Abstract
The main purpose of this in situ hybridization study was to investigate mRNA expression of three bone/cartilage matrix components (perlecan, DMP1, and MEPE) in developing primary (tibial) and secondary (condylar) cartilage. Perlecan mRNA expression was first detected in newly formed chondrocytes in tibial cartilage at E13.0, but this expression decreased in hypertrophic chondrocytes at E14.0. In contrast, at E15.0, perlecan mRNA was first detected in the newly formed chondrocytes of condylar cartilage; these chondrocytes had characteristics of hypertrophic chondrocytes, which confirmed the previous observation that progenitor cells of developing secondary cartilage rapidly differentiate into hypertrophic chondrocytes. DMP1 mRNA was detected in many chondrocytes within the lower hypertrophic cell zone in tibial cartilage at E14.0. In contrast, DMP1 mRNA expression was only transiently detected in a few chondrocytes of condylar cartilage at E15.0. Thus, DMP1 may be less important in the developing condylar cartilage than in the tibial cartilage. Another purpose of this study was to test the hypothesis that MEPE may be a useful marker molecule for cartilage. MEPE mRNA was not detected in any chondrocytes in either tibial or condylar cartilage; however, MEPE immunoreactivity was detected throughout the cartilage matrix. Western immunoblot analysis demonstrated that MEPE antibody recognized two bands, one of 67 kDa and another of 59 kDa, in cartilage-derived samples. Thus MEPE protein may gradually accumulate in the cartilage, even though mRNA expression levels were below the limits of detection of in situ hybridization. Ultimately, we could not designate MEPE as a marker molecule for cartilage, and would modify our original hypothesis.
Key words: Mandibular condylar cartilage, perlecan, DMP1; MEPE, in situ hybridization
Introduction
Mandibular condylar cartilage is embryologically classified as secondary cartilage.1-3 Secondary cartilage is somewhat different from primary cartilage; for example, it differs in origin, time of its appearance, and pattern of expression for several matrix components and growth factors.1-6 Previously, we reported that mouse condylar cartilage derives from alkaline phosphatase (ALP)-positive progenitor cells of the periosteum that is continuous with the ossifying mandible that forms before the condylar cartilage.7 Meanwhile, extracellular matrix (ECM) components are involved in organogenesis of bone or cartilage, and often utilized as marker molecules for each organ.3 From this point of view, we investigated expression patterns of bone/cartilage matrix components, including collagen types I, II, X, aggrecan, bone sialoprotein (BSP), and osteopontin (OPN) (secreted phosphoprotein-1:Spp-1).7-9 These previous studies confirm that expression patterns of matrix components in developing condylar cartilage differ from those in limb bud cartilage (representative primary cartilage), and they demonstrate that progenitor cells of condylar cartilage rapidly differentiate into hypertrophic chondrocytes. In the present in situ hybridization study, we focused on three other bone/cartilage matrix components, which involved cartilage formation and mineralization, to further elucidate structural features of developing condylar cartilage and limb bud cartilage.
Perlecan is a large heparan sulfate proteoglycan that is present in basement membranes and other extracellular matrices.10-13 It has a wide variety of functions in normal development and disease processes; these roles include binding to other extracellular proteins, growth factors, and cell membrane receptors.14 Additionally, perlecan-deficient mice display severe chondrodysplasia, which includes disorganized collagen fibrils and poor glycosaminoglycans, and this phenotype revealed that perlecan is essential for normal cartilage formation.15,16 However, transition of perlecan mRNA expression in the mesenchymal cell condensation to the initial formation of cartilage in normal mice has not been reported for either secondary cartilage or primary cartilage. Thus, investigation of perlecan mRNA expression would clarify further structural features of both types of cartilage.
DMP1 (dentin matrix protein 1) was first identified as a product of a rat pulp incisor cDNA library; it belongs to SIBLING (small integrin-binding ligand, N-linked glycoprotein) protein family, and previously it was thought to be dentin-specific.17 Subsequently, high levels of DMP1 mRNA were detected in osteocytes and lower levels were seen in other mineralized or unmineralized tissues such as dental pulp, brain, and submandibular glands.18-22 Regarding cartilage, DMP1 mRNA is expressed in hypertrophic chondrocytes in fetal limb bud cartilage at E16.0, and this expression in hypertrophic chondrocytes persisted throughout later embryonic and postnatal development, indicating involvement of this molecule in cartilage mineralization.23 Based on immunohistochemistry, DMP1 protein localization is complicated; for example, full-length DMP1 (108 kDa) and two proteolytic fragments (a 37-kDa N-terminal fragment and a 57-kDa C-terminal fragment) have different localization patterns in two different types of cartilage: limb bud cartilage24 and postnatal rat mandibular condylar cartilage.25 Thus, investigation of DMP1 mRNA expression during the initial stages of cartilage formation would clarify features of both types of cartilage in terms of mineralization.
MEPE (matrix extracellular phosphoglycoprotein), like DMP1, is a member of the SIBLING family and a bone marker protein expressed in osteocytes.26,27 MEPE is also expressed in odontoblasts, regenerating tissues after a fracture, and tumor tissue.28,29 Stain et al.30 recently reported that MEPE mRNA is expressed in hypertrophic chondrocytes in the growth plate of 3-week-old mice. However, expression of MEPE mRNA during the initial stages of cartilage formation was not investigated in any type of cartilage. Thus, the main purpose of this in situ hybridization study was to examine the expression of perlecan, DMP1, and MEPE mRNA in the developing mandibular condylar cartilage and the developing limb bud cartilage. In addition, we previously reported that MEPE immunoreactivity was clearly evident throughout the matrix of Meckel’s cartilage in mice, rats, and humans; 31 consequently, we hypothesized that MEPE may be a useful marker molecule for cartilage. Thus another purpose of this study is to further test this hypothesis by means of in situ hybridization, immunohistochemistry, and western immunoblot analysis.
Materials and Methods
Animals
A total of 10 pregnant ICR mice, E12.0-18.0 (8 a.m. on the day of the vaginal plug was designated as E0), and five male 3-weeks-old mice were used for this study. Each animal was housed in facilities approved by Tokyo Medical and Dental University. Our animal-use protocol and experimental system were approved by the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University (No. 0150011A).
