Abstract
Hepatitis C virus (HCV) is a major health problem worldwide. Early detection of the infection will help better management of the infected cases. The monoclonal antibodies (mAb) of mice are predominantly used for the immunodiagnosis of several viral, bacterial, and parasitic antigens. Serological detection of HCV antigens and antibodies provide simple and rapid methods of detection but lack sensitivity specially in the window phase between the infection and antibody development. Human mAb are used in the immunotherapy of several blood malignancies, such as lymphoma and leukemia, as well as for autoimmune diseases. In this review article, we will discuss methods of mouse and human monoclonal antibody production. We will demonstrate the role of mouse mAb in the detection of HCV antigens as rapid and sensitive immunodiagnostic assays for the detection of HCV, which is a major health problem throughout the world, particularly in Egypt. We will discuss the value of HCV-neutralizing antibodies and their roles in the immunotherapy of HCV infections and in HCV vaccine development. We will also discuss the different mechanisms by which the virus escape the effect of neutralizing mAb. Finally, we will discuss available and new trends to produce antibodies, such as egg yolk-based antibodies (IgY), production in transgenic plants, and the synthetic antibody mimics approach.
Keywords: Hepatitis C virus, Monoclonal antibodies, Immunodiagnosis, Immunotherapy
Core tip: The monoclonal antibodies (mAb) of mice are predominantly used for the immunodiagnosis of several viral, bacterial, and parasitic antigens. Human mAb are used in the immunotherapy of several blood malignancies, such as lymphoma and leukemia, as well as for autoimmune diseases. In this review, we discuss methods of mouse and human monoclonal antibody production. We will demonstrate the role of mouse mAb in the detection of hepatitis C virus (HCV) antigens as rapid and sensitive immunodiagnostic assays. We will also discuss the role of HCV-neutralizing antibodies in the immunotherapy of HCV infections and in HCV vaccine development.
INTRODUCTION
Monoclonal antibodies (mAb or moAb) are monospecific antibodies that have the ability to bind to the same epitope[1]. These antibodies are made by homogeneous hybrid cells (B cells) that are each clones of the same origin parent cell. Polyclonal antibodies, on the other hand, are made of several different immune cells (B cells). Hybridomas are hybrid cell lines that are generated through the fusion of an antibody-producing B cell with a myeloma (B cell cancer) cell. Myeloma cells are characterized by the ability to grow in tissue cultures and the absence of antibody chain synthesis. All antibodies produced by these hybrid cells (using hybridoma technology) are of a single specificity, and therefore mAb. The establishment of cell lines producing mAb was first reported by Köhler et al[2] in 1975.
The hepatitis C virus (HCV) is a major health problem worldwide. According to the estimates of the World Health Organization, this virus infects more than 180 million people across the world (representing 2%-3% of the world’s total population)[3,4]. HCV (genotype 4) is one of the major health issues in Egypt, infecting 22% of the country’s general population[3,5-7]. HCV is a small-enveloped, single-stranded RNA virus belonging to the family Flaviviridae. The genome encodes a single polyprotein that is co- and post-translationally processed into four structural and six non-structural proteins[4,8]. This is done by different cellular- and viral-encoded proteases. The envelope glycoproteins E1 and E2 are two structural proteins located on the surface of the HCV, and hence play a crucial role in HCV entry into hepatocytes. Their presence on the surface of the virus makes these two proteins, particularly E2, a major target for HCV antibody neutralization and interaction with host cellular receptors[3,8].
PRODUCTION OF MOUSE MAB
Mouse mAb by hybridoma technology
In 1975, Köhler et al[2] developed a hybridoma method for the production of mAb. The persistence of antibody-producing cells via their fusion with tumor cells may be an obvious procedure today, but at the time, this procedure was regarded as a key innovation that would allow for the unlimited yield of a specific antibody molecule. In 1984, Köhler et al[2] were awarded the Nobel Prize in Physiology or Medicine “for theories concerning the specificity in development and control of the immune system and the discovery of the principle for production of mAb. While mAb are now long-established as vital research products, their therapeutic use requires further development, particularly in terms of the humanization of mouse antibodies and recombinant productivity protocols. Several hundreds of mAb are currently under evaluation for the treatment of a broad range of conditions and use within a variety of therapeutics on the market[9]. The principle production of mouse mAb by hybridoma is shown in Figure 1. The different types and applications of mAb as diagnostic and therapeutic applications are presented in the Figure 2.
Figure 1.
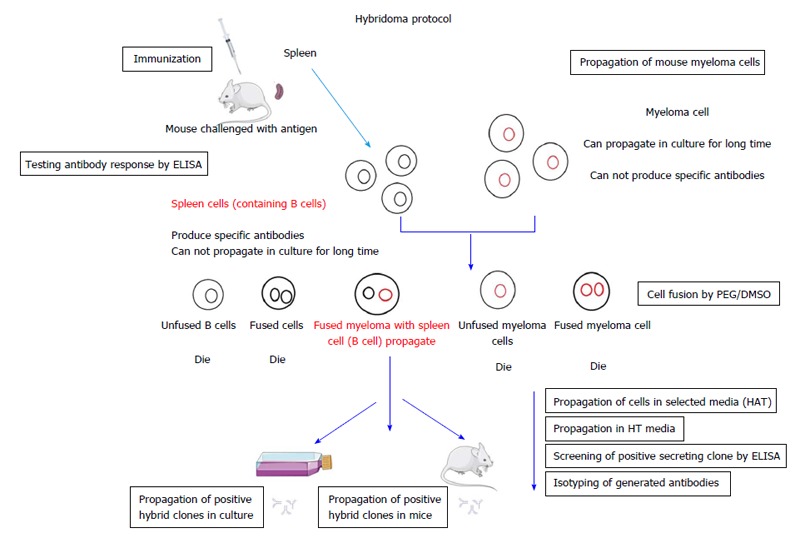
Diagrammatic procedure of the production of mouse monoclonal antibodies by hybridoma technology. ELISA: Enzyme-linked immunosorbent assay; PEG: Polyethylene glycol; DMSO: Dimethyl sulfoxide; HAT: Hypoxanthine-aminopterin-thymidine; HT: Hypoxanthine thymidine.
Figure 2.
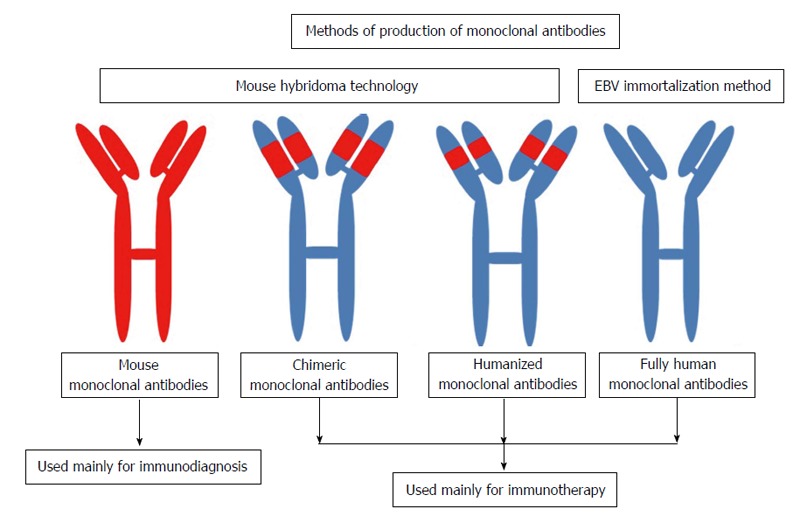
Diagrammatic presentations showing the applications of mouse and human monoclonal antibodies and their methods of production. EBV: Epstein-Barr virus.
Hybridoma cell production has conventionally been performed via cell fusion between spleen cells (B cell source) and myeloma cell lines by chemical fusion techniques using for example polyethylene glycol (PEG). A recent publication by Kandušer et al[10] in 2014, however, describes another technique for cell fusion based on electrofusion. This technique is superior to the PEG method due to its high fusion efficiency. Kato et al[11] have stated yet another technique that involves CpG oligodeoxynucleotide (CpG ODN) for cell activation prior to electrofusion. Kato et al[11] reported that CpG ODN stimulation not only increases fusion efficiency but also the number of antibody-producing cells, leading to an increased number of positive clones obtained.
Rat and rabbit mAb can be produced by the hybridoma technology using rat and rabbit spleen cells, respectively. A recent study[12] generated rat hybridoma clones via the cell fusion of immunized rat spleen cells with mouse myeloma SP2/0 cells and screened the generated antibodies using recombinant mouse CXCL4 and rhCXCL4. This study concluded that the CXCL4 signaling pathway is a potential therapeutic target in numerous diseases including cancer. In addition, Zhang et al[13] used rabbit hybridoma to produce highly sensitive rabbit mAb targeting an emerging cell surface in mesothelioma and other solid tumors (Mesothelin). They concluded that the generated rabbit mAb may be promising candidates for monitoring and treating mesothelioma and other mesothelin-expressing cancers.
