Abstract
Background: Populations in north central China are at high risk for oesophageal squamous cell carcinoma (ESCC) and gastric cancer (GC), and genetic variation in epigenetic machinery genes and pathways may contribute to this risk.
Methods: We used the adaptive multilocus joint test to analyse 192 epigenetic genes involved in chromatin remodelling, DNA methylation and microRNA biosynthesis in 1942 ESCC and 1758 GC cases [1126 cardia (GCA) and 632 non-cardia adenocarcinoma (GNCA)] and 2111 controls with Chinese ancestry. We examined potential function of risk alleles using in silico and expression quantitative trait loci (eQTLs) analyses.
Results: Suggestive pathway-based associations were observed for the overall epigenetic (P-valuePATH = 0.034) and chromatin remodelling (P-valuePATH = 0.039) pathways with risk of GCA, but not GC, GNCA or ESCC. Overall, 37 different epigenetic machinery genes were associated with risk of one or more upper gastrointestinal (UGI) cancer sites (P-valueGENE < 0.05), including 14 chromatin remodelling genes whose products are involved in the regulation of HOX genes. We identified a gastric eQTL (rs12724079; rho = 0.37; P = 0.0006) which regulates mRNA expression of ASH1L. Several suggestive eQTLs were also found in oesophageal (rs10898459 in EED), gastric cardia (rs7157322 in DICER1; rs8179271 in ASH1L), and gastric non-cardia (rs1790733 in PPP1CA) tissues.
Conclusions: Results of our analyses provide limited but suggestive evidence for a role of epigenetic gene variation in the aetiology of UGI cancer.
Keywords: Epigenetics, chromatin remodelling, DNA methylation, microRNA, oesophageal squamous cell carcinoma, gastric cancer, gastric cardia, gastric non-cardia, SNP, gene-based, pathway-based
Introduction
Gastric cancer (GC) is the third and oesophageal cancer (EC) is the sixth leading causes of cancer mortality in the world.1 Despite marked declines in recent decades, around half of the total GC and EC deaths in the world occur in China.1 Populations in north central China are at particularly high risk for both of these cancers. GCs here occur primarily in the cardia, and essentially all ECs are oesophageal squamous cell carcinomas (ESCC).2 Although Helicobacter pylori (H. pylori) infection is causally associated with GC and environmental risk factors (e.g. cigarette smoking and alcohol drinking) are recognized aetiological factors for ESCC in the West,3,4 the attributable fractions for these factors vary substantially by geographical region and ethnicity.5,6 Familial aggregation studies7 and germline mutations among familial cases provide primary evidence of genetic predisposition for upper gastrointestinal (UGI) cancers,8 and genome-wide association studies (GWAS) have identified around 20 genetic loci associated with risk of ESCC9–15 or GC11,16,17 in East Asians.
It is now well recognized that epigenetic regulation of gene expression by DNA methylation, chromatin remodelling and histone modifications, and non-coding RNAs is opening new and exciting insights into novel molecular mechanisms in carcinogenesis. Furthermore, there is growing evidence to suggest intricate interplay among epigenetic regulators and mechanisms. For example, more than 100 microRNAs have been reported to be affected by epigenetic aberrations through DNA methylation and/or histone modifications.18,19 Also, changes in DNA methylation occur in the presence of histone modifications and chromatin remodelling,20 which can either activate or suppress gene expression including microRNA expression in a tissue-specific manner and in different types of cancers.20–22 Collectively, these observations suggest that an intricate interplay exists across epigenetic machinery genes or mechanisms.
Previous studies related germline variants in DNA methyltransferase and microRNA biosynthesis genes to risk of a number of different cancers, including gastric, lung, liver, bladder, oesophageal, kidney, and head and neck cancers.23–33 However, to date, no comprehensive examination of the relation of genetic variants in established epigenetic machinery genes to risk of UGI cancer has been performed, nor has a mechanism that translates these variants into cancer risk been delineated. Therefore, an examination of genetic variants from a comprehensive list of epigenetic machinery genes may uncover novel genes and pathways that contribute to UGI cancer susceptibility.
In the present study we used genome-wide association study (GWAS) data11 to examine the relation of variation in epigenetic machinery genes and pathways to risk of ESCC and GC. We carried out bioinformatics analyses of SNP-containing regions using available ENCODE and NIH Roadmap Epigenomics project data. Further, we integrated gene expression data from normal tissues with genotype data from our GWAS participants, and used data from the NIH Genotype-Tissue Expression project (GTEx) portal to explore the functional relevance of our findings through expression quantitative trait loci (eQTLs).
Methods
Study population
This study reports analyses of data from a GWAS of ESCC and GC conducted among ethnic Chinese; full details have been described elsewhere.11 Briefly, participants were drawn from two studies/projects, the Shanxi UGI Genetics Project (Shanxi) and the Nutrition Intervention Trials (NIT). The Shanxi Project was conducted between 1997 and 2007 and had several components including a case-control study and a case-only study (tumour/nontumour study). Case-control and case-only studies were conducted contemporaneously, and the catchment area, enrolment process, information gathering and biological specimen collections were essentially identical. All cases were incident, histologically confirmed ESCC and GC cases who underwent surgical resection at the Shanxi Cancer Hospital/Institute in Taiyuan. Cases were individually matched to controls on age (±5 years), sex and neighbourhood for the case-control portion.34 The NITs were randomized, double-blind, placebo-controlled trials initiated in the mid 1980s that tested the effect of multiple vitamin and mineral combinations on the incidence and mortality of UGI cancers. NIT subjects included in this GWAS were drawn from the cohort of trial participants who participated in a 1999/2000 blood survey which collected blood for DNA. All incident and histologically confirmed ESCC and GC cases identified from blood survey participants followed through to the end of 2007 were included in this GWAS analysis. Controls were selected as a case-cohort from an age- and sex-stratified randomly sampled sub-cohort.35 Both projects in this GWAS collected self-reported risk factor information from interviews, obtained written informed consent from all participants and were approved by institutional review boards. The NCI Special Studies Institutional Review Board also approved the overall GWAS.
