Abstract
Most accounts of human cognitive architectures have focused on computational accounts of cognition while making little contact with the study of anatomical structures and physiological processes. A renewed convergence between neurobiology and cognition is well underway. A promising area arises from the overlap between systems/cognitive neuroscience on the one side and the discipline of network science on the other. Neuroscience increasingly adopts network tools and concepts to describe the operation of collections of brain regions. Beyond just providing illustrative metaphors, network science offers a theoretical framework for approaching brain structure and function as a multi-scale system comprised of networks of neurons, circuits, nuclei, cortical areas and systems of areas. This paper views large-scale networks at the level of areas and systems, mostly based on data from human neuroimaging, and how this view of network structure and function has begun to illuminate our understanding of the biological basis of cognitive architectures.
INTRODUCTION
The term “cognitive architecture” used to refer to concepts that were entirely the domain of cognitive or computer scientists (see Box 1) whose efforts to elucidate the rules behind human cognition (Fodor and Pylyshyn, 1988) made little of no reference to the underlying biological substrate – the human brain. Times have changed. A new picture of cognitive architecture has begun to emerge, as amply documented by the contributions to this Special Issue. Most “cognitive architectures” now are thought of as sets of brain regions that contribute to the performance of some set of related tasks, or a particular set of functions. Often these architectures are explicitly referred to as networks, for example the default mode network (Raichle et al., 2001), attention networks, etc (e.g. (Corbetta and Shulman, 2002)).
However, the meaning of the term “network” is highly variable. In many cases, the term network is informally applied to a simple collection of regions that is activated during a set of related fMRI imaging studies, without any explicit reference to connections between these regions. In contrast to this informal notion of networks as sets of regions stands the more formal definition of what constitutes a network, which is adopted in this article. A network is a set of pairwise relationships between the elements of a system – formally represented as a set of edges that link a set of nodes. Neurobiological networks come at many levels of scale from cell-specific metabolic or regulatory pathways inside of neurons to interactions between systems of cortical areas and sub cortical nuclei (see figure 1). At each level (neurons, neuronal circuits and populations, systems) different kinds of networks with importantly different properties are present. At each of these levels, it is important not just to understand how the individual elements work, but to understand the sets of pairwise relations that put the elements into the context of the larger interconnected system (Sporns, 2011). With some exceptions, cognitive architectures mostly involve structures and mechanisms at this highest level of analysis (Sejnowski and Churchland, 1989). For this article, we would like to focus at these highest levels, with a view to understanding networks that relate to all or much of the brain. We would like to explore large-scale architectural principles and properties that encompass the more specific architectures discussed in other articles in this issue.
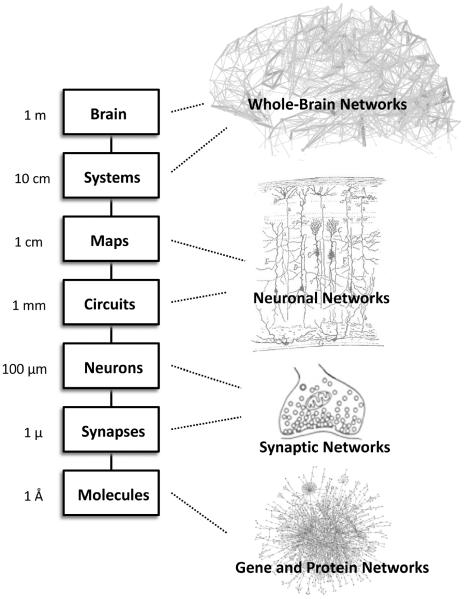
Figure 1.
Schematic representation of levels of structure within the nervous system. The large scale analyses discussed in the paper focus on the levels of areas/maps and systems, but network ideas clearly extend down to the level of neuronal circuits and populations, individual neurons and synapses, as well as genetic regulatory and protein interaction networks. Adapted from a similar illustration in (Churchland and Sejnowski, 1992; Sejnowski and Churchland, 1989).
Approaching large-scale brain networks
The bulk of the article will entail looking at some of the concepts and results coming from taking an explicitly network perspective to brain organization in two related types of studies.
We first turn to work that has aimed to elucidate the anatomical networks upon which all functional activity unfolds. Anatomical networks provide the skeleton that constrains the passage of neuronal signaling and information that is crucial for shaping our thoughts, understanding and actions.
A second major way in which many brain network studies have been studied is through correlated fluctuations of the functional MRI BOLD signal (cf (Power et al., 2014)). These studies often observe these correlations without any explicit task, forming so-called resting state “functional connectivity” (RSFC). This work began with the important observation that, even at rest, fluctuations of the fMRI BOLD signal correlate in anatomically specific ways across the brain. For example, many regions that relate to motor function are strongly correlated with one another in the absence of any task. The organization of RSFC has been demonstrated to provide insight into common functional relationships between many brain regions beyond the motor system. The second main section of the article explores some basic observations and properties that these studies have provided.
In the final section we explore the relationship between structural and functional networks which we think is fundamental for understanding the biological mechanisms that underpin cognitive architectures (see Box 2). While recent work has uncovered some relationships between these two types of brain networks, , many aspects of how structural connections constrain functional networks, and how these constraints play out on multiple time scales, remain incompletely understood. Integrative studies of networks across structure and function are an important goal for the future, and we end our article with charting some tentative footsteps down this path.
ANATOMICAL NETWORKS
The search for anatomical principles of neurocognitive networks has a long history, extending at least as far back as the 19th century marked by the development of new histological methods and new ideas about the localization of brain function. Deeply rooted in this tradition is the view that human cognition relies upon an intricately connected cortical architecture that underpins its various functional capacities. The fundamental idea that cognitive architecture has a structural foundation remains valid today.
Insights from Non-Human Primates
Preceding the recent expansion of studies utilizing fMRI methodology in humans, the biological foundations of cognition were mainly explored from the vantage point of large-scale anatomy and cellular physiology in model organisms such as nonhuman primates. These classic approaches have led to the formulation of candidate principles for the organization of neurocognitive networks that continue to influence our modern view. Key principles include functional specialization, distributed networks, segregated processing streams, cortical hierarchy and convergence zones. Functional specialization was articulated as principle of brain organization in the work of Semir Zeki (Zeki and Shipp, 1988) building on the finding that cortical regions maintained unique sets of afferent and efferent projections, later termed “cortical fingerprints” (Passingham et al., 2002). Distributed circuits, exemplified in the work of Patricia Goldman-Rakic and Vernon Mountcastle, consisted of sets of brain regions and interconnecting pathways that collectively performed a specific perceptual or cognitive function (Goldman-Rakic, 1988). Processing streams, for example the ventral and dorsal visual cortex, combined serial/hierarchical arrangements of regions with functional specialization. Cortical hierarchy was postulated based on projection patterns in collated anatomical data (Felleman and Van Essen, 1991), and was one of the hallmarks of Marcel Mesulam's seminal proposal for a cortical architecture that consisted of nested shells of areas ranging from unimodal sensory and motor regions to an inner core of transmodal or multimodal areas (Mesulam, 1990). A related idea was that of cortical convergence zones (Damasio, 1989), representing anatomical elements with key roles in binding and cross-referencing distributed sources of information.