RT-PCR
Mandibular condylar anlage or formed condylar cartilage was meticulously microdissected from embryonic mice at specific points within the E14.0-E16.0 window. ISOGEN (Nippon Gene, Tokyo, Japan) was used to extract total RNA from condylar cartilage of E15.0 and E16.0 and condylar anlage from E14.0. Primer Script RT reagent Kit and gDNA Eraser (Takara Bio Inc., Shiga, Japan) were used for RT-PCR to synthesize cDNA. SybrGreen Premix EX Taq II (Takara Bio Inc, Shiga, Japan) and a Light Cycler 480 Real-Time PCR System (Roche Diagnostics, Tokyo, Japan) were used for all real-time PCR amplification. Each primer sequence is listed in Table 1. The PCR conditions were as follows: initial denaturing at 95°C for 30 s followed by 40 cycles of denaturing at 95°C for 5 s, annealing at 60°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 7.5 min. β-actin mRNA levels were quantified as internal controls. Relative expression levels were calculated using the Δ(ΔCt) method. All data are expressed as the mean ± SD. The data analyses were performed with one-way analysis of variance for several group comparisons, and significance was determined using Tukey’s comparison test. A P-value of <0.01 was considered statistically significant. We examined three different samples for each embryonic day.
Table 1.
Primer sets for real-time RT-PCR.
| cDNA | Primer sequence (5'→3') | Size |
|---|---|---|
| Perlecan | Forward: AGCCGCAGCAAGGACTTCA Reverse: CGTCTACTTGGATGGAGCCTCT |
174 bp |
| DMP1 | Forward: AGCTGGGCTACATTGCTTTGG Reverse: TCCAGGCTTTGCTACTGTGGAAC |
155 bp |
| MEPE | Forward: GAGTCGTCATCCAGTGGGAGTTC Reverse: AGGTCTGTTGGCTTGCTCAGTTC |
171 bp |
| Aggrecan | Forward: AGTGGATCGGTCTGAATGACAGG Reverse: AGAAGTTGTCAGGCTGGTTTGGA |
105 bp |
| β-actin | Forward: CATCCGTAAAGACCTCTATGCCAAC Reverse: ATGGAGCCACCGATCCACA |
171 bp |
DMP1, dentin matrix protein 1; MEPE, matrix extracellular phosphoglycoprotein.
Tissue preparation
At each time point, pregnant mice were killed by cervical dislocation under ether anesthesia, after which each fetal mouse was killed by cervical dislocation. The heads and lower limbs were removed and then immersed in 4% paraformaldehyde (0.1 M phosphate buffer, pH 7.4) for 1 day at 4°C. Paraffin embedded serial sections (5 µm) were cut in the coronal plane (perpendicular to the sagittal plane) and parallel to the long axis of the condylar process of the mandible or longitudinally to the tibia. Sections were stained with 0.1% toluidine blue (0.1 M phosphate buffer, pH 7.4) for histological observations.
In situ hybridization
For in situ hybridization of perlecan, DMP1, and MEPE, total RNA was extracted from limb cartilage and bone; cDNA encoding mouse perlecan (NM_008305.3), DMP1 (NM_016779.2), and MEPE (NM_053172.2), were then synthesized via RT-PCR. Each primer sequence is listed in Table 2. A representative PCR product for each gene was subcloned into a pCRII vector (Strategene, La Jolla, CA, USA); antisense and sense RNA probes were then synthesized for each gene. The Riboprobe in vitro Transcription System (Promega, Madison, WI) was used to label each probe with 35S-UTP. In situ hybridization with 35S-UTP labeled probes was performed as previously described.32-34 Sections were dipped in emulsion (NTB, Kodak, Rochester, NY, USA) after hybridization and RNAase treatment, and then exposed for 2 weeks at 4°C for autoradiography. We extended exposure period up to 4wk to confirm enhanced signals would appear, but they were not recognized. Sense probes were used for negative control samples. We examined three different samples for each embryonic day, and the same results were obtained each time.
Table 2.
Primer sets for RNA probes.
| cDNA | Primer sequence (5'→3') | Size |
|---|---|---|
| Perlecan | Forward: CCGGGCTCTGGTAAATTTTA Reverse: CTGCCATCCCTGCAGTCTTC |
791 bp |
| DMP1 | Forward: GTGCTCTCCCAGTTGCCAGATA Reverse: TGATGAGGACTCCACAGACAC |
373 bp |
| MEPE | Forward: CTCCAGCAAAGCTGAAGT Reverse: GACAGCAGCGAGTCGTCATC |
611 bp |
Immunohistochemistry
A rat monoclonal antibody against perlecan (diluted 1:200 with PBS) was purchased from Seikagaku Corp (Tokyo, Japan). Rabbit polyclonal antibody against MEPE, LF-155 (diluted 1:200), was kindly supplied by Dr. Larry W. Fisher (Matrix Biology Unit, Craniofacial and Skeletal Disease Branch, NIDCR, NIH, Bethesda, MD, USA). LF-155 reacts only against COOH-terminal fragments of MEPE.35,36 For detection of DMP1, mouse monoclonal antibody raised against rat DMP1 57-kDa fragments was used (diluted 1:500). This monoclonal antibody specifically recognizes C-terminal fragments of DMP1.37 These antibodies were well characterized and used in our previous immunohistochemical studies.31,37,38
Before antibody addition, the sections were immersed in PBS and digested with testicular hyaluronidase (25 mg/mL in phosphate buffered saline; Sigma Chemical Co., St. Louis, MO, USA) to remove masking with hyaluronan and chondroitin sulphate chains for 3 h at 37°C. Sections for DMP1 immunohistochemistry were then immersed in mouse IgG blocking reagent from a MOM kit (Vector Laboratories, Burlingame, CA, USA) to block endogenous mouse IgG activity. The streptavidin-biotin method with a Histofine SAB kit (Nichirei, Tokyo, Japan) was then performed as previously described.7,31,38,39 The sections were treated with diaminobenzidine to visualize protein localization. Negative control sections were incubated with normal rabbit or mouse or rat IgG instead of the primary antibodies. Sections were observed after counter-staining with hematoxylin.
Protein extraction from bone and cartilage
Under anesthesia with diethyl ether and intraperitoneal injection of pentobarbital sodium (30 mg/kg body weight) (Kyoritu, Tokyo, Japan), bone of limbs, ribs, vertebra, and calvaria were dissected from 3-weeks-old mice and homogenized. The samples (2.8 g wet wt) were stirred for 72 h at 4°C in 30 mL of an E-extraction buffer (4 M guanidine HCl, 50 mM sodium acetate, 0.5 M Tetrasodium EDTA, pH 7.4) that contained a protease-inhibitor cocktail (Sigma-Aldrich).40 After centrifugation, the supernatant was removed and the remaining residues were stirred in the same buffer for another 72 h. The solution was again centrifuged, the supernatant removed and mixed with the first extract. The solution was dialyzed with 0.5 M Sodium Acetate (pH 7.4) containing protease inhibitors for 24 h at 4°C. Precipitated insoluble proteins (presumably collagen) were removed by centrifugation, and the supernatant was again dialyzed with dH2O for another 48 h and freeze-dried (bone-derived samples). Meanwhile, to collect large amounts of pure cartilage without bone, tracheal cartilage (6 mg wet wt) was dissected and stirred in 6 mL of an G-extraction buffer (4 M guanidine HCl, 50 mM sodium acetate, pH 7.4) containing protease inhibitors cocktail.40 The following steps were performed as described for bone-derived samples. Other tracheal cartilage specimens were fixed and embedded in paraffin, then subjected to immunohistochemistry to confirm whether postnatal tracheal cartilage also shows MEPE immunoreactivity.
