Abstract
The release of myxoma virus (MYXV) and Rabbit Haemorrhagic Disease Virus (RHDV) in Australia with the aim of controlling overabundant rabbits has provided a unique opportunity to study the initial spread and establishment of emerging pathogens, as well as their co-evolution with their mammalian hosts. In contrast to MYXV, which attenuated shortly after its introduction, rapid attenuation of RHDV has not been observed. By studying the change in virulence of recent field isolates at a single field site we show, for the first time, that RHDV virulence has increased through time, likely because of selection to overcome developing genetic resistance in Australian wild rabbits. High virulence also appears to be favoured as rabbit carcasses, rather than diseased animals, are the likely source of mechanical insect transmission. These findings not only help elucidate the co-evolutionary interaction between rabbits and RHDV, but reveal some of the key factors shaping virulence evolution.
Keywords: transmission, virulence, adaptive evolution, biological control, rabbit, calicivirus, emerging disease, genetic resistance
Introduction
A high level of pathogen virulence is a hallmark of many emerging diseases that severely impact human, animal and crop health. Accordingly, understanding the factors that select and maintain high virulence is of fundamental importance to the management and control of emerging diseases (1, 2). While most newly emerging diseases are considered harmful and efforts are made to reduce their spread, Australia has used two highly virulent viral pathogens as tools to control an introduced invasive vertebrate pest species – the European rabbit (Oryctolagus cuniculus). Wild rabbits were successfully introduced into Australia for hunting in 1859. They quickly spread through the southern half of the continent (3), and within a few decades became a major environmental and economic pest (4, 5). Large scale fences and conventional control methods such as poisoning and warren destruction had some local success in agricultural areas. However, broad scale control was not possible until the introduction of the two viral biocontrol agents, myxoma virus (MYXV) in 1950 and Rabbit Haemorrhagic Disease Virus (RHDV) in 1995. For ongoing effective biological control of a pest species such as the rabbit, sustained high pathogen virulence is essential, but virulence often appears to diminish as the emerging virus adapts to the target species or environment (6) and/or the target species develops genetic resistance.
The first virus used to control European rabbits in Australia, MYXV (family Poxviridae), has provided one of the best studied parasite-host co-evolutionary systems (7), and has become the classic example for the trade-off theory of virulence evolution (8, 9). Initially, MYXV caused case fatality rates of up to 99%, but within a few years rabbits began to develop genetic resistance and attenuated strains of MYXV were observed, although these were never dominant and quickly disappeared (10). In the field, virus strains with relatively moderate virulence persisted and became dominant. Although the highly virulent Lausanne strain of MYXV was repeatedly re-released, it failed to establish in the field (11). It has been proposed that the predominance of MYXV strains with intermediate levels of virulence indicates the need for the virus to produce enough viral particles in rabbit skin lesions to maximise transmission by mosquitoes or fleas, while not being so virulent that the rabbit died before this transmission could occur (12) Notably, parallel adaptive processes were observed on the European continent following the introduction of MYXV there (7, 13).
With the attenuation of MYXV and developing genetic host resistance, rabbit populations in Australia began to recover. Consequently, in 1991 RHDV (genus Lagovirus, family Caliciviridae), a recently described emerging and highly virulent pathogen of rabbits (14) was imported to assess its suitability as an additional biocontrol agent for rabbits (12). RHDV subsequently escaped from quarantine in 1995 and was subsequently officially released in 1996, spreading across the southern half of the Australian continent, resulting in declines of up to 95% in rabbit numbers in many areas (15–17). In contrast to MYXV, no rapid decline of virulence was observed in RHDV following its release. Although natural outbreaks of RHDV still occur regularly in most Australian rabbit populations, there is evidence that rabbit numbers are increasing again (18, 19), and recent studies in which rabbits were challenged with low doses of RHDV show that in some areas rabbits are developing resistance to infection (20). Other studies suggest that differential binding preferences of RHDV to carbohydrate structures on mucosal surfaces can convey partial resistance to infection with certain strains of RHDV in both Australian and European wild rabbits (21). This raises important questions about the trajectory of RHDV and rabbit co-evolution, including the expression of virulence. In particular, is the virus evolving to maintain relatively high virulence in response to higher host resistance?
One of the rabbit populations identified as having a moderate level of resistance to RHDV (20) is located at the Turretfield Research Centre in South Australia (22). A high proportion of rabbits from this site were resistant to low dose infection, as determined by a high survival rate and the absence of seroconversion in some rabbits following experimental challenge. However, these rabbits were not resistant to lethal disease because all those that became infected died, albeit with unusually prolonged survival times (20). On-going regular monitoring of the rabbit population has been carried out at the Turretfield site since the release of RHDV in 1996, and a steady increase in rabbit numbers has been observed there since 2006 (22). Despite the reported presence of genetic resistance at Turretfield and the increase in rabbit numbers, RHDV outbreaks still occur regularly and at increasing frequency, compatible with an ongoing evolutionary ‘arms race’ between rabbits and RHDV. Importantly, virus samples have been collected from rabbits found dead during each RHDV outbreak and stored in laboratory freezers. These archived samples provide an excellent opportunity to study virus-host co-evolution. This analysis is assisted by the fact that all field isolates of RHDV in Australia are derived from a single founding strain used for biocontrol, the Czech CAPM V-351 strain (herein denoted CAPM V-351) (23), in contrast to Europe where multiple viral strains co-circulate.
