Abstract
Background: Epigenetic mechanisms such as altered DNA methylation have been suggested to play a role in autism, beginning with the classical association of Prader-Willi syndrome, an imprinting disorder, with autistic features.
Objectives: Here we tested for the relationship of paternal sperm DNA methylation with autism risk in offspring, examining an enriched-risk cohort of fathers of autistic children.
Methods: We examined genome-wide DNA methylation (DNAm) in paternal semen biosamples obtained from an autism spectrum disorder (ASD) enriched-risk pregnancy cohort, the Early Autism Risk Longitudinal Investigation (EARLI) cohort, to estimate associations between sperm DNAm and prospective ASD development, using a 12-month ASD symptoms assessment, the Autism Observation Scale for Infants (AOSI). We analysed methylation data from 44 sperm samples run on the CHARM 3.0 array, which contains over 4 million probes (over 7 million CpG sites), including 30 samples also run on the Illumina Infinium HumanMethylation450 (450K) BeadChip platform (∼485 000 CpG sites). We also examined associated regions in an independent sample of post-mortem human brain ASD and control samples for which Illumina 450K DNA methylation data were available.
Results: Using region-based statistical approaches, we identified 193 differentially methylated regions (DMRs) in paternal sperm with a family-wise empirical P-value [family-wise error rate (FWER)] <0.05 associated with performance on the Autism Observational Scale for Infants (AOSI) at 12 months of age in offspring. The DMRs clustered near genes involved in developmental processes, including many genes in the SNORD family, within the Prader-Willi syndrome gene cluster. These results were consistent among the 75 probes on the Illumina 450K array that cover AOSI-associated DMRs from CHARM. Further, 18 of 75 (24%) 450K array probes showed consistent differences in the cerebellums of autistic individuals compared with controls.
Conclusions: These data suggest that epigenetic differences in paternal sperm may contribute to autism risk in offspring, and provide evidence that directionally consistent, potentially related epigenetic mechanisms may be operating in the cerebellum of individuals with autism.
Keywords: Epigenetics, DNA methylation, sperm, autism risk
Key Messages.
In a high-risk autism spectrum disorder (ASD) birth cohort, paternal sperm genome-wide DNA methylation patterns are associated with 12-month ASD-related phenotypes.
Observations were validated in a partially independent set of fathers using a second method of DNA methylation assessment.
Regions of altered sperm DNA methylation in fathers associated with infant ASD phenotypes included overlapping genes previously associated with Prader-Willi syndrome.
A significant subset of sperm DMRs associated with ASD phenotypes showed directionally consistent methylation changes in post-mortem cerebellar tissue of ASD patients compared with controls.
Introduction
Autism spectrum disorder (ASD) is a complex neurodevelopmental disorder characterized by deficits in communication and social behaviours, as well as stereotypic movements.1 Recent estimates from the Autism and Developmental Disabilities Monitoring (ADDM) Network report the prevalence of ASD among 8-year-old US children to be 1 in 68.2 The recurrence rate of ASD among children with an older affected sibling is estimated at 18.7% [95% confidence interval (CI) = 13.3% to 25.5%] in the Baby Siblings Research Consortium,3 representing 10-fold enrichment over the general population. Neuroanatomy evidence,4,5,6 teratogen exposure studies7,8 and very early life behaviour differences9 suggest that ASD neurobiology likely begins during the in utero period. Intervention research demonstrates early identification and diagnosis lead to better treatment outcomes for children with ASD,10 thus identification of prenatal or early life ASD risk factors are critical to disease prevention or mitigation.
The aetiology of ASD is incompletely characterized, and recent heritability estimates suggest approximately equal contribution from genetic and environmental risk factors.11,12 Several large genome-wide studies have identified de novo and inherited genetic risk factors for ASD,13,14 yet there appears to be high genetic heterogeneity and much of the overall ASD risk has not yet been explained.15 Similarly, evidence of prenatal toxicant risk factors for ASD is growing, but is inconsistent to date.16,17 Parental characteristics may be associated with ASD risk; numerous studies have observed associations between advanced paternal age,18 maternal age19 or both.20 Parental age, particularly paternal age, has been implicated in de novo genetic findings for ASD21,22 and has been associated with brain expression of particular genes in offspring.23 Other paternal risk factors might also be important, however. In vitro fertilization by intracytoplasmic sperm injection is associated with an increased frequency of autism that is specific for method, with a 4.6-fold increased risk after surgical sperm extraction and > 9-fold if also associated with preterm birth.24 Consideration of combined genetic, environmental and demographic factors is likely to yield further insights into ASD aetiology.
Epigenetics, defined as genomic modifications heritable during cell division other than DNA sequence per se, such as DNA methylation (DNAm), represents an important intersection of genetic and environmental influences that may be particularly relevant for ASD. DNAm alterations have been observed in multiple regions of post-mortem brains of individuals with ASD compared with controls25 and between blood samples from discordant ASD twins.26 Epigenetic changes to the developing fetus may underscore risk of ASD,27 and nutritional deprivation can impact on the male germline with epigenetic changes to the offspring.28
In the current project, we examine genome-wide DNAm in paternal semen biosamples obtained from an ASD enriched-risk birth cohort to estimate associations between sperm DNAm and prospective ASD development, using a 12-month ASD symptoms assessment. We used two DNAm measurement approaches with complementary types of information. We lastly examined regions showing association with early autism symptoms in an independent sample of post-mortem human brain ASD and control samples.25
Methods
Study sample
Semen samples (n = 44) were obtained from fathers of families enrolled in the Early Autism Risk Longitudinal Investigation (EARLI), an autism enriched-risk pregnancy cohort that focuses on the prenatal and early life periods of newborns who have biological siblings already diagnosed with an ASD.29 For diseases such as ASD that demonstrate familial aggregation, disease-specific familial enriched-risk cohort studies represent an attractive alternative to large birth cohorts.30 In enriched-risk studies, subjects are over-sampled for increased familial risk, increasing power to detect associations between risk factors and disease.29 The EARLI study is comprised of four data collection sites (Drexel University, Johns Hopkins University, University of California, Davis, and Kaiser Permanente Division of Research) and includes prospective collection of biological samples from multiple family members along with environmental, questionnaire, interview and clinical assessment data collected at multiple time points throughout the pregnancy and through age 3 years of the newborn sibling.29 The EARLI study was reviewed and approved by Human Subjects Institutional Review Boards (IRBs) from each of the four study sites.
