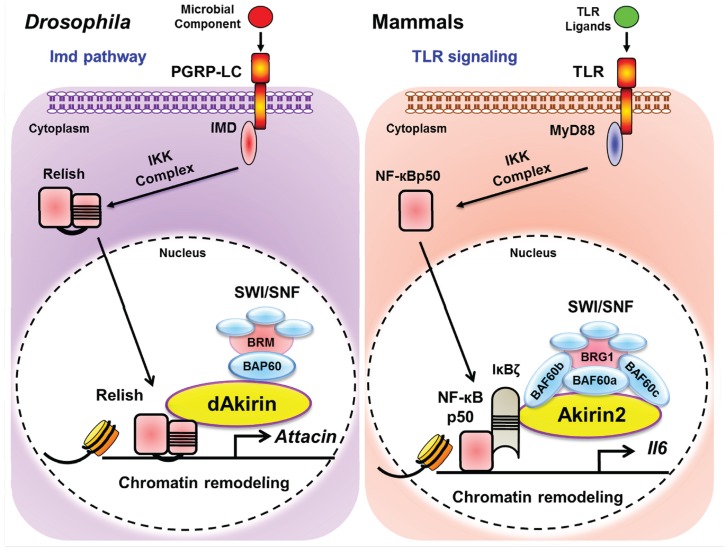
Figure 3.
Evolutionary conservation of chromatin remodeling in inflammatory gene expression. (Left Panel) The Drosophila melanogaster Imd pathway. Peptidoglycan (PGN) from Gram-negative bacteria is sensed by PGRP-LC, which activate Imd. Imd recruits IKK signaling complex, which phosphorylates Relish, triggering its cleavage. The ankyrin repeats of Relish remain in the cytoplasm, and the “REL” moiety translocates to the cytoplasm to activate transcription of target genes. Akirin forms a complex with Bap60 (component of BRM complex) and facilitate the recruitment of whole complex to the promoters of its target genes. (Right Panel) The mammalian TLR signaling pathway. Activation of TLRs by their respective ligands leads to signaling that ultimately activates NF-κB. Similar to the process of the Imd pathway, IKK complex phosphorylates an inhibitory regulator of NF-κB, IκBα, resulting in release of NF-κB for translocation to the nucleus and activation of gene expression. p50 subunit of NF-κB interacts with the pre-formed complex of IκB-ζ-Akirin2-BAF60 (component of Brg1 complex) and facilitates the recruitment of SWI/SNF complex to the promoters of its target genes for their transcription.