Given the major biological differences between RHDV and MYXV it might be expected that these viruses would follow different evolutionary trajectories. In particular, MYXV possesses a large double-stranded DNA genome with multiple genes that may act as virulence determinants, whereas RHDV is a small, rapidly-replicating RNA virus with likely a limited set of mutations controlling virulence (24, 25). In addition, MYXV relies on insect-mediated transmission with virus particle uptake from lesions on live animals (7), while RHDV transmission can be by direct contact with virus particles passed via mucous membranes (26) or by scavenging insects feeding on carcasses, in turn facilitating long distance transmission of the virus (23).
In this study we used the offspring of wild rabbits from Turretfield to examine the short-term evolution of virulence in recent RHDV isolates taken from the same sampling site. Rabbits were collected from the field following the 2007 spring RHDV outbreak, and three recent virus isolates from the same year (2007), the previous year (2006) and the most recent virus sample available at the time of testing (2009) were compared with the original CAPM V-351 release strain. Three key parameters of virulence were assessed: case fatality rates, disease duration (time to death), and the amount of virus progeny produced in infected animals.
Materials and Methods
Ethics Statement
All research on animals was conducted in strict accordance with the guidelines of the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (7th Edition). Transport and breeding of wild rabbits and the infection studies were conducted under Queensland Community Access Animal Ethics Committee approval CA 2007/10/220, and every effort was made to minimize animal suffering.
Turretfield site and rabbit population
Turretfield is an agricultural research station approximately 50 km north of Adelaide, South Australia (34° 33′ 00″ S, 138° 49′ 47″ E) (22). The site is regularly searched for rabbit carcases or sign of rabbits dying in warrens as indicated by blowflies (Calliphoridae and Sarcophagidae), their maggots or meat-ants (Iridomyrmex sp.) emerging from burrows or the smell of decaying carcases in and around warrens. Liver, heart, kidney or bone marrow from large leg bones are sampled from any carcase found and frozen at −30° C as soon as possible.
Preparation of virus isolates for challenge tests
The field isolates used came from three livers of individual rabbits found freshly dead and verified by standard PCR as having died from RHD. These were collected during winter/spring outbreaks of RHDV in 2006, 2007 and 2009 (strains denoted TUR06, TUR07 and TUR09, respectively). The inoculum to which they were compared was a commercially available inoculum prepared from the CAPM V-351 virus that escaped quarantine in 1995 and was released in 1996.
To standardise doses for each of the four isolates, given their different origins, preparations and storage times, each was passaged through a domestic rabbit and, upon death (within 48 hours), livers were stored frozen (−20°C) for seven days and subsequently used to produce inoculums. Equal-sized (1 cm3) sections of liver were macerated in 5 mL of PBS and made up into equal volumes in PBS (50 mL). These stocks were then used to challenge rabbits. While the viral load in livers of rabbits that succumb to RHDV infection is independent of the inoculation dose (21), and genome copy numbers are not an exact measure for the number of infectious particles, quantitative reverse transcription-PCR (qRT-PCR) analysis was done to confirm that all animals received a high virus dose: each preparation contained at least 2.5 × 108 genome copies per dose (CAPM V-351: 2.50 ×108; TUR06: 7.42 ×109; TUR07: 1.95 ×109; TUR09: 3.01 ×109).
Preparation of wild rabbits for challenge
Four male and seven female rabbits were live trapped from the Turretfield site within one month of the 2007 RHD outbreak. These were housed at the Robert Wicks Pest Animal Research Centre, Inglewood, Queensland. Four females (seronegative to RHDV, to reduce the risk of young carrying temporary maternal antibodies) were randomly chosen and randomly paired to the four males (each seropositive to RHDV) and housed in a temperature controlled animal house (20 ± 1°C, day/night cycle 12hr / 12hr) in large cages in an environment where freedom from exposure to RHDV and non-pathogenic RCV-A1(27) could be assured. Presence of this non-pathogenic virus could have resulted in erroneous experimental results because it partially protects rabbits against the full effects of RHDV (28, 29)
All rabbits were fed commercial pellets daily, carrots twice weekly and water ad libitum. From these four pairs, 80 offspring were reared and held until more than 13 weeks of age when infection and mortality rates are no longer influenced by age related resilience (30). Prior to challenge, the young adults were assigned to four groups of 20, to distribute animals of each sex and known parentage equally.
For experimental challenge, rabbits were individually housed in plastic boxes (610 × 410 × 400 mm) with insect-proof gauze lids and wire mesh floors over absorbent litter. These were held in a climate-controlled room (22°C ± 1°C, 50% RH, 12hr /12hr day/night cycle). Water and food (commercial rabbit pellets and fresh carrot) were provided ad libitum.
Monitoring and sampling of experimentally infected rabbits
Before challenge, a blood sample (0.5 mL) was taken from an ear of each rabbit to confirm freedom from exposure to RHDV and the non-pathogenic rabbit calicivirus RCV-A1. To initiate the experiment, the twenty rabbits in each group received 1 mL of the appropriate virus inoculums (i.e. CAPM V-351, TUR06, TUR07 or TUR09) administered orally using a tuberculin syringe (without needle) placed through the diastema onto the back of the tongue.
Rabbits were subsequently checked at eight hour intervals (nominally 0700, 1500 and 2300 hours) to minimise disturbance while still allowing time of death to be calculated and blood samples to be collected. Upon death, rectal temperature and a blood and liver sample were taken. Rectal temperature was used to more precisely calculate time of death within the previous 8 hours as described previously (20). Rabbits surviving to 14 days post-inoculation were euthanized. Blood and liver samples were collected from all rabbits and stored frozen at −20°C for later serological and qRT-PCR analysis. Serum samples were tested for RHDV antibodies using a suite of ELISAs (31).