PRODUCTION OF FULLY HUMAN MAB
There are several methods for the production of human mAb, such as phase display, transgenic mice, humanized mouse mAb, and immortalization by Epstein-Barr virus (EBV). In this review, we focus on the production of fully human mAb by EBV immortalization. Human mAb (hMAb) provide novel ways for probing the B cell repertoire of various health and disease issues. Several difficulties have been encountered in the development of the hMAb, including cell line instability, low levels of specific antibody secretion, and poor cloning potency, particularly when using lymphoblastoid cells[14]. Martin et al[15] reported that the immortalization of B lymphocytes by EBV is a time consuming methodology for antibody production. EBV infects B cells via their CD21 receptors, which then transforms the B cells into lymphoblastoid cell lines that produce antibodies, representing the humoral immune response in vivo. Based on the type of parent cell, the generated antibodies target either an infective agent or a tumor cell, which makes them suitable therapeutic candidates against these diseases.
Compared to other antibody manufacturing techniques, the immortalization of B cells stands as the best technique, as the generation of fully human antibodies from the immortalized human B lymphocyte repertoire does not require immunization[16,17]. The first B lymphocyte immortalization experiment was performed by culturing B cells in the presence of a herpes virus obtained from a marmoset lymphocyte cell line B95-8[18,19]. Numerous changes to this procedure have since been tried, yet the immortalization and B lymphocyte rates remain inefficient. Several successful mAbs have been generated with this technique for use against different pathogens; Schieffelin et al[20] report that human mAb against dengue virus envelope were generated by the EBV transformation of B cells from patients after two years of naturally-acquired dengue virus infection. These antibodies were found to have completely different cross-reactivity, and neutralizing patterns. Schieffelin et al[20] and others[21,22] have used CpG2006 as stimulators for EBV immortalization. Furthermore, antibodies neutralizing the SARS corona virus and cytomegalovirus have been successfully created via the introduction of the polyclonal B lymphocyte activator CpG2006 into the B lymphocyte immortalization method and by B lymphocyte activation before EBV infection, respectively[21,22]. Moreover, Fraussen et al[23] report that seeding low B lymphocyte numbers per well serves to limit bias toward the advantageous outgrowth of fast-growing immortalized B cells including interleukin-2 (IL2) and CpG 2006.
Siemoneit et al[24] used herpes virus immortalization for the production of human organism antibodies to target HCV core macromolecules. In doing this, they revealed the establishment of two vegetative cell lines secreting human mAb to the 22-kD nucleocapsid macromolecule (core, p22) of HCV. Siemoneit et al[24] isolated B lymphocytes from an anti-HCV-positive donor and immortalized them by EBV infection. Two of the cell colonies were fused with the (mouse/human) heteromyeloma cell line K6H6/B5. The generated fused hybridomas produced antibodies of the IgG1/kappa (U1/F10) and the IgM/kappa (Ul/F11) isotype and were found to specifically react with the recombinant core macromolecule p22. Recently, Steinitz[25] reported that EBV has the in vitro ability to immortalize nearly all human B lymphocytes, which allows for the isolation of monoclonal cell lines that secrete specific human mAb.
The method of immortalization by EBV allows for the production of human mAb of different classes (IgM, IgG, IgA, and IgE) from any individual. Human mAb produced by EBV immortalization resemble the supplies of antibody molecules derived from the lymphocytes of blood donors. Therefore, Steinitz[25] concluded that human mAb are promising reagents of passive immunization for several diseases, such as cancer, and viral or bacterial infections. In our recently published paper[26], we aimed to produce human cell lines by manufacturing neutralizing human mAb for use against the envelope E1/E2 macromolecule of HCV. For this, we used two methods for the EBV immortalization of CD22+ cells taken from HCV-positive patients: (1) immortalization with 100% EBV only; and (2) immortalization by 30% EBV and CPG2006 with IL2 (Figure 3). Our results indicated that cell stimulators play a role in the production of antibodies, with immortalization by 100% EBV only producing large clones compared to immortalization by 30% EBV and CpG2006 with IL2. The antibody levels, as measured by enzyme-linked immunosorbent assay (ELISA), showed high optical density in cells immortalized with 30% EBV and stimulants CpG2006 and IL2. Our results also indicated that the immortalization of low B cell numbers by 30% EBV, CPG2006, and IL2 was introduced with high efficiency and reproducibility leading to immediate generation of single clones that produced mAb. The specificity of the generated mAbs was confirmed by screening them against linear synthetic peptides (as epitopes) derived from HCV E1 (a.a 315-323) and two synthetic peptides derived from HCV E2 (a.a 412-419 and a.a 517-531) using in-house, ELISA-based optimized assays, all of which were studied by several investigators[27-29]. Fifteen clones secreting human immunoglobulin G against HCV E1/E2 protein were isolated. The ELISA results showed that one antibody was binding to the E2 peptide (a.a 517-531), while two antibodies were binding to the HCV E2 peptide (a.a 412-419). The three generated antibodies (IgG3, one antibody, and IgG2, two antibodies) showed high neutralization activity against HCVpp. We therefore concluded that these antibodies may be useful for the passive immunotherapy of HCV infections, particularly for HCV-positive liver transplantation patients.
Figure 3.
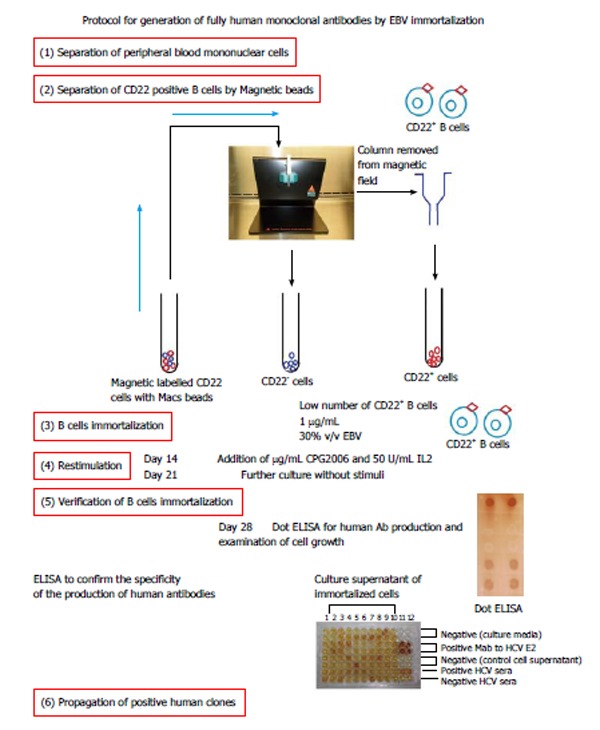
Generation of fully human monoclonal antibodies by Epstein-Barr virus immortalization. EBV: Epstein-Barr virus; HCV: Hepatitis C virus; IL2: Interleukin-2; ELISA: Enzyme-linked immunosorbent assay.
USING MOUSE MAB FOR THE DETECTION OF HCV ANTIGEN(S) AS DIAGNOSTIC MARKERS
The detection of anti-HCV antibodies involves a simple, inexpensive, and quick test, yet this test has a low sensitivity in the first six to eight weeks of infection, or given the presence of several clinical conditions, such as chronic immunosuppression or hemodialysis[30]. A recent study[31] has shown that the proteins of Core and E2 genes within the antigenic regions of a local HCV-3a genotype can be used to develop highly sensitive, specific, and economical diagnostic assays for the detection of HCV infection. The recombinant antigen showed 100% reactivity against HCV-infected sera, with no cross reactivity in the HCV-negative sera. The authors therefore concluded that a mixture of Core and E2 antigens is potentially valuable for HCV-Ab detection[31].
The disadvantage of HCV diagnosis via the detection of HCV-Abs is the inability to distinguish between active and past infection. Given this, HCV-Ag detection is preferable, especially during the window phase of HCV infection. Recently, Florea et al[32] investigated the diagnosis of an HCV infection based on a HCV core Ag detection assay and found both a good correlation between the core Ag results with the HCV RNA viral load and very high specificity. Furthermore, Chevaliez et al[33] evaluated the clinical performance of the Architect HCV-Ag assay in terms of the detection and quantification of HCV core antigens in patients with chronic HCV genotype 1-6 infections. They concluded that the Architect HCV-Ag assay is highly specific, easy to perform, and represents a valuable screening, diagnostic, and monitoring tool.
Several investigators have demonstrated evidence of HCV antigens in liver tissue[34-37] serum samples[38,39] and plasma samples[40]. We reported the detection of HCV NS4 antigen in the sera of infected HCV patients using the Dot ELISA technique as a rapid assay[41]. The assay developed was able to detect the HCV target antigen in 95% of the serum samples from HCV-infected individuals with a specificity of 97% compared to the sera of uninfected individuals, using reverse transcription polymerase chain reaction results as a reference. This antigen-detection-based method showed high positive predictive values (99%) and negative predictive values (90%). The added advantage of the assay is that it was able to detect HCV target antigens in the sera of patients during the window phase (negative for HCV-Ab and positive for HCV-RNA), as well as in the sera of both low and high HCV-RNA viral loads. The authors showed that their developed assay was highly sensitive and specific for HCV antigen detection, and that it could be applied for the mass screening of HCV infection.