Electrophoresis and Western immunoblotting
Samples of bone- or cartilage-derived protein (~100 ng) were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in12.5% (W/V) gel. SDS-PAGE was done under reducing conditions, and protein bands were transferred to Immobilon-P membranes with Horizblot from ATTO (Tokyo, Japan). The membranes were then immersed in a solution of 3% bovine serum albumin in PBS for 1 h at RT to block any non-specific reactions. They were then reacted with mouse monoclonal antibody that was against a 57-kDa fragment of rat DMP1 (diluted 1:2000 with PBS) or rabbit polyclonal antibody against MEPE, LF-155 (diluted 1:1000), in PBS at 4°C overnight. The membranes were then incubated in biotin-labeled anti-mouse IgG+IgA+IgM (diluted 1:10 with PBS) or biotin-labeled anti-rabbit IgG (diluted 1:10) from HISTOFINE SAB kits for 1 h at 37°C, and subsequently incubated in peroxidase-labelled streptoavidin (diluted 1:10 with PBS) for 30 min at RT. Membranes were then washed in PBS and next treated with AEC (Nichirei) to detect any immunoreactivity. The membranes generated to detect MEPE were treated with ECL detection system (GE Healthcare UK Ltd., Little Chalfont, UK) and exposed to soft X-ray film.
Results
RT-PCR analysis of perlecan, DMP1, and MEPE mRNA in developing mandibular condylar cartilage
Based on RT-PCR analysis, aggrecan mRNA was barely evident in the condylar anlagen at E14.0. Aggrecan mRNA expression levels had increased modestly by E15.0 and were markedly upregulated by E16.0 (Figure 1a). Perlecan mRNA expression on E14.0 was similar to that on E15.0, but levels increased 2-fold by E16.0 (Figure 1b). In contrast, neither DMP1 nor MEPE mRNAs was evident from E14.0 to E16.0 (Figure 1 c,d).
Figure 1.
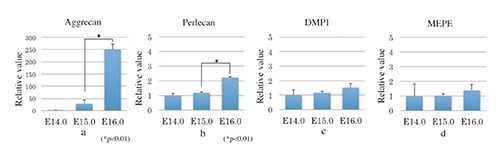
Real-time RT-PCR analysis of perlecan, DMP1, and MEPE mRNA in condylar cartilage. a) Aggrecan mRNA was barely evident in the condylar anlagen at E14.0; aggrecan mRNA expression levels had increased modestly by E15.0 and were markedly upregulated by E 16.0. b) Perlecan mRNA expression on E14.0 was similar to that on E15.0, but levels increased 2-fold by E16.0. c, d) Neither DMP1 nor MEPE mRNAs was evident from E14.0 to E16.0. Data are presented as means ± SD (n=3).
In situ hybridization in developing mandibular condylar cartilage
Mandibular bone that contained many osteocytes was evident on E18.0 (Figure 2a). DMP1 mRNA was expressed in many of these osteocytes (Figure 2b); in contrast, MEPE mRNA was expressed in fewer of these mandibular bone osteocytes (Figure 2c). These expression patterns were similar to patterns seen in previous studies,18,26 indicating probes for DMP1 and MEPE could be reliably used for this in situ hybridization study. At E14.0, the anlage of the future mandibular condylar process (designated the condylar anlage) was recognizable in the posterior position of the ossifying mandible, which comprised osteoid-like tissue. The condylar anlage consisted of a mesenchymal cell condensation, as previously described.6-9,33 Cartilage formation indicated by matrix metachromasia was not observed in the anlage (Figure 3a). Perlecan mRNA was not detected in the anlage at this stage (Figure 3b). DMP1 mRNA was slightly expressed in cells in the osteoid-like tissue, but not in the condylar anlage (Figure 3c). MEPE mRNA expression was not detected in the condylar anlage or in the osteoid-like tissue (Figure 3d). At E15.0, a metachromatically stained matrix, which indicated that the condylar cartilage had formed, was first detected in posterior to the ossifying mandible (Figure 3e). Several chondrocytes in this newly formed cartilage showed marked hypertrophy. The bone collar had formed around the cartilage (Figure 3e). Perlecan mRNA expression was detected in newly formed chondrocytes (Figure 3f). DMP1 mRNA was expressed in the osteogenic cells of the bone collar and of the mandibular bone as well as in a few chondrocytes (Figure 3g). MEPE mRNA expression was barely detected both in the cartilage and the mandibular bone (Figure 3h). At E16.0, the condylar cartilage had increased in length, especially the hypertrophic cell zone; each zone had become clearly identifiable and distinct. In this study, condylar cartilages were classified according to the system developed by Luder et al.:41 fibrous cell (articular cell) zone, polymorphic cell zone, flattened cell zone, and hypertrophic cell zone (Figure 3i). Perlecan mRNA was expressed in chondrocytes from flattened cell zone to upper hypertrophic cell zone, but perlecan mRNA expression was reduced in the lower hypertrophic cell zone (Figure 3j). At E16.0, DMP1 mRNA was expressed in osteogenic cells of the bone collar, but not in chondrocytes (Figure 3k). MEPE mRNA expression was detected in a few osteocytes in mandibular bone, as described in Figure 2b (data not shown), but not in the bone collar or in the cartilage (Figure 3l).
Figure 2.
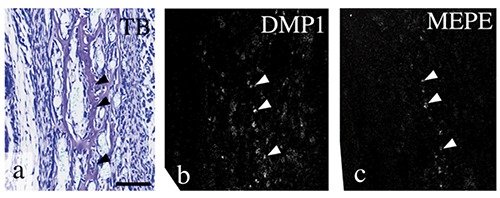
A higher magnification of mandibular bone at E18.0. Toluidine blue staining (a) and in situ hybridization for DMP1 (b) and MEPE (c). Mandibular bone contained many osteocytes (arrowheads in a). DMP1 mRNA was expressed in many of these osteocytes (arrowhead in b), but MEPE mRNA was expressed in fewer of them (arrowheads in c). Scale bars: 100 µm.
Figure 3.