The samples analysed within this project were collected from biological fathers of infant siblings using a home-collection semen kit distributed at the time of enrolment, typically in the first or second trimester of pregnancy. Semen collection kits with instructions were mailed to the father of the current pregnancy prior to the first prenatal visit. The goal was to collect the semen sample as close to the time of the family’s enrolment in EARLI as possible, but samples were accepted any time during the study. Paternal demographic information was collected via the EARLI paternal interview (or, if unavailable, from the maternal interview). Paternal age for each biosample was calculated by subtracting the father’s date of birth from the date of sample collection.
A total of 44 sperm DNA samples were available for CHARM DNA methylation (DNAm) analysis, and an overlapping validation set of 30 samples with DNAm assessment via the Illumina 450 K platform (see methods below) was available. All 44 individuals had 12-month developmental assessment data from the child available.
Phenotype assessment
EARLI siblings participated in extensive neurodevelopmental phenotyping at regular intervals during development, including clinician assessment and parent report. The 12-month clinical assessment included the Autism Observation Scale for Infants (AOSI) tool,31 an 18-item clinician-administered, semi-structured, play-based observation that is targeted for high-risk infants. It assesses eye contact, visual tracking, imitation, atypical sensory behaviour and social babbling (among other behaviours), many of which have been associated with later ASD.32 AOSI scores at 12 months have been shown to predict Autism Diagnostic Observation Schedule (ADOS) classification at 249 and 36 months33 and are thus used in this analysis as an early quantitative indicator of ASD-related phenotype in at-risk siblings.
Laboratory analyses
Sample processing and DNA extraction
Semen samples were frozen upon collection as per the EARLI collection kit instructions and then shipped (either same or next day) directly to the Johns Hopkins Biosample Repository (JHBR) for storage (-80°C) until processing. Genomic DNA for the 44 samples used in CHARM analyses and for 26 of the samples used in 450 K analyses was extracted from semen samples using a QIAgen QIAsymphony automated workstation with the Blood 1000 protocol of the DSP DNA Midi kit (Cat. No. 937255, Qiagen, Valencia, CA) as per manufacturer’s instructions. For the additional four samples measured via 450 K analyses, DNA was extracted at the Johns Hopkins School of Medicine Center for Epigenetics using a version of the Epicentre MasterPure DNA Purification Kit (Cat. No. MCD85201).
CHARM DNA methylation measurement
Genome-wide sperm DNA methylation was measured using the Comprehensive High-throughput Arrays for Relative Methylation assay (CHARM).34 Genomic sperm DNA (4 µg) was sheared on a HydroShear Plus (DigiLab, Marlborough, MA), digested with McrBC, gel-purified, labelled and hybridized to arrays as described.34 The CHARM 3.0 array (Roche NimbleGen, Madison, WI) includes over 4 million probes and covers over 7 million CpGs arranged into probe groups (consecutive probes are within 300 bp of each other). These arrays include probes covering all annotated and non-annotated promoters and microRNA sites in addition to features present in the original CHARM method.34
Illumina 450K DNA methylation measurement
DNAm for 30 overlapping paternal semen samples was measured via the Illumina Infinium HumanMethylation450 BeadChip assay (Illumina, San Diego, CA). Genomic DNA (1 µg) was processed by the Johns Hopkins University SNP Center using the automated Infinium workflow. Technical control samples, including technical replicates of liver and placenta, and spike-in samples (here, 0%, 50% and 100% methylated commercial DNA) were repeated across plates to ensure consistent high-quality data. Note these samples were not included in any subsequent analyses of semen.
Statistical analyses
Data pre-processing
CHARM raw data were pre-processed using the CHARM package (v.2.8.0)35 in R (version 3.0.3). Briefly, probe-level percentage DNAm estimates were obtained by first removing background signal using anti-genomic background probes, followed by normalization using control probes (loess for within arrays, followed by quantile between arrays). After normalization, we excluded background, control and repetitive probe groups, yielding 3 811 046 total probes per array for each of the 44 discovery samples. Surrogate variable analysis (SVA) was then performed on these percentage methylation estimates to estimate latent factors influencing DNAm levels which typically represent ‘batch’ effects.36 We estimated the number of surrogate variables (SVs) to include in our statistical models using the Buja and Eyuboglu (‘be’) algorithm, which identifies how many latent variables are present in the data.37 Then the SVA algorithm estimates these SVs which are adjusted for as confounders in downstream differential methylation bumphunting analysis.
Validation 450 K raw data were pre-processed using the minfi package (v.1.10.1) in R (version 3.1.0).38 Data were stratified quantile normalized and SVA was performed on the percentage methylation estimates (beta scale) to adjust for potential batch effects.
Age and developmental assessment associations
Differences in AOSI score across demographic and technical/laboratory variables were compared using F-tests. Bivariate associations between 12-month AOSI scores and ordinal variables were calculated using linear model trend tests. Similar tests were also performed to assess the degree of association between SVs and demographic/laboratory variables. Spearman rank correlations were estimated between paternal age and offspring 12-month AOSI scores.
Regional DNAm discovery
Regions of CHARM DNAm differences by infant AOSI score were identified using the ‘bump hunting’ approach previously developed for the CHARM platform,39 adjusting for estimated surrogate variables. The statistical model treated AOSI as the outcome of interest, and adjusted for paternal age and 10 surrogate variables (described above) as confounders. This model is applied to every high-quality probe on the microarray to identify the adjusted linear effect of AOSI on DNA methylation levels. Regions of differential methylation were identified by smoothing the linear effects of AOSI on DNA methylation, and then thresholding these smoothed statistics across all probes using the 99.9th percentile as a cutoff, as previously described in Jaffe et al. 2012.39 P-values for each DMR were calculated from a genome-wide empirical distribution of null statistics generated using a linear model bootstrapping approach across 10 000 permutations as described in Jaffe et al.39 We identified DMRs with a genome-wide family-wise empirical P-value [family-wise error rate (FWER)] < 0.05. Note that the DMRs are ranked prior to the permutation procedure, which is used to identify the threshold that controls for the target genome-wide FWER. Mean methylation values across the DMR for each sample were also plotted by AOSI score and Spearman rank correlation coefficients estimated.
We performed post hoc sensitivity analyses to assess the potential influence of skewed or extreme AOSI scores and the influence of race on identified DMRs. First, to account for right-skewed AOSI scores, a natural log(AOSI+1) transformation was used in regressions against the average methylation per DMR, for each DMR with P < 0.05. We also considered a dichotomous outcome comparing the highest quartile of AOSI scores (> 10) with the lowest three quartiles. These provide comparisons of effect size magnitude and direction between the discovery DMRs and these alternative forms of AOSI modelling. Bump-hunting models were also reanalysed adjusting for principal components of race (PC1 and PC2 from ancestry analysis of GWAS SNP data) and beta coefficients compared between this race-adjusted model and the primary discovery bump-hunting results.