In two reports[41,42], Attallah et al[41] describe their use of ELISA for the detection of HCV-NS4 and assessing the diagnostic performance of the assay in 883 chronic HCV patients. Taking quantitative HCV-RNA as a gold standard for HCV diagnosis, areas under the receiver operating characteristics (ROC) curves (AUC) were used to assess the diagnostic accuracy of ELISA for HCV-NS4. Attallah et al[42] identified HCV-NS4 to be at 27 kDa using Western blot. The areas under the ROC curves (AUC) in HCV-NS4 detection were 0.95 among patients with different pathological states of liver disease, with 0.93 for liver fibrosis, 0.95 for liver cirrhosis, and 0.98 for hepatocellular carcinoma (HCC). The mean ± SD (ng/mL) of HCV-NS4 in liver fibrosis was 94.2 ± 55.6, in liver cirrhosis was 99.3 ± 64.8, and in HCC was 124.9 ± 70.3. The authors therefore concluded that HCV-NS4 antigen detection using ELISA is a reliable test to confirm HCV infection. We established a hybrid cell line that produced mouse mAb targeting HCV E1 a.a 315-323[43]. We also produced mouse mAb targeting HCV E1/E2 and used them in the ELISA as a diagnostic assay for HCV infection (unpublished data). Our results showed a sensitivity of 80% and a specificity 96%.
HCV neutralizing antibodies
HCV cellular receptors are targets for HCV neutralization: Many HCV neutralizing antibodies target E1 and E2 HCV glycoproteins. However, an alternative strategy to preventing HCV entry may be achieved by targeting host receptors, such as CD81 and SR-BI. These antibodies block viral-host receptor interaction. A large number of broad spectrum, anti-HCV, host-targeting antivirals (HTAs) have been developed that trigger the innate immune system. Examples of these anti-HCV compounds include anti-SR-BI and toll-like receptor agonists[3,8]. The mechanism of action in this group of compounds involves the inhibition of certain cellular factors that are crucial to the HCV lifecycle. One of the main advantages of HTAs is that they act on host factors that are of a much lower rate of mutation[3]. Therefore, we are going to discuss primarily cellular receptors required for the attachment of HCV together with their involvement in the neutralization process.
CD81 receptor: The tetraspanin CD81 (26 kDa) is an integral membrane unglycosylated protein. CD81 is reported to possess several functions, such as signal transduction, cell activation, and cell adhesion. Moreover, in prior studies uding HCVpp and HCVcc systems, it has been confirmed that the CD81 receptor plays a major role in HCV cell entry[4]. A large number of broadly anti-HCV neutralizing antibodies block CD81 interaction with the HCV envelope glycoprotein E2. Indeed, it has been previously shown that anti-CD81 mAb inhibit the entry of both HCVcc and HCVpp into the Huh-7 cell line[44,45]. Resolving the crystal structure of CD81 complexed with E2 protein[44,45] has revealed that CD81 binds the HCV envelope glycoprotein E2 within certain specific amino acid residues (i.e., 412-423, 432-447, 480-493, 528-535, and 544-551). K04, a recently generated anti-human CD81 monoclonal antibody, was shown to have a broad-spectrum antiviral action in the prevention and treatment of HCV infection[46].
Lipoprotein receptor scavenger receptor BI: The scavenger receptor class B type I [scavenger receptor BI (SR-BI)] is highly expressed in hepatocytes. This receptor functions as a lipoprotein receptor that mediates cholesteryl ester selective uptake from high density lipoproteins[47]. The SR-BI receptor can bind both high density lipoproteins (HDL) and low density lipoproteins[8,47]. SR-BI has been previously shown to mediate interactions of E2-CD81. It has been suggested[45,48,49] that the SR-BI receptor interacts with HCV glycoprotein E2 hypervariable region 1 [hypervariable region-1 (HVR1), the first 27 amino acids in E2]. In line with this hypothesis, HVR1 deletion has been shown to inhibit E2-SR-BI interaction and to reduce HCV infectivity[50-52]. Indeed, it has been shown that HVR1 facilitates the interaction between HDL and SR-BI, which inhibits the neutralization of both HCVpp and HCVcc. HCV infection in cell cultures has also been shown to be inhibited with antibodies against SR-BI, confirming the crucial role of SR-BI in HCV cell entry[47,53,54]. Lastly, it was recently indicated that anti-SR-BI antibodies inhibit the HCV infection of different genotypes, both in cell cultures and humanized mice[55].
Other HCV receptors: Aside from SR-BI and CD81, other receptors including claudin-1 (CLDN1) and occludin (OCLN) compose the tight junction factors[56]. The tight junction multiprotein complex is comprised of four types of transmembrane proteins: Claudins, occludins, junction associated molecules, and the coxsackie virus B adenovirus receptors[57-59]. It is still unclear how the CLDN1 and OCLN inhibit HCV cell entry. Anti-claudin-1 antibodies neutralize HCV infectivity via inhibiting the interaction between CD81 and claudin-1 receptors, which is important to the viral entry process[58,59]. Occludin has been reported to co-precipitate with the HCV E2 glycoprotein. However, unlike CD81 and SR-BI and claudin-1 cellular receptors, no virus-specific neutralizing antibodies for occludin have been identified thus far[60].
HCV-NEUTRALIZING EPITOPES AND ANTIBODIES
The identification of the various mechanisms involved in immune protection is an important step in designing an HCV vaccine. The production of neutralizing antibodies by adaptive immune systems following vaccination has been prior demonstrated as a key strategy for protection against human viruses[61]. In the case of the HCV, a viral infection can persist even in the presence of a broadly neutralizing antibody, in many cases leading to chronic infection[44,62]; this can result in liver fibrosis, cirrhosis, and even eventually lead to hepatocellular carcinoma, causing death.
Anti-HCV antibodies can be targeted against structural and nonstructural protein epitopes (classified as either linear or conformational)[63,64]. The envelope glycoproteins E1 and E2 are considered major targets for neutralizing antibodies, as they are present on the surface of the HCV virus. Consequently, the development of an HCVpp system using unmodified HCV E1 and E2 envelope glycoproteins has allowed researchers to achieve remarkable progress in the study of neutralizing antibodies. However, little is understood so far as to the structure of the HCV envelope glycoproteins and how they interact with neutralizing antibodies[65].
Despite the fact that the HCV E1 glycoprotein is more conserved than E2, it has been proposed that E1 is of lower immunogenicity and hence a more difficult target for neutralizing antibodies[47,66]. However, two mAb against HCV glycoprotein E1 (IGH 505 and IGH 526) have been identified[27]. Both antibodies neutralize HCVpp bearing the E1 envelope glycoprotein of genotypes such as 1a, 1b, 4a, 5a, and 6a. These antibodies work within the region of E1 amino acids comprised of amino acids 313 to 327[27]. In addition to these two antibodies, the H-111 antibody has been reported to neutralize expressed E1 proteins from genotypes 1a, 1b, 2b, and 3a via binding to the 192YEVRNVSGVYH211 region of E1[67,68].
Anti-E2 human conformational-dependent HCV antibodies targeting E2 in HCVpp cell culture systems have been used to identify E2 epitopes. Due to such studies, three different regions of the E2 HCV glycoprotein E2 have been identified[50]. These regions include the E2 HVR1, the E2 HVR2, and the CD81 binding region of E2 and the C-terminus of HVR1[69]. The HCV glycoprotein HVR1 is a major target of neutralizing antibodies. This region is crucial for the virus, as it plays an important role in the HCV virus binding and entry process[50,70]. The physicochemical properties of the residues of HVR1 and its conformation are both highly conserved among various species despite the sequence variability of HVR1. It has been suggested that the sequence variability of E2 HVR1 is driven by the antibody selection of immune-escape variants[71]. Stable HVR1 sequences associated with resolved infection have also been reported, with HVR1 sequence change being suggested as one of the reasons for persistent HCV infection. This suggests that the most important target of antibody response to the HCV E2 glycoprotein is HVR1 (Table 1 for other examples)[72].
Table 1.
