Abstract
Despite the high frequency of asymptomatic carriage of bacterial pathogens, we understand little about the bacterial molecular genetic underpinnings of this phenomenon. To obtain new information about the molecular genetic mechanisms underlying carriage of group A Streptococcus (GAS), we performed whole-genome sequencing of GAS strains recovered from a single individual during acute pharyngitis and subsequent asymptomatic carriage. We discovered that compared to the initial infection isolate, the strain recovered during asymptomatic carriage contained three single nucleotide polymorphisms, one of which was in a highly conserved region of a gene encoding a sensor kinase, liaS, resulting in an arginine-to-glycine amino acid replacement at position 135 of LiaS (LiaSR135G). Using gene replacement, we demonstrate that introduction of the carrier allele (liaSR135G) into a serotype-matched invasive strain increased mouse nasopharyngeal colonization and adherence to cultured human epithelial cells. The carrier mutation also resulted in a reduced ability to grow in human blood and reduced virulence in a mouse model of necrotizing fasciitis. Repair of the mutation in the GAS carrier strain restored virulence and decreased adherence to cultured human epithelial cells. We also provide evidence that the carrier mutation alters the GAS transcriptome, including altered transcription of GAS virulence genes, providing a potential mechanism for the pleiotropic phenotypic effects. Our data obtained using isogenic strains suggest that the liaSR135G mutation in the carrier strain contributes to the transition from disease to asymptomatic carriage and provides new information about this poorly described regulatory system in GAS.
INTRODUCTION
Human bacterial pathogens, including Neisseria meningitidis (1), Staphylococcus aureus (2), Streptococcus pneumoniae (3), and Streptococcus pyogenes (group A Streptococcus [GAS]) (4), are frequently carried asymptomatically. However, in contrast to the relatively sophisticated knowledge regarding how bacterial pathogens cause disease, we understand little of the processes used by bacteria to persist on mucosal surfaces in the absence of symptoms. Asymptomatic carriage by or colonization of a susceptible host is a key step in the development of a myriad of diseases caused by bacterial pathogens. Inhibiting the ability of a pathogen to colonize a host may severely hamper its ability to subsequently cause disease. For example, vaccination against S. pneumoniae reduces the potential for disease by interfering with the organism's ability to colonize mucosal surfaces (5). In contrast, no effective GAS vaccine exists, and the ability of current candidate GAS vaccine formulations to prevent colonization is unknown. Thus, enhancing our understanding of the mechanisms used to colonize human hosts is paramount to reducing the incidence of disease caused by bacterial pathogens.
GAS is an ideal model organism for the study of bacterial asymptomatic carriage. GAS causes a broad range of diseases in humans, including severe invasive diseases, such as necrotizing fasciitis (flesh-eating disease) and streptococcal toxic shock syndrome, and mild infections, such as impetigo and pharyngitis (6). In addition, GAS is carried asymptomatically in the throats of healthy individuals. Between 5 and 15% in children carry GAS asymptomatically (7), but this rate of carriage exceeds the incidence of any disease caused by GAS. For example, in the United States, the rate of invasive disease caused by GAS ranges from 3 to 4/100,000 people per year (8). Despite the high rate of asymptomatic carriage and in contrast to the multitude of studies examining GAS virulence, we have only a rudimentary understanding of the bacterial molecular mechanisms that contribute to GAS asymptomatic carriage.
Gene regulation is critical for GAS and other bacterial pathogens in the transition from initial colonization to disease or to respond to host selective pressures. Gene regulation in GAS uses several two-component regulatory systems (TCSs) consisting of a membrane-anchored sensor kinase, used for sensing environmental stimuli, that upon activation phosphorylates an associated cytoplasmic transcriptional response regulator (9). The CovRS regulatory system is the best-described TCS in GAS (10, 11). Inactivation of CovRS alters approximately 10% of the GAS transcriptome and derepresses or increases the expression of several virulence genes (10–12). Small mutations affecting CovRS regulation may play a crucial role in the transition to an invasive disease state. Studies of paired superficial and invasive isolates from the same patient suggest that altered gene regulation through covRS mutation may facilitate GAS breaching of epithelial barriers and cause invasive disease (13, 14). Extensive whole-genome investigations with serotype M3 (15) and M1 (16) invasive GAS strains also found high frequencies of mutation in covRS. In contrast to CovRS, relatively few studies of the other TCSs in GAS have been conducted, which means that their contribution to GAS disease or asymptomatic carriage is unknown.
The GAS carrier state has been described to be an enigma for decades. However, exceedingly few studies examining the ability of GAS carrier strains to colonize mucosal surfaces and cause disease exist. One hypothesis is that changes in the physiologic state of the organism are key to the development of the carrier state (17) and that the transition from a disease-causing state to a carrier state is facilitated by mutation. Very early studies clearly demonstrated that the longer that GAS was carried in the human throat the less likely it was that those individuals would transmit disease (17, 18). A decreased risk of transmission was also associated with a decreased bacterial burden in the human upper respiratory tract (19). Recently, using a mouse nasopharyngeal colonization model, it was shown that a decreased bacterial burden was associated with single nucleotide mutations in GAS carrier strains that eliminated capsule production (20). Despite relatively modest decreases in bacterial burden, the carrier mutations resulted in large decreases in virulence. Similar patterns have been observed in GAS carrier strains with mutations affecting expression of the stand-alone regulator Mga (21) and the surface protein SclA (22). Taken together, these data indicate that it is undoubtedly the case that multiple genetic pathways contribute to GAS carriage, making individual mutations rare events. That is, the frequency with which an individual mutation would occur in a carrier population is exceedingly low, given the multiple routes to a common phenotype. Importantly, relatively small genetic changes in GAS carrier strains result in distinct phenotypes depending on the host niche, revealing new information about the biology of this important pathogen (20–22).
Herein, we describe a mutation in the gene encoding the sensor kinase, liaS, of the three-component system (3CS) LiaFSR in a GAS carrier strain. Using allelic exchange, we provide evidence that the mutation in liaS contributes to the transition from disease to persistence by increasing the ability or the organism to adhere to and persist on mucosal surfaces and decreasing its virulence. Furthermore, the carrier mutation significantly alters global gene transcription, including the transcription of known virulence genes. Little is known regarding the LiaFSR 3CS in GAS, and our studies provide critical new information on GAS regulation of virulence, in addition to new pathways leading to GAS carriage.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table S1 in the supplemental material. MGAS10870 is a serotype M3 GAS strain isolated in 2002 from an individual with a soft tissue infection in Ontario, CA (23). The genome of strain MGAS10870 has been sequenced (23). MGAS23412 was isolated from an individual with GAS pharyngitis, and MGAS23431 was isolated from the same individual 63 days later during asymptomatic carriage (20). The genomes of MGAS23412 and MGAS23431 have been sequenced (20). All GAS strains were grown in Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY; Difco Laboratories) unless otherwise specified. Additional details of the experimental growth conditions are provided in the supplemental material.