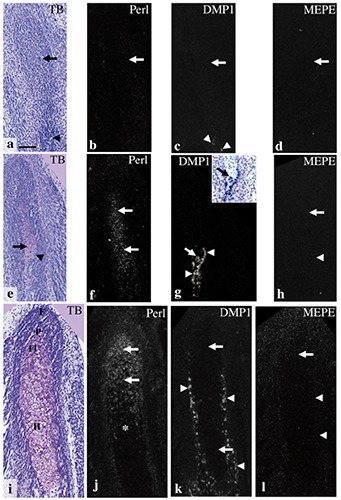
Condylar anlage/cartilage in coronal plane at E14.0 (a-d), E15.0 (e-h), and E16.0 (i-l). Toluidine blue staining (a,e,i), and in situ hybridization for perlecan (b,f,j), DMP1 (c,g,k), and MEPE (d,h,l). Inset in (g) shows a bright field image of identical section stained with toluidine blue. a) Although the ossifying mandible was recognizable (arrowhead), metachromasia was not observed in the condylar anlage (arrow). b-d) Perlecan, DMP1, and MEPE mRNA expression were not detected in the anlage (arrows in b-d), but DMP1 mRNA was slightly expressed in cells in the osteoid-like tissue (arrowheads in c). e) A metachromatically stained matrix was first detected in the anlage (arrow); note that newly formed chondrocytes showed a considerable hypertrophy, and the bone collar had formed around the cartilage (arrowhead). f) Perlecan mRNA expression was detected in newly formed chondrocytes (arrows). g) DMP1 mRNA was expressed in the osteogenic cells of the bone collar (arrowheads) as well as in a few chondrocytes (arrows). h) MEPE mRNA expression was barely detected both in the cartilage (arrow) and the bone collar (arrowhead). i) Cell zones in the condylar cartilage had become distinct: fibrous cell (articular cell) zone (F), polymorphic cell zone (P), flattened cell zone (Fl), and hypertrophic cell zone (H). j) Perlecan mRNA was expressed in chondrocytes from flattened cell zone to upper hypertrophic cell zone (arrows), but reduced in the lower hypertrophic cell zone (*). k) DMP1 mRNA was expressed in osteogenic cells of the bone collar (arrowheads), but not in chondrocytes (arrows). l) MEPE mRNA expression was not detected in the bone collar (arrowheads) or in the cartilage (arrow). Scale bars: 100 µm.
In situ hybridization in developing tibia
At E12.0, the anlage of the future tibia consisted of a mesenchymal cell condensation (Figure 4a). At this stage, in situ hybridization for perlecan, DMP1, and MEPE failed to show any positive reaction in this condensation (Figure 4 b-d). At E13.0, the anlage of the future tibia exhibited metachromasia by toluidine blue staining, indicating initial cartilage formation; this cartilage was designated tibial cartilage (Figure 4e); Perlecan mRNA was expressed in chondrocytes throughout this cartilage (Figure 4f), but DMP1 and MEPE mRNA was not detected in tibial cartilage at this stage (Figure 4 g,h). At E14.0, chondrocyte hypertrophy was first observed in the diaphysis of the tibial cartilage. At this stage, the hypertrophic cell zone could be divided into two zones: the upper hypertrophic cell zone, which contained relatively large hypertrophic cells, and the lower hypertrophic cell zone, which was located in the center of diaphysis and contained relatively small hypertrophic chondrocytes (Figure 4i). Perlecan mRNA was strongly expressed in chondrocytes throughout the entire proliferation and maturation zone, but expression was weaker in the hypertrophic cell zone (Figure 4j). DMP1 mRNA was expressed in osteogenic cells of the bone collar and in many hypertrophic chondrocytes of the lower hypertrophic cell zone (Figure 4k). MEPE mRNA expression was not detected at this stage (Figure 4l). Negative controls using sense probes showed no positive reactions at any stage examined (data not shown). Expression patterns of mRNA for three molecules were summarized in Figure 5.
Figure 4.
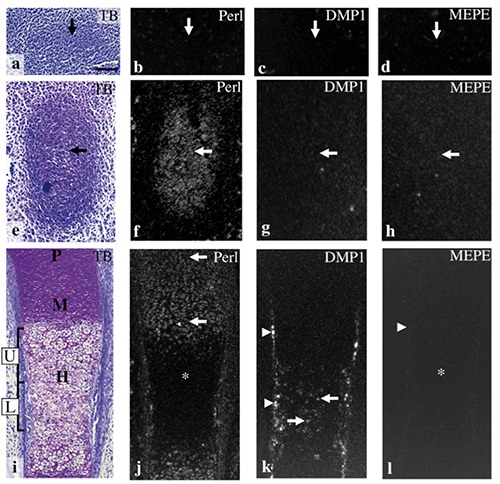
The anlage of the future tibia at E12.0 (a-d), and tibial cartilage at E13.0 (e-h), E14.0 (i-l). Toluidine blue staining (a,e,i) and in situ hybridization for perlecan (b,f,j), DMP1 (c,g,k), and MEPE (d,h,l). a) The anlage of the future tibia consisted of a mesenchymal cell condensation (arrow). b-d) Perlecan, DMP1, and MEPE mRNA were not expressed in the condensation (arrows). e) A metachromatically stained matrix was first detected in the anlage of the future tibia (arrow). f) Perlecan mRNA was expressed in chondrocytes throughout the cartilage (arrow). g,h) DMP1 and MEPE mRNA were not detected in tibial cartilage (arrows). i) Proliferative (P), maturation (M), and hypertrophic cell zone (H) had become clearly identifiable in the cartilage, and hypertrophic cell zone could be divided into two zones: the upper hypertrophic cell zone (U) and the lower hypertrophic cell zone (L). j) Perlecan mRNA was expressed in chondrocytes throughout the entire proliferative and maturation cell zone (arrows), but expression was weaker in the hypertrophic cell zone (*). k) DMP1 mRNA was expressed in the osteogenic cells of the bone collar (arrowheads) and in chondrocytes of the lower hypertrophic cell zone (arrows). l) MEPE mRNA expression was not detected both in the bone collar (arrowhead) or in the cartilage (*). Scale bars: 100 µm.
Figure 5.
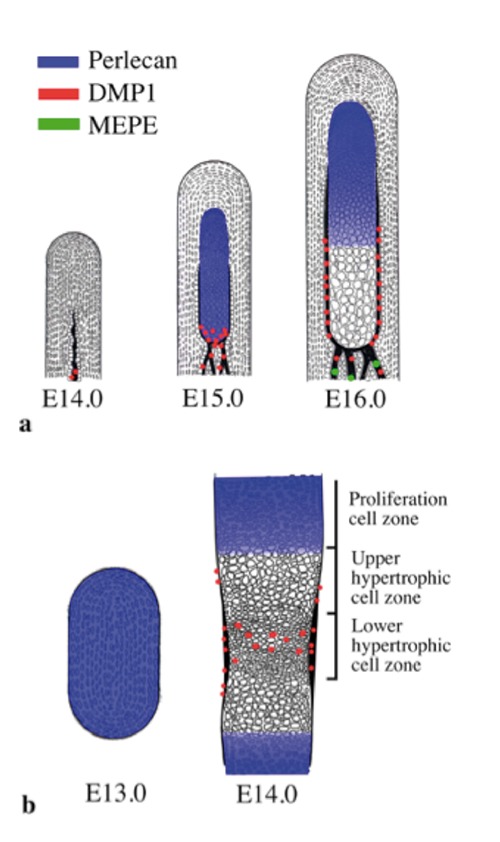
Schematic representations of the expression pattern of perlecan, DMP1, and MEPE based on the present findings. a) Condylar anlage/cartilage at E14.0 to E16.0. b) Tibial cartilage at E13.0 and E14.0.