Statistical analysis
Mortality differences between the four isolates were measured using Fisher’s Exact (32) tests to determine which isolates differed significantly from the others. Survival times were analysed using a Mann-Whitney U test (32). As mortality and survival time are linked, Kaplan-Meier survival analysis (33) was used to further show differences between the four isolates. Differences in virus titres in deceased rabbits were analysed using the Mann-Whitney U test. Fisher’s Exact tests and Mann-Whitney U tests were run using the R program (34).
Quantitative PCR (qRT-PCR)
RNA was prepared from liver tissue using the QIAGEN RNeasy kit according to the instructions of the supplier. Primers and standards for RHDVqRT-PCR have been described previously (29). The BioRad iScript One Step RT-PCR kit and a BioRad CFX Real time PCR machine were used for amplification according to the instructions of the manufacturer. Each 10μl reaction contained a final concentration of 0.3 μM of each primer. Cycle conditions used were 10 min at 50 °C, followed by 42 repeats of a two-step cycle with 10s at 95 °C and 40 s at 63 °C, and signal acquisition for 10 s at 78 °C. All samples were analysed in triplicates.
Genome sequencing
Oligonucleotides used for sequencing are listed in Supplementary Table S1. First strand cDNA was prepared as described previously (27) and used for amplification. Cycle conditions were 95°C for 3 min, and 42 repeats of 95°C for 15 sec, 50°C for 30 sec and 72°C for 90 sec, using the Invitrogen Platinum Tag polymerase according to the instructions of the manufacturer. Amplified fragments were cut and purified using the QIAquick Gel Extraction Kit (QIAGEN) protocol. Custom Sanger sequencing was carried out at the Biological Resources Facility of the Australian National University. Sequences were then assembled using BioEdit, version 7.0.1 (35). All sequences generated here have been submitted to GenBank and assigned accession numbers (KF594474-6, KJ606058-9). A full list of all the strains used and their GenBank accession numbers is shown in Supplementary Table S2.
Evolutionary analysis
To determine the evolutionary relationships of the three Turretfield viruses (TUR06, TUR07, TUR09) in relation to other strains of RHDV we collated from GenBank the complete genome sequences of 37 strains of RHDV. Sequence alignment of the total data set was undertaken using MUSCLE (36) assuming default parameters. This resulted in a final alignment of 40 sequences, 7032 nucleotides (nt) in length.
The phylogenetic relationships among these sequences were determined using the maximum likelihood (ML) method available in the PhyML program (37). This analysis incorporated the GTR+Γ model of nucleotide substitution (parameters estimated from the empirical data) and SPR branch-swapping. The support for individual nodes was determined using 1000 bootstrap replicate ML trees, this time employing NNI branch-swapping in PhyML. All amino acid substitutions were mapped onto the ML tree using the parsimony algorithm available in the PAUP* package (37).
To determine the selection pressures acting on RHDV, and particularly those from Turretfield, we employed a variety of methods available in the HyPhy package (38) and based on the ML tree inferred above. All analyses compared the relative numbers of nonsynonymous (dN) and synonymous (dS) substitutions per site (i.e. the ratio dN/dS). To determine the selection pressures acting on individual amino acid sites we utilized the Single Likelihood Ancestor Counting (SLAC) and Fast Unbiased Bayesian Approximation (FUBAR) methods (39), while the Branch-Site REL method (40) was used to estimate selection pressures on individual branches.
Serology
Enzyme Linked Immuno Sorbent Assays (ELISAs) for IgM, IgA and IgG subclass antibodies, as well as a competition ELISA were used to detect antibodies to RHDV as previously described (31). The absence of RCV-A1 antibodies was confirmed using a highly specific blocking ELSIA (41). All 80 animals used in this study were free from antibodies to the non-pathogenic calicivirus RCV-A1 prior to RHDV inoculation.
Results
Phylogenetic relationships of Turretfield viruses
To place the evolution of TUR06, TUR07 and TUR09 in the context of global RHDV we performed a phylogenetic analysis of the complete genome sequences of 40 RHDV strains sampled from diverse localities. This revealed that the three Turretfield viruses formed a well-supported monophyletic group that was most closely related to other viruses sampled from Australia and New Zealand, and which was ultimately derived from the introduced Czech CAPM V-351 strain (Figure 1). A monophyletic group of Australasian viruses was previously observed in an analysis of viral capsid gene sequences (VP60) (23). Also of note was that the two Turretfield viruses of the highest virulence and most recent sampling – TUR07 and TUR09 – clustered together to the exclusion of the lower virulence TUR06 strain (see below).
Figure 1.
Maximum likelihood (ML) tree showing the phylogenetic relationships among 40 complete genomes of RHDV. All branches are scaled according to the number of nucleotide substitutions per site, and relevant bootstrap values >90% are denoted by asterisks. The monophyletic grouping of RHDV from Australia and New Zealand, derived from the imported Czech CAPM V-351 strain, are shaded grey, while the three Turretfield viruses (TUR06, TUR07 and TUR09) are shown in bold italics. The tree is mid-point rooted for purposes of clarity only. The insert shows the mean dN/dS value and the amino acid (AA) changes on those branches associated with the evolution of TUR06, TUR07 and TUR09. The single site associated with positive selection across RHDV has a whole – amino acid residue 12 in the p16 protein – fixes a mutation on the branch leading to the three Turretfield viruses and is colored red.
Experimental infections
All three field isolates (TUR06, TUR07, TUR09) caused higher mortality rates compared to the original CAPM V-351 strain. The mortality rates also increased in the more recent field isolates, from 75% in the CAPM V-351 to 90% in the TUR06 isolate, and 100% in the most recent TUR07 and TUR09 isolates. Mortality rates differed significantly between the four virus isolates (Fisher’s Exact test p=0.014), and the CAPM V-351 isolate caused significantly lower mortality than the TUR07 and TUR09 isolates (Table 1).
Table 1.