Hepatitis C virus neutralizing antibodies
| Neutralizing antibody | Epitope | Specificity | Escape mutants | Ref. |
| IGH505 | E1 (313-326) linear | Cross-reactive | NA | [27] |
| IGH526 | E1 (313-326) linear | Cross-reactive | NA | [27] |
| H-111 | E1 (192-202) linear | Cross-reactive | NA | [68] |
| 95-2 | E2 (412-423) linear | Cross-reactive | NA | [65] |
| HCV-1 | E2 (412-423) linear | Cross-reactive | NA | [65] |
| HC-1 | E2 (523-540) conformational | Cross-reactive | No escape | [69,125] |
| 3/11 | E2 (412-423) linear | Cross-reactive | N415Y, N415D, N417S, G418D | [75] |
| 3C7 | E2 (396-407) | H strain | NA | [126] |
| AP33 | E2 (412-423) linear | Cross-reactive | N415Y, N415D, N417S, G418D | [75] |
| CBH7 | E2 conformational | Cross-reactive | NA | [74] |
| CBH5 | E2 (523-540) conformational | Cross-reactive | NA | [68,127] |
| CBH2 | E2 (425-447), E2 (523-540) conformational | Cross-reactive | D431G, A439E | [68,125,128] |
| CBH-8C | E2 conformational | Cross-reactive | NA | [129] |
| CBH-8E | E2 conformational | Cross-reactive | NA | [129] |
| CBH-11 | E2 conformational | Cross-reactive | NA | [129] |
| CBH-17 | E2 conformational | Cross-reactive | NA | [129] |
| CBH4B | E2 conformational | Cross-reactive | NA | [129] |
| CBH4D | E2 conformational | Cross-reactive | NA | [129] |
| CBH4G | E2 conformational | Cross-reactive | NA | [129] |
| 9/27 | E2 conformational | H strain | NA | [130] |
| A8 | E2 (523-540) conformational | Cross-reactive | NA | [127] |
| J6.36 | E2 Partially conformational | J6 strain | NA | [131] |
| HC-11 | E2 (425-447) | NA | L438F | [69,125] |
| E2 (523-540) conformational | ||||
| Fab e20 | E2 (523-540) conformational | Cross-reactive | NA | [132] |
| Fab e137 | E2 (412-423) | Cross-reactive | NA | [73] |
| E2 (523-540) conformational | ||||
| 1:7 | E2 (523-540) conformational | Cross-reactive | NA | [127] |
| AR3A | E2 (394-424), E2 (437-447) | Cross-reactive | NA | [76] |
| E2 (523-540) conformational | ||||
| AR3B | E2 (394-424) | Cross-reactive | NA | [76] |
| E2 (437-447) | ||||
| E2(523-540) conformational | ||||
| AR3C | E2 (394-424), E2(437-447) | Cross-reactive | NA | [76] |
| E2 (523-540) conformational | ||||
| AR3D | E2 (394-424), E2 (437-447) | Cross-reactive | NA | [76] |
| E2 (523-540) conformational | ||||
| AP213 | E2 (396-407) partially conformational | Gla strain | NA | [133] |
| H77.39 | E2 (384-520) linear | Cross-reactive | NA | [131] |
| H35 | E2-conformational | Poorly cross-reactive | NA | [134] |
| H48 | E2-conformational | Poorly cross-reactive | NA | [134] |
| 2/69a | E2 (436-443) linear | NA | NA | [135] |
| 9/86a | E2 conformational | NA | NA | [130] |
| 6/1a | E2 (464-471) linear | NA | NA | [130] |
| 9/75 | E2 (524-531) linear | NA | NA | [130] |
| 6/53 | E2 (544-551) linear | NA | NA | [130] |
| 6/16 | E2 (384-391) linear | NA | NA | [130] |
| 1/39 | E2 (432-443) linear | NA | NA | [130] |
| 6/41a | E2 (480-493) linear | NA | NA | [130] |
| 11/20c | E2 (436-447) linear | NA | NA | [130] |
| ALP98 | E2 (644-651) linear | Cross-reactive | NA | [136] |
| ALP1 | E2 (647-658) linear | NA | NA | [136] |
NA: Not available.
EXAMPLES OF HCV-NEUTRALIZING ANTIBODIES
Epitopes can be classified into two main groups (linear or conformational epitopes). Various viral epitopes that are targeted by neutralizing antibodies have been identified and characterized. Two human mAb, HCV1 and 95-2, have been identified as successfully neutralizing HCVpp belonging to different genotypes (i.e., 1a, 1b, 2b, 3a, and 4a). In addition, a highly conserved linear epitope in E2 (amino acids 412 to 423) has been reported as recognized by HCV1 and 95-2 mAb[65]. Moreover, e137, a human monoclonal Fab, has been shown to bind to the HCV E2 glycoproteins of all HCV genotypes, with the exception of HCV genotype 5. Furthermore, it has been confirmed that this epitope interacts with highly conserved residues in all HCV genotypes, such as T416, W529, and D535[73]. CBH-5 has also shown itself to be capable of neutralizing all examined genotypes (genotypes 1-6). It was revealed that two of the amino acids comprising the epitope of CBH-5 are crucial for E2-CD81 interaction, which suggests direct competition between CBH-5 and CD81 to bind with the HCV E2 glycoprotein[74]. AP33 is another broadly neutralizing mouse monoclonal antibody that has been shown to neutralize all genotypes (i.e., 1a, 1b, 2a, 2b, 3a, 4, 5, and 6). This antibody recognizes a highly conserved epitope in HCV glycoprotein E2 (amino acids 412 to 423)[75]. The high conservation of its epitope may have resulted in the broadly neutralizing activity of this antibody. Finally, AR3B displays a broadly neutralizing human antibody activity. AR3B neutralizing antibodies have been shown to protect against viremia in an infected mouse model[76].
NEUTRALIZING ANTIBODIES’ MECHANISMS OF ACTION
The lack of cell culture-based assays to measure and quantify HCV activity has long hindered the study of the role of neutralizing antibodies in HCV infections. The mechanism of action in the antibody neutralization process still remains unclear. Several mechanisms through which neutralizing antibodies interfere with different stages of the HCV life cycle have been suggested[77,78], including immune aggregation and the blocking the attachment of the virion to the viral receptor which inhibits HCV infection. In addition to these mechanisms, neutralizing antibodies have been reported to interfere with other stages of the HCV life cycle following the binding process, such as the penetration of the virus through the cell membrane via host entry factors. Neutralizing antibodies may also prevent conformational changes important for the fusion of the virus to a host cell[77,78].
VIRAL MECHANISMS FOR EVADING NEUTRALIZING ANTIBODIES
HCV has developed the following different mechanisms to evade neutralizing antibody responses[79]: (1) the association of HCV with low density lipoproteins[47]; (2) the association of HCV with certain glycans can play a role in shielding important neutralizing epitopes of E1 and E2 envelope glycoproteins[80]; (3) the production of interfering antibodies, which may interfere with anti-HCV neutralizing antibodies[81]; and (4) direct cell-to-cell transmission of the HCV virus[82]. HCV typically employs a combination of these mechanisms together at the same time.
Lipoproteins
It is reported that poorly infectious HCV particles linked to immunoglobulins have been separated from the higher density fractions of chronic HCV plasma samples, with highly infectious particles found in lower density fractions, indicating the effect of lipoproteins on the infectivity of HCV[83]. The binding of HCV to lipoproteins may facilitate HCV uptake by liver cells[47,84]. This facilitated viral entry is mediated through interaction between the ApoB and SR-BI receptors, which enable HCV to escape recognition via different HCV neutralizing antibodies. This suggests that lipoproteins could possibly play a role in protection against antibody-mediated neutralization by masking the epitopes of the viral surface glycoproteins[85,86].
HCV genetic diversity
The high variability of the HCV genome provides the virus with an effective escape strategy from antibody neutralization[87,88]. HCV is classified into seven different genotypes, with each genotype including a number of subtypes that are roughly 25% different at the nucleotide level. The loss of the proofreading ability of HCV NS5B polymerase increases the mutation rate of the virus to one nucleotide per replication cycle[89]. This results in the evolution of HCV into many quasispecies with different though closely related nucleotide sequences even within the same patient, which may help the virus to escape from the human immune response[54,90]. HCV likely escapes the effects of the immune system because immune responses develop over weeks and HCV replicates on a timespan of days[91]. Interestingly, the sequence variation occurs mainly in the HCV E2 HVR1. HVR1 can remain with no change in its genome sequence for roughly two years given the absence of neutralizing antibodies. Given the virus’s exposure to neutralizing antibodies, the HCV genomic RNA sequence undergoes several adaptive mutations[89].
Glycans
Glycans associated with HCV envelope proteins protect the virus from neutralization by various antibodies via shielding crucial epitopes, especially in the E2 glycoprotein. With eleven N-glycosylation sites, E2 is the most glycosylated protein of the E1-E2 heterodimer, whereas the E1 protein contains only four sites. These fifteen glycans in total were reported to play a role in the entry of HCV into host cells. Several glycans (E2N1, E2N2, E2N3, E2N4, E2N6, E2N8, E2N9, E2N10, an E2N11) on E2 were found to limit the accessibility of neutralizing epitopes on E2[92]. These nine glycosylation sites were found to be conserved across all genotypes[80], which may indicate their importance as an HCV escape mechanism[92].
Direct viral transmission from cell to cell
Direct transmission between cells helps HCV to evade both innate and adaptive immune systems. Cell-to-cell spread has been found to occur in other viral families, such as the herpes virus, retroviruses, and rhabdoviruses[93]. Direct cell-to-cell transmission is more efficient in spreading the virus in host, as it allows the virus to escape neutralizing antibodies. Furthermore, direct cell-to-cell transmission has been shown to be CD81 independent. Lastly, both CLDN1 and OCLDN cell receptors were reported to play a role in cell-to-cell transmission[94,95].
AVAILABLE AND NEW TRENDS IN ANTIBODY PRODUCTION
The high cost of production of antibodies by hybridoma or the humanization of mouse antibodies, page displays, or transgenic mice has led to the emergence of new trends to produce antibodies, such as egg-yolk-based antibody (IgY) production within transgenic plants and synthetic antibody mimics.