Immunohistochemistry in developing mandibular condylar cartilage and tibial cartilage
In developing condylar cartilage at E14.0, perlecan immunoreactivity was evident in Meckel’s cartilage, but not in the anlage (Figure 6a). DMP1 immunoreactivity was weak but definite in the osteoid-like tissue, but it was not evident in the condylar anlage or Meckel’s cartilage (Figure 6b). MEPE immunoreactivity was not detected in the condylar anlage, but it was evident in the osteoid-like tissue and in Meckel’s cartilage (Figure 6c). At E15.0, perlecan immunoreactivity was clearly detected in newly formed cartilage matrix (Figure 6d); DMP1 immunoreactivity was evident in the bone collar, but not in the cartilage matrix (Figure 6e); MEPE immunoreactivity was evident in the bone collar and in the cartilage matrix (Figure 6f). At E16.0, perlecan immunoreactivity was detected throughout the cartilage matrix (Figure 6g); DMP1 immunoreactivity was detected in the bone collar, but not in the cartilage matrix (Figure 6h); MEPE immunoreactivity was detected in the bone collar and throughout the cartilage matrix (Figure 6i). In developing limb bud cartilage at E12.0, immunohistochemistry for perlecan, DMP1, and MEPE failed to show any positive reaction in the in the tibial anlage (Figure 6 j-l). At E13.0, perlecan and MEPE immunoreactivity was evident throughout the cartilage matrix (Figure 6 m,o); DMP1 immunoreactivity was not detected at this stage (Figure 6n). At E14.0, perlecan immunoreactitivity was detected throughout the cartilage matrix (Figure 6p); DMP1 immunoreactivity was detected in the bone collar and in the lower hypertrophic cell zone (Figure 6q); MEPE immunoreactivity was detected in the bone collar and throughout the cartilage matrix (Figure 6r). In maxillary region at E16.0, MEPE immunoreactivity was detected in some bone lacunae, but not in the oral epithelium in the same section (Figure 6 s,t), supporting the specificity of the antibody used in the present study.
Figure 6.
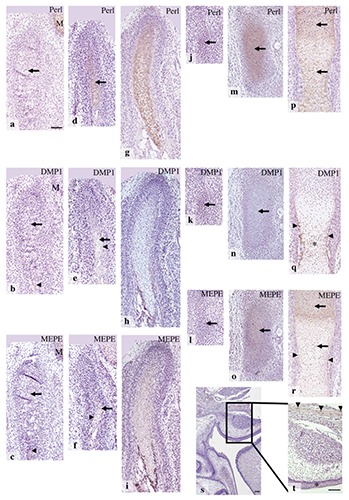
Condylar anlage/cartilage at E14.0 (a-c), E15.0 (d-f), E16.0 (g-i), and maxillary region at E16.0 (s,t); t) is the higher magnification of rectangular area of (s). The anlage of future tibia at E12.0 (j-l), and tibial cartilage at E13.0 (m-o), and E14.0 (p-r). Immunohistochemistry for perlecan (a,d,g,j,m,p), DMP1 (b,e,h,k,n,q), and MEPE (c,f,i,l,o,r,s,t). a) Perlecan immunoreactivity was evident in Meckel’s cartilage (M), but not in the anlage (arrow). b) DMP1 immunoreactivity was weak but definite in the osteoid-like tissue (arrowhead), but not in the condylar anlage (arrow) or Meckel’s cartilage (M). c) MEPE immunoreactivity was not detected in the condylar anlage (arrow), but it was evident in the osteoid-like tissue (arrowhead) and in Meckel’s cartilage (M). d) Perlecan immunoreactivity was clearly detected in newly formed cartilage matrix (arrow). e) DMP1 immunoreactivity was evident in the bone collar (arrowhead), but not in the cartilage matrix (arrow). f) MEPE immunoreactivity was evident in the bone collar (arrowhead) and in the cartilage matrix (arrow). g-i) Immunostaining patterns were similar to those of E15.0. j-l) Perlecan, DMP1, and MEPE immunoreactivity were not detected in the mesenchymal cell condensation (arrows). m-o) Perlecan and MEPE immunoreactivity was evident in the matrix throughout the cartilage matrix (arrows in m and o), but DMP1 immunoreactivity was not detected (arrow in n). p) Perlecan immunoreactitivity was detected throughout the cartilage matrix (arrows). q) DMP1 immunoreactivity was detected in the bone collar (arrowheads) and in the lower hypertrophic cell zone (*). r-t) MEPE immunoreactivity was detected in the bone collar (arrowheads in r), throughout the cartilage (arrows in r), and in some bone lacunae (arrowheads in t), but not in the oral epithelium (asterisk in t). Scale bars; a-r) 100 µm; s,t) 80 µm.
Western immunoblotting for DMP1 and MEPE in bone- or cartilage-derived samples
Tracheal cartilage from 3-week-old animals exhibited metachromasia throughout the cartilage (Figure 7a). DMP1 immunoreactivity was not detected in the cartilage matrix (Figure 7b), but MEPE immunoreactivity was clearly recognized in the cartilage matrix, as seen in fetal cartilages (Figure 7c).
Figure 7.
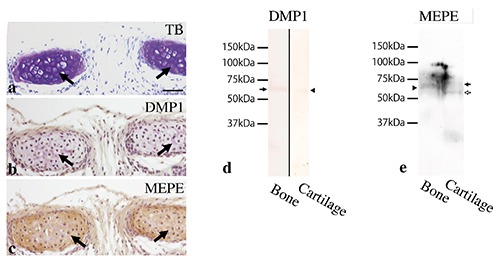
Tracheal cartilage at 3 weeks old (a-c) and Western immunoblotting for DMP1 and MEPE in bone- or cartilage- derived samples (d,e). a) Tracheal cartilage matrix exhibited metachromasia by toluidine blue staining (arrows). (b) DMP1 immunoreactivity was not detected in the cartilage matrix (arrows). c) MEPE immunoreactivity was clearly recognized in the cartilage matrix (arrows). d) DMP1 antibody clearly recognized a strong protein band of 65 kDa in bone-derived samples (arrow), but a weaker band in cartilage-derived samples (arrowhead). e) MEPE antibody recognized a protein band of 62 kDa in bone-derived samples (arrowhead) and two bands, one of 67 kDa (black arrow) and another of 59 kDa (white arrow) in cartilage-derived samples. Scale bars: 50 µm.