More recent work has shed new light on some of these classic concepts, by applying data-driven and quantitative analytic tools from graph theory and network science to neurocognitive systems. For example, analysis of the topology of projections among a subset of regions in macaque cortex has shown that unique sets of inputs and outputs, especially those made over longer distances convey functional specificity (Markov et al., 2013). Cortical hierarchies, while not always arranged in strictly serial order, define gradients of progressively more complex physiological properties in sensory and motor systems. Processing streams, for example those in visual cortex, may correspond to network modules or communities that are defined by the topology of interregional projections in cerebral cortex. Diverse and widespread anatomical connections have repeatedly been described as the defining feature of transmodal/multimodal areas, e.g. the various subdivisions of the prefrontal cortex (Markov et al., 2013)or the precuneus (Parvizi et al., 2006). The network embedding of these regions renders them candidate network hubs, putative focal points that are important for attracting and dispersing a diverse set of neural signals (van den Heuvel and Sporns, 2013). Another prominent network feature is based on a high density of anatomical linkages among hub nodes, which are often seen as forming a core (Hagmann et al., 2008) or rich club (van den Heuvel and Sporns, 2011). Overall, modern network-based studies and analyses validate most classic anatomical principles and advance a coherent framework for the topology of neurocognitive systems that is rooted within the larger context of network science.
Going forward, the continued exploration of the anatomical basis of cognitive networks will benefit from the development of more sensitive quantitative methods for estimating the geometry and topology of cortical projection systems. Invasive labeling and tract-tracing technologies are evolving towards more comprehensive (Bota et al., 2015) and high-throughput (e.g. (Oh et al., 2014)) detection of interregional pathways. Despite their status as “gold standards” in connectional anatomy, these methods also have some methodological limitations, as they require aggregating data across many individuals (often without tracking gender, age, or hemispheric location) and do not capture inter-individual variability.
Structural Networks in the Human Brain
In recent years, much work on the structural basis of human cognitive networks has relied on reconstructions of anatomical networks derived from diffusion imaging and tractography. Methodological advantages of this approach are partly complementary to the limitations of invasive tract tracing studies – for example, whole-brain coverage in neuroimaging allows construction of complete anatomical networks from single individuals and hence the potential systematic assessment of individual differences and heritability. However, diffusion imaging also suffers from numerous limitations and biases in data acquisition and computational reconstruction of connectivity. These include the complete lack of gray-matter connections, an inability to determine directionality or physiological efficacy, and uncertain measures of connection strength or magnitude (Fornito et al., 2013). Considerable efforts are underway to further improve diffusion imaging acquisition and signal deconvolution. In addition, computational inference of anatomical pathways with noninvasive imaging is undergoing continuing development – one promising avenue is the introduction of model-based global tractography approaches (Pestilli et al., 2014). Cross-validation between tract-tracing and diffusion data continues to be invaluable for verifying key features of human anatomical networks. In model organisms, such cross-validation has led to mixed results, with some studies reporting significant mismatches (Thomas et al., 2014) and others finding significant convergence and overlap (Calabrese et al., 2015), with gains in reliability and sensitivity that depend on the selection of optimal tractography parameters and parcellation schemes (Chen et al., 2015).
Over the past few years, a large number of studies have attempted to reconstruct whole-brain (or at least cortical) network maps in humans and reported a number of significant features of network topology (Sporns, 2013, 2014). These network features include unique connectivity fingerprints, a high density of triangles (high clustering) and short path length, densely connected network communities or modules, and skewed degree distributions characterized by a small set of regions that maintain a large set of diverse connections. Several studies have reported these regions to comprise portions of the superior and lateral frontal cortex, and portions of medial parietal cortex, the cingulate and the insula. In addition to their high degree of connectivity, these regions have been found to be mutually densely interconnected (van den Heuvel and Sporns, 2011), paralleling the high density of connections among network hubs found in model organisms (Bota et al., 2015; Rubinov et al., 2015; Shih et al., 2015; Towlson et al., 2013)). It is worth noting that, while the methods for reconstructing human anatomical networks continue to evolve, there is strong convergence between humans and other species across very different anatomical measurement techniques with respect to prominent features of large-scale network topology (e.g. clustering, modules, hubs, core). This convergence raises the possibility that common architectural themes are the result of common driving forces shaping anatomical networks.
Factors Shaping Anatomical Networks
One of the most enduring observations in anatomical connectivity is an overabundance and high density of short-range projections and, as a consequence, a high propensity for neighboring brain regions to be anatomically linked (Averbeck and Seo, 2008; Young, 1992). These findings suggest that the layout of anatomical projections is largely determined by spatial constraints, minimizing or at least conserving wiring length and volume as well as conduction delays. Recent studies have expanded on these views, for example proposing an exponential distance rule as the key generative factor for inter-regional projections (Ercsey-Ravasz et al., 2013). However, distance or wiring length alone can neither account for all observed topological features of anatomical brain networks (Kaiser and Hilgetag, 2006), nor can it predict specific patterns of (often long-distance) couplings among areas that share high degree (van den Heuvel and Sporns, 2011) and common cytoarchitectonic patterns (Barbas, 2015). Thus, generative principles for anatomical networks likely comprise a combination of factors, including connectional geometry and cost as well as aspects of topology and microstructure, competing as part of an economic trade-off between low cost and efficient performance (Bullmore and Sporns, 2012). The search for generative principles that can explain the arrangement of network elements in nervous systems may eventually provide insights regarding the evolutionary origin of cognitive architectures. It appears that the anatomical substrate is subject to severe and ultimately inviolable constraints that force a trade-off between the expense of material, space and energy on the one side, and computational performance on the other. This trade-off places sharp boundaries around the subsets of architectures that can be physically realized and, at the same time, are biologically viable. A corollary of this perspective is that existing cognitive architectures may be optimally negotiating a trade-off among multiple design constraints but may also fall well short of theoretical limits on any one dimension, i.e. combine suboptimal cost with suboptimal performance.
This last point reinforces the importance of considering the biological implementation (for example, in the topology of anatomical networks) as inseparably linked with the more abstract level of neural computation – a point that runs counter to David Marr's classic notion of separable levels of analysis (Marr, 1982). Instead, structure (implementation) and function (computation) appear inseparable. Anatomical networks define the space of what is functionally possible (Avena-Koenigsberger et al., 2015)– their structure imposes strong constraints on patterns of neural signaling and dynamics, effectively shrinking an impossibly large space of functional network configurations to a lower-dimensional manifold that defines an envelope of possible functional interactions. This envelope is expressed in spontaneous and task-evoked fluctuations in functional connectivity which in turn define functional networks.
FUNCTIONAL CORRELATION NETWORKS
As stated in the Introduction, one approach to large-scale brain networks is through the use of RSFC. At this time, there is considerable consternation by many about what correlations really represent in these very “unconstrained” situations. However, there is little question that observations of RSFC, both at the group and individual level, can show high levels of reliability and reproducibility. Indeed, many of the observations of network data from RSFC persist across different types of “rest” (eyes open vs. eyes closed (McAvoy et al., 2012) vs. light anesthesia, early stages of sleep (Horovitz et al., 2009; Larson-Prior et al., 2011) etc). Further, many of the overall network relationships appear to persist across task states (Cole et al., 2014). Nonetheless, tasks do produce perturbations on the underlying networks present during rest, (Cole et al., 2014; Davison et al., 2015) and deep sleep and deep anesthesia (Heine et al., 2012) also produce clear disruptions of the functional architecture of RSFC.