Generation of isoallelic mutants of MGAS10870 and MGAS23431.
The plasmids and primers used in this study are listed in Table S2 in the supplemental material. We used a previously described procedure for generating the mutants MGAS10870liaSR135G (whose genome carries liaS with an R-to-G change at position 135 [liaSR135G]) and MGAS23431liaSWT (whose genome carries the liaS wild type [WT]) (20, 21). The sequences of all mutants were confirmed using Sanger sequencing (Applied Biosystems). Details of the methods used for mutant generation are provided in the supplemental material.
MIC determination.
MICs were determined using broth microdilution in a 96-well plate. Serial dilutions of nisin (Sigma-Aldrich), polymyxin B (Sigma-Aldrich), LL-37 (AnaSpec), colistin (Sigma-Aldrich), bacitracin (Sigma-Aldrich), daptomycin (Sigma-Aldrich), and vancomycin (Sigma-Aldrich) were performed in THY. Strains were grown to mid-exponential (ME) phase (optical density at 600 nm [OD600], ≈0.5) in THY, and subsequently, 20 μl of culture was added to 180 μl of THY (with and without antibiotic) in triplicate in a 96-well optically clear plate. The plates were incubated at 37°C and scanned every 30 min for 8 h using a plate reader (BioTek Synergy H1). The lowest concentration at which growth was inhibited compared to the growth on plates lacking antibiotic was considered the MIC. Assays were performed in triplicate with a minimum of 2 biological replicates.
Mouse nasopharyngeal and intramuscular infection.
All animal experiments were conducted under a protocol approved by the Houston Methodist Research Institute Institutional Animal Care and Use Committee. A total of 30 2-week old female CD1 mice (Harlan Laboratories) were inoculated intranasally with 5 × 107 CFU of the appropriate GAS strain in 50 μl phosphate-buffered saline (PBS). Mouse throats were swabbed prior to inoculation to document the absence of beta-hemolytic bacteria and daily thereafter for a total of 14 days. For intramuscular infection, mice were inoculated in the right hind limb with 5 × 106 CFU of the appropriate GAS strain in 100 μl PBS. Infected mice were assessed for histopathology, and bacteria were enumerated at 24 and 72 h postinfection as previously described (24). All mice were observed and sacrificed when they reached a condition near mortality, determined using predefined criteria (24).
Cultured human epithelial cell adherence assays.
Adherence to cultured human epithelial cells was carried out as previously described (20). Approximately 1 × 107 CFU GAS (multiplicity of infection, ≈10) grown to mid-exponential phase was added to 8 replicate wells previously seeded with HaCaT cells, and the plate was rocked briefly and incubated for 2 h at 37°C. See the supplemental material for experimental details on the adherence assays. Percent adherence was calculated by dividing the number of CFU recovered by the number of CFU in the original inoculum.
Ex vivo bactericidal assays in human blood.
Growth in whole human blood was conducted under a Houston Methodist Research Institute Institutional Review Board experimental protocol and performed as described by Lancefield (25). Blood from a minimum of two, healthy, nonimmune adult donors was used for each experiment. Details about the bactericidal assays in human blood are given in the supplemental material. Multiplication factors were calculated by dividing the number of CFU per milliliter obtained after 3 h of incubation by the number of CFU per milliliter in the initial inoculum.
RNA isolation and qRT-PCR analysis.
Growth of GAS strains, RNA isolation, cDNA synthesis, and quantitative real-time PCR (qRT-PCR) analysis were performed as previously described (21). See the supplemental material for experimental details. The primers and probes used in the transcript analyses are given in Table S2 in the supplemental material. All reactions were performed in triplicate using RNA purified from at least three biological replicates.
RNA sequencing (RNA-seq).
GAS strains were grown in THY, and cells were harvested by centrifugation at either ME or early stationary (ES) phase of growth. RNA isolation, RNA purification, rRNA depletion, and the development of adapter-tagged cDNA libraries were performed as previously described (26). cDNA libraries were run on an Illumina HiSeq 2000 to obtain 51-bp reads, and the reads were subsequently mapped to the reference serotype M3 genome using the CLC Genomics Workbench, version 8 (Qiagen). Pairwise comparisons of 1,672 coding regions (the total number after exclusion of tRNA- and rRNA-encoding regions) were carried out following normalization using empirical analysis of differentially expressed genes (genes for which the change in expression was >1.5-fold with a P value of <0.05 after applying Bonferroni's correction, which was considered to be a statistically significant difference in expression).
Statistics.
A two-tailed t test (unequal variance) was used to compare multiplication factors between strains grown in human blood, adherence of strains to epithelial cells, and gene transcript levels. Kruskal-Wallis analysis of variance (ANOVA) was used to compare rates of nasopharyngeal colonization between strains. A two-way ANOVA with correction for multiple comparisons (Bonferroni) was used to compare the in vitro growth of the strains. A log rank test was used to compare survival, and a Mann-Whitney U test was performed to compare bacterial CFU counts following intramuscular infection in mice. A P value of less than 0.05 was considered significant for all statistical tests.
RESULTS
Identification of a GAS carrier strain with a single nucleotide mutation in liaS.