Western immunoblotting indicated that DMP1 antibody clearly recognized a strong protein band of 65 kDa in bone-derived samples, but a weaker band in cartilage-derived samples (Figure 7d). MEPE antibody recognized a protein band of 62 kDa in bone-derived samples and two bands, one of 67 kDa and another of 59 kDa, in cartilage-derived samples (Figure 7e).
Discussion
Perlecan
In the present study, we first reported transition of perlecan mRNA expression from mesenchymal cell condensation to initial cartilage formation in normal limb bud cartilage using in situ hybridization. Perlecan mRNA expression was not detected in mesenchymal cell condensations at E12.0; it was first detected in the newly formed cartilage at E13.0. At E14.0, perlecan mRNA expression was weak in hypertrophic chondrocytes of the diaphysis. Meanwhile, perlecan immunoreactivity was evident throughout the tibial cartilage; these findings indicated that cells developing into mature chondrocytes actively synthesized perlecan, but the hypertrophic chondrocytes had downregulated perlecan synthesis. This discordance between mRNA expression and protein accumulation was similar to the discordance noted for aggrecan localization in the tibial cartilage.42
In the condylar cartilage, perlecan mRNA expression was not detected in the E14.0 anlage, which consisted of mesenchymal cell condensation: it was first detected in newly formed cartilage at E15.0. We have previously described that, as newly formed chondrocytes can simultaneously express collagen type I, II, X, and aggrecan, progenitor cells of condylar cartilage rapidly differentiate into hypertrophic chondrocytes; notably, this process is different from that in limb bud cartilage.6,7,9 Our current results indicated that newly formed hypertrophic chondrocytes can also express perlecan mRNA, supporting our previous theory. At E16.0, perlecan mRNA expression was reduced in lower hypertrophic cell zone, although immunoreactivity was detected throughout the cartilage matrix. These results are consistent with results previously described for aggrecan expression,9 and realtime PCR in the present study demonstrated that perlecan mRNA showed a similar expression pattern to aggrecan mRNA. Thus, these results indicated that perlecan expression is concomitant with aggrecan expression during the process of condylar cartilage formation. Thus, we believe that perlecan is a component of an essential cartilage matrix, as perlecan gene-knockout studies have indicated.15
DMP1
DMP1 is primarily expressed in mineralized tissues such as osteocytes and odontoblasts.17,43 DMP1-deficient mice displayed severe deletion of bone and dentin mineralization and an abnormal growth plate, suggesting that DMP1 play a role in mineralization. Notably, Feng et al.23 reported that DMP1 mRNA was highly expressed in hypertrophic chondrocytes within the diaphysis of limb bud cartilage at E16.0 and in the growth plate of 8-weeks-old mice. Meanwhile, in limb bud cartilage, cartilage marker molecules further divide the hypertrophic cell zone into upper hypertrophic cell zone and lower hypertrophic cell zone, and the latter expresses OPN as well as type X collagen.8,44 During initial formation of the hypertrophic cell zone in limb bud cartilage (before bone marrow cavity formation), both OPN and BSP mRNAs are expressed in the lower hypertrophic cell zone (mineralizing cartilage core), although the expression of BSP mRNA is weaker than that of OPN.8,45 In the present study, hypertrophic cell zone in the tibial cartilage was first observed at E14.0, and DMP1 mRNA expression was detected in lower hypertrophic cell zone; this pattern was similar to the pattern of OPN mRNA. Thus, DMP1 is potentially another marker molecule for lower hypertrophic cell zone in limb bud cartilage.
In newly formed condylar cartilage at E15.0, BSP mRNA is more extensively expressed than OPN mRNA; this pattern differed from that in limb bud cartilage at the corresponding stage.8 In the present study, DMP1 mRNA was detected in a few chondrocytes in newly formed condylar cartilage at E15.0; this pattern was similar to that of OPN. DMP1 mRNA expression was, however, hardly detected at E16.0. Thus DMP1 expression in hypertrophic chondrocytes was only transient, and DMP1 may not have important roles in developing condylar cartilage, compared with OPN and BSP. Meanwhile, intense DMP1 mRNA expression was observed both in newly formed bone collar around the diaphysis of tibial cartilage and around newly formed condylar cartilage, indicating DMP1 seems to be involved in the formation of bone collar in both types of cartilage.
In vitro findings suggest that the full-length DMP1 inhibits mineralization, but N-terminal and C-terminal fragments promote hydroxyapatite formation.46,47 Maciejewska et al.48,49 demonstrated that, in the growth plate of long bones in newborn rats, N-terminal DMP1 fragments were predominantly localized in the matrix of the proliferation and hypertrophic cell zone, while the C-terminal fragments were primarily localized in the hypertrophic cell zone and mineralization front, indicating the C-terminal fragments are involved in cartilage mineralization. Based on our western blot analysis, a protein band of 65 kDa reacted with anti-DMP1 antibody in bone-derived samples, confirming that this antibody can recognize C-terminal fragments of DMP1. Based on immunohistochemistry, C-terminal fragments were recognized in the newly formed lower hypertrophic cell zone in the tibial cartilage at E14.0; this pattern was identical to that of DMP1 mRNA expression. These results were similar to previous finding for the postnatal rat growth plate,48,49 and we reasoned that C-terminal fragments may also be involved in initial mineralization of limb bud cartilage.
Regarding the mandibular condylar cartilage, the C-terminal fragments were mainly localized in the nuclei of chondroblastic and hypertrophic layers and cartilage-bone interface in postnatal rat condylar cartilage.25 In the present study, immunoreactivity from C-terminal fragments was barely evident in developing condylar cartilage; this finding was not consistent with previous findings25 or with our findings for tibial cartilage. These differences were due to differences in the developmental stages and species examined, and they suggested that roles of C-terminal fragments in the developing condylar cartilage may not be as important as those in the limb bud cartilage. This is also supported by results of in situ hybridization described above.
MEPE
MEPE is predominantly expressed in osteocytes.26,50,51 In situ hybridization in the present study confirmed this observation, indicating that the probes used were reliable markers for in situ hybridization of this molecule. MEPE-deficient mice display an increase in bone mass and enhancement of mineralization; conversely, MEPE-overexpressing mice had wider epiphyseal growth plates and expansion of primary cancellous bone.50,52 Thus, MEPE may be involved in mineralization of cartilage matrix, as indicated by a report on MEPE mRNA expression in hypertrophic chondrocytes of the postnatal murine growth plate.30 Nevertheless, the exact role of MEPE in cartilage remains unclear. Our previous study31 shows that MEPE immunoreactivity is evident in Meckel’s cartilage in fetal mice and rats and in condylar cartilage in human fetuses, and we hypothesized that MEPE may be useful as a marker molecule for cartilage.