Egg yolk antibodies (IgY)
Providing passive immunity to chicks, IgY is passively transmitted to egg yolks to help safeguard a chick against infection until its own immune response can be developed. IgYs are functionally similar to IgG in mammals, and intensive research has been conducted on the utilization of IgY for passive immunization[96]. The transient activity of passive antibodies increases the need for large-scale production due to their frequent administration. Hen eggs are an excellent source of antibodies for passive immunization due to their being a non-invasive means of production and the large production capability of a chicken[97]. The IgY collected from egg yolks can yield eighteen times more antibodies than the serum obtained from rabbits without sacrificing the animal[98]. An average hen can lay roughly 325 eggs a year. Given that an egg can produce 60-150 mg of IgY[99], one hen can produce 20-40 g of antibodies a year, with 2%-10% of the antibodies being antigen specific[100].
Additional advantages of using IgYs to combat infections in the human body include the ease of isolating egg yolk antibodies and the absence of interaction accruing between IgYs and the Fc receptors of mammals, which can initiate an inflammatory reaction and fail to induce complement activation in mammals[101]. In addition, chicken antibodies produce a different antibody repertoire and identify different epitopes than the antibodies produced by mammals[102]. Furthermore, the large-scale industrial production of eggs makes the process of IgY production technically efficient. IgY has been used effectively against several human and veterinary viruses, such as the bovine rotavirus[103], infectious bursal disease virus[104], human influenza virus H1N1[105,106], rabies virus[107], bovine leukemia Virus[108], rabbit hemorrhagic disease virus[109], bovine respiratory syncytial virus[110], avian reovirus[111], norovirus[112], hepatitis A virus[113], and the white spot syndrome virus[114].
The use of genetic engineering technologies to produce chicken mAb (mIgYs) will enhance the utility of IgY antibodies[115]. The combination of the high specificity and homogeneity of the mAbs and the unique features of chicken antibodies provide additional features to mIgYs. Very few studies have investigated the generation and utility of mIgYs with respect to prion immunogen[116]. The use of antibody engineering technologies facilitates the production of mIgY, while the phage display technique provides the features of a large, diverse library, as well as an efficient selection procedure[117].
Antibody expression in plants
The first trial of the production of antibodies in plants was carried out in 1989 in transgenic plants. Plant expression systems have several advantages in terms of the production of antibodies: First, these systems cost less compared to other conventional expression systems. Second, with regard to safety, they do not contain mammalian viruses or pathogens and do not produce endotoxins. Furthermore, as reported by Xu et al[118], the antibodies can be applied parenterally, topically, or orally. Castilho et al[119] and Olinger et al[120] report that, with antibody plant expression systems, it is possible to produce antibodies with desired glycoforms, and that glyco-engineered plants have a much higher degree of glycan homogeneity. In a recent review article by Virdi et al[121], they report that the production of antibodies in edible tissues would allow for oral passive immunization of the mucosal surface of the stomach. The use of technology together with the natural capacity of plant tissues to collect complex antibodies will enable in the enrichment of the antibody market. A review by Virdi et al[121] also showed the role of plants as an adaptable expression system for passive immunotherapy. Nianiou et al[122] showed the production of antibodies against HCV core gene in transgenic tobacco plants. The resultant HCV core antigen was found to be immunoreactive not only with polyclonal and mAb, but also with human sera positive for HCV-infected patients. Therefore, the authors prospected that the use of a plant-based HCV vaccine could be possible. Recently, Iranian scientists Mohammadzadeh et al[123] designed a highly codon-optimized HCV core protein gene for the construction of an effective plant expression system for the production of HCV core proteins with antigenic properties in an Iranian Jafarabadi-cultivar tobacco plant. The authors concluded that, through the use of a gene optimization strategy that uses vectors based on HCV and the suppression of plant-derived, gene-silencing effects, an effective expression system including the HCV core proteins of tobacco plants with antigenic immunogenic characteristics may be possible.
Synthetic antibody mimics
Recently, McEnaney et al[124] of the Yale University lab have crafted the first synthetic molecules (synthetic antibody mimics) that possess both the targeting abilities and functions of natural antibodies. The synthetic antibody mimics (SyAMs) attach themselves simultaneously to disease cells and immune fighting cells, performing a similar action to natural human antibodies. McEnaney et al[124] showed these molecules to be synthetic organic compounds that are approximately one-twentieth the size of natural antibodies. The authors report that their new SyAMs are thermally stable and can be administered orally. Furthermore, the authors report that the SyAMs have the potential to be used in treatments for cancer and other diseases, such as human immunodeficiency virus and various bacterial diseases[124]. We believe that these synthetic antibody mimics will open new areas of research and practice in the field of immunotherapy.
Footnotes
P- Reviewer: Lasfar A, Youssef SS S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
Conflict-of-interest statement: The authors declare no conflict of interests for this article.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: April 21, 2015
First decision: July 17, 2015
Article in press: September 8, 2015
References
- 1.Tansey EM, Catterall PP. Monoclonal antibodies: a witness seminar in contemporary medical history. Med Hist. 1994;38:322–327. doi: 10.1017/s0025727300036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 3.Baugh JM, Garcia-Rivera JA, Gallay PA. Host-targeting agents in the treatment of hepatitis C: a beginning and an end? Antiviral Res. 2013;100:555–561. doi: 10.1016/j.antiviral.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akazawa D, Date T, Morikawa K, Murayama A, Miyamoto M, Kaga M, Barth H, Baumert TF, Dubuisson J, Wakita T. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81:5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doss W, Shiha G, Hassany M, Soliman R, Fouad R, Khairy M, Samir W, Hammad R, Kersey K, Jiang D, et al. Sofosbuvir plus ribavirin for treating Egyptian patients with hepatitis C genotype 4. J Hepatol. 2015;63:581–585. doi: 10.1016/j.jhep.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Breban R, Doss W, Esmat G, Elsayed M, Hellard M, Ayscue P, Albert M, Fontanet A, Mohamed MK. Towards realistic estimates of HCV incidence in Egypt. J Viral Hepat. 2013;20:294–296. doi: 10.1111/j.1365-2893.2012.01650.x. [DOI] [PubMed] [Google Scholar]
- 7.Jansson J, Wilson DP. Feasible HCV targets in Egypt. Lancet Glob Health. 2014;2:e687. doi: 10.1016/S2214-109X(14)70323-7. [DOI] [PubMed] [Google Scholar]
- 8.Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A, et al. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34–43. doi: 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madorsky Rowdo FP, Baron A, Urrutia M, Mordoh J. Immunotherapy in Cancer: A Combat between Tumors and the Immune System; You Win Some, You Lose Some. Front Immunol. 2015;6:127. doi: 10.3389/fimmu.2015.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandušer M, Ušaj M. Cell electrofusion: past and future perspectives for antibody production and cancer cell vaccines. Expert Opin Drug Deliv. 2014;11:1885–1898. doi: 10.1517/17425247.2014.938632. [DOI] [PubMed] [Google Scholar]
- 11.Kato M, Sasamori E, Chiba T, Hanyu Y. Cell activation by CpG ODN leads to improved electrofusion in hybridoma production. J Immunol Methods. 2011;373:102–110. doi: 10.1016/j.jim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Wu M, Gao J, Wang X, Zhang Y, Zhu S, Yu Y, Han W. Generation and Characterization of a New Monoclonal Antibody Against CXCL4. Monoclon Antib Immunodiagn Immunother. 2015;34:110–115. doi: 10.1089/mab.2014.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YF, Phung Y, Gao W, Kawa S, Hassan R, Pastan I, Ho M. New high affinity monoclonal antibodies recognize non-overlapping epitopes on mesothelin for monitoring and treating mesothelioma. Sci Rep. 2015;5:9928. doi: 10.1038/srep09928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mondelli MU, Cerino A. Production of human monoclonal antibodies to hepatitis C virus and their characterization. Methods Mol Med. 1999;19:451–461. doi: 10.1385/0-89603-521-2:451. [DOI] [PubMed] [Google Scholar]