Thus real questions exist about what these RSFC fluctuations represent. They appear to be constrained by the underlying anatomical relationships, but overall they clearly do not duplicate anatomical relationships (see the next section). Very strong functional correlations can be found between brain regions that demonstrably are not linked by any direct (one-step) anatomical connections. For example, functional correlations can be found between left motor cortex and right cerebellum (Buckner et al., 2011), two structures that are multiple steps away from one another in anatomical terms (figure 2). The eccentric representations of primary visual cortex in left and right hemispheres also clearly correlate, also without the presence of direct anatomical connections (Vincent et al., 2007).
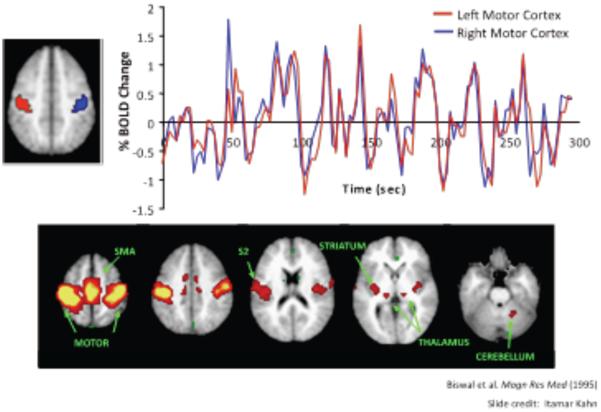
Figure 2.
The top half of the figure shows two regions of interest from motor cortex in red and blue on a brain image. On the right are resting time courses from these two regions showing high correlation across several minutes. This is resting state functional correlation (RSFC). The bottom panel shows regions of the brain that have high correlation with the red ROI from above. Each of these regions is nominally related to motor and somatosensory processing suggesting that RSFC illuminates regions that are functionally related. Note also that the right cerebellar response in the rightmost brain section is several anatomical steps from the ROI.
The fact that RSFC does not represent single step anatomical correlations unfortunately call into question the appropriateness of some otherwise very useful network tools that are based on representations of paths and path lengths (referring to the number of steps between two locations). While some measures based on paths, including global efficiency and some centrality measures, are widely used in functional network studies they must be interpreted with caution. Path-based analyses in correlation networks may be useful for inferring hierarchical relationships and multi-step associations among the brain's functional systems. However, interpretations that make direct reference to information flow or communication along functional connections may be inappropriate in light of the complex and indirect way that correlation networks relate to network paths in the underlying anatomical networks. Neural signals are passed along anatomical paths, and functional connections emerge as a result.
Hence, instead of interpreting functional connections as direct links between two brain regions, a more reasonable supposition is that an RSFC correlation represents composite (perhaps a weighted sum) of the functional relationships along many or all of the anatomical paths that exist between the two regions, e.g. see (Power et al., 2014) for review. Importantly, these correlations reflect not only the anatomical presence, but also the synaptic efficiencies of these connections. Thus functional relationships can vary not only as a result of the structural arrangements of paths but also as a result of changes in the synaptic efficiencies along these paths. For example, performance of a task a large number of times can increase the correlation between commonly activated regions e.g. (Lewis et al., 2009; Mackey et al., 2011). Thus an interesting way to think about the generic correlation structure across the brain is that it represents a very high-level statistical representation of historical coactivation between regions constrained (but not fully determined) by the underlying anatomy.
Early studies using RSFC used a seed-based approach. This technique took a seed or contiguous collection of voxels, and looked at how all the rest of the voxels in the brain correlated with the seed (Biswal et al., 1995). In many cases, the correlation patterns seem to represent functionally related regions. Notably, in 2003, a study by (Greicius et al., 2003) showed that placing a seed in the posterior cingulate region related to the default mode network revealed a set of correlations that looked very much like the set of regions comprising the default mode network, much as seeding the connections from left motor cortex reveals much of motor systems. The default mode network had previously been described as a set of regions that have the unusual property of decreasing their activity whenever a subject goes into many different kinds of active task states. This study unleashed a torrent of further studies with similar outcomes. Different attentional systems were defined using different seeds, and these networks again showed that resting correlation patterns followed coactivation patterns during tasks.
Community detection
The success of this piecemeal approach at describing local relationships of “neighbors” (and the difficulties in putting these different “neighborhoods” together) encouraged several different groups to attempt descriptions of all or much of the brain in terms of sets of modularized correlation relationships. In network science, the attempt to find underlying group structure in a large-scale network is termed community detection. A community is a set of objects or nodes (in this case, brain regions) that maintains denser and stronger relations among themselves than with members of other communities.
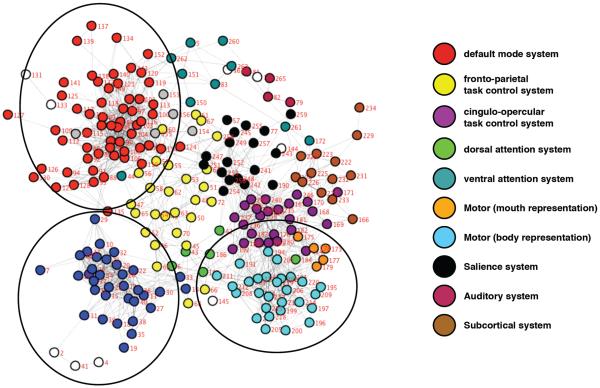
Using quite different techniques, similar community or cluster structures have been described by several groups (e.g., see figure 3) (Damoiseaux et al., 2006; Power et al., 2011; Sorg et al., 2007; Yeo et al., 2011). Further, the community structure itself has a very high degree of face plausibility. In many cases, it replicated the neighborhoods found using seed-based techniques, which in turn had replicated coactivation relationships found in task-based fMRI studies. For example, the “attentional” networks found in figure 4 had striking similarity to the dorsal and ventral attention systems of (Corbetta and Shulman, 2002), and the frontoparietal and cingulo/opercular systems of (Dosenbach et al., 2007) (see also executive control and salience systems of (Seeley et al., 2007)).
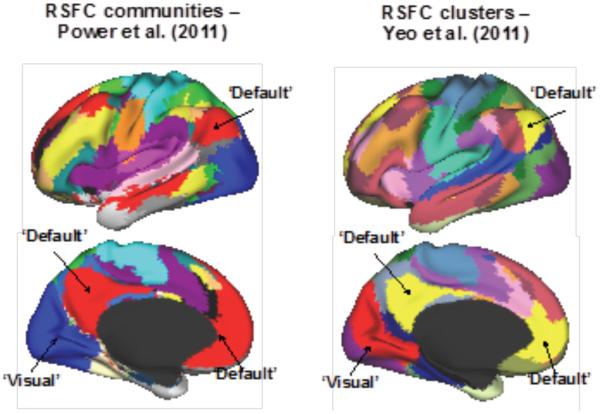
Figure 3.