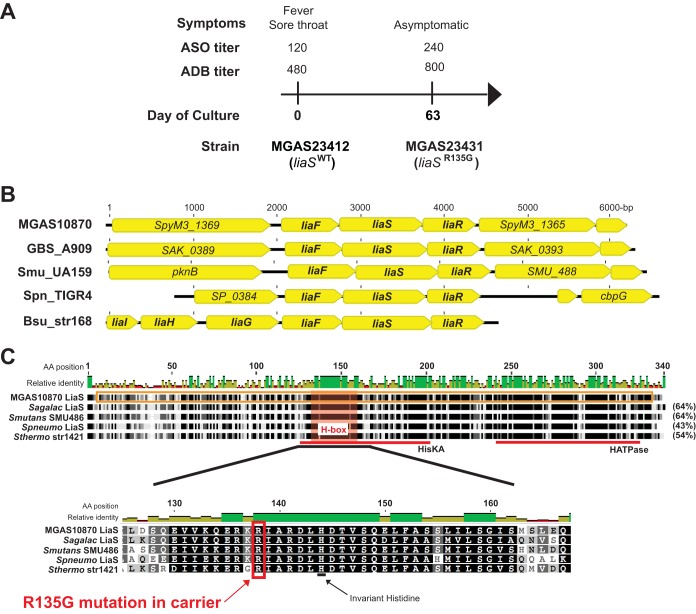
We previously reported the complete genome sequence of serotype M3 GAS strains isolated serially over time from the throats of human subjects (20). Examination of the genome sequences of strains recovered from one patient revealed that the strain recovered during asymptomatic carriage (day 63) contained three single nucleotide polymorphisms (SNPs) compared with the sequence of the original infecting isolate (day 0): one in SpyM3_1367, one in SpyM3_0244, and one in SpyM3_0499. The SNP in SpyM3_1367, encoding a putative sensor kinase, resulted in an amino acid change from arginine (R) to glycine (G) at position 135 (R135G) in the putative protein. Importantly, at the time of isolation of the initial infecting isolate, signs and symptoms of pharyngitis (e.g., sore throat, fever) were present. The patient subsequently showed an immunological response to GAS infection with an increase in both serum anti-streptolysin O (ASO) and anti-DNase B (ADB) titers at day 63, further suggesting bona fide GAS pharyngitis at day 0 (Fig. 1A). The carrier strain isolated at day 63 differed from the initial infecting isolate by only three polymorphisms in the entire genome, strongly suggesting that the carrier strain is a lineal descendant of the original strain and not a different strain causing a new infection. Previous analyses of over 200 serotype M3 disease-causing GAS strains (15) did not identify polymorphisms in SpyM3_1367, suggesting that the mutation in this locus may be unique to carrier strains.
FIG 1.
A mutation in liaS develops during asymptomatic carriage and is predicted to affect protein function. (A) Timing of strain collection, symptoms, and selected laboratory values of a patient with serotype M3 GAS pharyngitis. Anti-streptolysin O (ASO) and anti-DNase B (ADB) serum titers are given above the timeline. (B) Comparison of the liaFSR operons from serotype M3 GAS (MGAS10870), Streptococcus agalactiae strain A909 (GBS_A909), Streptococcus mutans strain UA159 (Smu_UA159), Streptococcus pneumoniae strain TIGR4 (Spn_TIGR4), and Bacillus subtilis strain 168 (Bsu_str168). (C) Homology of the LiaS (SpyM3_1367) proteins from GAS (MGAS10870 LiaS), Streptococcus agalactiae (Sagalac LiaS), Streptococcus mutans (Smutans SMU486), Streptococcus pneumoniae (Spneumo LiaS), and Streptococcus thermophilus (Sthermo str1421). The percent identity to the MGAS10870 LiaS is given in parentheses on the right of the alignment. The highly conserved regions representing the highly conserved histidine kinase A (HisKA) and histidine kinase-type ATPase (HATPase) catalytic domains are indicated with red lines. The H box harboring the invariant histidine residue is magnified, and the position of the amino acid (AA) change in the GAS carrier strain is shown.
The inferred protein encoded by SpyM3_1367 has homology (29% amino acid identity) to the sensor kinase, LiaS, of a three-component system (3CS), LiaFSR, in Bacillus subtilis (27). BLAST analysis of SpyM3_1367 revealed a high degree of conservation (>99% protein identity) among sequenced GAS serotypes and homology to the 3CS sensor kinase LiaS from other Firmicutes (Fig. 1B and C). Based on the protein homology and the conservation of a genome location adjacent to the putative genes for LiaF and LiaR (Fig. 1B), we propose that SpyM3_1367 henceforth be referred to as liaS. The amino acid change found in the carrier strain (R135G) is in a very highly conserved region adjacent to the invariant histidine (H box) (28) of the putative sensor kinase (Fig. 1C). Thus, we hypothesized that the mutation identified in liaS contributes to the transition to asymptomatic carriage in GAS.
The carrier strain LiaSR135G increases antibiotic susceptibility.
The Lia (lipid II-interacting antibiotics) 3CS was originally identified in Bacillus subtilis as part of a regulatory network responding to cell wall stress induced by antibiotics, such as bacitracin and cationic antimicrobial peptides (CAMPs) (29). Homologous systems have also been described in other members of the Firmicutes family, including the human pathogens Listeria monocytogenes (30), Staphylococcus aureus (31), Streptococcus agalactiae (32), and Streptococcus pneumoniae (33). Studies of the Lia 3CS have reported a response to antibiotics that interferes with the lipid II cycle (29, 31, 32). Thus, we hypothesized that if the carrier mutation alters the LiaS function, we would observe increased susceptibility to lipid II-interfering antibiotics. To directly test this hypothesis, we generated a mutant strain (MGAS10870liaSR135G) that differed from parental invasive serotype M3 strain MGAS10870 by only the carrier liaS allele. No significant difference in the growth rate between the parental and mutant strains was observed (see Fig. S1A and B in the supplemental material). However, mutant strain MGAS10870liaSR135G exhibited a moderate increase in chain length in late exponential phase compared to that of parental strain MGAS10870 (see Fig. S1C in the supplemental material). The phenotype of an increase in chain length in the mutant was no longer apparent in stationary phase.
We next tested the strains for susceptibility to several antibiotics. Consistent with our hypothesis of altered LiaS function in the carrier strain, we observed an increased susceptibility of the mutant strain to bacitracin, polymyxin B, colistin, daptomycin, and nisin compared to that of the parental strain but no difference in susceptibility to vancomycin (Table 1). Thus, the liaSR135G carrier mutation increases susceptibility to antimicrobial agents targeting cell envelope synthesis and maintenance, consistent with the findings of previous studies of LiaFSR (29, 31, 32).
TABLE 1.
MICs of selected antimicrobials
| Strain | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|
| Vancomycin | Bacitracin | Polymyxin B | Colistin | Daptomycin | Nisin | LL-37 | |
| MGAS10870 | 0.5 | 0.025 | 20 | 50 | 2 | 2 | 175 |
| MGAS10870liaSR135G | 0.5 | 0.0125 | 10 | 25 | 1 | 1 | 125 |
The carrier allele liaSR135G confers an increased ability to colonize and persist in the mouse nasopharynx in vivo.