Gene enrichment analyses
We tested for enrichment of genes within 10 kb of DMRs with FWER < 0.05 based on Gene Ontology (Biological Processes database)40 using the hypergeometric test41 restricted to gene sets with at least four members. We used the GOstats R Bioconductor package to compare genes mapped to top DMRs (FWER < 0.05) to all genes on the CHARM array that also have an Entrez ID as background.
Cross-platform validation
We attempted to validate DMRs discovered via CHARM for AOSI score using overlapping genomic coverage on the 450 K array in a partially overlapping set of paternal samples. Within these subsets of genomic sites, linear regression was first used to test the relationship between single-site 450 K DNAm and AOSI score. Statistical models were adjusted for surrogate variables estimated from the data of only overlapping samples (n = 30 samples with seven SVs for AOSI score). Spearman correlation tests were used to calculate the correlation between effect estimates from CHARM and 450K.
Comparison with autism brain data
We downloaded publicly available Illumina 450 K data from post-mortem human brain tissue on 40 samples (19 ASD and 21 controls) across three brain regions (frontal cortex, temporal cortex and cerebellum) from GEO dataset: GSE53162.25 Illumina 450 K data were normalized as above with the FlowSorted.DLPFC.450 K Bioconductor dataset to estimate cellular composition in each sample.42 We calculated mean differences and resulting t-statistics and P-values within each brain region comparing cases to controls using the limma Bioconductor package,43 and considered probes with differential methylation with P < 0.05 marginally significant for the cross-tissue comparison analysis.
Results
Study sample characteristics
Infant sibling performance on the 12-month AOSI for children of the fathers examined in the CHARM discovery sample ranged from 0 to 18. Fathers were predominantly White and non-Hispanic (73%) and their ages ranged between 27 and 51 years (Table 1). Sibling AOSI scores were not associated with paternal age (ρ = −0.04), education level or family income, though higher AOSI scores were observed at the Drexel study site.
Table 1.
Bivariate associations of AOSI score with demographic and laboratory variables for the EARLI study population
| Total 12-month AOSI score (N = 44) |
CHARM (N = 44) | Subset also with 450 K (N = 30) | ||
|---|---|---|---|---|
| Variable | Mean (SD) | P-value | N (%) | N (%) |
| Age quartile | ||||
| 27–32.7 | 6.5 (4.5) | 12 (27.3) | 9 (30) | |
| 32.8–36 | 5 (5.8) | 10 (22.7) | 7 (23.3) | |
| 36.1–40.3 | 5.7 (4) | 11 (25) | 10 (33.3) | |
| 40.2–51.2 | 5.4 (3.7) | 0.88 | 11 (25) | 4 (13.3) |
| Race, ethnicity | ||||
| Black, Hispanic | 7 (NA) | 1 (2.3) | 1 (3.3) | |
| Black, non-Hispanic | 4.7 (2.5) | 3 (6.8) | 2 (6.7) | |
| Other, Hispanic | 7.5 (0.7) | 2 (4.5) | 1 (3.3) | |
| Other, non- Hispanic | 5 (4.2) | 2 (4.5) | 1 (3.3) | |
| White, Hispanic | 3 (NA) | 1 (2.3) | 1 (3.3) | |
| White, non- Hispanic | 5.4 (4.5) | 32 (72.7) | 22 (73.3) | |
| White, other | 12 (8.5) | 0.63 | 2 (4.5) | 2 (6.7) |
| Missing | 4 (NA) | 1 (2.3) | - | |
| Study site | ||||
| Drexel | 8.5 (5.5) | 14 (31.8) | 10 (33.3) | |
| Johns Hopkins | 3.6 (2.8) | 15 (34.1) | 9 (30) | |
| Kaiser | 4.3 (3.8) | 7 (15.9) | 6 (20) | |
| UC Davis | 5.9 (2.4) | 0.014 | 8 (18.2) | 5 (16.7) |
| Paternity status | ||||
| Proband and sibling | 5.6 (4.6) | 37 (84.1) | 24 (80) | |
| Sibling only | 5.9 (3.2) | 0.91 | 7 (15.9) | 6 (20) |
| Paternal education | ||||
| High school graduate or less | 5.5 (2.1) | 8 (18.2) | 5 (16.7) | |
| Some college | 8.2 (6.2) | 9 (20.5) | 6 (20) | |
| Bachelor's degree | 4.8 (3.8) | 13 (29.5) | 9 (30) | |
| Graduate degree | 4.9 (4.3) | 0.27 | 14 (31.8) | 10 (33.3) |
| Annual income | ||||
| < US$49,999 | 5.6 (2.3) | 9 (20.5) | 6 (20) | |
| $50,000–$74,999 | 7.6 (7.2) | 5 (11.4) | 4 (13.3) | |
| $75,000–$99,999 | 5.5 (4.8) | 13 (29.5) | 10 (33.3) | |
| >=$100,000 | 5.4 (4.2) | 0.79 | 17 (38.6) | 10 (33.3) |
| Shear date | ||||
| 1 | 5.9 (4.5) | 9 (20.5) | - | |
| 2 | 5.1 (4.4) | 18 (40.9) | - | |
| 3 | 6.2 (4.4) | 0.77 | 17 (38.6) | - |
| Shear machine (HS) | ||||
| 2 | 5.6 (5.2) | 19 (43.2) | - | |
| 3 | 5.9 (3.8) | 22 (50) | - | |
| 4 | 4.7 (4.2) | 0.90 | 3 (6.8) | - |
| CHARM gel | ||||
| 1 | 0 (NA) | 1 (2.3) | - | |
| 2 | 5.2 (3.2) | 4 (9.1) | - | |
| 3 | 8 (5.1) | 4 (9.1) | - | |
| 4 | 7 (7.4) | 4 (9.1) | - | |
| 5 | 3.3 (1.5) | 3 (6.8) | - | |
| 6 | 2 (0) | 2 (4.5) | - | |
| 7 | 4.2 (3.9) | 4 (9.1) | - | |
| 8 | 8 (4) | 3 (6.8) | - | |
| 9 | 4.5 (3.5) | 2 (4.5) | - | |
| 10 | 8.3 (8.5) | 3 (6.8) | - | |
| 11 | 7.2 (4.9) | 4 (9.1) | - | |
| 12 | 3 (1) | 3 (6.8) | - | |
| 13 | 7.8 (2.2) | 4 (9.1) | - | |
| 14 | 3.7 (1.5) | 0.65 | 3 (6.8) | - |
| Gel location | ||||
| Bottom middle | 2 (NA) | 1 (2.3) | - | |
| Bottom left | 4.6 (4.5) | 12 (27.3) | - | |
| Bottom right | 6.9 (2.9) | 8 (18.2) | - | |
| Top left | 5.8 (5.6) | 12 (27.3) | - | |
| Top right | 6.2 (3.8) | 0.71 | 11 (25) | - |
| Hybridization date | ||||
| 1 | 5 (4.5) | 20 (45.5) | 20 (66.7) | |
| 2 | 6.2 (4.3) | 0.39 | 24 (54.5) | 8 (26.7) |
| 3 | - | - | 2 (6.7) | |
NA, not applicable.