Two versions of community detection are presented in figure 3. On the left is the layout of communities from a network science infomap community detection algorithm on group RSFC data. On the right is the layout of communities from a clustering approach. Notice the high spatial similarity from the two versions. Adapted from figures in Power et al 2011 and Yeo et al. 2011 (Power et al., 2011; Yeo et al., 2011)
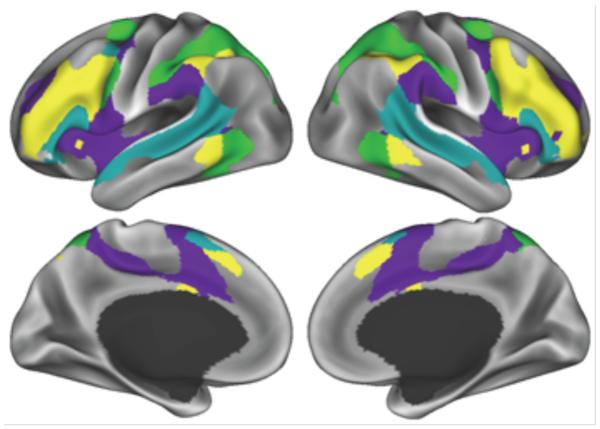
Figure 4.
The isolated layout of some frontal and parietal communities. These spatial layouts are recognizable as the cingulo-opercular (purple) and fronto-parietal (yellow) systems from fMRI studies of Dosenbach, 2007 (Dosenbach et al., 2007) and the dorsal and ventral attention systems, based on fMRI studies of Corbetta and Shulman, 2002 (Corbetta and Shulman, 2002).
Some of the communities that were found in these studies had not been previously described. In some cases, the new communities identified regions that had previously described coactivation relationships. For example, a set of parietal regions were described, near default mode regions that shared specific coactivation relationships across memory encoding and retrieval tasks, and have come to be termed, perhaps inauspiciously, the parietal encoding retrieval network (PERN) (light blue regions on midline of Power communities, and gray regions in Yeo clusters in figure 3). Other communities were somewhat surprising in their configuration. For example, somatic motor regions were broken into regions related to the mouth representation and to the rest of the body separately. In further exploring this distinction, it was found that the mouth representation community more closely correlated with an auditory community than the body representation did (Power et al., 2011). This suggests the tantalizing speculation that the strong relationships created between oral and aural processing in language leads, perhaps by way of Hebbian plasticity, to this somewhat unintuitive result.
Several studies have taken a more formal approach to the question of the relationship of underlying correlation patterns at rest with what is seen during tasks. Meta-analyses of large databases recording task-evoked cortical activation patterns allow the description of co-activation patterns estimated across a large number of cognitive tasks. These co-activation patterns often show significant overlap with RSFC clustering. The cluster structure of these patterns may also be used to extract relations among different cognitive tasks and domains, a step towards creating a data-driven ontology of cognitive states (e.g., (Smith et al., 2009)). Such ontology would further reinforce the notion that the capacities of human cognition have their roots in the network architecture of the human brain.
Large-scale network science tools also allow interpretation beyond the simple presence of separate communities. One way is by creating a representation of how the different communities relate to one another, not in an anatomical space, but in network space. A popular network visualization tool treats each of the correlations as a mechanical spring with a spring constant proportional to the correlation strength. A repulsive force is placed on all the regions/nodes of the network, and the springs between the nodes pull the entire network into a new energy-minimizing configuration. As can be seen in figure 5, spring embedding places the more enclosed processing type networks, the blue visual system and cyan and brown motor systems, on the edges of the network. The more control-related attentional systems reside more centrally, as if they would be expected to relate to other systems more broadly. Interestingly, the default mode “network” sits at the edge, more like a “processing” than a control system (see (Power et al., 2014) for more discussion).
Figure 5.
A spring embedded representation of 264 region RSFC from Power 2011(Power et al., 2011). This pulls anatomically disparate members of systems together in a “network space” (see text for description of spring-embedding. Important to note is that the circles emcompass visual (blue), motor (cyan and brown) and default systems (red) that are located along the edge of the network, while control-related and attentional systems are more centrally located.
Relation to cognitive architectures
All of this leads to some interesting functional questions. The community-oriented studies have suggested that there may be somewhere between one and two dozen different systems in resting correlation. It is our supposition that several of these will relate to the more specific cognitive architectures that are addressed in the other articles in this issue. On the other hand, humans seem to have amazing behavioral flexibility with a myriad of well-defined sets of functional capabilities. One can imagine that relatively separable RSFC systems should be associated with these different kinds of capabilities. So as we go through the other chapters in this Special Issue, one might be interested in mapping say a “reward system” onto one of our communities. What if this doesn't happen?
One explanation might be that network communities that manifest in the resting state just do not faithfully represent specific cognitive or behavioral functional distinctions. But the many counterexamples already explored here at least somewhat argue against a strong dissociation. A second explanation is that there may be further breakdowns within the dozen or so “coarse-grained” systems detected in most resting-state studies that represent more “fine-grained” functional distinctions. This, of course, can be explored in future studies. A third and more interesting possibility is that many kinds of task distinctions that may be important to us as humans do not represent statistically useful descriptions of overall coactivation in the life of the individual, and hence are not expressed in aggregate RSFC patterns.
To this end, let us look at a set of regions consistently activated during reading tasks that in some cases have been identified as the “reading network” e.g. ((Dehaene et al., 2010; Fiez and Petersen, 1998; Perfetti and Bolger, 2004). These include a region that has come to be called the “visual word form area” in extrastriate cortex, regions in the angular and supramarginal gyri, and others. When examining resting state among these regions, as well as viewing these regions inside of large-scale networks, their mutual RSFC correlations are unremarkable or non-existent. It is not that these regions do not have strong relationships at all (Vogel et al., 2013). In fact these regions seem to be very much parts of other quite coherent networks, particularly the dorsal attention system (Vogel et al., 2012), see figure 6 So what's going on here?
Figure 6.
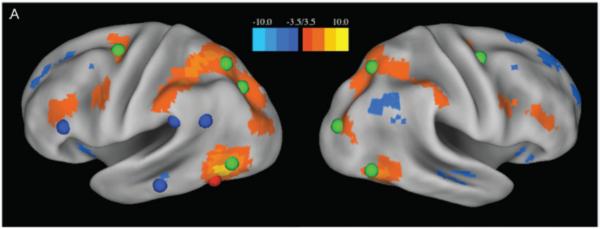
The upper panels show RSFC (color bar patches across cortex) from the putative visual word form area (pVWFA)(red sphere) (Adapted from (Vogel et al., 2012)). “Reading regions” from metanalysis are shown in blue, and dorsal attention network areas are shown in green. The RSFC from pVWFA are almost exclusively related to dorsal attention regions and avoid members of the task-based reading network.