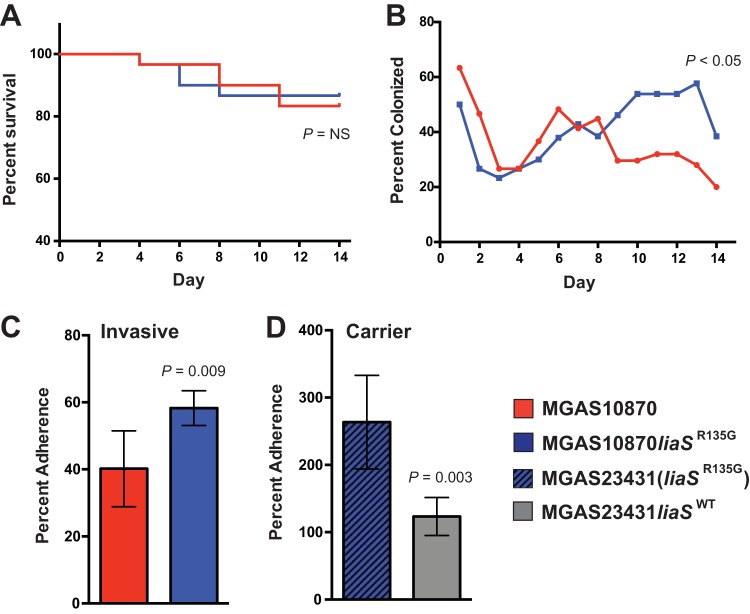
Previous studies have suggested that GAS carrier strains are characterized by an increased ability to adhere to mucosal surfaces and decreased virulence (20, 22). We hypothesized that the carrier allele liaSR135G may alter the ability of GAS strains to adhere to and colonize mucosal surfaces. To test this hypothesis, we inoculated mice intranasally with either the parental invasive strain or MGAS10870liaSR135G and compared the rates of bacterial recovery from the mouse nasopharynx. No significant differences in survival between mice inoculated with the parental strain and mice inoculated with the mutant strain were observed (Fig. 2A). However, beginning on day 8, MGAS10870liaSR135G was recovered from mice at a significantly higher rate than the MGAS10870 parental strain (Fig. 2B), suggesting that MGAS10870liaSR135G has an increased ability to colonize the mouse nasopharynx. This observation stands in stark contrast to the findings of previous studies of carrier strain mutations, where decreases in mouse nasopharyngeal colonization were described (20–22).
FIG 2.
Carrier allele liaSR135G increases mouse nasopharyngeal colonization and in vitro adherence. (A) Kaplan-Meier survival curve following nasopharyngeal infection with either the invasive (liaSWT; red line) or the mutant (blue line) strain. P values were determined by the log-rank test. (B) Rates of bacterial recovery from mice (n = 30) following inoculation of the invasive (liaSWT; red line) or the mutant (blue line) strain. P values were determined using Kruskal-Wallis repeated-measures ANOVA. (C and D) Cultured epithelial cell (HaCaT) adherence assay using the invasive strain and the derived mutant (C) or the carrier strain [MGAS23431(liaSR135G)] and the repaired mutant (MGAS23431liaSWT) (D). Error bars represent SEMs, and P values were determined using a t test (unequal variance). NS, not significant.
In vitro adherence to cultured human epithelial cells is increased in strains with liaSR135G.
To further characterize the ability of the carrier mutant to colonize mucosal surfaces, we performed assays of in vitro adherence to cultured human epithelial cells. Based on the increased ability of strain MGAS10870liaSR135G to colonize the nasopharynx of mice, we hypothesized that the carrier allele would confer an increased ability to adhere to cultured human epithelial cells. Consistent with the differences in mouse colonization observed, MGAS10870liaSR135G had significantly increased adherence to HaCaT cells compared to parental strain MGAS10870 (Fig. 2C). As a further test of the hypothesis, we repaired the mutation present in carrier strain MGAS23431 recovered on day 63 (Fig. 1A) and compared the ability of both the carrier [MGAS23431(liaSR135G)] and the mutant (MGAS23431liaSWT) strains to adhere to cultured human epithelial cells. In line with the hypothesis that liaSR135G contributes to increased adherence, the parental carrier strain [MGAS23431(liaSR135G)] had significantly increased cell adherence compared to the repaired mutant strain (MGAS23431liaSWT) (Fig. 2D).
Strains with the carrier liaSR135G allele have decreased virulence in an in vivo mouse model of GAS invasive disease.
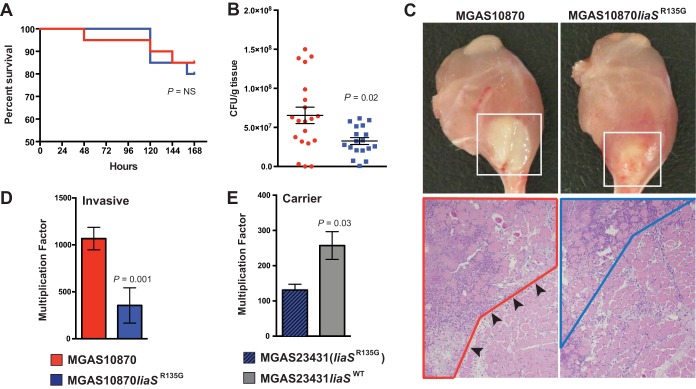
GAS carrier strains have significantly reduced virulence in mice compared to invasive strains of the same serotype (22, 34). We hypothesized that the carrier mutation, liaSR135G, results in decreased virulence in in vivo models of GAS disease. We tested the virulence of the parental invasive strain and mutant strain MGAS10870liaSR135G in a mouse model of necrotizing fasciitis. We observed similar rates of survival for mice infected with the parental strain and the MGAS10870liaSR135G strain (Fig. 3A). However, at 72 h significantly fewer numbers of CFU were recovered from mice infected with MGAS10870liaSR135G than from mice infected with the parental strain (Fig. 3B). Visual inspection of mouse limbs at 72 h postinfection showed a smaller abscess-like lesion after infection with MGAS10870liaSR135G (Fig. 3C, top). The histopathology at 24 h postinfection correlated with the visual appearance (Fig. 3C, bottom). The parental invasive strain produced severe necrotizing fasciitis and myositis, while the amount of necrosis caused by strain MGAS10870liaSR135G was decreased and more viable tissue was present, consistent with the smaller lesion size.
FIG 3.