Methylation data quality assessment
Total AOSI did not vary by CHARM DNA shearing date (P-value = 0.77), HydroShear machine (P-value = 0.9), CHARM gel (P-value = 0.65), sample location in CHARM gel (P-value = 0.71) or CHARM hybridization date (P-value = 0.39). Supplementary Table 1 (available as Supplementary data at IJE online) shows the degree of association between CHARM variables to each of the estimated surrogate variables.
Illumina 450 K between-plate correlation Pearson coefficients of unnormalized DNAm between technical control replicates ranged from 0.848 to 0.996 with a mean [standard deviation (SD)] of 0.963 (0.041). DNA samples were hybridized to Illumina arrays across three dates and AOSI score did vary by hybridization date (P-value = 0.06). Supplementary Table 2 (available as Supplementary data at IJE online) shows the degree of association between 450 K variables to each of the estimated surrogate variables.
DNAm and AOSI score
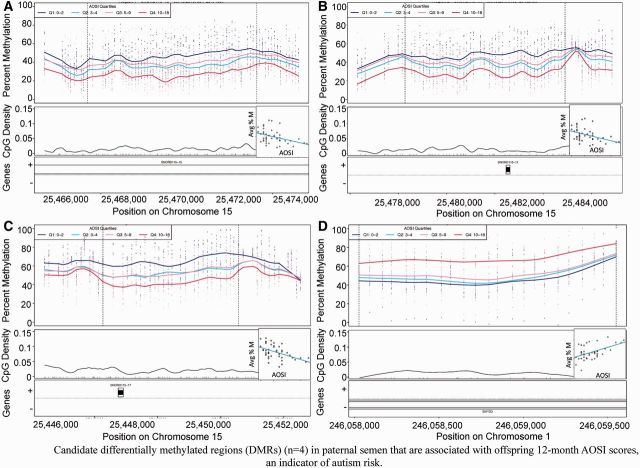
We identified 2605 candidate DMRs using our bump-hunting method in paternal sperm associated with child AOSI score at 12 months and, after permutation analysis, the top193 DMRs had genome-wide P < 0.05. The top 10 ranked DMRs are shown in Table 2, and Supplementary Table 3 (available as Supplementary data at IJE online) includes the complete list of 193 DMRs. Methylation plots by AOSI score for all 193 DMRs are shown in Supplementary Figure 1 (available as Supplementary data at IJE online). We highlight the top four regions where DNAm associates linearly (genome-wide P = 0.001) with AOSI score in Figure 1, with the average DNAm across the DMR shown in each inset. Figure 1a shows hypomethylation with increased AOSI score on chromosome 15 overlapping SNORD115-15 where the highest quartile of AOSI scores (≥ 10) corresponded to 27.5% average sperm methylation compared with 46.9% average methylation for the lowest quartile (<3). Similarly, Figure 1b and c shows hypomethylation with increased AOSI scores on chromosome 15 covering SNORD115-11 and SNORD115-17, respectively, with differences in regional average methylation between the highest and lowest AOSI quartiles of 19.7% and 22.6%, respectively. Figure 1d illustrates hypermethylation with increased AOSI scores on chromosome 1 overlapping SMYD3, with 68.9% average methylation for the highest AOSI score quartile compared with 47.3% average methylation of the lowest quartile.
Table 2.
Candidate differentially methylated regions (DMRs) associated with 12-month total AOSI score, an indicator of autism risk
| Rank | Chr | Start | End | # Probes | Average MethylationQ1 | Avg.Meth.Q4 | Q4-Q1 difference | Smoothed effect estimate | FWER* P-value | Gene symbol | Location relative to gene | Distance from transcription start site |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 15 | 25467684 | 25471631 | 51 | 0.469 | 0.275 | −0.194 | −0.0130 | 0 | SNORD115-15 | Inside intron | 16 276 |
| 2 | 15 | 25479 139 | 25482453 | 44 | 0.478 | 0.281 | −0.197 | −0.0137 | 0 | SNORD115-11 | Covers | 0 |
| 3 | 15 | 25448132 | 25450278 | 27 | 0.652 | 0.426 | −0.226 | −0.0161 | 0 | SNORD115-17 | Covers | 0 |
| 4 | 1 | 246058026 | 246059550 | 23 | 0.473 | 0.689 | 0.216 | 0.0181 | 0 | SMYD3 | Inside intron | 521160 |
| 5 | 7 | 100678945 | 100682385 | 32 | 0.643 | 0.500 | −0.143 | −0.0126 | 0 | MUC17 | Inside exon | 15582 |
| 6 | 12 | 114527770 | 114529240 | 21 | 0.485 | 0.252 | −0.233 | −0.0192 | 0 | RBM19 | Upstream | 123595 |
| 7 | 4 | 89679863 | 89681417 | 23 | 0.625 | 0.379 | −0.245 | −0.0173 | 0 | FAM13A | Covers exon(s) | 62984 |
| 8 | 4 | 22389344 | 22391462 | 31 | 0.559 | 0.439 | −0.119 | −0.0124 | 0 | GPR125 | Covers exon(s) | 126209 |
| 9 | 4 | 10167294 | 10169448 | 32 | 0.490 | 0.353 | −0.137 | −0.0119 | 0 | WDR1 | Upstream | 48722 |
| 10 | 15 | 25445204 | 25447276 | 28 | 0.665 | 0.492 | −0.174 | −0.0132 | 1.00E-04 | SNORD115-17 | Covers | 0 |
Chr, Chromosome
*Genome-wide family-wise empirical P-value.