Reading is clearly very important to humans – in fact it is a task that you, the reader, are currently engaged in and presumably spend a lot of time exercising. However, it also appears that the regions that are commonly utilized in reading tasks are also utilized for many other different kinds of tasks (Price and Devlin, 2003; Vogel et al., 2012). Thus the statistical nature of the relationships specific to reading may not represent the “day jobs” of many of the involved regions. This suggests that our behavioral flexibility may depend on our ability to usefully configure sets of regions for specific tasks, and that these configurations are not necessarily representative of the baseline way that those regions are “normally” conjoined. Reading appears to be a very interesting example of this – it involves breaking baseline network coherence to create task-specific new networks bound by new sets of dynamic relationships. This immediately implies that the relation between (relatively stable) anatomical networks and (highly dynamic) functional networks is bound to be a complex issue.
RELATIONS ETWEEN ANATOMICAL AND FUNCTIONAL NETWORKS
Structure/Function Relations
So far, we have examined cognitive architectures separately from a structural/anatomical and functional/physiological perspective. A more complete understanding of the biological foundations of cognition requires considering their interaction – the emergence of functional brain activity and dynamics on top of structural networks, as well as the continued modification of structural networks that results from activity-dependent modulation and plasticity.
Functional Networks Emerging on Structural Networks
Patterns of structural and resting-state functional connectivity exhibit significant relationships in both nonhuman primates (Vincent et al., 2007) and humans ((Hagmann et al., 2008; Hermundstad et al., 2013). Components of functionally coherent resting-state networks are anatomically interconnected (Greicius et al., 2009), structural and functional connection strengths are significantly correlated (Honey et al., 2009), and the path structure linking indirectly connected node pairs is partly predictive of the strength of functional couplings (Adachi et al., 2012). Nonlinear dynamic simulations of spontaneous neural activity (Ghosh et al., 2008; Gollo et al., 2015; Honey et al., 2007) as well as formally simple generative models based on the topology of anatomical connections can create synthetic patterns of functional connectivity that resemble empirical resting-state (Abdelnour et al., 2014; Goni et al., 2014; Misic et al., 2015) (figure 7). These and other findings strongly suggest that anatomical connectivity plays an important role in shaping the patterns of functional connectivity that characterize long-time averages of spontaneous BOLD fluctuations. Recent work on relations between anatomical patterns and EEG functional connectivity further strengthens this idea (Chu et al., 2015). Structure is predictive of function in other domains as well. For example, in line with findings coming from nonhuman primates (see above), anatomical connectivity patterns can predict functional specialization of brain regions in the fusiform gyrus (Saygin et al., 2012) and in other portions of human temporal cortex (Gomez et al., 2015).
Figure 7.
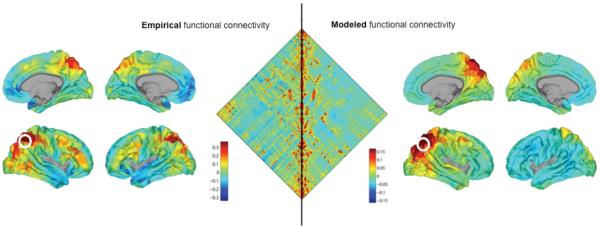
A network-based computational model of functional connectivity (Goni et al., 2014). The left side of the figure represents empirical RSFC, while the right side represents results from a model of functional connectivity based on the network architecture of structural connectivity. In the middle, the left triangular half of the plot shows a functional connectivity (crosscorrelation) map of 500 parcels comprising the right cortical hemisphere (Hagmann et al., 2008). The right triangular half of the plot shows modeled or predicted functional connectivity derived from the computational model. The model was based entirely on network measures of communication applied to the underlying structural connectivity (connectome) matrix. The two halves of the plot are significantly correlated (R = 0.60). The outer plots (left and right) show examples of a seed-based cross-correlation map (with the seed placed in the superior parietal cortex) projected onto the lateral and medial surface of the cortex. Plots on the left depict a correlation map from empirical data (Hagmann et al., 2008). Plots on the right depict model predictions (R = 0.55).
Despite strong and reproducible relations between structural and functional networks, their topology diverges in a number of important ways. A root cause for this divergence is a fundamental difference in the nature of structural and functional connectivity. Structural connections represent direct anatomical linkages and define (within fine-grained parcellations) relatively sparse networks. Functional connections express (in their most common usage) the similarity of BOLD time courses and define correlation networks that are dense due to transitive closure (Zalesky et al., 2012). Transitivity refers to the mathematical fact that individual correlations in functional connectivity are generally interdependent such that changes in individual correlations often propagate across the network. Transitive closure implies that this basic property of correlation networks induces topological structure such as triadic closure or clustering, an effect that should be accounted for in appropriate statistical comparisons and null models (Zalesky et al., 2012). As a result of these and other features, network measures must be interpreted differently across the two domains of structure and function (Power et al., 2014; Sporns, 2014). For example, while it is appropriate to use node degree as a defining feature of network hubs in structural networks, “functional node degree” is confounded by the properties (e.g. transitivity) of networks built from pairwise cross-correlations. Node participation, i.e. the diversity of its functional connections relative to a module partition offers a more robust approach (Power et al., 2014). As different measures for functional connectivity (e.g. partial correlations or directed information flow) come into play, interpretation of network measures must remain sensitive to the nature of what is expressed in the edge weights and their topology.
Earlier in this Perspective, we discussed the important roles of resting-state or intrinsic connectivity networks as functional building blocks of cognitive architecture. It is therefore an important question to investigate how patterns of structural connectivity relate to the partitions defined by these functional building blocks. In general, structural and functional networks do not simply “line up” across domains – for example, structural network communities do not, for the most part, correspond to functional communities. Nevertheless, anatomical nodes and connections are organized in ways that relate to functional partitions. For example, several studies have shown that high-degree (anatomical) brain regions or network hubs are widely dispersed around the brain, across lateral and superior frontal cortex, parietal cortex and the insula, among others, and they are densely interconnected, a hallmark of “rich club organization” (van den Heuvel and Sporns, 2011). Comparison to functional modules has shown that these structural hubs are also widely dispersed among resting-state networks, and that their interconnections may be important for communication between such networks (van den Heuvel and Sporns, 2013). The latter point about the “importance of weak links” (Granovetter, 1973) (defined as links that connect different communities) has been reinforced by other studies that have shown significant anatomical links spanning functional modules (Gallos et al., 2012).
While structural and functional networks are clearly related, no structurally-based computational model has thus far succeeded in capturing all of the variance observed in functional brain recordings. Likely causes for this shortfall are complex physiological underpinnings of the BOLD response and its temporal fluctuations, the lack of important information on the directionality and physiological strength of pathways, as well as biases and noise corrupting both structural and functional data acquisition. While the latter may be partly addressable through future methodological refinements, current technology does not allow direct noninvasive observations of neural processes in the human brain that combines both spatio-temporal precision and whole-brain coverage. Extremely promising work addressing this gap in knowledge is underway in model organisms, where whole-brain connectomics and large-scale functional recordings are likely to converge soon, offering unprecedented glimpses of highly resolved network structure/function relationships.