Statistical analysis of the three key virulence parameters in RHDV.
| Virus isolate comparison | Mortality Rates | Survival Time* | Virus titres* | ||
|---|---|---|---|---|---|
|
| |||||
| p-value | W | p-value | W | p-value | |
| TUR06 vs TUR07 | 0.4872 | 318.5 | < 0.0001 | 188 | 0.8150 |
| TUR06 vs TUR09 | 0.4872 | 285.5 | 0.0019 | 91.5 | 0.0097 |
| TUR06 vs CAPM V-351 | 0.4075 | 83.5 | 0.0620 | 190 | 0.0467 |
| TUR07 vs TUR09 | 1.0000 | 147.5 | 0.1389 | 106.5 | 0.0114 |
| TUR07 vs CAPM V-351 | 0.0471 | 27.5 | < 0.0001 | 194 | 0.1424 |
| TUR09 vs CAPM V-351 | 0.0471 | 44.5 | 0.0004 | 249 | 0.0010 |
| All field isolates vs CAPM V-351 | 155.5 | 0.0001 | 237 | 0.0069 | |
| TUR09 vs all other isolates | 811 | 0.0005 | |||
| All strains against each other | 0.0142 | ||||
only survival times and virus titres of rabbits that succumbed to the infection were analysed. Virus titres are expressed as copy numbers/mg liver, as determined by qRT-PCR.
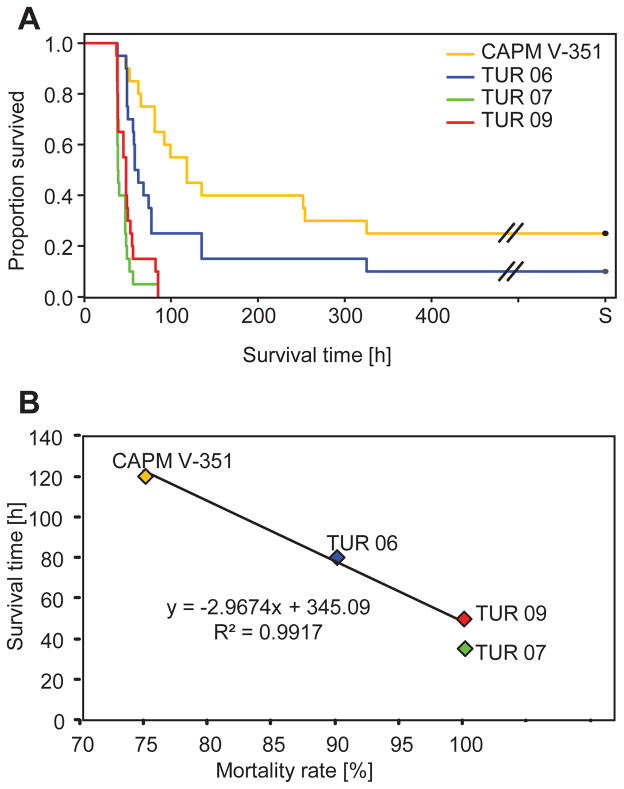
Survival times of infected rabbits also differed significantly between the virus isolates (Mann-Whitney U test W=155.5, p=0.0001, Table 1). They were highest in rabbits infected with the CAPM V-351 isolate (121.27 h ± 22.35), and became progressively shorter in the TUR06 (81.17 h ± 15.68) and TUR09 isolates (50.35 h ± 3.50). Notably, the TUR07 isolate had the shortest average survival times of all isolates tested (44.45 h ± 2.49). The differences between CAPM V-351 and TUR07 and TUR09, as well as the differences between TUR06 and both the TUR07 and TUR09 isolates, were statistically significant (Table 1). Kaplan-Meier analysis illustrates increasingly shorter survival times and higher mortality rates for the more recent isolates (Figure 2A). The two parameters were closely associated (y = −2.967× + 345.07, R2 = 0.9917) (Figure 2B).
Figure 2.
A: Kaplan Meier Survival analysis of rabbits infected with different virus isolates. S: Surviving rabbits were euthanized 14 d.p.i.B: Increasingly shorter survival times and higher mortality rates for the more recent isolates are strongly associated.
QRT-PCR analysis of the livers of dead rabbits showed that average virus loads increased in rabbits infected with the more recent isolates and were highest in TUR09 (Figure 3). For the two isolates TUR09 and TUR06 the highest virus titres were obtained in animals that died within the first 70h following inoculation. Three of four rabbits (across all groups) that died more than 250 h post infection still had virus titres of >10 × 109 copies per mg liver tissue. Rabbits infected with CAPM V-351 had the lowest average viral loads (4.15 × 108). Infection with TUR06 resulted in increased average viral loads in the liver (4.74 × 108) compared to CAPM V-351, while the most recent isolate, TUR09, had the highest average viral loads in the liver (1.12 × 109). Average virus loads in rabbits infected with TUR07 (6.12 × 108), were comparable to TUR06; however, the variance within the TUR07 group was very high and some individuals that died within the first 48 hours post infection had very low virus titres, suggesting that TUR07 may have killed some rabbits before they had developed very high virus loads in the liver. Although differences in median RNA copy numbers per mg liver tissue in non-surviving rabbits were within an order of magnitude, there were significant differences between treatment groups (Table 1). The mean copy numbers of rabbits infected with the TUR09 isolate were significantly higher compared to all other treatment groups. The difference between the median virus titres of TUR06 and CAPM V-351 was also significant, but the difference between TUR07 and the CAPM V-351 strain was not (Table 1) due to the rabbits that died quickly with low viral loads. In this context it needs to be noted that the inoculum preparations used for infection were not identical in terms of their genome equivalents. This caveat notwithstanding, a very high dose was applied in all cases. The viral load in livers of rabbits that succumb to RHDV infection is independent of the inoculation dose (21), and differences in mortality rates and time to death are only noted when very low doses of virus are applied (limiting dilution to titrate ID50).