Carrier allele liaSR135G decreases virulence in in vivo and ex vivo models of GAS disease. (A) Kaplan-Meier survival curve of mice (n = 15) after intramuscular infection with the parental invasive (red line) or mutant (blue line) strain. (B) The recovery of each bacterial strain (numbers of CFU) from mice (n = 20) at 72 h postinfection is plotted as the mean ± SEM. P values were determined by the Mann-Whitney U test. Red symbols, invasive strain; blue symbols, mutant strain. (C) Visual (top; 72 h postinoculation) and microscopic (bottom; 24 h postinoculation; magnification, ×4) inspection of mouse limbs. MGAS10870 caused a large necrotic lesion centered at the inoculation site (white box). Microscopic examination revealed extensive areas of necrosis (area with red outline) that breached fascial planes (black arrowheads) to cause severe muscle destruction with numerous nonviable (ghost) cells. MGAS10870liaSR135G caused a comparatively smaller lesion (white box) with less severe fascial and muscle necrosis (area with blue outline). (D) Bactericidal assays of the parental invasive strain and the isoallelic mutant strain. (E) Bactericidal assays of the carrier strain [MGAS23431(liaSR135G)] and the repaired mutant (MGAS23431liaSWT). The error bars in panels D and E represent SEMs, and P values were determined by the t test (unequal variance).
The liaSR135G carrier allele results in a decreased ability to grow ex vivo in human blood.
In the one previous study to examine LiaS in GAS, a serotype M1 GAS liaS deletion mutant had a reduced ability to survive in human blood (35). Thus, we next hypothesized that the carrier mutation in liaS in serotype M3 GAS would decrease the ability to grow in human blood. We tested the ability of parental strain MGAS10870 and the derived mutant strain to survive in human bactericidal assays. Compared to the parental invasive strain (with liaSWT), MGAS10870liaSR135G had a significantly reduced ability to multiply in human blood (Fig. 3D). To further test the hypothesis, we compared the abilities of the MGAS23431 carrier strain recovered on day 63 [MGAS23431(liaSR135G)] (Fig. 1A) and the MGAS23431liaSWT mutant to grow in human blood. Again, consistent with liaSR135G reducing virulence, we observed a significantly increased ability of MGAS23431liaSWT to multiply in human blood compared to that of the carrier strain MGAS23431 with the liaSR135G mutation (Fig. 3E). Taken together, the data on virulence for mice in vivo and growth in human blood ex vivo suggest that the carrier mutation liaSR135G decreases virulence in GAS.
liaSR135G significantly alters the GAS transcriptome in vitro.
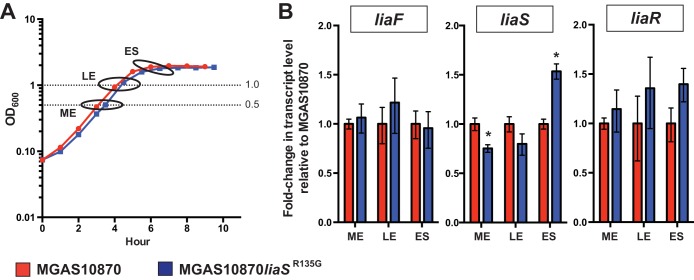
The data presented thus far demonstrate that the carrier allele liaSR135G confers an increased ability to adhere to and persist on mucosal surfaces and decreased virulence. Inasmuch as liaS is part of a 3CS whose homologues are known to regulate global gene transcription in other Gram-positive pathogens (32), we hypothesized that the phenotypic differences observed in strain MGAS10870liaSR135G are the result of an altered transcriptome, including altered transcription of GAS virulence genes. Prior to global transcript analyses, we first assayed the transcript levels of liaF, liaS, and liaR at three stages of growth in vitro in parental invasive strain MGAS10870 using qRT-PCR (Fig. 4A). Gene transcripts were detected in all phases of growth assayed. We compared the transcript levels between the parental and mutant strains at mid-exponential (ME), late exponential (LE), and early stationary (ES) phases of growth. We observed modest but significant differences in the levels of liaS transcripts from MGAS10870liaSR135G and the parental wild-type strain at the ME and ES phases of growth (Fig. 4B). In contrast, no significant differences in liaR or liaF transcript levels were observed between MGAS10870liaSR135G and MGAS10870.
FIG 4.
Carrier allele liaSR135G minimally alters the levels of the liaF, liaS, and liaR transcripts. (A) Growth curves of strains indicating the time points used for transcript analyses (ovals). Dotted lines, growth at ME phase (OD600, 0.5) and LE phase (OD600, 1.0). The ES phase was 2 h after the LE phase. (B) TaqMan analysis of liaF, liaS, and liaR transcripts in the parental and isoallelic mutant strains at ME, LE, and ES phases. Error bars represent SEMs, and P values were determined by the t test (unequal variance). *, P < 0.05.
Few studies have defined the global regulation by LiaFSR in Gram-positive human pathogens. Most recently, comparison of a wild-type strain and an liaR deletion strain in group B Streptococcus (GBS; Streptococcus agalactiae) found altered expression of >200 genes, including genes encoding known and putative virulence genes (32). To test our hypothesis that the carrier mutation alters GAS global gene regulation, we compared the transcriptomes of parental strain MGAS10870 and the isoallelic mutant MGAS10870liaSR135G using RNA-seq. Differential gene expression was compared at both ME and ES phases of growth, as modest but significant differences in liaS transcript levels were identified by qRT-PCR (Fig. 4B).
Compared to the levels of expression in the wild-type parental strain, 111 genes (6.6% of the genome) were significantly differentially expressed (>1.5-fold, P < 0.05 after Bonferroni correction) in MGAS10870liaSR135G in the ME phase of growth (Table 2; see also Table S3 in the supplemental material). Of the 111 differentially expressed genes, 52 were significantly upregulated and 59 were significantly downregulated. Of note, decreased d-alanylation of teichoic acids is known to reduce virulence in GAS (36, 37), and we observed significantly reduced transcription from the operon encoding the genes (dltABCD) necessary for d-alanylation of teichoic acids in MGAS10870liaSR135G compared to their levels of transcription in the parental strain (Table 2; see also Table S3 in the supplemental material). A summary of the most highly up- and downregulated genes is presented in Table 2.
TABLE 2.