Network Dynamics, Flexibility and Reconfiguration
In most previous studies, relations between structural and functional networks have been most evident when considering long-time averages of correlations among spontaneous or resting-state fluctuations. Over shorter time periods, however, structure/function relations diminish (Van Dijk et al., 2010). One possible interpretation of the latter finding is that shorter observation periods undersample the set of dynamic patterns that jointly contribute to the long-term average of resting-state functional connectivity. The implication is that functional connectivity is “dynamic”, i.e. its spatial pattern changes over time (Hutchison et al., 2013). In line with this view, some theoretical and computational models have suggested that spontaneous dynamics in the brain occurs “near criticality”, a dynamic regime characterized by ongoing noisy fluctuations and a rich repertoire of brain states (Deco et al., 2011; Haimovici et al., 2013). Others have suggested that observed fluctuations may result, more broadly, from processes that confer “dynamic instability” (Breakspear, 2002; Friston et al., 2012). In general, criticality and dynamic instability both suggest that noise-driven fluctuations should result in short-term deviations from the long-term average pattern of functional connectivity.
In empirical resting-state fMRI studies, the status of time-dependent functional connectivity is still in flux. Some studies, usually carried out on the temporal evolution of windowed patterns of functional connectivity, have suggested that a subset of functional connections exhibits non-stationary fluctuations in magnitude (Zalesky et al., 2014) and others have provided evidence that functional connectivity passes through a restricted set of network states with distinct topology and community structure (Allen et al., 2014). Important and difficult methodological issues involve demonstrating the robustness of clustering methods used for deriving families of network states, inherent limitations in diagnosing network-wide transient connectivity states on the basis of mutually dependent sets of pairwise correlations, the uncertain level of persistence of such states in conservatively configured (e.g. phase-randomized) null models, and the unknown nature of neurobiological mechanisms driving state transitions. Multimodal studies as well as studies in model organisms (Keilholz, 2014; Tagliazucchi and Laufs, 2015) will likely help to clarify the physiological origin of fluctuating rs-fMRI functional connectivity. It should be noted that fast non-stationary fluctuations in spontaneous task-free brain connectivity are well documented in the EEG/MEG and neurophysiological literature (Breakspear, 2002; Ioannides, 2007). For example, long-term recordings of spontaneous network patterns with intracranial EEG revealed a core of persistent functional connections as well as a set of connections that are consistently more variable and metastable (Kramer et al., 2011). More recently, rapid transitions (on the order of 100–200 ms) among transient brain states resembling cortical resting-state networks have been observed in MEG recordings of resting brain activity (Baker et al., 2014).
In addition to these fast responses of functional connectivity in response to sensory perturbations and momentary shifts in cognitive demands, structural and functional networks underpinning human cognitive architecture are also changing more slowly in the course of learning and plasticity. Resting-state functional networks were sensitively remodeled in the course of a visual perceptual learning task (Lewis et al., 2009) and the acquisition of a complex motor skill was found to be associated with changes in the modular organization of fMRI functional networks (Bassett et al., 2011). Taken together these findings demonstrate that the networks underlying human cognitive architecture partly reflect individual experience and skill acquisition. This view is supported by numerous studies that have shown that individual differences in neurocognitive networks (structural and functional) can be predictive of individual differences in cognitive and behavioral performance.
CONCLUSIONS
The emerging picture is one in which dynamic processes of neuronal signaling and communication play out on an intricate web of anatomical projections. The resulting interplay between structure and function renders brain networks capable of both robust computational performance and flexible adaptive response – a cognitive architecture that as a “network of networks” maintains consistent, recognizable and reproducible topology across individuals, and yet retains many additional degrees of freedom for context, stimulus and task-dependent reconfiguration. Different networks make different contributions – while some may be more heavily engaged in domain-specific (e.g. visual, motor) processes, others may be more important for integrating multimodal information, or for task switching and control. Importantly, the view that emerges is one where the elementary building blocks of cognitive architecture are networks, not regions or individual neurons. We believe that this network-centric perspective provides a fruitful basis for how to understand the biological basis of human cognitive architecture.
BOX 1: Current Status of the Field.
The classic notion of “cognitive architecture” postulated the basic idea that human cognition is a computational process carried out as a series of operations on symbolic representations. This view explicitly embraced functionalism, which implies that cognition can be studied and understood without much (if any) reference to its biological basis.
In parallel, understanding of the neural bases of human cognition was materially advanced through the mechanistic study of neurocognitive circuits in non-human primates and the application of noninvasive imaging technology in the human brain. An enduring achievement was the discovery of task-specific activations of specific neuronal populations and localized brain regions aided by the development of statistical tools for mass-univariate region-based analyses.
Today, ROI-based analyses are increasingly complemented by an alternative perspective, based on the notion that cognitive function emerges from the dynamics of extended cortical and subcortical networks. Unlike classic “neural nets”, these networks have a distinct anatomical basis in the brain's structural connectivity (the connectome) and manifest through coherent fluctuations in neural activity at rest as well as distributed patterns of activation in task states.
Network approaches are appealing because they (i) transcend local and global function, as connectivity simultaneously accounts for regional differences (segregation) and interregional signaling and communication (integration); (ii) can provide a common framework for describing both endogenously and exogenously driven neuronal activity, and their mutual relations; and (iii) they can be applied across spatial scales from neurons to regions, and even across different data domains from genes to neural dynamics to social interactions.
Current challenges for network approaches include the development of novel data acquisition and analytic methodologies that can cope with the ever-increasing volume and complexity of “big data”. Mapping cognition to the brain will increasingly rely on sophisticated multivariate statistical algorithms involving clustering, module detection and other dimension reduction approaches. In future the growing application of “data-driven” machine learning or pattern recognition approaches could substantially benefit from added constraints coming from the rich tradition of cognitive anatomy.
BOX 2: Future Directions.
We need a more complete and more accurate view of the anatomical underpinnings of cognitive architectures. In the case of the human brain this will require the development of more reliable tools for noninvasive imaging of anatomical connections, as well as rigorous cross-validation with more invasive histological or imaging approaches in non-human primates. We need progress towards more consistent and biologically motivated quantification of the geometry, strengths and efficacies of anatomical pathways.
We need increasingly accurate functional parcellations, both within groups as well as within individuals. The overarching goal behind these efforts is to more clearly define the functional building blocks from which large-scale brain networks are configured. Key challenges here are related to the quality of functional imaging data, excluding systematic biases and sources of noise, as well as deploying sophisticated data analysis techniques that can reveal network communities across scales, down to the level of individual brain regions.
We need new approaches for mapping brain networks engaged in specific cognitive tasks that can capture their rapid reconfiguration and dynamic functional connectivity. Current methods for creating functional connectivity maps with fMRI are limited in terms of their temporal resolution, and they cannot reveal the direction of information flow.
We need more systematic assessment of individual variability in brain structure and function as a basis for revealing biological mechanisms that drive individual differences in behavior and cognition. Going beyond work that aims at creating “population averages” of brain networks, mapping the anatomical and functional networks of individuals will be essential for revealing the network basis of their specific cognitive capabilities and styles.
We need more accurate and more powerful computational models of dynamic brain activity. Such models will be indispensable for understanding the complex patterns of signaling and communication within and between brain networks. Also, such models will be able to inform empirical research by generating predictions about the network structures and dynamic relationships that are most important for maintaining cognitive function.