Figure 3.
Viral RNA copy numbers in liver tissue were quantified using qRT-PCR. Virus load is expressed as genome copies per mg tissue. B: Cumulative virus load of rabbits that died within a specified time. The cumulative virus load is the product of the number of rabbits that died and the viral genome copy number in their liver. Grey bars indicate the cumulative load of all dead rabbits, black bars the cumulative load of all rabbits that died within the first 60 hours, before the onset of adaptive immune responses.
Seven animals survived the experimental infections, five in the group infected with the CAPM V-351 strain, and two in the TUR06 group. Of these survivors, only one animal infected with the CAPM V-351 strain had seroconverted and produced antibodies against RHDV 14 days after the challenge (Table 2). Notably, initial seroconversion (IgM and a positive competition ELISA) was also found in two rabbits that did not survive the challenge, one of which was infected with the CAPM V-351 isolate and one with TUR07. These animals had prolonged survival times that were amongst the highest in their group (135h and 85h, respectively, data not shown). The four rabbits that died more than 10 days post infection had no detectable RHDV antibodies.
Genome-scale selection pressures and amino acid replacements in Turretfield viruses
To help determine the genetic basis to virulence evolution in the Turretfield viruses we performed a series of evolutionary analyses. Across the data set as a whole, three amino acid residues were found to be positively selected according to the FUBAR program (posterior probability >0.95): sites 3, 12 and 597, which are located in the first non-structural protein p16 (aa 3 and aa 12) and the viral helicase (aa 597), respectively. Of these, sites 3 and 12 were confirmed at the p-value < 0.05 significance level in the SLAC program. Interestingly, the branch leading to TUR06, TUR07 and TUR09 is characterized by a T to A amino acid replacement at aa 12 that that has evolved independently four times across the RHDV phylogeny and shown here to be subject to positive selection (insert, Figure 1), although it’s function is uncertain. In contrast, the Branch-Site REL method revealed no evidence for positive selection in the lineages involving TUR06, TUR07 and TUR09 (nor across the phylogeny as a whole at p < 0.05) with the range of dN/dS values among these three viruses (0.033 – 0.062) lower than the phylogeny-wide mean of 0.074 (insert, Figure 1).
Also of note were the 44 amino acid changes fixed in the evolutionary history of the Turretfield viruses (i.e. that distinguish these lineages from their closest relative in the sample; Figure 1, insert). Notably, among these mutations, 18 (41%) occur within the first non-structural protein p16 (aa 1-144), a protein of yet unknown function, which represents only 6% of the 2344 amino acid residue alignment of ORF1. In addition, all three Turretfield viruses share an amino substitution in the V1-loop (residues 2066 to 2075) compared to CAPM V-351: from SASYTGSNAT to SASYPGNNAT. This region has been suggested to play a key role in virus-host interaction, both as a major immuno-dominant epitope and for receptor recognition (42). Finally, it is notable that the higher virulence TUR07 and TUR09 strains are characterized by clustered mutations at residues 134, 136, and 139, all of which are located in p16.
Discussion
Finding suitable model systems to study the mechanisms of disease emergence is essential to understand and ultimately predict the virulence patterns of novel infections and reduce their impacts. In Australia, two biocontrol agents were released into naïve rabbit populations, creating unique opportunities to study the changes in pathogen virulence as a virus establishes itself in the population. In addition, the two biocontrol viruses provide examples of the two main mechanisms of disease emergence. While MYXV allows insights into how the virulence of a large DNA virus may evolve following a species jump (10), RHDV represents a small, rapidly evolving RNA virus that has likely emerged from a previously non-pathogenic group of viruses by mutation (25, 43, 44) and is apparently maintaining high levels of virulence.
Cooke and Berman (2000) showed that CAPM V-351 killed 22 of 24 unselected, nonresistant Australian wild rabbits, with survival times averaging 72.5 hr for orally inoculated rabbits (and BDC, pers comm.). However, increasing resistance would mediate such severity. Herein, we show that the virulence of recent field isolates has increased in genetically resistant rabbits compared to the CAPM V-351 strain originally released. In particular, isolates collected in 2006, 2007 and 2009 show a general trend towards causing higher mortality rates, shorter survival times, increased replication speed and higher virus loads in the livers of rabbits succumbing to the disease (Figure 2). This evolutionary trend is in stark contrast to MYXV, which may in part reflect their different mechanisms of transmission. MXYV is mechanically transmitted by biting or blood sucking arthropod vectors such as fleas and mosquitoes. In this case a reduction of virulence is thought to maximise transmission, such that strains of intermediate virulence with reduced mortality rates and prolonged disease became dominant because they produced enough virus in skin lesions for a sufficient period to allow uptake by biting insects (10). In contrast, RHDV can be transmitted both orally by fomites or direct contact between rabbits (26), and passively via insect vectors, with the latter likely responsible for the escape of RHDV from quarantine in 1995 (45). While fomite/contact transmission is likely to play an important role in transmission during virus outbreaks (46), long distance transmission of RHDV occurs through flies that scavenge on decomposing carcasses, or fresh carcasses opened by predators, and then transmit the virus passively by landing on mucous membranes of rabbits directly, or by leaving faeces or regurgita on pasture that is subsequently ingested by rabbits (45, 47, 48). Hence, MYXV requires vectors to bite a live, diseased animal. However, in the case of RHDV the carcass of a dead rabbit appears to be the main source of virus for transmission in the field, enabling its spread between distant rabbit populations that are not directly connected
The amount of tissue taken up and transmitted by flies that have fed on rabbit carcasses is likely to be very small (47). In this context, the speed with which the most recent field isolates of RHDV in this study killed rabbits is striking and suggests that the virus may be selected to avoid adaptive immune responses. The rabbit’s immune response to RHDV is rapid and the first IgM antibodies can be detectable as early as 50 to 60 h.p.i. (49), and are reliably detected at 72 h.p.i. (49). Cooke and Berman reported that the levels of intact virus detected with virus-capture ELISA in rabbit livers reached maximal values at about 50 h (50), indicating that with the onset of the adaptive immune response virus titres start to decline. RHDV recovered from rabbit tissues 144 hpi were not infectious, possibly due to the formation of immune complexes or degradation of viral particles (51). Thus, it is likely that selection will favour those RHDV variants that can multiply most efficiently in less than 60 hours, as these will have the highest number of infectious viral particles in the liver (Figure 3B). Indeed, the average survival time of rabbits infected with the most recent TUR07 and TUR09 isolates was 44 and 50 h, respectively, and viral titres were highest at this time as determined by qRT-PCR. In contrast, only 15% of the CAPM V-351 rabbits died within 60 hours, compared to 77% mortality in the 60 rabbits infected with the three recent field isolates, and the virus titre in the livers of the CAPM V-351 infected rabbits was consistently lower. In the field this would significantly decrease the likelihood of insect transmission for CAPM V-351. This is consistent with the observation that despite several hundred deliberate releases for local rabbit control over the past 18 years, CAPM V-351 was never able to reestablish and replace circulating field strains, although a local reduction in rabbit numbers was often achieved (23).