Most highly differentially expressed genes in MGAS10870liaSR135G versus MGAS10870 at ME phase of growth
| Regulation | Locus taga | Gene name | Fold change | P valueb |
|---|---|---|---|---|
| Downregulated | SpyM3_1799 | spxA2 | −5.4 | <1.0E−23 |
| SpyM3_0411 | −3.3 | <1.0E−23 | ||
| SpyM3_1713 | trpG | −2.7 | 5.8E−09 | |
| SpyM3_0993 | dltB | −2.6 | 1.0E−11 | |
| SpyM3_0364 | −2.5 | 5.6E−11 | ||
| Upregulated | SpyM3_1409 | sdn | 2.7 | 2.8E−12 |
| SpyM3_0535 | 2.7 | 5.0E−12 | ||
| SpyM3_0077 | 3.0 | 4.8E−11 | ||
| SpyM3_1666 | 3.2 | 1.4E−09 | ||
| SpyM3_0068 | 4.7 | 4.4E−23 |
Locus tag defined in the serotype M3 strain MGAS315 reference genome.
P value after Bonferroni correction.
Transcriptome comparisons at ES phase revealed 127 significantly differentially expressed genes (7.6% of the genome), with 82 genes being upregulated and 45 genes being downregulated in MGAS10870liaSR135G compared to their levels of expression in the parental strain (Table 3; see also Table S4 in the supplemental material). We previously reported that a mutation decreasing the level of transcription of the gene encoding the stand-alone regulator Mga contributed to a carrier phenotype (21). Similarly, compared to the level of expression in the parental strain, we observed a significant downregulation of mga in the carrier mutant MGAS10870liaSR135G (see Table S4 in the supplemental material). The gene emm3, encoding M protein and regulated by Mga (38), was downregulated in ES but did not meet the statistical criteria for inclusion (−1.6, P = 0.056).
TABLE 3.
Most highly differentially expressed genes in MGAS10870liaSR135G versus MGAS10870 at ES phase of growth
| Regulation | Locus taga | Gene name | Fold change | P valueb |
|---|---|---|---|---|
| Downregulated | SpyM3_1799 | spxA2 | −3.8 | <1.0E−38 |
| SpyM3_0985 | malG | −2.4 | 1.9E−07 | |
| SpyM3_0014 | −2.4 | 1.1E−08 | ||
| SpyM3_0984 | malF | −2.3 | 2.0E−08 | |
| SpyM3_0375 | rnc | −2.3 | 3.7E−05 | |
| Upregulated | SpyM3_0617 | pyrE | 5.4 | 5.2E−25 |
| SpyM3_0578 | fruR | 5.4 | 7.0E−32 | |
| SpyM3_0579 | fruK | 5.6 | 1.5E−34 | |
| SpyM3_0561 | carA | 5.6 | 1.9E−38 | |
| SpyM3_0562 | carB | 6.2 | 2.7E−29 |
Locus tag defined in the serotype M3 strain MGAS315 reference genome.
P value after Bonferroni correction.
A total of 32 genes (28.8% of genes differentially expressed in ME phase and 25.2% of genes differentially expressed in ES phase) were similarly differentially expressed between the two phases of growth (see the bold font in Tables S3 and S4 in the supplemental material). Uncharacterized or hypothetical proteins accounted for 31% (10/32) of these shared genes. Common to both phases of growth was the upregulation in MGAS10870liaSR135G of the gene speB, encoding the secreted cysteine protease. The most highly differentially expressed gene at both phases of growth was spxA2 (SpyM3_1799; Tables 2 and 3), which encodes an RNA polymerase binding protein conserved in many Gram-positive bacteria (39). In summary, the carrier mutation identified in liaS significantly and distinctly altered the GAS transcriptome at multiple phases of growth in vitro, and the patterns of differential gene expression are consistent with the observed phenotype of the carrier mutant.
DISCUSSION
The mechanisms used by GAS and other bacterial pathogens to persist in the human throat in the absence of symptoms are poorly understood. Recent longitudinal studies of bacterial isolates obtained from chronic infections have identified similar patterns of in vivo adaptation. That is, mutations predicted to negatively affect virulence were frequently identified in chronic infections with Pseudomonas aeruginosa (40, 41), Burkholderia dolosa (42), and Burkholderia pseudomallei (43). In addition to studies in GAS (20, 34), comparison of the genome sequences of disease-causing and carrier strains of Staphylococcus aureus (44) and Neisseria meningitidis (45) suggests a role for small genetic changes in the transition from disease to carriage. Our line of investigation represents one of the only ones to compare the genome sequences of GAS disease-causing and carrier strains from the same individual and improves upon previous investigations by linking carrier-specific mutations in the transition from disease to asymptomatic carriage.
The current study expands our knowledge of GAS carriage. Using precise gene replacement, we conclusively demonstrate that the mutation in liaS identified during asymptomatic human carriage contributes to both decreased virulence and increased adherence. This finding is consistent with that of previous studies in which mutations identified in GAS carrier strains led to similar phenotypes (20–22). However, in contrast to the findings of previous GAS carrier studies, the presence of the liaSR135G carrier mutation increased the ability to recover GAS from the mouse nasopharynx, which perhaps suggests an advantage for persistence in the human throat.
In addition to providing new data on GAS carriage, our findings represent one of the few studies connecting the LiaFSR 3CS to virulence. A single study with a serotype M1 GAS strain showed that deletion of liaS results in decreased survival in human blood and in a mouse subcutaneous model of infection (35). However, complete restoration of the wild-type phenotype was not achieved with complementation, and no follow-up investigations ascribing a mechanism to the observed findings have been published. Two studies with S. pneumoniae link LiaFSR to virulence. In the first, signature-tagged mutagenesis (STM) revealed LiaF to be essential for murine pneumonia (46). Deletion of liaS in another study led to increased pilus expression (i.e., negative regulation) but decreased virulence in a mouse intranasal infection model (33). More recently, deletion of the gene encoding the LiaR response regulator in GBS decreased pilus expression and virulence in mouse models of sepsis and pneumonia (32).
The liaSR135G polymorphism is the first mutation identified in a human GAS carrier strain that has been found to alter global gene regulation. Transcriptome analysis strongly suggests that this carrier mutation affects protein function. Our finding that between 6% and 7% of GAS genes were differentially expressed is similar to the finding in GBS, in which approximately 8% of the genes were differentially expressed in an liaR deletion mutant (32). However, in contrast to investigations into LiaFSR in GBS (32) and S. pneumoniae (33), pilus gene expression was not significantly affected by the liaS carrier mutation in our analyses. It is important to note that this carrier mutation is predicted to alter but not eliminate LiaS protein function. Thus, it is plausible that deletion and thus a complete loss of function of liaS (or liaR) in GAS would affect pilus or other gene expression not observed with the carrier mutation alone.