We need better understanding of the neurobiological mechanisms that determine switches or transitions between cognitive states. While convergent lines of evidence suggest that such transitions are associated with reconfigurations in functional brain networks, we know very little about what the underlying causes. One possibility is that specific network nodes are responsible for triggering transitions in global network states. An alternative possibility is that switches reflect metastable transitions in brain dynamics. Understanding the mechanisms behind network transitions is crucial for moving the field beyond mere description and towards prediction and control.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Network approaches to brain function have begun to illuminate how structural and functional connectivity support cognition and behavior. The authors present an overview of how networks inform theories of cognitive architectures and discuss future issues in the field.
References
- Abdelnour F, Voss HU, Raj A. Network diffusion accurately models the relationship between structural and functional brain connectivity networks. Neuroimage. 2014;90:335–347. doi: 10.1016/j.neuroimage.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Osada T, Sporns O, Watanabe T, Matsui T, Miyamoto K, Miyashita Y. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb Cortex. 2012;22:1586–1592. doi: 10.1093/cercor/bhr234. [DOI] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2014;24:663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avena-Koenigsberger A, Goni J, Sole R, Sporns O. Network morphospace. Journal of the Royal Society Interface. 2015;12 doi: 10.1098/rsif.2014.0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck BB, Seo M. The statistical neuroanatomy of frontal networks in the macaque. PLoS Comput Biol. 2008;4:e1000050. doi: 10.1371/journal.pcbi.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AP, Brookes MJ, Rezek IA, Smith SM, Behrens T, Probert Smith PJ, Woolrich M. Fast transient networks in spontaneous human brain activity. Elife. 2014;3:e01867. doi: 10.7554/eLife.01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas H. General Cortical and Special Prefrontal Connections: Principles from Structure to Function. Annu Rev Neurosci. 2015;38:269–289. doi: 10.1146/annurev-neuro-071714-033936. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A. 2011;108:7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bota M, Sporns O, Swanson LW. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci U S A. 2015;112:E2093–2101. doi: 10.1073/pnas.1504394112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear M. Nonlinear phase desynchronization in human electroencephalographic data. Hum Brain Mapp. 2002;15:175–198. doi: 10.1002/hbm.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:2322–2345. doi: 10.1152/jn.00339.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Cofer G, Qi Y, Johnson GA. A Diffusion MRI Tractography Connectome of the Mouse Brain and Comparison with Neuronal Tracer Data. Cereb Cortex. 2015 doi: 10.1093/cercor/bhv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Liu T, Zhao Y, Zhang T, Li Y, Li M, Zhang H, Kuang H, Guo L, Tsien JZ, Liu T. Optimization of large-scale mouse brain connectome via joint evaluation of DTI and neuron tracing data. Neuroimage. 2015;115:202–213. doi: 10.1016/j.neuroimage.2015.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CJ, Tanaka N, Diaz J, Edlow BL, Wu O, Hamalainen M, Stufflebeam S, Cash SS, Kramer MA. EEG functional connectivity is partially predicted by underlying white matter connectivity. Neuroimage. 2015;108:23–33. doi: 10.1016/j.neuroimage.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland PS, Sejnowski TJ. The Computational Brain. MIT Press; Cambridge: 1992. [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nature Reviews Neuroscience. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- Damasio AR. The brain binds entities and events by multiregional activation from convergence zones. Neural Comput. 1989;1:123–132. [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison EN, Schlesinger KJ, Bassett DS, Lynall ME, Miller MB, Grafton ST, Carlson JM. Brain network adaptability across task states. PLoS Comput Biol. 2015;11:e1004029. doi: 10.1371/journal.pcbi.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Dehaene S, Pegado F, Braga LW, Ventura P, Nunes Filho G, Jobert A, Dehaene-Lambertz G, Kolinsky R, Morais J, Cohen L. How learning to read changes the cortical networks for vision and language. Science. 2010;330:1359–1364. doi: 10.1126/science.1194140. [DOI] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RAT, Fox MD, Snyder AZ, Vincent JL, Raichle ME, et al. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci U S A. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercsey-Ravasz M, Markov NT, Lamy C, Van Essen DC, Knoblauch K, Toroczkai Z, Kennedy H. A predictive network model of cerebral cortical connectivity based on a distance rule. Neuron. 2013;80:184–197. doi: 10.1016/j.neuron.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fiez JA, Petersen SE. Neuroimaging studies of word reading. Proceedings of the National Academy of Sciences. 1998;95:914–921. doi: 10.1073/pnas.95.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor JA, Pylyshyn ZW. Connectionism and cognitive architecture: a critical analysis. Cognition. 1988;28:3–71. doi: 10.1016/0010-0277(88)90031-5. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage. 2013;80:426–444. doi: 10.1016/j.neuroimage.2013.04.087. [DOI] [PubMed] [Google Scholar]
- Friston K, Breakspear M, Deco G. Perception and self-organized instability. Front Comput Neurosci. 2012;6:44. doi: 10.3389/fncom.2012.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallos LK, Makse HA, Sigman M. A small world of weak ties provides optimal global integration of self-similar modules in functional brain networks. Proc Natl Acad Sci U S A. 2012;109:2825–2830. doi: 10.1073/pnas.1106612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Rho Y, McIntosh AR, Kotter R, Jirsa VK. Noise during rest enables the exploration of the brain's dynamic repertoire. PLoS Comput Biol. 2008;4:e1000196. doi: 10.1371/journal.pcbi.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Topography of cognition: Parallel distributed networks in primate association cortex. Annu Rev Neurosci. 1988;11:137–156. doi: 10.1146/annurev.ne.11.030188.001033. [DOI] [PubMed] [Google Scholar]
- Gollo LL, Zalesky A, Hutchison RM, van den Heuvel M, Breakspear M. Dwelling quietly in the rich club: brain network determinants of slow cortical fluctuations. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J, Pestilli F, Witthoft N, Golarai G, Liberman A, Poltoratski S, Yoon J, Grill-Spector K. Functionally defined white matter reveals segregated pathways in human ventral temporal cortex associated with category-specific processing. Neuron. 2015;85:216–227. doi: 10.1016/j.neuron.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni J, van den Heuvel MP, Avena-Koenigsberger A, Velez de Mendizabal N, Betzel RF, Griffa A, Hagmann P, Corominas-Murtra B, Thiran JP, Sporns O. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014;111:833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovetter MS. The Strength of Weak Ties. American Journal of Sociology. 1973;78:1360–1380. [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovici A, Tagliazucchi E, Balenzuela P, Chialvo DR. Brain organization into resting state networks emerges at criticality on a model of the human connectome. Phys Rev Lett. 2013;110:178101. doi: 10.1103/PhysRevLett.110.178101. [DOI] [PubMed] [Google Scholar]