Our work also suggests that selection for virus strains with increased virulence that rapidly replicate to very high titres in the liver could partially off-set developing resistance to RHDV infection. Although the mechanisms for genetic resistance to RHDV are not completely understood, rabbits express a variety of different histo-blood group antigen (HBGA) phenotypes on their intestinal tissues that differ in their ability to bind different RHDV strains, leading to partial resistance to infection (21). However, these protective effects were only observed at low virus doses, and overcome when high challenge doses were used. Although the HBGA phenotypes of the Turretfield rabbits used in this experiment were not known, resistance of rabbits from the same location to infection with low doses of RHDV was demonstrated previously (20) and six of the seven rabbits surviving the challenge infections in this our study did not seroconvert to RHDV. This suggests that these rabbits also avoided productive infections and that the mechanism contributing to genetic resistance in the Turretfield rabbits similarly leads to less efficient uptake of virus, and perhaps slows virus spread in those animals that did become infected.
Despite this apparent evolutionary trend toward higher virulence, the causative viral mutations are unclear, although our sequence analysis suggests that any positive selection within the three Turretfield viruses was likely limited to a small number of amino acid sites. Interestingly, we noted a marked clustering of amino acid changes in the first non-structural protein (p16), including a number of putatively positively selected sites. However, both the phenotypic and fitness effects of these mutations are unclear, and the function of this protein has not yet been described. Studies are currently underway aimed at characterising the function of the RHDV non-structural proteins and which will enable us to make the central link with genotype and phenotype. In addition, the three recent field isolates share an amino acid substitution on an area of the capsid protein that has been suggested to play a key role in virus-host interactions (42). Hence, there are a variety of mutations that could underpin the virulence evolution observed here, including at receptor binding/virus entry or virus replication level.
Case fatality rates of 100% for the recent field isolates may seem unusually high. However, overall mortality rates of the strains tested in this study will depend on the presence and proportion of young rabbits in the population. In the case of RHDV young rabbits become infected with the virus but are innately resistant to lethal disease, likely due to their immature immune system (52). This resistance is almost 100% in four week old rabbits but gradually lost until they become fully susceptible at 12 weeks of age. Maternal antibodies can prolong the period of reduced susceptibility but are not essential in this age-related resistance (30). Importantly, young rabbits survive infection with a strong adaptive immune response providing lifelong immunity to RHDV (30). These animals are then likely to be recruited into the immune breeding population, and the offspring they produce form the next generation of susceptible hosts.
The combination of high environmental stability of the infectious viral particles (53), the innate resistance of young rabbits and high fecundity of the species that allows populations to recover quickly, as well as the ability of the virus to travel considerable distances on insect vectors provides a resilient system that has allowed RHDV to maintain high levels of virulence over an 18 year period. The rabbit population densities sufficient to sustain this equilibrium have been much lower compared to pre-RHDV years, and this has provided some much needed relief for the continent’s biodiversity (5). However, a recent increase in rabbit numbers (4) highlights the need for further studies into RHDV-host co-evolution. Such studies also need to take into account factors such as climate variables that can influence rabbit breeding patterns and vector availability, as well as the complex interactions of the two other viruses endemic to Australia’s rabbits – MYXV and the partially protective non-pathogenic calicivirus RCV-A1 (28).
Understanding the mechanisms and circumstances that allow a pathogen to develop and sustain high levels of virulence is not only crucial to ensure optimal gains are made from RHDV as a biological control agent in Australia, but represents a powerful model system for emerging pathogens in general. RNA viruses cause the majority of emerging or re-emerging diseases, which has been attributed to their high evolutionary rates and rapid replication (54). Importantly, RHDV can also be studied experimentally in its natural host. Further investigation of RHDV virulence evolution will provide insights into both the RHDV-rabbit interaction and virus-host co-evolution in general.