Differential expression of several GAS virulence genes provides potential mechanisms for the pleiotropic effect of the liaSR135G carrier mutation. For example, in ME phase of growth, we observed decreased expression of the dlt operon (dltABCD), encoding the enzymes necessary for d-alanylation of lipoteichoic acids (LTAs) (47, 48). Modification of LTAs with d-alanine residues occurs in many Gram-positive bacteria, including S. aureus, GBS, and GAS (49). The LiaFSR 3CS has also been shown to positively regulate the dlt operon in Streptococcus gordonii (50). Mutants lacking the ability to form d-alanyl-LTA in GAS have increased susceptibility to cationic antimicrobial peptides (CAMPs), increased killing by human neutrophils, and reduced growth in human blood (36, 37). In addition to the decreased expression of the dlt operon, expression of the gene encoding the stand-alone regulator Mga was decreased in ES phase of growth in the carrier mutant, as was that of emm, although the decreased expression of emm did not reach statistical significance. Of note, GAS mutants lacking the ability to form d-alanyl-LTA have also been reported to have decreased amounts of cell wall-associated M protein (36). Furthermore, deletion of mga in serotype M2 and M49 GAS strains resulted in increased adherence to and internalization of cultured human epithelial cells (51). GAS carrier mutations that decrease mga and emm transcript levels have also been identified (21), suggesting an important role of Mga and M protein in the transition from disease to carriage. Specifically, decreased Mga and M-protein levels appear to be associated with carriage, a finding that may make an M-protein-based vaccine strategy suboptimal due to a lack of sterilizing immunity.
We also observed significantly increased speB transcript levels in the carrier mutant at both phases of growth studied. The SpeB cysteine protease secreted by GAS plays a significant role in GAS virulence (52). However, evidence also suggests that SpeB may promote colonization through a tight association with the bacterial cell surface and mediation of adhesion to the extracellular matrix component laminin (53). Increased adhesion to laminin has also been associated with a carrier mutation restoring function to the streptococcal collagen-like protein SclA (22). Thus, the sum of the evidence suggests that the liaSR135G carrier mutation contributes to the transition from disease to carriage by altering the expression of multiple GAS virulence genes. The precise mechanism leading to altered gene expression and the subsequent pleiotropic phenotypic effects requires further investigation.
Our study is limited in that we examined only the contribution of the liaSR135G mutation to the GAS carrier phenotype. In principle, it is possible that one or both of the other two polymorphisms identified in the carrier strain also contribute to the carrier phenotype and the transition from disease to carriage. However, the phenotype of the isogenic mutant strain MGAS10870liaSR135G recapitulates the carrier strain phenotype of increased adherence and decreased virulence (Fig. 2 and 3). Likewise, repair of the liaSR135G mutation in the carrier strain restored virulence and decreased adherence (Fig. 2D and 3E). Thus, the data support the conclusion that the liaSR135G carrier mutation is the major contributor to the carrier phenotype in this strain.
In summary, a unifying theme in GAS carriage research is beginning to emerge and may serve as a paradigm for carriage research with other human pathogens. Whole-genome comparisons of human disease and carrier strains reveal multiple independent pathways toward a carrier phenotype of persistence and decreased virulence. A recurring theme is that independent genetic changes that alter cell surface components and, thus, the host-microbe interaction are central to the carrier phenotype. Importantly, study of the carriage state may expose previously unrecognized virulence mechanisms leading to new control and prevention strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Concepcion Cantu for assistance with the animal studies and E. Kaplan and D. Johnson for providing the clinical information for the carrier subject studied.
A.R.F. is supported by a grant from the Harold Amos Medical Faculty Development Program of the Robert Wood Johnson Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00656-15.
REFERENCES
- 1.Yazdankhah SP, Caugant DA. 2004. Neisseria meningitidis: an overview of the carriage state. J Med Microbiol 53:821–832. doi: 10.1099/jmm.0.45529-0. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 3.Weiser JN. 2010. The pneumococcus: why a commensal misbehaves. J Mol Med 88:97–102. doi: 10.1007/s00109-009-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan EL. 1980. The group A streptococcal upper respiratory tract carrier state: an enigma. J Pediatr 97:337–345. doi: 10.1016/S0022-3476(80)80178-8. [DOI] [PubMed] [Google Scholar]
- 5.Dagan R, Patterson S, Juergens C, Greenberg D, Givon-Lavi N, Porat N, Gurtman A, Gruber WC, Scott DA. 2013. Comparative immunogenicity and efficacy of 13-valent and 7-valent pneumococcal conjugate vaccines in reducing nasopharyngeal colonization: a randomized double-blind trial. Clin Infect Dis 57:952–962. doi: 10.1093/cid/cit428. [DOI] [PubMed] [Google Scholar]
- 6.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaikh N, Leonard E, Martin JM. 2010. Prevalence of streptococcal pharyngitis and streptococcal carriage in children: a meta-analysis. Pediatrics 126:e557–e564. doi: 10.1542/peds.2009-2648. [DOI] [PubMed] [Google Scholar]
- 8.O'Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, Van Beneden C. 2007. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000-2004. Clin Infect Dis 45:853–862. doi: 10.1086/521264. [DOI] [PubMed] [Google Scholar]
- 9.Sitkiewicz I, Musser JM. 2006. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group A Streptococcus. Infect Immun 74:1339–1351. doi: 10.1128/IAI.74.2.1339-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horstmann N, Sahasrabhojane P, Suber B, Kumaraswami M, Olsen RJ, Flores A, Musser JM, Brennan RG, Shelburne SA III. 2011. Distinct single amino acid replacements in the control of virulence regulator protein differentially impact streptococcal pathogenesis. PLoS Pathog 7:e1002311. doi: 10.1371/journal.ppat.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Treviño J, Perez N, Ramirez-Peña E, Liu Z, Shelburne SA III, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun 77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia AF, Abe LM, Erdem G, Cortez CL, Kurahara D, Yamaga K. 2010. An insert in the covS gene distinguishes a pharyngeal and a blood isolate of Streptococcus pyogenes found in the same individual. Microbiology 156:3085–3095. doi: 10.1099/mic.0.042614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores AR, Sahasrabhojane P, Saldaña M, Galloway-Peña J, Olsen RJ, Musser JM, Shelburne SA. 2014. Molecular characterization of an invasive phenotype of group A Streptococcus arising during human infection using whole genome sequencing of multiple isolates from the same patient. J Infect Dis 209:1520–1523. doi: 10.1093/infdis/jit674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shea PR, Beres SB, Flores AR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Ayers SD, Webb P, Willey BM, Low DE, Musser JM. 2011. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc Natl Acad Sci U S A 108:5039–5044. doi: 10.1073/pnas.1016282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A 111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wannamaker LW. 1954. The epidemiology of streptococcal infection, p 157–175. In McCarty M. (ed), Streptococcal infections. Columbia University Press, New York, NY. [Google Scholar]