- Heine L, Soddu A, Gomez F, Vanhaudenhuyse A, Tshibanda L, Thonnard M, Charland-Verville V, Kirsch M, Laureys S, Demertzi A. Resting state networks and consciousness: alterations of multiple resting state network connectivity in physiological, pharmacological, and pathological consciousness States. Front Psychol. 2012;3:295. doi: 10.3389/fpsyg.2012.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermundstad AM, Bassett DS, Brown KS, Aminoff EM, Clewett D, Freeman S, Frithsen A, Johnson A, Tipper CM, Miller MB, et al. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc Natl Acad Sci U S A. 2013;110:6169–6174. doi: 10.1073/pnas.1219562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey C, Kotter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Braun AR, Carr WS, Picchioni D, Balkin TJ, Fukunaga M, Duyn JH. Decoupling of the brain's default mode network during deep sleep. Proc Natl Acad Sci U S A. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, et al. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannides AA. Dynamic functional connectivity. Curr Opin Neurobiol. 2007;17:161–170. doi: 10.1016/j.conb.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput Biol. 2006;2:e95. doi: 10.1371/journal.pcbi.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilholz SD. The neural basis of time-varying resting-state functional connectivity. Brain Connect. 2014;4:769–779. doi: 10.1089/brain.2014.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MA, Eden UT, Lepage KQ, Kolaczyk ED, Bianchi MT, Cash SS. Emergence of persistent networks in long-term intracranial EEG recordings. J Neurosci. 2011;31:15757–15767. doi: 10.1523/JNEUROSCI.2287-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior LJ, Power JD, Vincent JL, Nolan TS, Coalson RS, Zempel J, Snyder AZ, Schlaggar BL, Raichle ME, Petersen SE. Modulation of the brain's functional network architecture in the transition from wake to sleep. Prog Brain Res. 2011;193:277–294. doi: 10.1016/B978-0-444-53839-0.00018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Baldassarre A, Committeri G, Romani GL, Corbetta M. Learning sculpts the spontaneous activity of the resting human brain. Proc Natl Acad Sci USA. 2009;106:17558–17563. doi: 10.1073/pnas.0902455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey AP, Hill SS, Stone SI, Bunge SA. Differential effects of reasoning and speed training in children. Dev Sci. 2011;14:582–590. doi: 10.1111/j.1467-7687.2010.01005.x. [DOI] [PubMed] [Google Scholar]
- Markov NT, Ercsey-Ravasz M, Lamy C, Ribeiro Gomes AR, Magrou L, Misery P, Giroud P, Barone P, Dehay C, Toroczkai Z, et al. The role of long-range connections on the specificity of the macaque interareal cortical network. Proc Natl Acad Sci U S A. 2013;110:5187–5192. doi: 10.1073/pnas.1218972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr D. Vision. W.H. Freeman and Company; New York: 1982. [Google Scholar]
- McAvoy M, Larson-Prior L, Ludwikow M, Zhang D, Snyder AZ, Gusnard DL, Raichle ME, d'Avossa G. Dissociated mean and functional connectivity BOLD signals in visual cortex during eyes closed and fixation. JNeurophysiol. 2012;108:2363–2372. doi: 10.1152/jn.00900.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM. DUPLICATE-Large-scale neurocognitive networks and distributed processing for attention, language, and memory. Ann Neurol. 1990;28:597–613. doi: 10.1002/ana.410280502. [DOI] [PubMed] [Google Scholar]
- Misic B, Betzel RF, Nematzadeh A, Goni J, Griffa A, Hagmann P, Flammini A, Ahn YY, Sporns O. Cooperative and Competitive Spreading Dynamics on the Human Connectome. Neuron. 2015;86:1518–1529. doi: 10.1016/j.neuron.2015.05.035. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvizi J, Van Hoesen GW, Buckwalter J, Damasio A. Neural connections of the posteromedial cortex in the macaque. Proc Natl Acad Sci U S A. 2006;103:1563–1568. doi: 10.1073/pnas.0507729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kotter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3:606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Perfetti CA, Bolger DJ. The brain might read that way. Scientific Studies of Reading. 2004;8:293–304. [Google Scholar]
- Pestilli F, Yeatman JD, Rokem A, Kay KN, Wandell BA. Evaluation and statistical inference for human connectomes. Nat Methods. 2014;11:1058–1063. doi: 10.1038/nmeth.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. Studying brain organization via spontaneous fMRI signal. Neuron. 2014;84:681–696. doi: 10.1016/j.neuron.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CJ, Devlin JT. The myth of the visual word form area. Neuroimage. 2003;19:473–481. doi: 10.1016/s1053-8119(03)00084-3. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Ypma RJ, Watson C, Bullmore ET. Wiring cost and topological participation of the mouse brain connectome. Proc Natl Acad Sci U S A. 2015;112:10032–10037. doi: 10.1073/pnas.1420315112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygin ZM, Osher DE, Koldewyn K, Reynolds G, Gabrieli JD, Saxe RR. Anatomical connectivity patterns predict face selectivity in the fusiform gyrus. Nat Neurosci. 2012;15:321–327. doi: 10.1038/nn.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejnowski TJ, Churchland PS. Brain and Cognition. In: Posner M, editor. Foundations of Cognitive Science. MIT Press; Cambridge: 1989. p. 888. [Google Scholar]
- Shih CT, Sporns O, Yuan SL, Su TS, Lin YJ, Chuang CC, Wang TY, Lo CC, Greenspan RJ, Chiang AS. Connectomics-based analysis of information flow in the Drosophila brain. Curr Biol. 2015;25:1249–1258. doi: 10.1016/j.cub.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Muhlau M, Calhoun VD, Eichele T, Laer L, Drzezga A, Forstl H, Kurz A, Zimmer C, Wohlschlager AM. Selective changes of resting-state networks in individuals at risk for Alzheimer's disease. ProcNatlAcadSciUSA. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Networks of the brain. MIT Press; Cambridge, Mass: 2011. [Google Scholar]
- Sporns O. Network attributes for segregation and integration in the human brain. Curr Opin Neurobiol. 2013;23:162–171. doi: 10.1016/j.conb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. Multimodal imaging of dynamic functional connectivity. Front Neurol. 2015;6:10. doi: 10.3389/fneur.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A. 2014;111:16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towlson EK, Vertes PE, Ahnert SE, Schafer WR, Bullmore ET. The rich club of the C. elegans neuronal connectome. J Neurosci. 2013;33:6380–6387. doi: 10.1523/JNEUROSCI.3784-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: Theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Vogel AC, Church JA, Power JD, Miezin FM, Petersen SE, Schlaggar BL. Functional network architecture of reading-related regions across development. Brain Lang. 2013;125:231–243. doi: 10.1016/j.bandl.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel AC, Petersen SE, Schlaggar BL. The left occipitotemporal cortex does not show preferential activity for words. Cereb Cortex. 2012;22:2715–2732. doi: 10.1093/cercor/bhr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MP. Objective analysis of the topological organization of the primate cortical visual system. Nature. 1992;358:152–155. doi: 10.1038/358152a0. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore E. On the use of correlation as a measure of network connectivity. Neuroimage. 2012;60:2096–2106. doi: 10.1016/j.neuroimage.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. Proc Natl Acad Sci U S A. 2014;111:10341–10346. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeki S, Shipp S. The functional logic of cortical connections. Nature. 1988;335:311–317. doi: 10.1038/335311a0. [DOI] [PubMed] [Google Scholar]