To test this hypothesis, we performed in situ hybridization with a 35S-UTP-labeled RNA probe for MEPE. No mRNA expression was detected in any chondrocytes in the tibial cartilage or the condylar cartilage, even when exposure period was extended up to 4 weeks; this finding was consistent with the results of our real-time PCR analysis. MEPE immunoreactivity was, however, detected throughout the cartilage matrix in Meckel’s, tibial, condylar, and tracheal cartilages, as we previously described. As negative controls never showed positive reactions, we believed that this immunoreactivity was not background staining. To determine whether this positive reaction was a non-specific reaction, we performed western immunoblotting with bone- and tracheal cartilage-derived samples. Previous western immunoblotting studies demonstrated that a protein band of 55 kDa in rat bone53 and 50 kDa in rat dental pulp36 were recognized as full-length MEPE.
In the present study, a protein band of 62 kDa in bone-derived samples reacted with antibody, indicating that this antibody could recognize full-length MEPE. The slight increase in molecular weight may be due to differences between species or post-translational modification, such as phosphorylation or glycosylation. In cartilage-derived samples, two bands of 67 kDa and 59 kDa were detected. Wang et al.36 identified three forms of MEPE protein, 50, 38, and 25 kDa in rat pulp tissue; these finding indicate that the low-molecular weight proteins are cleaved forms of MEPE. We cannot confirm whether these bands were products of post-translational modification or cleaved forms of MEPE, but believe that there are some proteins that the MEPE antibody can recognize and that the immunoreactivity in cartilage was not a non-specific reaction.
It is conceivable that MEPE protein accumulated gradually in the cartilage, even though the mRNA expression levels were consistently below the limits of detection of in situ hybridization. We cannot, however, confirm that MEPE is a useful marker molecule for cartilage, and we will modify our previous hypothesis. In addition, the exact role of MEPE in developing condylar/tibial cartilage still remains unclear, since MEPE immunoreactivity was evenly recognized throughout cartilage matrix.
Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research (No. 15K11005) from Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1.Beresford WA. Chondroid bone, secondary cartilage and metaplasia, p. 23-47. Baltimore: Urban & Schwarzenberg; 1981. [Google Scholar]
- 2.Vinkka H. Secondary cartilages in the facial skeleton of the rat. Proc Finn Dent Soc 1982;78(Suppl 7):1-137. [PubMed] [Google Scholar]
- 3.Hall BK. Bones and cartilage: developmental and evolutionary skeletal biology, p. 149-65. San Diego, London: Elsevier Academic Press; 2005. [Google Scholar]
- 4.Durkin JF, Heeley JD, Irving JT. The cartilage of the mandibular condyle. Oral Sci Rev 1973;2:29-99. [PubMed] [Google Scholar]
- 5.Hinton RJ. Genes that regulate morphogenesis and growth of the temporomandibular joint: a review. Dev Dyn 2014;243:864-74. [DOI] [PubMed] [Google Scholar]
- 6.Koyama E, Saunders C, Salhab I, Decker RS, Chen I, Um H, et al. Lubricin is required for the structural integrity and post-natal maintenance of TMJ. J Dent Res 2014;93:663-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibata S, Fukada K, Suzuki S, Yamashita Y. Immunohistochemistry of collagen types II and X, and enzyme-histochemistry of alkaline phosphatase in the developing condylar cartilage of the fetal mouse mandible. J Anat 1997;191:561-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata S, Fukada K, Suzuki S, Ogawa T, Yamashita Y. In situ hybridization and immunohistochemistry of bone sialoprotein and secreted phosphoprotein 1 (osteopontin) in the developing mouse mandibular condylar cartilage compared with limb bud cartilage. J Anat 2002;200:309-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukada K, Shibata S, Suzuki S, Ohya K, Kuroda T. In situ hybridisation study of type I, II, X collagens and aggrecan mRNAs in the developing condylar cartilage of fetal mouse mandible. J Anat 1999;195:321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noonan DM, Fulle A, Valente P, Cai S, Horigan E, Sasaki M, et al. The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein-receptor, and the neural cell adhesion molecule. J Biol Chem 1991;266:22939-47. [PubMed] [Google Scholar]
- 11.Iozzo RV, Cohen IR, Grässel S, Murdoch AD. The biology of perlecan: the multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem J 1994;302:625-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen BR. Life without perlecan has its problems. J Cell Biol 1999;147:909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeCarlo AA, Whitelock JM. The role of heparan sulfate and perlecan in boneregenerative procedures. J Dent Res 2006;85:122-32. [DOI] [PubMed] [Google Scholar]
- 14.Handler M, Yurchenco PD, Iozzo RV. Developmental expression of perlecan during murine embryogenesis. Dev Dyn 1997;210:130-45. [DOI] [PubMed] [Google Scholar]
- 15.Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat Genet 1999;23:354-8. [DOI] [PubMed] [Google Scholar]
- 16.Costell M, Gustafsson E, Aszódi A, Mörgelin M, Bloch W, Hunziker E, et al. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol 1999;147:1109-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George A, Sabsay B, Simonian PA, Veis A. Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J Biol Chem 1993;268:12624-30. [PubMed] [Google Scholar]
- 18.D’Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, et al. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res 1997;12:2040-9. [DOI] [PubMed] [Google Scholar]
- 19.MacDougall M. Refined mapping of the human dentin sialophosphoprotein (DSPP) gene within the critical dentinogenesis imperfecta type II and dentin dysplasia type II loci. Eur J Oral Sci 1998;106(Suppl 1):227-33. [DOI] [PubMed] [Google Scholar]
- 20.He G, George A. Dentin matrix protein 1 immobilized on type I collagen fibrils facilitates apatite deposition in vitro. J Biol Chem 2004;279:11649-56. [DOI] [PubMed] [Google Scholar]