Figure 1.
Methylation plots for the top four statistical DMRs (P < 1.0 x 10−4) identified using CHARM and 12-month AOSI score. (a) SNORD115-15, (b) SNORD115-15, (c) SNORD115-17, (d) SMYD3. Top panels show individual methylation levels at each probe by genomic position. Dotted vertical lines represent the boundaries of the DMR, and coloured lines represent the average methylation curve for samples grouped by quartiles of AOSI scores—the scores within each quartile are shown in the legend. Middle panel shows location of CpG dinucleotides (as black tick marks) and CpG density by genomic position (black curved line). Bottom panel shows location (boxes) and direction (+ or −) of refseq-annotated genes. Inset scatterplot reflects linear regression of average methylation across all probes within DMR per sample by AOSI 12-month score.
An inverse relationship between AOSI and DNAm was observed at 141 (73%) of these regions. These DMRs overlapped genes (87 were located within 10 kb of a gene) that were enriched for gene ontology biological processes of cellular movement, neurogenesis and neuronal development categories (Table 3). Sensitivity analysis involving linear regression on log-transformed AOSI scores [ln(AOSI+1)] yielded consistent results with the bump-hunting performed on the linear AOSI models described above—all 193 DMRs matched directionality with regression statistics and 144/193 regions (74.6%) had P < 0.05. Similarly, when performing linear regression using a model of the highest quartile of AOSI score relative to the other three quartiles, the results were also consistent with the primary analyses—all 193 DMRs matched directionality with regression statistics and 142/193 regions (73.6%) had P < 0.05. Results from bump-hunting analyses that adjusted for principal components of race were not substantially different from those reported. Beta coefficients for DMRs before and after race adjustment were highly correlated (ρ = 0.99, Supplementary Figure 3, available as Supplementary data at IJE online).
Table 3.
Gene ontology, biological processes enriched in differentially methylated regions associated with 12-month AOSI score
| Rank | GOBPID | P-value | Odds ratio | Expected Count | Count | Size | Term |
|---|---|---|---|---|---|---|---|
| 1 | GO:0006928 | 0.00027 | 3.55 | 5.0 | 14 | 1495 | Cellular component movement |
| 2 | GO:0040011 | 0.00035 | 3.61 | 4.5 | 13 | 1344 | Locomotion |
| 3 | GO:0030155 | 0.00045 | 6.86 | 1.0 | 6 | 297 | Regulation of cell adhesion |
| 4 | GO:0007155 | 0.00139 | 3.57 | 3.3 | 10 | 989 | Cell adhesion |
| 5 | GO:0022610 | 0.00143 | 3.56 | 3.3 | 10 | 993 | Biological adhesion |
| 6 | GO:0051056 | 0.00161 | 5.31 | 1.3 | 6 | 380 | Regulation of small gtpase mediated signal transduction |
| 7 | GO:0022008 | 0.00226 | 3.15 | 4.2 | 11 | 1242 | Neurogenesis |
| 8 | GO:0048011 | 0.00231 | 5.96 | 0.9 | 5 | 278 | Neurotrophin TRK receptor signaling pathway |
| 9 | GO:0038179 | 0.00242 | 5.89 | 0.9 | 5 | 281 | Neurotrophin signaling pathway |
| 10 | GO:0007626 | 0.00244 | 7.77 | 0.6 | 4 | 169 | Locomotory behavior |
| 11 | GO:0008543 | 0.00325 | 7.15 | 0.6 | 4 | 183 | Fibroblast growth factor receptor signaling pathway |
| 12 | GO:0044344 | 0.00462 | 6.46 | 0.7 | 4 | 202 | Cellular response to fibroblast growth factor stimulus |
| 13 | GO:0071774 | 0.00478 | 6.39 | 0.7 | 4 | 204 | Response to fibroblast growth factor |
| 14 | GO:0048699 | 0.00485 | 2.96 | 3.9 | 10 | 1173 | Generation of neurons |
| 15 | GO:0007264 | 0.00618 | 3.99 | 1.7 | 6 | 499 | Small gtpase mediated signal transduction |
| 16 | GO:0007399 | 0.00647 | 2.52 | 6.2 | 13 | 1848 | Nervous system development |
| 17 | GO:0051235 | 0.00653 | 5.83 | 0.7 | 4 | 223 | Maintenance of location |
| 18 | GO:0007173 | 0.00663 | 5.80 | 0.8 | 4 | 224 | Epidermal growth factor receptor signaling pathway |
| 19 | GO:0009894 | 0.00687 | 3.47 | 2.3 | 7 | 675 | Regulation of catabolic process |
| 20 | GO:0007265 | 0.00695 | 5.72 | 0.8 | 4 | 227 | Ras protein signal transduction |
| 21 | GO:0038127 | 0.00706 | 5.70 | 0.8 | 4 | 228 | ERBB signaling pathway |
| 22 | GO:0007268 | 0.00726 | 3.44 | 2.3 | 7 | 682 | Synaptic transmission |
| 23 | GO:0007411 | 0.00748 | 4.47 | 1.2 | 5 | 367 | Axon guidance |
| 24 | GO:0097485 | 0.00748 | 4.47 | 1.2 | 5 | 367 | Neuron projection guidance |
| 25 | GO:0051674 | 0.00802 | 3.37 | 2.3 | 7 | 695 | Localization of cell |
| 26 | GO:0050790 | 0.00841 | 2.50 | 5.7 | 12 | 1688 | Regulation of catalytic activity |
| 27 | GO:0030182 | 0.00891 | 2.84 | 3.6 | 9 | 1081 | Neuron differentiation |
GOBPID, Gene Ontology Biological Processes ID.
