levels of Shiga toxin 2 production by Escherichia coli O157:H7 strains. Appl Environ Microbiol 76:474–482. doi: 10.1128/AEM.01288-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellor GE, Besser TE, Davis M, Beavis B, Jung W, Smith HV, Jennison AV, Doyle CJ, Chandry PS, Gobius KS, Fegan N. 2013. Multilocus genotype analysis of Escherichia coli O157 isolates from Australia and the United States provides evidence of geographic divergence. Appl Environ Microbiol 79:5050–5058. doi: 10.1128/AEM.01525-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Laing CR, Steele M, Ziebell K, Johnson R, Benson A, Taboada E, Gannon VPJ. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8:121. doi: 10.1186/1471-2164-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyoda S, Manning SD, Seto K, Kimata K, Isobe J, Etoh Y, Ichihara S, Magita Y, Ogata K, Honda M, Kubota T, Kawano K, Matsumoto K, Kudaka J, Asai N, Yabata J, Tominaga K, Terajima J, Morita-Ishihara T, Izumiya H, Ogura Y, Saitoh T, Iguchi A, Kobayashi H, Hara-Kudo Y, Ohnishi M, EHEC Working Group in Japan. 2014. Phylogenetic clades 6 and 8 of enterohemorrhagic Escherichia coli O157:H7 with particular stx subtypes are more frequently found in isolates from hemolytic uremic syndrome patients than from asymptomatic carriers. Open Forum Infect Dis 1:ofuD61. doi: 10.1093/ofid/ofu061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DL, James CE, Sergeant MJ, Yaxian Y, Saunders JR, McCarthy AJ, Allison HE. 2007. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J Bacteriol 189:7223–7233. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis 181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- 26.Loś JM, Loś M, Węgrzyn A, Węgrzyn G. 2010. Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol Med Microbiol 58:322–329. doi: 10.1111/j.1574-695X.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- 27.McGannon CM, Fuller CA, Weiss AA. 2010. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob Agents Chemother 54:3790–3798. doi: 10.1128/AAC.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamage SD, Strasser JE, Chalk CL, Weiss AA. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect Immun 71:3107–3115. doi: 10.1128/IAI.71.6.3107-3115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto H, Nakai W, Yajima N, Fujibayashi A, Higuchi T, Sato K, Matsushiro A. 1999. Sequence analysis of Stx2-converting phage VT2-Sa shows a great divergence in early regulation and replication regions. DNA Res 6:235–240. doi: 10.1093/dnares/6.4.235. [DOI] [PubMed] [Google Scholar]
- 30.Smith DL, Rooks DJ, Fogg PCM, Darby AC, Thomson NR, McCarthy AJ, Allison HE. 2012. Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics 13:311. doi: 10.1186/1471-2164-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laing CR, Zhang Y, Gilmour MW, Allen V, Johnson R, Thomas JE, Gannon VPJ. 2012. A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS One 7:e37362. doi: 10.1371/journal.pone.0037362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basta M, Karmali M, Lingwood C. 1989. Sensitive receptor-specified enzyme-linked immunosorbent assay for Escherichia coli verocytotoxin. J Clin Microbiol 27:1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Islam MR, Ogura Y, Asadulghani M, Ooka T, Murase K, Gotoh Y, Hayashi T. 2012. A sensitive and simple plaque formation method for the Stx2 phage of Escherichia coli O157:H7, which does not form plaques in the standard plating procedure. Plasmid 67:227–235. doi: 10.1016/j.plasmid.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Eaton KA, Friedman DI, Francis GJ, Tyler JS, Young VB, Haeger J, Abu-Ali G, Whittam TS. 2008. Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect Immun 76:3054–3063. doi: 10.1128/IAI.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamage SD, Patton AK, Strasser JE, Chalk CL, Weiss AA. 2006. Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect Immun 74:1977–1983. doi: 10.1128/IAI.74.3.1977-1983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centers for Disease Control and Prevention. 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb Mortal Wkly Rep 55:1045–1046. [PubMed] [Google Scholar]
- 37.Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer F-D, Boelter J, Petersen H, Gottschalk G, Daniel R. 2011. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol 193:883–891. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 39.Toshima H, Yoshimura A, Arikawa K, Hidaka A, Ogasawara J, Hase A, Masaki H, Nishikawa Y. 2007. Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl Environ Microbiol 73:7582–7588. doi: 10.1128/AEM.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. 2009. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun 77:783–790. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iversen H, Lindbäck T, L'Abée-Lund TM, Roos N, Aspholm M, Stenfors Arnesen L. 2015. The gut bacterium Bacteroides thetaiotaomicron influences the virulence potential of the enterohemorrhagic Escherichia coli O103:H25. PLoS One 10:e0118140. doi: 10.1371/journal.pone.0118140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Appleyard RK. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]