Acknowledgments
We thank L. Capucci for the provision of RHDV ELISA reagents, S. Haboury, J. Wright, D. George and M. Drewer for expert technical assistance, David Peacock for assistance with sample and rabbit acquisition, Greg Mutze for assistance with capture of rabbits and helpful comments on the draft manuscript, David Aster and Andrew Granzotto for maintaining and breeding rabbits at Inglewood, and Nina Schwensow for helpful comments. This project is supported the ARC (grant DP140103362). ECH is supported by an NHMRC Australia Fellowship and by NIH grant R01 AI093804-01A1. PE’s postgraduate studies were supported by the Invasive Animals Cooperative Research Centre.
Footnotes
Competing Interests
The authors declare they have no competing interests.
References
- 1.Frank SA. Models of parasite virulence. Quart Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 2.Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson B. Calicivirus, myxoma virus and the wild rabbit in Australia: a tale of three invasions. In: Smith GLIW, McCauley J, Rowlands DJ, editors. New Challenges to Health: the Threat of Virus Infection. 60. Cambridge University Press; Cambridge, UK: 2001. pp. 67–87. [Google Scholar]
- 4.Cooke B, Chudleigh P, Simpson S, Saunders G. The economic Benefits of the biological control of rabbits in Australia, 150–2011. Australian Econ Hist Rev. 2013;53:1–17. [Google Scholar]
- 5.Cooke BD. Rabbits: manageable environmental pests or participants in new Australian ecosystems? Wildlife Res. 2012;39:279–289. [Google Scholar]
- 6.Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 7.Fenner F, Ratcliffe FN. Myxomatosis. Cambridge University Press; 1965. [Google Scholar]
- 8.Levin BR. The evolution and maintenance of virulence in microparasites. Emerg Infect Dis. 1996;2:93–102. doi: 10.3201/eid0202.960203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.May RM, Anderson RM. Epidemiology and genetics in the coevolution of parasites and hosts. Proc Roy Soc Lond B. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. [DOI] [PubMed] [Google Scholar]
- 10.Kerr PJ. Myxomatosis in Australia and Europe: a model for emerging infectious diseases. Antiviral Res. 2012;93:387–415. doi: 10.1016/j.antiviral.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Berman D, Kerr PJ, Stagg R, van Leeuwen BH, Gonzalez T. Should the 40-year-old practice of releasing virulent myxoma virus to control rabbits (Oryctolagus cuniculus) be continued? Wildlife Res. 2006;33:549–556. [Google Scholar]
- 12.Fenner F, Fantini B. Biological control of vertebrate pests: the history of myxomatosis, an experiment in evolution. Biological control of vertebrate pests: the history of myxomatosis, an experiment in evolution. 1999:xii, 339. [Google Scholar]
- 13.Kerr PJ, Ghedin E, Depasse JV, Fitch A, Cattadori IM, Hudson PJ, Tscharke DC, Read AF, Holmes EC. Evolutionary history and attenuation of myxoma virus on two continents. PLoS Pathog. 2012;8:e1002950. doi: 10.1371/journal.ppat.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu SJ, Xue HP, Pu BQ, Qian NH. A new viral disease in rabbits. Animal Husb Vet Med. 1984;6:253–255. [Google Scholar]
- 15.Bowen Z, Read J. Population and demographic patterns of rabbits (Oryctolagus cuniculus) at Roxby Downs in arid South Australia and the influence of rabbit haemorrhagic disease. Wildlife Res. 1998;25:655–662. [Google Scholar]
- 16.Kovaliski J. Monitoring the spread of rabbit hemorrhagic disease virus as a new biological agent for control of wild European rabbits in Australia. J Wildl Dis. 1998;34:421–428. doi: 10.7589/0090-3558-34.3.421. [DOI] [PubMed] [Google Scholar]
- 17.Mutze G, Cooke B, Alexander P. The initial impact of rabbit hemorrhagic disease on European rabbit populations in South Australia. J Wildl Dis. 1998;34:221–227. doi: 10.7589/0090-3558-34.2.221. [DOI] [PubMed] [Google Scholar]
- 18.McPhee SR, Butler KL, Kovaliski J, Mutze G, Capucci L, Cooke BD. Antibody status and survival of Australian wild rabbits challenged with rabbit haemorrhagic disease virus. Wildlife Res. 2009;36:447–456. [Google Scholar]
- 19.Sandell PR. Implications of rabbit haemorrhagic disease for the short-term recovery of semi-arid woodland communities in north-west Victoria. Wildlife Res. 2002;29:591–598. [Google Scholar]
- 20.Elsworth PG, Kovaliski J, Cooke BD. Rabbit haemorrhagic disease: are Australian rabbits (Oryctolagus cuniculus) evolving resistance to infection with Czech CAPM 351 RHDV? Epidemiol Infect. 2012;140:1972–1981. doi: 10.1017/S0950268811002743. [DOI] [PubMed] [Google Scholar]
- 21.Nystrom K, Le Gall-Recule G, Grassi P, Abrantes J, Ruvoen-Clouet N, Le Moullac-Vaidye B, Lopes AM, Esteves PJ, Strive T, Marchandeau S, Dell A, Haslam SM, Le Pendu J. Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog. 2011;7:e1002188. doi: 10.1371/journal.ppat.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peacock DE, Sinclair RG. Longevity record for a wild European rabbit (Oryctolagus cuniculus) from South Australia. Australian Mammol. 2009;31:65–66. [Google Scholar]
- 23.Kovaliski J, Sinclair R, Mutze G, Peacock D, Strive T, Abrantes J, Esteves PJ, Holmes EC. Molecular epidemiology of Rabbit Haemorrhagic Disease Virus in Australia: when one became many. Mol Ecol. 2014;23:408–420. doi: 10.1111/mec.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes EC. What can we predict about viral evolution and emergence? Curr Opin Virol. 2013;3:180–184. doi: 10.1016/j.coviro.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerr PJ, Kitchen A, Holmes EC. Origin and phylodynamics of rabbit hemorrhagic disease virus. J Virol. 2009;83:12129–12138. doi: 10.1128/JVI.01523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morisse JP, Le Gall G, Boilletot E. Hepatitis of viral origin in Leporidae: introduction and aetiological hypotheses. Rev Sci Tech. 1991;10:269–310. [PubMed] [Google Scholar]