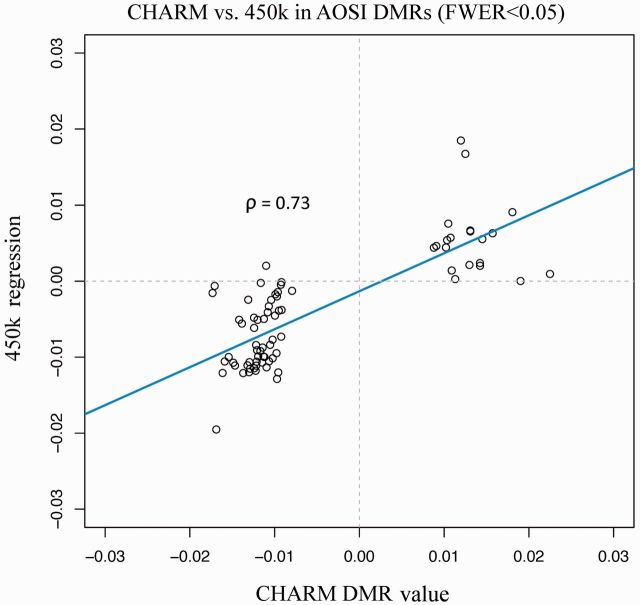
Among these 193 AOSI-associated DMRs, we extracted Illumina 450 K probes located within 500 bp of the region boundaries. This was possible for75 (38.9%) CHARM-discovered DMRs (see Supplementary Table 4, available as Supplementary data at IJE online) represented by 244 probes on the 450 K array. The direction of association between AOSI and DNAm was consistent with CHARM results for 74 (98.7%) of these 75 regions (Figure 2).
Figure 2.
Validation of CHARM AOSI DMR values using 450K data from overlapping samples (n = 30).The CHARM DMR value corresponds to the smoothed effect estimate at each probe, plotted against the single-site regression coefficients from 450 K data for each corresponding DMR. Spearman rank correlation coefficient is also included.
AOSI-associated DMRs in post-mortem brain
A subset of the AOSI-associated DMRs identified in semen also shows directionally consistent effects in post-mortem human brain samples in a study comparing patients with ASD with unaffected controls. The AOSI DMRs were most consistent comparing cases and controls in the cerebellum, where 21 of 75 of AOSI-associated DMRs contained at least one Illumina 450 K probe that was differentially methylated comparing autism with controls at P-value < 0.05; 18 of these were directionally consistent with the sperm AOSI score association data (e.g. DNAm positively associated with both AOSI score in semen and in ASD brains). This cross-tissue consistency was not present in the cortical brain regions—in the frontal cortex, only 12 AOSI DMRs were significant, of which 3 were directionally consistent, and in the temporal cortex, only 10 AOSI DMRs were significant, of which 2 were directionally consistent. Surprisingly, 15 of 18 cerebellum-consistent AOSI DMRs were overlapping genes in the SNORD families (Supplementary Table 5, available as Supplementary data at IJE online).
Discussion
In summary, we observed a strong relationship between DNA methylation in sperm from fathers and 12-month ASD-related phenotype in infant siblings within an autism enriched-risk birth cohort. The degree of methylation change was quite substantial compared with recent reports for other epigenome-wide association studies (EWAS): 6.2–30.7% difference between top and bottom quartiles of AOSI score among the 193 DMRs with FWER < 0.05. These AOSI-associated DMRs showed enrichment for genes involved in neurogenesis and more general neuronal development.
We focused on 12-month infant developmental assessment as a measure of early ASD-related phenotype. Total scores on the AOSI administered to high-risk infants at 12 months of age has shown high inter-rater reliability (0.93), test-retest reliability (0.61) and correlation with ASD diagnosis at age 3 years.31 Because of the unusual nature of this prospective high-risk cohort, direct replication is difficult. However, an independent data set of autism and control brains showed overlap with the AOSI-associated DMRs, with 18 of 21 genes directionally consistent in methylation values.
How might these DMRs contribute to ASD? The sperm contributes half of the genome to somatic cells, and after germline reprogramming (except for generating germ cells in the next generation), this would likely persist into brain development.44 The fact that these genes were enriched for neurological function is consistent with this idea. Remarkably, 15 members of the small nucleolar RNA, C/D Box (SNORD-115) genes were in this overlapping set. This gene cluster lies within the Prader-Willi critical region of chromosome 15 and are considered candidate mechanistic targets for the disorder,45,46 a canonical imprinted Mendelian disorder with autism features.47 Gene dosage alterations in this region are also associated with ASD.48,49 The data reported here suggest that paternal epigenetic changes of this gene cluster confer risk of ASD among offspring, at least among those with an older affected sibling. Molecularly, aberrant imprinting, as seen in Prader-Willi syndrome, would not explain these results as the paternal allele is normally expressed.
The association with the cerebellum is intriguing. A case report of twins discordant for ASD showed cerebellar anatomical changes in the affected twin,49 and some neuro-imaging studies have shown changes in the cerebellum in autism, or autistic features in children with cerebellar defects.50,51
One potential limitation of the study is that DNA was extracted from semen rather than purified sperm, so we can not absolutely rule out some partial cell type confounding. However, DNA methylation has been shown to be highly uniform regardless of the quality of sperm subpopulations.52,53 Thus, we did not adjust for sperm quality or subpopulations. Also, there has been recent increased awareness of possible bias in enrichment statistics when gene size is not considered.54 We found no difference between gene size (P = 0.055), coding length (P = 0.79) and GC content (P = 0.1) comparing DMRs with FWER < 0.05 (n = 193) with those with FWER > 0.05 (n = 2412). Further, the average gene length was actually shorter for AOSI DMRs in our results (144 kb vs 178 kb). Additionally, the CHARM design features sets of probe groups, each with very evenly spaced probes (56–90 bp), which was designed solely to target regions of at least moderate CpG density in the genome in an annotation-agnostic manner. Note this is different from the more promoter-focused designs, where probe density has been shown to bias gene set analysis. The CHARM method is less quantitative at individual loci, due to the use of a restriction enzyme and how signal is measured. However, we have previously shown that it is more powerful for identifying relative changes in DNAm at the region level.55
The prevalence of autism spectrum disorder (ASD) has been increasing in recent years, but even the current estimate of 1–2%2 makes study of a general population prospective pregnancy cohort not feasible for ASD. For diseases such as ASD that demonstrate familial aggregation, disease-specific enriched-familial risk cohort studies are a commonly employed design.30 There is a long history of enriched-risk (family-based) cohort studies in epidemiology29,56,57,58 to examine risk factors for rare diseases. These studies are not designed to be generalizable to the general population, though targeted results from an enriched-risk cohort can be replicated in a representative population to test generalizability. Nevertheless, in our study, fathers of second children with high and low AOSI scores are from the same underlying EARLI enriched-risk population, and thus effect estimates of association are not distorted due to selection bias, but are in fact internally valid.
In summary, we have identified relatively large inter-individual differences in paternal sperm DNAm that associate with later 12-month ASD-related phenotype in their biological offspring, among a sample of autism enriched-risk infant siblings. Many of these regions of paternal sperm DNAm were also associated with ASD in cerebellum brain samples. Further, the genes implicated are enriched for neurodevelopment and include regions implicated in Prader-Willi syndrome. This unique set of biosamples is comparatively small, given the nature of the biosample type and required perinatal collection timing in an enriched-risk cohort. Thus it is important to examine these findings in larger samples to confirm these associations and explore mechanistic implications.