- 21.Terasawa M, Shimokawa R, Terashima T, Ohya K, Takagi Y, Shimokawa H. Expression of dentin matrix protein 1 (DMP1) in non-mineralized tissues. J Bone Miner Metab 2004;22:430-8. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Xie X, Wang X, Sun Y, Liu P, Chen L, et al. Differential expression and localization of dentin matrix protein 1 (DMP1) fragments in mouse submandibular glands. J Mol Histol 2013;44:231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng JQ, Zhang J, Dallas SL, Lu Y, Chen S, Tan X, et al. Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. J Bone Miner Res 2002;17:1822-31. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Ma S, Zhou J, Yamoah AK, Feng JQ, Hinton RJ, et al. Distribution of small integrin-binding ligand, N-linked glycoproteins (SIBLING) in the articular cartilage of the rat femoral head. J Histochem Cytochem 2010;58:1033-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Gandhi V, Prasad M, Yu W, Wang X, Zhu Q, et al. Distribution of small integrin-binding ligand, N-linked glycoproteins (SIBLING) in the condylar cartilage of rat mandible. Int J Oral Maxillofac Surg 2010;39:272-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi M, Kamiya N, Ito K, Takagi M. In situ localization and in vitro expression of osteoblast/osteocyte factor 45 mRNA during bone cell differentiation. Histochem J 2002;34:255-63. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Chen L, Jahangiri A, Chen B, Wu Y, Chuang HH, et al. Expression and processing of small integrin-binding ligand N-linked glycoproteins in mouse odontoblastic cells. Arch Oral Biol 2008;53:879-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, Huang S, Miclau T, Helms JA, Colnot C. Mepe is expressed during skeletal development and regeneration. Histochem Cell Biol 2004;121:493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Brown TA, Zhou J, Xiao ZS, Awad H, Guilak F, et al. Role of matrix extracellular phosphoglycoprotein in the pathogenesis of X-linked hypophosphatemia. J Am Soc Nephrol 2005;16:1645-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staines KA, Mackenzie NC, Clarkin CE, Zelenchuk L, Rowe PS, MacRae VE, et al. MEPE is a novel regulator of growth plate cartilage mineralization. Bone 2012;51:418-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata S, Sakamoto Y, Baba O, Qin C, Murakami G, Cho BH. An immunohistochemical study of matrix proteins in the craniofacial cartilage in midterm human fetuses. Eur J Histochem 2013;57:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata S, Yokohama-Tamaki T. An in situ hybridization study of Runx2, Osterix, and Sox9 in the anlagen of mouse mandibular condylar cartilage in the early stages of embryogenesis. J Anat 2008;213:274-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata S, Fukuoka H, Sato R, Abe T, Suzuki Y. An in situ hybridization study of the IGF system in developing condylar cartilage of the fetal mouse mandible. Eur J Histochem 2012;56:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata S, Sato R, Murakami G, Fukuoka H, Rodríguez-Vázquez JF. Origin of mandibular condylar cartilage in mice, rats and humans: periosteum or separate blastema? J Oral Biosci 2013;55:208-16. [Google Scholar]
- 35.Gluhak-Heinrich J, Pavlin D, Yang W, MacDougall M, Harris SE. MEPE expression in osteocytes during orthodontic tooth movement. Arch Oral Biol 2007;52:684-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang HG, Kawashima N, Iwata T, Xu J, Takahashi S, Sugiyama T, et al. MEPE activated by furin promotes pulpal cell adhesion. J Dent Res 2011;90:529-34. [DOI] [PubMed] [Google Scholar]
- 37.Baba O, Qin C, Brunn JC, Wygant JN, McIntyre BW, Butler WT. Colocalization of dentin matrix protein 1 and dentin sialoprotein at late stages of rat molar development. Matrix Biol 2004;23:371-9. [DOI] [PubMed] [Google Scholar]
- 38.Shibata S, Baba O, Oda T, Yokohama-Tamaki T, Qin C, Butler WT, et al. An immunohistochemical and ultrastructural study of the pericellular matrix of uneroded hypertrophic chondrocytes in the mandibular condyle of aged c-src-deficient mice. Arch Oral Biol 2008;53:220-30. [DOI] [PubMed] [Google Scholar]
- 39.Shibata S, Morita T, Yokohama-Tamaki T, Murakami G, Cho BH. An immunohistochemical study of matrix components in early-stage vascular canals within mandibular condylar cartilage in midterm human fetuses. Anat Rec 2015;298:1560-71. [DOI] [PubMed] [Google Scholar]
- 40.Takeushi Y, Matsumoto T, Ogata E, Shishiba Y. Isolation and characterization of proteoglycans synthesized by mouse osteoblastic cells in culture during the mineralization process. Biochem J 1990;266:15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luder HU, Leblond CP, von der Mark K. Cellular stages in cartilage formation as revealed by morphometry, radioautography and type II collagen immunostaining of the mandibular condyle from weanling rats. Am J Anat 1988;182:197-214. [DOI] [PubMed] [Google Scholar]
- 42.Shibata S, Fukada K, Imai H, Abe T, Yamashita Y. In situ hybridization and immunohistochemistry of versican, aggrecan and link protein, and histochemistry of hyaluronan in the developing mouse limb bud cartilage. J Anat 2003;203:425-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyosawa S, Shintani S, Fujiwara T, Ooshima T, Sato A, Ijuhin N, et al. Dentin matrix protein 1 is predominantly expressed in chicken and rat osteocytes but not in osteoblasts. J Bone Miner Res 2001;16:2017-26. [DOI] [PubMed] [Google Scholar]
- 44.Nomura S, Wills AJ, Edwards DR, Heath JK, Hogan BL. Developmental expression of 2ar (osteopontin) and SPARC (osteonectin) RNA as revealed by in situ hybridization. J Cell Biol 1988;106:441-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sommer B, Bickel M, Hofstetter W, Wetterwald A. Expression of matrix proteins during the development of mineralized tissues. Bone 1996;19:371-80. [DOI] [PubMed] [Google Scholar]
- 46.Tartaix PH, Doulaverakis M, George A, Fisher LW, Butler WT, Qin C, et al. In vitro effects of dentin matrix protein-1 on hydroxyapatite formation provide insights into in vivo functions. J Biol Chem 2004;279:18115-20. [DOI] [PubMed] [Google Scholar]
- 47.Gericke A, Qin C, Sun Y, Redfern R, Redfern D, Fujimoto Y, et al. Different forms of DMP1 play distinct roles in mineralization. J Dent Res 2010;89:355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maciejewska I, Qin D, Huang B, Sun Y, Mues G, Svoboda K, et al. Distinct compartmentalization of dentin matrix protein 1 fragments in mineralized tissues and cells. Cells Tissues Organs 2009;189:186-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maciejewska I, Cowan C, Svoboda K, Butler WT, D'Souza R, Qin C. The NH2-terminal and COOH-terminal Fragments of Dentin Matrix Protein 1 (DMP1) Localize Differently in the Compartments of Dentin and Growth Plate of Bone. J Histochem Cytochem 2009; 57:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, et al. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem 2003;278:1998-2007. [DOI] [PubMed] [Google Scholar]
- 51.Nampei A, Hashimoto J, Hayashida K, Tsuboi H, Shi K, Tsuji I, et al. Matrix extracellular phosphoglycoprotein (MEPE) is highly expressed in osteocytes in human bone. J Bone Miner Metab 2004;22:176-84. [DOI] [PubMed] [Google Scholar]
- 52.David V, Martin A, Hedge AM, Rowe PS. Matrix extracellular phosphoglycoprotein (MEPE) is a new bone renal hormone and vascularization modulator. Endocrinology. 2009;150:4012-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Midura RJ, Midura SB, Su X, Gorski JP. Separation of newly formed bone from older compact bone reveals clear compositional differences in bone matrix. Bone 2011;49:1365-74. [DOI] [PMC free article] [PubMed] [Google Scholar]