- 27.Strive T, Wright JD, Robinson AJ. Identification and partial characterisation of a new lagovirus in Australian wild rabbits. Virol. 2009;384:97–105. doi: 10.1016/j.virol.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Strive T, Elsworth P, Liu JN, Wright JD, Kovaliski J, Capucci L. The non-pathogenic Australian rabbit calicivirus RCV-A1 provides temporal and partial cross protection to lethal Rabbit Haemorrhagic Disease Virus infection which is not dependent on antibody titres. Vet Res. 2013;44:51. doi: 10.1186/1297-9716-44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strive T, Wright J, Kovaliski J, Botti G, Capucci L. The non-pathogenic Australian lagovirus RCV-A1 causes a prolonged infection and elicits partial cross-protection to rabbit haemorrhagic disease virus. Virol. 2010;398:125–134. doi: 10.1016/j.virol.2009.11.045. [DOI] [PubMed] [Google Scholar]
- 30.Robinson AJ, So PTM, Muller WJ, Cooke BD, Capucci L. Statistical models for the effect of age and maternal antibodies on the development of rabbit haemorrhagic disease in Australian wild rabbits. Wildlife Res. 2002;29:663–671. [Google Scholar]
- 31.Cooke BD, Robinson AJ, Merchant JC, Nardin A, Capucci L. Use of ELISAs in field studies of rabbit haemorrhagic disease (RHD) in Australia. Epidemiol Infect. 2000;124:563–576. doi: 10.1017/s0950268899003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. 3. W.H. Freeman & Co; New York: 1995. [Google Scholar]
- 33.Kaplan EL, Meier P. Nonparametric-Estimation from Incomplete Observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 34.Team RCD. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. http://www.R-project.org/, Vienna, Austri. [Google Scholar]
- 35.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 36.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates; Sunderland, Ma: 2003. [Google Scholar]
- 38.Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 39.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. A random effects branch-site model for detecting episodic diversifying Selection. Mol Biol Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Kerr PJ, Strive T. A sensitive and specific blocking ELISA for the detection of rabbit calicivirus RCV-A1 antibodies. Virol J. 2012;9:182. doi: 10.1186/1743-422X-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Xu FT, Liu JS, Gao BQ, Liu YX, Zhai YJ, Ma J, Zhang K, Baker TS, Schulten K, Zheng D, Pang H, Sun F. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog. 2013;9:e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hicks AL, Duffy S. One misdated sequence of rabbit hemorrhagic disease virus prevents accurate estimation of its nucleotide substitution rate. BMC Evol Biol. 2012;12:74. doi: 10.1186/1471-2148-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinnear M, Linde CC. Capsid gene divergence in rabbit hemorrhagic disease virus. J Gen Virol. 2010;91:174–181. doi: 10.1099/vir.0.014076-0. [DOI] [PubMed] [Google Scholar]
- 45.Cooke BD, Fenner F. Rabbit haemorrhagic disease and the biological control of wild rabbits, Oryctolagus cuniculus, in Australia and New Zealand. Wildlife Res. 2002;29:689–706. [Google Scholar]
- 46.Matthaei MKP, Read AJ, Hick P, Haboury S, Wright JD, Strive T. Comparative quantitative monitoring of rabbit haemorrhagic disease viruses in rabbit kittens. Virol. doi: 10.1186/1743-422X-11-109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asgari S, Hardy JRE, Sinclair RG, Cooke BD. Field evidence for mechanical transmission of rabbit haemorrhagic disease virus (RHDV) by flies (Diptera : Calliphoridae) among wild rabbits in Australia. Virus Res. 1998;54:123–132. doi: 10.1016/s0168-1702(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 48.McColl KA, Merchant JC, Hardy J, Cooke BD, Robinson A, Westbury HA. Evidence for insect transmission of rabbit haemorrhagic disease virus. Epidemiol Infect. 2002;129:655–663. doi: 10.1017/s0950268802007756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbieri I, Lavazza A, Brocchi E, Konig M, Capucci L. Proc. 1st Int. Symp. Calicivirus ESVV; Reading, UK. 1997. pp. 182–193. [Google Scholar]
- 50.Cooke BD, Berman D. Effect of inoculation route and ambient temperature on the survival time of rabbits, Oryctolagus cuniculus (L.), infected with Rabbit Haemorrhagic Disease Virus. Wildlife Research. 2000;27:137–142. [Google Scholar]
- 51.Pages Mante A. Consideraciones tecnicas de la sueroterapia y de la profilaxis vacunal en la enfermedad hemorragica virica del conejo (R.H.D.V.) Medicina Veterinaria. 1989;6:285–291. [Google Scholar]
- 52.Marques RM, Teixeira L, Aguas AP, Ribeiro JC, Costa-e-Silva A, Ferreira PG. Immunosuppression abrogates resistance of young rabbits to Rabbit Haemorrhagic Disease (RHD) Veterinary research. 2014:45. doi: 10.1186/1297-9716-45-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henning J, Meers J, Davies PR, Morris RS. Survival of rabbit haemorrhagic disease virus (RHDV) in the environment. Epidemiology and Infection. 2005;133:719–730. doi: 10.1017/s0950268805003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmes EC. The comparative genomics of viral emergence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1742–1746. doi: 10.1073/pnas.0906193106. [DOI] [PMC free article] [PubMed] [Google Scholar]