Supplementary Data
Supplementary data are available at IJE online.
Funding
Funding for this work was provided by NIH R01 ES017646 (PIs MDF and APF), NIH R01 ES16443 (CJN) and Autism Speaks.
Supplementary Material
Acknowledgements
We thank Stacey Cayetano, Samantha Bragan and David Kelly of JHBR for EARLI sample processing/handling and for DNA extractions, and Roxann Ashworth of CIDR for coordinating the methylation assays.
Conflict of interest: None declared.
References
- 1.King BH, Navot N, Bernier R, Webb SJ. Update on diagnostic classification in autism. Current opinion in psychiatry. 2014 Mar;27(2):105–9. PubMed PMID: 24441420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. Prevalence of autism spectrum disorder among children aged 8 years - Autism and Developmental Disabilities Monitoring network, 11 sites, USA, 2010. MMWR Surveillance Summaries. 2014;63(SS02):1–21. [PubMed] [Google Scholar]
- 3.Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011 Sep;128(3):e488–95. PubMed PMID: 21844053. Pubmed Central PMCID: 3164092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005 Apr-May;23(2-3):183–7. PubMed PMID: 15749244. Epub 2005/03/08. eng. [DOI] [PubMed] [Google Scholar]
- 5.Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: developmental anomalies of the cranial nerve motor nuclei. J Comp Neurol. 1996 Jun 24;370(2):247–61. PubMed PMID: 8808733. Epub 1996/06/24. eng. [DOI] [PubMed] [Google Scholar]
- 6.Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, et al. A clinicopathological study of autism. Brain. 1998 May;121 (Pt 5):889–905. PubMed PMID: 9619192. Epub 1998/06/10. eng. [DOI] [PubMed] [Google Scholar]
- 7.Stromland K, Nordin V, Miller M, Akerstrom B, Gillberg C. Autism in thalidomide embryopathy: a population study. Dev Med Child Neurol. 1994 Apr;36(4):351–6. PubMed PMID: 8157157. Epub 1994/04/01. eng. [DOI] [PubMed] [Google Scholar]
- 8.Williams G, King J, Cunningham M, Stephan M, Kerr B, Hersh JH. Fetal valproate syndrome and autism: additional evidence of an association. Dev Med Child Neurol. 2001 Mar;43(3):202–6. PubMed PMID: 11263692. [PubMed] [Google Scholar]
- 9.Zwaigenbaum L, Bryson S, Rogers T, Roberts W, Brian J, Szatmari P. Behavioral manifestations of autism in the first year of life. Int J Dev Neurosci. 2005 Apr-May;23(2-3):143–52. PubMed PMID: 15749241. Epub 2005/03/08. eng. [DOI] [PubMed] [Google Scholar]
- 10.Tonge BJ, Bull K, Brereton A, Wilson R. A review of evidence-based early intervention for behavioural problems in children with autism spectrum disorder: the core components of effective programs, child-focused interventions and comprehensive treatment models. Current opinion in psychiatry. 2014 Mar;27(2):158–65. PubMed PMID: 24452070. [DOI] [PubMed] [Google Scholar]
- 11.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA: the journal of the American Medical Association. 2014 May 7;311(17):1770–7. PubMed PMID: 24794370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of general psychiatry. 2011 Nov;68(11):1095–102. PubMed PMID: 21727249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, State MW, et al. The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012 Dec 20;76(6):1052–6. PubMed PMID: 23259942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Current opinion in genetics & development. 2012 Jun;22(3):229–37. PubMed PMID: 22463983. [DOI] [PubMed] [Google Scholar]
- 15.Miles JH. Autism spectrum disorders–a genetics review. Genetics in medicine: official journal of the American College of Medical Genetics. 2011 Apr;13(4):278–94. PubMed PMID: 21358411. [DOI] [PubMed] [Google Scholar]
- 16.Herbert MR. Contributions of the environment and environmentally vulnerable physiology to autism spectrum disorders. Current opinion in neurology. 2010 Apr;23(2):103–10. PubMed PMID: 20087183. [DOI] [PubMed] [Google Scholar]
- 17.Hertz-Picciotto I. Environmental risk factors in autism: Results from large-scale epidemiologic studies. In: Amaral DG, Dawson G, Geschwind D, editors. Autism spectrum disorders . New York: Oxford University Press; 2011. p. 827–62. [Google Scholar]
- 18.Hultman CM, Sandin S, Levine SZ, Lichtenstein P, Reichenberg A. Advancing paternal age and risk of autism: new evidence from a population-based study and a meta-analysis of epidemiological studies. Mol Psychiatry. 2011 Dec;16(12):1203–12. PubMed PMID: 21116277. Epub 2010/12/01. eng. [DOI] [PubMed] [Google Scholar]
- 19.Sandin S, Hultman CM, Kolevzon A, Gross R, MacCabe JH, Reichenberg A. Advancing maternal age is associated with increasing risk for autism: a review and meta-analysis. Journal of the American Academy of Child and Adolescent Psychiatry. 2012 May;51(5):477–86 e1. PubMed PMID: 22525954. [DOI] [PubMed] [Google Scholar]
- 20.Idring S, Magnusson C, Lundberg M, Ek M, Rai D, Svensson AC, et al. Parental age and the risk of autism spectrum disorders: findings from a Swedish population-based cohort. International journal of epidemiology. 2014 Feb;43(1):107–15. PubMed PMID: 24408971. [DOI] [PubMed] [Google Scholar]
- 21.Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell reports. 2014 Oct 9;9(1):16–23. PubMed PMID: 25284784. Pubmed Central PMCID: 4194132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 May 10;485(7397):246–50. PubMed PMID: 22495309. Pubmed Central PMCID: 3350576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith RG, Fernandes C, Kember R, Schalkwyk LC, Buxbaum J, Reichenberg A, et al. Transcriptomic changes in the frontal cortex associated with paternal age. Molecular autism. 2014;5(1):24 PubMed PMID: 24655730. Pubmed Central PMCID: 3998024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandin S, Nygren KG, Iliadou A, Hultman CM, Reichenberg A. Autism and mental retardation among offspring born after in vitro fertilization. JAMA: the journal of the American Medical Association. 2013 Jul 3;310(1):75–84. PubMed PMID: 23821091. [DOI] [PubMed] [Google Scholar]