- 15.Martin DR, Marlowe RL, Ahearn JM. Determination of the role for CD21 during Epstein-Barr virus infection of B-lymphoblastoid cells. J Virol. 1994;68:4716–4726. doi: 10.1128/jvi.68.8.4716-4726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 17.Meijer PJ, Andersen PS, Haahr Hansen M, Steinaa L, Jensen A, Lantto J, Oleksiewicz MB, Tengbjerg K, Poulsen TR, Coljee VW, et al. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J Mol Biol. 2006;358:764–772. doi: 10.1016/j.jmb.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 18.Rosén A, Gergely P, Jondal M, Klein G, Britton S. Polyclonal Ig production after Epstein-Barr virus infection of human lymphocytes in vitro. Nature. 1977;267:52–54. doi: 10.1038/267052a0. [DOI] [PubMed] [Google Scholar]
- 19.Steinitz M, Klein G, Koskimies S, Makel O. EB virus-induced B lymphocyte cell lines producing specific antibody. Nature. 1977;269:420–422. doi: 10.1038/269420a0. [DOI] [PubMed] [Google Scholar]
- 20.Schieffelin JS, Costin JM, Nicholson CO, Orgeron NM, Fontaine KA, Isern S, Michael SF, Robinson JE. Neutralizing and non-neutralizing monoclonal antibodies against dengue virus E protein derived from a naturally infected patient. Virol J. 2010;7:28. doi: 10.1186/1743-422X-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funaro A, Gribaudo G, Luganini A, Ortolan E, Lo Buono N, Vicenzi E, Cassetta L, Landolfo S, Buick R, Falciola L, et al. Generation of potent neutralizing human monoclonal antibodies against cytomegalovirus infection from immune B cells. BMC Biotechnol. 2008;8:85. doi: 10.1186/1472-6750-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraussen J, Vrolix K, Martinez-Martinez P, Losen M, Meulemans E, De Baets MH, Stinissen P, Somers V. A novel method for making human monoclonal antibodies. J Autoimmun. 2010;35:130–134. doi: 10.1016/j.jaut.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siemoneit K, da Silva Cardoso M, Wölpl A, Koerner K, Subanek B. Isolation and epitope characterization of human monoclonal antibodies to hepatitis C virus core antigen. Hybridoma. 1994;13:9–13. doi: 10.1089/hyb.1994.13.9. [DOI] [PubMed] [Google Scholar]
- 25.Steinitz M. Production of human monoclonal antibodies by the epstein-barr virus method. Methods Mol Biol. 2014;1060:111–122. doi: 10.1007/978-1-62703-586-6_6. [DOI] [PubMed] [Google Scholar]
- 26.Tabll A, El Abd Y, El Din NG, El Shenawy R, Abdelhafez TH, El Awady M, El-Mohamady H, Viazov S. Establishment of human clones producing neutralizing human monoclonal antibodies to the envelope E1/E2 protein of hepatitis C virus by EBV immortalization of immune CD22+ B cells. Hum Antibodies. 2013;22:55–65. doi: 10.3233/HAB-140271. [DOI] [PubMed] [Google Scholar]
- 27.Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, Union A, Faulk KN, Bukh J, Emerson SU, Purcell RH. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J Virol. 2008;82:966–973. doi: 10.1128/JVI.01872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci USA. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauss S, Thomas B, Juergen R, Christoph S, Heiner W. Hepatology- A Clinical Textbook. 4th ed. Düsseldorf, Germany: Flying publisher; 2013. [Google Scholar]
- 31.Ali A, Nisar M, Idrees M, Rafique S, Iqbal M. Expression of Hepatitis C Virus Core and E2 antigenic recombinant proteins and their use for development of diagnostic assays. Int J Infect Dis. 2015;34:84–89. doi: 10.1016/j.ijid.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Florea D, Neaga E, Nicolae I, Maxim D, Popa M, Otelea D. Clinical usefulness of HCV core antigen assay for the management of patients with chronic hepatitis C. J Gastrointestin Liver Dis. 2014;23:393–396. doi: 10.15403/jgld.2014.1121.234.chcv. [DOI] [PubMed] [Google Scholar]
- 33.Chevaliez S, Soulier A, Poiteau L, Bouvier-Alias M, Pawlotsky JM. Clinical utility of hepatitis C virus core antigen quantification in patients with chronic hepatitis C. J Clin Virol. 2014;61:145–148. doi: 10.1016/j.jcv.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka T, Lau JY, Mizokami M, Orito E, Tanaka E, Kiyosawa K, Yasui K, Ohta Y, Hasegawa A, Tanaka S. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J Hepatol. 1995;23:742–745. doi: 10.1016/0168-8278(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 35.Yap SH, Willems M, Van den Oord J, Habets W, Middeldorp JM, Hellings JA, Nevens F, Moshage H, Desmet V, Fevery J. Detection of hepatitis C virus antigen by immuno-histochemical staining: a histological marker of hepatitis C virus infection. J Hepatol. 1994;20:275–281. doi: 10.1016/s0168-8278(05)80069-8. [DOI] [PubMed] [Google Scholar]
- 36.González-Peralta RP, Fang JW, Davis GL, Gish RG, Kohara M, Mondelli MU, Urdea MS, Mizokami M, Lau JY. Significance of hepatic expression of hepatitis C viral antigens in chronic hepatitis C. Dig Dis Sci. 1995;40:2595–2601. doi: 10.1007/BF02220447. [DOI] [PubMed] [Google Scholar]
- 37.Sansonno D, Cornacchiulo V, Iacobelli AR, Di Stefano R, Lospalluti M, Dammacco F. Localization of hepatitis C virus antigens in liver and skin tissues of chronic hepatitis C virus-infected patients with mixed cryoglobulinemia. Hepatology. 1995;21:305–312. [PubMed] [Google Scholar]
- 38.Tanaka E, Kiyosawa K, Matsumoto A, Kashiwakuma T, Hasegawa A, Mori H, Yanagihara O, Ohta Y. Serum levels of hepatitis C virus core protein in patients with chronic hepatitis C treated with interferon alfa. Hepatology. 1996;23:1330–1333. doi: 10.1053/jhep.1996.v23.pm0008675147. [DOI] [PubMed] [Google Scholar]
- 39.Aoyagi K, Ohue C, Iida K, Kimura T, Tanaka E, Kiyosawa K, Yagi S. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–1808. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masalova OV, Atanadze SN, Samokhvalov EI, Petrakova NV, Kalinina TI, Smirnov VD, Khudyakov YE, Fields HA, Kushch AA. Detection of hepatitis C virus core protein circulating within different virus particle populations. J Med Virol. 1998;55:1–6. doi: 10.1002/(sici)1096-9071(199805)55:1<1::aid-jmv1>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 41.Attallah AM, Ismail H, Tabll AA, Shiha GE, El-Dosoky I. A novel antigen detection immunoassay for field diagnosis of hepatitis C virus infection. J Immunoassay Immunochem. 2003;24:395–407. doi: 10.1081/IAS-120025777. [DOI] [PubMed] [Google Scholar]
- 42.Attallah AM, Omran MM, Nasif WA, Ghaly MF, El-Shanshoury Ael-R, Abdalla MS, Sharada HM, Farid K, El-Shony W, Moussa el-SM, et al. Diagnostic performances of hepatitis C virus-NS4 antigen in patients with different liver pathologies. Arch Med Res. 2012;43:555–562. doi: 10.1016/j.arcmed.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Tabll AA, Moustafa RI, El Abd YS, Bader El Din NG, El-Shenawy R, Yousef H, Hussein M, Dawood RM, Omran MH, El-Awady MK. Mouse monoclonal antibody towards e1 specific epitope blocks viral entry and intracellular viral replication in vitro. J Immunoassay Immunochem. 2014;35:60–73. doi: 10.1080/15321819.2013.792831. [DOI] [PubMed] [Google Scholar]
- 44.Allander T, Drakenberg K, Beyene A, Rosa D, Abrignani S, Houghton M, Widell A, Grillner L, Persson MA. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J Gen Virol. 2000;81:2451–2459. doi: 10.1099/0022-1317-81-10-2451. [DOI] [PubMed] [Google Scholar]
- 45.Cocquerel L, Kuo CC, Dubuisson J, Levy S. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J Virol. 2003;77:10677–10683. doi: 10.1128/JVI.77.19.10677-10683.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji C, Liu Y, Pamulapati C, Bohini S, Fertig G, Schraeml M, Rubas W, Brandt M, Ries S, Ma H, et al. Prevention of hepatitis C virus infection and spread in human liver chimeric mice by an anti-CD81 monoclonal antibody. Hepatology. 2015;61:1136–1144. doi: 10.1002/hep.27603. [DOI] [PubMed] [Google Scholar]
- 47.Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J, et al. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285–18295. doi: 10.1074/jbc.M602706200. [DOI] [PubMed] [Google Scholar]
- 48.Cocquerel L, Voisset C, Dubuisson J. Hepatitis C virus entry: potential receptors and their biological functions. J Gen Virol. 2006;87:1075–1084. doi: 10.1099/vir.0.81646-0. [DOI] [PubMed] [Google Scholar]
- 49.Boo I, teWierik K, Douam F, Lavillette D, Poumbourios P, Drummer HE. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidine residues of hepatitis C virus glycoprotein E1 and E2. Biochem J. 2012;443:85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- 50.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter HJ, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callens N, Ciczora Y, Bartosch B, Vu-Dac N, Cosset FL, Pawlotsky JM, Penin F, Dubuisson J. Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein e2 contribute to virus entry. J Virol. 2005;79:15331–15341. doi: 10.1128/JVI.79.24.15331-15341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mondelli MU, Cerino A, Segagni L, Meola A, Cividini A, Silini E, Nicosia A. Hypervariable region 1 of hepatitis C virus: immunological decoy or biologically relevant domain? Antiviral Res. 2001;52:153–159. doi: 10.1016/s0166-3542(01)00180-2. [DOI] [PubMed] [Google Scholar]
- 53.von Hahn T, Lindenbach BD, Boullier A, Quehenberger O, Paulson M, Rice CM, McKeating JA. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology. 2006;43:932–942. doi: 10.1002/hep.21139. [DOI] [PubMed] [Google Scholar]