- 18.Kuttner AG, Krumwiede E. 1944. Observations on the epidemiology of streptococcal pharyngitis and the relation of streptococcal carriers to the occurrence of outbreaks. J Clin Invest 23:139–150. doi: 10.1172/JCI101477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamburger M, Green MJ, Hamburger VG. 1945. The problem of the dangerous carrier of hemolytic streptococci. II. Spread of infection by individuals with strongly positive nose cultures who expelled large numbers of hemolytic streptococci. J Infect Dis 79:68–81. [DOI] [PubMed] [Google Scholar]
- 20.Flores AR, Jewell BE, Olsen RJ, Shelburne SA III, Fittipaldi N, Beres SB, Musser JM. 2014. Asymptomatic carriage of group A Streptococcus is associated with elimination of capsule production. Infect Immun 82:3958–3967. doi: 10.1128/IAI.01788-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flores AR, Olsen RJ, Wunsche A, Kumaraswami M, Shelburne SA III, Carroll RK, Musser JM. 2013. Natural variation in the promoter of the gene encoding the Mga regulator alters host-pathogen interactions in group A Streptococcus carrier strains. Infect Immun 81:4128–4138. doi: 10.1128/IAI.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flores AR, Jewell BE, Versalovic EM, Olsen RJ, Bachert BA, Lukomski S, Musser JM. 2015. Natural variant of collagen-like protein A in serotype M3 group A Streptococcus increases adherence and decreases invasive potential. Infect Immun 83:1122–1129. doi: 10.1128/IAI.02860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. 2010. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A 107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen RJ, Sitkiewicz I, Ayeras AA, Gonulal VE, Cantu C, Beres SB, Green NM, Lei B, Humbird T, Greaver J, Chang E, Ragasa WP, Montgomery CA, Cartwright J Jr, McGeer A, Low DE, Whitney AR, Cagle PT, Blasdel TL, DeLeo FR, Musser JM. 2010. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A 107:888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lancefield RC. 1959. Persistence of type-specific antibodies in man following infection with group A streptococci. J Exp Med 110:271–292. doi: 10.1084/jem.110.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. 2015. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res 43:418–432. doi: 10.1093/nar/gku1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascher T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett 264:133–144. doi: 10.1111/j.1574-6968.2006.00444.x. [DOI] [PubMed] [Google Scholar]
- 28.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 30.Fritsch F, Mauder N, Williams T, Weiser J, Oberle M, Beier D. 2011. The cell envelope stress response mediated by the LiaFSRLm three-component system of Listeria monocytogenes is controlled via the phosphatase activity of the bifunctional histidine kinase LiaSLm. Microbiology 157:373–386. doi: 10.1099/mic.0.044776-0. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. [DOI] [PubMed] [Google Scholar]
- 32.Klinzing DC, Ishmael N, Dunning Hotopp JC, Tettelin H, Shields KR, Madoff LC, Puopolo KM. 2013. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology 159:1521–1534. doi: 10.1099/mic.0.064444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosch JW, Mann B, Thornton J, Sublett J, Tuomanen E. 2008. Convergence of regulatory networks on the pilus locus of Streptococcus pneumoniae. Infect Immun 76:3187–3196. doi: 10.1128/IAI.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci U S A 103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichikawa M, Minami M, Isaka M, Tatsuno I, Hasegawa T. 2011. Analysis of two-component sensor proteins involved in the response to acid stimuli in Streptococcus pyogenes. Microbiology 157:3187–3194. doi: 10.1099/mic.0.050534-0. [DOI] [PubMed] [Google Scholar]
- 36.Cox KH, Ruiz-Bustos E, Courtney HS, Dale JB, Pence MA, Nizet V, Aziz RK, Gerling I, Price SM, Hasty DL. 2009. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes. PLoS One 4:e5366. doi: 10.1371/journal.pone.0005366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. 2005. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol 187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caparon MG, Scott JR. 1987. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci U S A 84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol 186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. 2011. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, Keim P, Pearson T, Currie BJ. 2013. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 4(4):e00388–13. doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Miller RR, Godwin H, Knox K, Everitt RG, Iqbal Z, Rimmer AJ, Cule M, Ip CL, Didelot X, Harding RM, Donnelly P, Peto TE, Crook DW, Bowden R, Wilson DJ. 2012. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A 109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoen C, Blom J, Claus H, Schramm-Gluck A, Brandt P, Muller T, Goesmann A, Joseph B, Konietzny S, Kurzai O, Schmitt C, Friedrich T, Linke B, Vogel U, Frosch M. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A 105:3473–3478. doi: 10.1073/pnas.0800151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heaton MP, Neuhaus FC. 1992. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J Bacteriol 174:4707–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. 1995. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 270:15598–15606. [DOI] [PubMed] [Google Scholar]
- 49.Percy MG, Grundling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 50.McCormick NE, Halperin SA, Lee SF. 2011. Regulation of d-alanylation of lipoteichoic acid in Streptococcus gordonii. Microbiology 157:2248–2256. doi: 10.1099/mic.0.048140-0. [DOI] [PubMed] [Google Scholar]
- 51.Fiedler T, Kreikemeyer B, Sugareva V, Redanz S, Arlt R, Standar K, Podbielski A. 2010. Impact of the Streptococcus pyogenes Mga regulator on human matrix protein binding and interaction with eukaryotic cells. Int J Med Microbiol 300:248–258. doi: 10.1016/j.ijmm.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Carroll RK, Musser JM. 2011. From transcription to activation: how group A Streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol 81:588–601. doi: 10.1111/j.1365-2958.2011.07709.x. [DOI] [PubMed] [Google Scholar]
- 53.Hytonen J, Haataja S, Gerlach D, Podbielski A, Finne J. 2001. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol Microbiol 39:512–519. doi: 10.1046/j.1365-2958.2001.02269.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.