- 25.Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2014 Aug;19(8):862–71. PubMed PMID: 23999529. Pubmed Central PMCID: 4184909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2014 Apr;19(4):495–503. PubMed PMID: 23608919. Pubmed Central PMCID: 3906213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaSalle JM. Epigenomic strategies at the interface of genetic and environmental risk factors for autism. Journal of human genetics. 2013 Jul;58(7):396–401. PubMed PMID: 23677056. Pubmed Central PMCID: 3955092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014 Aug 15;345(6198):1255903 PubMed PMID: 25011554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newschaffer CJ, Croen LA, Fallin MD, Hertz-Picciotto I, Nguyen DV, Lee NL, et al. Infant siblings and the investigation of autism risk factors. Journal of neurodevelopmental disorders. 2012;4(1):7 PubMed PMID: 22958474. Pubmed Central PMCID: 3436647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopper JL. Disease-specific prospective family study cohorts enriched for familial risk. Epidemiologic perspectives & innovations: EP+I. 2011;8(1):2 PubMed PMID: 21352566. Pubmed Central PMCID: 3055804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryson SE, Zwaigenbaum L, McDermott C, Rombough V, Brian J. The Autism Observation Scale for Infants: scale development and reliability data. Journal of autism and developmental disorders. 2008 Apr;38(4):731–8. PubMed PMID: 17874180. [DOI] [PubMed] [Google Scholar]
- 32.Jones EJ, Gliga T, Bedford R, Charman T, Johnson MH. Developmental pathways to autism: a review of prospective studies of infants at risk. Neuroscience and biobehavioral reviews. 2014 Feb;39:1–33. PubMed PMID: 24361967. Pubmed Central PMCID: 3969297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Sasson A, Habib S, Tirosh E. Feasibility and validity of early screening for identifying infants with poor social-communication development in a well-baby clinic system. Journal of pediatric nursing. 2014 May-Jun;29(3):238–47. PubMed PMID: 24333238. [DOI] [PubMed] [Google Scholar]
- 34.Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM). Haines JL, et al. Current Protocols In Human Genetics 2010. Apr;Chapter 20:Unit 20 1 1-19. PubMed PMID: 20373514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aryee MJ, Wu Z, Ladd-Acosta C, Herb B, Feinberg AP, Yegnasubramanian S, et al. Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostatistics. 2011 Apr;12(2):197–210. PubMed PMID: 20858772. Pubmed Central PMCID: 3062148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS genetics. 2007 Sep;3(9):1724–35. PubMed PMID: 17907809. Pubmed Central PMCID: 1994707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buja A EN. Remarks on parallel analysis. Multivariate Behav Res. 1992;27:509–40. [DOI] [PubMed] [Google Scholar]
- 38.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014 May 15;30(10):1363–9. PubMed PMID: 24478339. Pubmed Central PMCID: 4016708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. International journal of epidemiology. 2012 Feb;41(1):200–9. PubMed PMID: 22422453. Pubmed Central PMCID: 3304533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao Y, Van Auken K, Li D, Arighi CN, McQuilton P, Hayman GT, et al. Overview of the gene ontology task at BioCreative IV. Database: the journal of biological databases and curation. 2014;2014. PubMed PMID: 25157073. Pubmed Central PMCID: 4142793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007 Jan 15;23(2):257–8. PubMed PMID: 17098774. [DOI] [PubMed] [Google Scholar]
- 42.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smyth GK. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor: Springer; 2005. p. 397–420. [Google Scholar]
- 44.Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005 Aug;6(8):633–42. PubMed PMID: 16136654. [DOI] [PubMed] [Google Scholar]
- 45.Bortolin-Cavaille ML, Cavaille J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic acids research. 2012 Aug;40(14):6800–7. PubMed PMID: 22495932. Pubmed Central PMCID: 3413130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girardot M, Cavaille J, Feil R. Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenetics: official journal of the DNA Methylation Society. 2012 Dec 1;7(12):1341–8. PubMed PMID: 23154539. Pubmed Central PMCID: 3528689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veltman MW, Craig EE, Bolton PF. Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatric genetics. 2005 Dec;15(4):243–54. PubMed PMID: 16314754. [DOI] [PubMed] [Google Scholar]
- 48.Bolton PF, Veltman MW, Weisblatt E, Holmes JR, Thomas NS, Youings SA, et al. Chromosome 15q11-13 abnormalities and other medical conditions in individuals with autism spectrum disorders. Psychiatric genetics. 2004 Sep;14(3):131–7. PubMed PMID: 15318025. [DOI] [PubMed] [Google Scholar]
- 49.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008 Oct 16;455(7215):919–23. PubMed PMID: 18923514. [DOI] [PubMed] [Google Scholar]
- 50.Wegiel J, Flory M, Kuchna I, Nowicki K, Ma S, Imaki H, et al. Stereological study of the neuronal number and volume of 38 brain subdivisions of subjects diagnosed with autism reveals significant alterations restricted to the striatum, amygdala and cerebellum. Acta neuropathologica communications. 2014 Sep 18;2(1):141 PubMed PMID: 25231243. Pubmed Central PMCID: 4177256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker EB, Stoodley CJ. Autism spectrum disorder and the cerebellum. International review of neurobiology. 2013;113:1–34. PubMed PMID: 24290381. [DOI] [PubMed] [Google Scholar]
- 52.Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, et al. Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PloS one. 2012;7(10):e44479 PubMed PMID: 23071498. Pubmed Central PMCID: 3467000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seshadri S, Flanagan B, Vince G, Lewis Jones DI. Leucocyte subpopulations in the seminal plasma and their effects on fertilisation rates in an IVF cycle. Andrologia. 2012 Dec;44(6):396–400. PubMed PMID: 22537602. [DOI] [PubMed] [Google Scholar]
- 54.Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics. 2013 Aug 1;29(15):1851–7. PubMed PMID: 23732277. [DOI] [PubMed] [Google Scholar]
- 55.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Research. 2008 May;18(5):780–90. PubMed PMID: WOS:000255504600010. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berg D, Marek K, Ross GW, Poewe W. Defining at-risk populations for Parkinson's disease: lessons from ongoing studies. Movement disorders: official journal of the Movement Disorder Society. 2012 Apr 15;27(5):656–65. PubMed PMID: 22508284. [DOI] [PubMed] [Google Scholar]
- 57.John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast cancer research: BCR. 2004;6(4):R375–89. PubMed PMID: 15217505. Pubmed Central PMCID: 468645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev 2007;16:2331–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.