- 54.Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 55.Vercauteren K, Van Den Eede N, Mesalam AA, Belouzard S, Catanese MT, Bankwitz D, Wong-Staal F, Cortese R, Dubuisson J, Rice CM, et al. Successful anti-scavenger receptor class B type I (SR-BI) monoclonal antibody therapy in humanized mice after challenge with HCV variants with in vitro resistance to SR-BI-targeting agents. Hepatology. 2014;60:1508–1518. doi: 10.1002/hep.27196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benedicto I, Molina-Jiménez F, Bartosch B, Cosset FL, Lavillette D, Prieto J, Moreno-Otero R, Valenzuela-Fernández A, Aldabe R, López-Cabrera M, et al. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J Virol. 2009;83:8012–8020. doi: 10.1128/JVI.00038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bekker V, Chanock SJ, Yeager M, Hutchinson AA, von Hahn T, Chen S, Xiao N, Dotrang M, Brown M, Busch MP, et al. Genetic variation in CLDN1 and susceptibility to hepatitis C virus infection. J Viral Hepat. 2010;17:192–200. doi: 10.1111/j.1365-2893.2009.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 59.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris HJ, Farquhar MJ, Mee CJ, Davis C, Reynolds GM, Jennings A, Hu K, Yuan F, Deng H, Hubscher SG, et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol. 2008;82:5007–5020. doi: 10.1128/JVI.02286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abrignani S, Houghton M, Hsu HH. Perspectives for a vaccine against hepatitis C virus. J Hepatol. 1999;31 Suppl 1:259–263. doi: 10.1016/s0168-8278(99)80413-9. [DOI] [PubMed] [Google Scholar]
- 62.Arthur RR, Hassan NF, Abdallah MY, el-Sharkawy MS, Saad MD, Hackbart BG, Imam IZ. Hepatitis C antibody prevalence in blood donors in different governorates in Egypt. Trans R Soc Trop Med Hyg. 1997;91:271–274. doi: 10.1016/s0035-9203(97)90070-5. [DOI] [PubMed] [Google Scholar]
- 63.Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709–1714. doi: 10.1084/jem.20050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci USA. 2003;100:14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol. 2009;83:12473–12482. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci USA. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haberstroh A, Schnober EK, Zeisel MB, Carolla P, Barth H, Blum HE, Cosset FL, Koutsoudakis G, Bartenschlager R, Union A, Depla E, Owsianka A, Patel AH, Schuster C, Stoll-Keller F, Doffoël M, Dreux M, Baumert TF. Neutralizing host responses in hepatitis C virus infection target viral entry at postbinding steps and membrane fusion. Gastroenterology. 2008;135:1719–1728.e1. doi: 10.1053/j.gastro.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 68.Keck ZY, Sung VM, Perkins S, Rowe J, Paul S, Liang TJ, Lai MM, Foung SK. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J Virol. 2004;78:7257–7263. doi: 10.1128/JVI.78.13.7257-7263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keck ZY, Li SH, Xia J, von Hahn T, Balfe P, McKeating JA, Witteveldt J, Patel AH, Alter H, Rice CM, et al. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol. 2009;83:6149–6160. doi: 10.1128/JVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anjum S, Wahid A, Afzal MS, Albecka A, Alsaleh K, Ahmad T, Baumert TF, Wychowski C, Qadri I, Penin F, et al. Additional glycosylation within a specific hypervariable region of subtype 3a of hepatitis C virus protects against virus neutralization. J Infect Dis. 2013;208:1888–1897. doi: 10.1093/infdis/jit376. [DOI] [PubMed] [Google Scholar]
- 71.Binder M, Kochs G, Bartenschlager R, Lohmann V. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology. 2007;46:1365–1374. doi: 10.1002/hep.21829. [DOI] [PubMed] [Google Scholar]
- 72.Falkowska E, Kajumo F, Garcia E, Reinus J, Dragic T. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J Virol. 2007;81:8072–8079. doi: 10.1128/JVI.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perotti M, Mancini N, Diotti RA, Tarr AW, Ball JK, Owsianka A, Adair R, Patel AH, Clementi M, Burioni R. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J Virol. 2008;82:1047–1052. doi: 10.1128/JVI.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Owsianka AM, Tarr AW, Keck ZY, Li TK, Witteveldt J, Adair R, Foung SK, Ball JK, Patel AH. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J Gen Virol. 2008;89:653–659. doi: 10.1099/vir.0.83386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJ, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology. 2006;43:592–601. doi: 10.1002/hep.21088. [DOI] [PubMed] [Google Scholar]
- 76.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 77.Barth H, Liang TJ, Baumert TF. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology. 2006;44:527–535. doi: 10.1002/hep.21321. [DOI] [PubMed] [Google Scholar]
- 78.Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 79.Wahid A, Dubuisson J. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat. 2013;20:369–376. doi: 10.1111/jvh.12094. [DOI] [PubMed] [Google Scholar]
- 80.Helle F, Goffard A, Morel V, Duverlie G, McKeating J, Keck ZY, Foung S, Penin F, Dubuisson J, Voisset C. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81:8101–8111. doi: 10.1128/JVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, Feinstone SM. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci USA. 2007;104:8449–8454. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Witteveldt J, Evans MJ, Bitzegeio J, Koutsoudakis G, Owsianka AM, Angus AG, Keck ZY, Foung SK, Pietschmann T, Rice CM, et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol. 2009;90:48–58. doi: 10.1099/vir.0.006700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carrick RJ, Schlauder GG, Peterson DA, Mushahwar IK. Examination of the buoyant density of hepatitis C virus by the polymerase chain reaction. J Virol Methods. 1992;39:279–289. doi: 10.1016/0166-0934(92)90101-i. [DOI] [PubMed] [Google Scholar]
- 85.Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J, Tercé F, Duverlie G, Rouillé Y, Dubuisson J. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology. 2012;55:998–1007. doi: 10.1002/hep.25501. [DOI] [PubMed] [Google Scholar]
- 86.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anjum S, Ali S, Ahmad T, Afzal MS, Waheed Y, Shafi T, Ashraf M, Andleeb S. Sequence and structural analysis of 3’ untranslated region of hepatitis C virus, genotype 3a, from pakistani isolates. Hepat Mon. 2013;13:e8390. doi: 10.5812/hepatmon.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdel-Hamid M, El-Daly M, Molnegren V, El-Kafrawy S, Abdel-Latif S, Esmat G, Strickland GT, Loffredo C, Albert J, Widell A. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol. 2007;88:1526–1531. doi: 10.1099/vir.0.82626-0. [DOI] [PubMed] [Google Scholar]
- 89.Chen S, Wang YM. Evolutionary study of hepatitis C virus envelope genes during primary infection. Chin Med J (Engl) 2007;120:2174–2180. [PubMed] [Google Scholar]
- 90.Larrubia JR, Moreno-Cubero E, Lokhande MU, García-Garzón S, Lázaro A, Miquel J, Perna C, Sanz-de-Villalobos E. Adaptive immune response during hepatitis C virus infection. World J Gastroenterol. 2014;20:3418–3430. doi: 10.3748/wjg.v20.i13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gal-Tanamy M, Keck ZY, Yi M, McKeating JA, Patel AH, Foung SK, Lemon SM. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc Natl Acad Sci USA. 2008;105:19450–19455. doi: 10.1073/pnas.0809879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goffard A, Callens N, Bartosch B, Wychowski C, Cosset FL, Montpellier C, Dubuisson J. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol. 2005;79:8400–8409. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat Rev Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 94.Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL, et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol. 2011;85:596–605. doi: 10.1128/JVI.01592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ciesek S, Westhaus S, Wicht M, Wappler I, Henschen S, Sarrazin C, Hamdi N, Abdelaziz AI, Strassburg CP, Wedemeyer H, et al. Impact of intra- and interspecies variation of occludin on its function as coreceptor for authentic hepatitis C virus particles. J Virol. 2011;85:7613–7621. doi: 10.1128/JVI.00212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kovacs-Nolan J, Mine Y. Egg yolk antibodies for passive immunity. Annu Rev Food Sci Technol. 2012;3:163–182. doi: 10.1146/annurev-food-022811-101137. [DOI] [PubMed] [Google Scholar]
- 97.Schade R, Zhang X-Y, Terzolo H. Use of IgY Antibodies in Human and Veterinary Medicine. In: Huopalahti R, López-Fandiño R, Anton M, Schade R, editors. Bioactive Egg Compounds: Springer Berlin Heidelberg; 2007. pp. 213–222. [Google Scholar]
- 98.Larsson A, Sjöquist J. Chicken IgY: utilizing the evolutionary difference. Comp Immunol Microbiol Infect Dis. 1990;13:199–201. doi: 10.1016/0147-9571(90)90088-b. [DOI] [PubMed] [Google Scholar]
- 99.Pauly D, Dorner M, Zhang X, Hlinak A, Dorner B, Schade R. Monitoring of laying capacity, immunoglobulin Y concentration, and antibody titer development in chickens immunized with ricin and botulinum toxins over a two-year period. Poult Sci. 2009;88:281–290. doi: 10.3382/ps.2008-00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tini M, Jewell UR, Camenisch G, Chilov D, Gassmann M. Generation and application of chicken egg-yolk antibodies. Comp Biochem Physiol A Mol Integr Physiol. 2002;131:569–574. doi: 10.1016/s1095-6433(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 101.Carlander D, Kollberg H, Wejåker PE, Larsson A. Peroral immunotherapy with yolk antibodies for the prevention and treatment of enteric infections. Immunol Res. 2000;21:1–6. doi: 10.1385/IR:21:1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gassmann M, Thömmes P, Weiser T, Hübscher U. Efficient production of chicken egg yolk antibodies against a conserved mammalian protein. FASEB J. 1990;4:2528–2532. doi: 10.1096/fasebj.4.8.1970792. [DOI] [PubMed] [Google Scholar]
- 103.Vega C, Bok M, Chacana P, Saif L, Fernandez F, Parreño V. Egg yolk IgY: protection against rotavirus induced diarrhea and modulatory effect on the systemic and mucosal antibody responses in newborn calves. Vet Immunol Immunopathol. 2011;142:156–169. doi: 10.1016/j.vetimm.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yousif AA, Mohammad WA, Khodeir MH, Zeid AZ, el-Sanousi AA, Saber MS, Reda IM. Oral administration of hyperimmune IgY: an immunoecological approach to curbing acute infectious bursal disease virus infection. Egypt J Immunol. 2006;13:85–94. [PubMed] [Google Scholar]
- 105.Nguyen HH, Tumpey TM, Park HJ, Byun YH, Tran LD, Nguyen VD, Kilgore PE, Czerkinsky C, Katz JM, Seong BL, et al. Prophylactic and therapeutic efficacy of avian antibodies against influenza virus H5N1 and H1N1 in mice. PLoS One. 2010;5:e10152. doi: 10.1371/journal.pone.0010152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang YE, Wen J, Zhao S, Zhang K, Zhou Y. Prophylaxis and therapy of pandemic H1N1 virus infection using egg yolk antibody. J Virol Methods. 2014;206:19–26. doi: 10.1016/j.jviromet.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 107.Motoi Y, Sato K, Hatta H, Morimoto K, Inoue S, Yamada A. Production of rabies neutralizing antibody in hen’s eggs using a part of the G protein expressed in Escherichia coli. Vaccine. 2005;23:3026–3032. doi: 10.1016/j.vaccine.2004.11.071. [DOI] [PubMed] [Google Scholar]
- 108.Martinez C, Gutierrez G, Alvarez I, Porta N, Lomonaco M, Wigdorovitz A, Chacana P, Trono K. Egg yolk antibodies (IgY) against Bovine Leukemia Virus. Retrovirology. 2014;11 Suppl 1:46. [Google Scholar]
- 109.Li ZX, Hu WD, Li BC, Li TY, Zhou XY, Zhang Z. Egg yolk IgY against RHDV capsid protein VP60 promotes rabbit defense against RHDV infection. Vet Immunol Immunopathol. 2014;157:97–104. doi: 10.1016/j.vetimm.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 110.Ferella A, Bellido D, Chacana P, Wigdorovitz A, Santos MJD, Mozgovoj MV. Chicken egg yolk antibodies against bovine respiratory syncytial virus neutralize the virus in vitro. Procedia in Vaccinology. 2012;6:33–38. [Google Scholar]
- 111.Jung KM, Bae EH, Jung YT, Kim JW. Use of IgY antibody to recombinant avian reovirus σC protein in the virus diagnostics. Acta Virol. 2014;58:108–113. doi: 10.4149/av_2014_02_108. [DOI] [PubMed] [Google Scholar]
- 112.Dai YC, Wang YY, Zhang XF, Tan M, Xia M, Wu XB, Jiang X, Nie J. Evaluation of anti-norovirus IgY from egg yolk of chickens immunized with norovirus P particles. J Virol Methods. 2012;186:126–131. doi: 10.1016/j.jviromet.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Paula VS, da Silva Ados S, de Vasconcelos GA, Iff ET, Silva ME, Kappel LA, Cruz PB, Pinto MA. Applied biotechnology for production of immunoglobulin Y specific to hepatitis A virus. J Virol Methods. 2011;171:102–106. doi: 10.1016/j.jviromet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 114.Lu Y, Liu J, Jin L, Li X, Zhen Y, Xue H, You J, Xu Y. Passive protection of shrimp against white spot syndrome virus (WSSV) using specific antibody from egg yolk of chickens immunized with inactivated virus or a WSSV-DNA vaccine. Fish Shellfish Immunol. 2008;25:604–610. doi: 10.1016/j.fsi.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 115.Zhang X, Chen H, Tian Z, Chen S, Schade R. Chicken monoclonal IgY antibody: a novel antibody development strategy. Avian Biology Research. 2010;3:97–106. [Google Scholar]
- 116.Shimamoto T, Nishibori N, Aosasa M, Horiuchi H, Furusawa S, Matsuda H. Stable production of recombinant chicken antibody in CHO-K1 cell line. Biologicals. 2005;33:169–174. doi: 10.1016/j.biologicals.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 117.Chen HX, He F, Sun Y, Luo Y, Qiu HJ, Zhang XY, Sutton BJ. Generation and characterization of chicken-sourced single-chain variable fragments (scFvs) against porcine interferon-gamma (pIFN-γ) J Immunoassay Immunochem. 2015;36:27–44. doi: 10.1080/15321819.2014.892511. [DOI] [PubMed] [Google Scholar]
- 118.Xu J, Dolan MC, Medrano G, Cramer CL, Weathers PJ. Green factory: plants as bioproduction platforms for recombinant proteins. Biotechnol Adv. 2012;30:1171–1184. doi: 10.1016/j.biotechadv.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 119.Castilho A, Bohorova N, Grass J, Bohorov O, Zeitlin L, Whaley K, Altmann F, Steinkellner H. Rapid high yield production of different glycoforms of Ebola virus monoclonal antibody. PLoS One. 2011;6:e26040. doi: 10.1371/journal.pone.0026040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Olinger GG, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci USA. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Virdi V, Depicker A. Role of plant expression systems in antibody production for passive immunization. Int J Dev Biol. 2013;57:587–593. doi: 10.1387/ijdb.130266ad. [DOI] [PubMed] [Google Scholar]
- 122.Nianiou I, Kalantidis K, Madesis P, Georgopoulou U, Mavromara P, Tsaftaris A. Expression of an HCV core antigen coding gene in tobacco (N. tabacum L.) Prep Biochem Biotechnol. 2008;38:411–421. doi: 10.1080/10826060802325667. [DOI] [PubMed] [Google Scholar]
- 123.Mohammadzadeh S, Khabiri A, Roohvand F, Memarnejadian A, Salmanian AH, Ajdary S, Ehsani P. Enhanced-Transient Expression of Hepatitis C Virus Core Protein in Nicotiana tabacum, a Protein With Potential Clinical Applications. Hepat Mon. 2014;14:e20524. doi: 10.5812/hepatmon.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McEnaney PJ, Fitzgerald KJ, Zhang AX, Douglass EF, Shan W, Balog A, Kolesnikova MD, Spiegel DA. Chemically synthesized molecules with the targeting and effector functions of antibodies. J Am Chem Soc. 2014;136:18034–18043. doi: 10.1021/ja509513c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Keck ZY, Saha A, Xia J, Wang Y, Lau P, Krey T, Rey FA, Foung SK. Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations. J Virol. 2011;85:10451–10463. doi: 10.1128/JVI.05259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cerino A, Meola A, Segagni L, Furione M, Marciano S, Triyatni M, Liang TJ, Nicosia A, Mondelli MU. Monoclonal antibodies with broad specificity for hepatitis C virus hypervariable region 1 variants can recognize viral particles. J Immunol. 2001;167:3878–3886. doi: 10.4049/jimmunol.167.7.3878. [DOI] [PubMed] [Google Scholar]
- 127.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MA. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci USA. 2007;104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keck ZY, Xia J, Cai Z, Li TK, Owsianka AM, Patel AH, Luo G, Foung SK. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J Virol. 2007;81:1043–1047. doi: 10.1128/JVI.01710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hadlock KG, Lanford RE, Perkins S, Rowe J, Yang Q, Levy S, Pileri P, Abrignani S, Foung SK. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74:10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Flint M, Dubuisson J, Maidens C, Harrop R, Guile GR, Borrow P, McKeating JA. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J Virol. 2000;74:702–709. doi: 10.1128/jvi.74.2.702-709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, Lemon SM, Ball JK, Bukh J, Evans MJ, et al. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol. 2011;85:7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mancini N, Diotti RA, Perotti M, Sautto G, Clementi N, Nitti G, Patel AH, Ball JK, Clementi M, Burioni R. Hepatitis C virus (HCV) infection may elicit neutralizing antibodies targeting epitopes conserved in all viral genotypes. PLoS One. 2009;4:e8254. doi: 10.1371/journal.pone.0008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vieyres G, Dubuisson J, Patel AH. Characterization of antibody-mediated neutralization directed against the hypervariable region 1 of hepatitis C virus E2 glycoprotein. J Gen Virol. 2011;92:494–506. doi: 10.1099/vir.0.028092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol. 2006;80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Clayton RF, Owsianka A, Aitken J, Graham S, Bhella D, Patel AH. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J Virol. 2002;76:7672–7682. doi: 10.1128/JVI.76.15.7672-7682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