Genotyping, quality control, and exclusion
DNAs were genotyped as part of the GWAS at the Cancer Genomic Research laboratory of the National Cancer Institute's Division of Cancer Epidemiology and Genetics as previously described.11 After the initial report, additional subjects from the same contributing studies were scanned on the same platform at the same facility. Both the initial and additional scan data underwent similar processing and quality control metrics. We excluded SNPs with minor allele frequency (MAF) < 5%, < 98% completion and < 95% concordance and a Hardy–Weinberg equilibrium P-value < 1E-6. Linkage disequilibrium (LD) was measured by pairwise r2 among controls using Haploview [http://www.broad.mit.edu/mpg/haploview/]. We excluded subjects whose SNP completion rates were < 94%, who had abnormal mean heterozygosity values (> 30 or < 25%), were gender discordant, or were an unexpected duplicate pair. Following all subject exclusions, data on a total of 5811 subjects were included in this analysis, including 1942 ESCCs, 1758 GCs (1126 cardia [GCA] and 632 non-cardia adenocarinoma [GNCA]) and 2111 controls. Data are available upon request from the NIH Data Access Committee [http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000361.v1.p1].
Gene and SNP selection in epigenetic pathway(s)
To obtain as comprehensive a list of epigenetic machinery genes as possible, we identified from the literature a priori genes which encode proteins associated with known epigenetic mechanisms (DNA methylation, histone modifications, chromatin remodelling and microRNA biosynthesis; collectively referred to here as epigenetic pathway genes).36–39 We searched PubMed for epigenetic genes and associated mechanisms/pathways using the terms: epigenetics, epigenetic modifications, epigenetic enzymes, epigenetic machinery genes, genes encoding epigenetic regulator, DNA methylation machinery, microRNA biosynthesis, histone modifications, chromatin enzymes, chromatin remodelling.
We extended the search and confirmation of gene data and associated pathways to five resources or pathway catalogues: BioCarta [http://www.biocarta.com/genes/index.asp], HumanCyc [http://humancyc.org/], the Kyoto Encyclopedia of Genes and Genomes database [http://www.genome.jp/kegg/], the NCI-Nature Curated database [http://pid.nci.nih.gov/download.shtml] and Reactome [http://www.reactome.org/]. We identified 198 autosomal epigenetic pathway genes and mapped SNPs including regions located 20 Kb upstream and 10 kb downstream of each coding gene (Human Genome Build 36 Hg18). We excluded two genes with no SNP data (TARBP2P and PCGF1), one gene which contained SNPs with MAFs < 5% (HDAC1) and four genes for which the proportion of subjects with at least one missing genotype (gene maximum missing rates) was higher than 10% [HDAC9, KDM4C and SMYE3 in all cancers; MBD5 in ESCC, GC and GCA]. After this, a total of 192 genes were included in the epigenetic pathway (Supplementary Table 1, available as Supplementary data at IJE online). To allow subpathway-based analyses, we further subdivided the genes into groups based on their gene regulation mechanism: chromatin remodelling (including histone modification, 142 genes), DNA methylation (15 genes) and microRNA biosynthesis (35 genes) pathways. After quality control processes, we included a total of 2181 unique SNPs in all epigenetic pathway genes for ESCC, 2181 SNPs for GC, 2173 SNPs for GCA and 2225 SNPs GNCA in the final analysis.
Statistical analyses
We carried out individual SNP-, gene- and pathway-based analyses for ESCC and GC overall in addition to the GC subsites of GCA and GNCA. SNP-based analyses were tested under the additive model, and per-allele odds ratios (ORs), 95% confidence intervals (CIs) and corresponding P-values (called P-valueSNP) were calculated using unconditional logistic regression with adjustment for age (10-year categories), sex, study (Shanxi or NIT) and the top 5 eigenvectors (to control for population stratification). After excluding SNPs with LD r2 ≥ 0.80 in controls, our Bonferroni-corrected threshold for SNP analyses was P = 3.1E-05 (the number of independent signals varied from 1563 to 1593 for ESCC, GC, GCA and GNCA).
Gene-based analyses were conducted using the AdaJoint method [http://dceg.cancer.gov/tools/analysis/adajoint]. The test statistic for gene-based analyses (P-valueGENE) accumulated the evidence of association from the best single SNP and the best pair of SNPs within the candidate gene.
The test statistics for pathway-level analyses (P-valuesPATH) were based on the adaptive rank truncated product (ARTP)40 method which combined P-valueGENE from the AdaJoint method across all relevant genes in the pathway. The P-values for gene-based analyses and pathway-based analysis were determined through 106 resampling steps.41 The resampling procedure was used to generate datasets under the null hypothesis while keeping the correlation among SNPs the same as in the observed dataset.
The Bonferroni-corrected significance threshold was P = 2.6E-04 (0.05/192 genes) for gene-based analyses and P = 0.013 (0.05/4 pathways) for pathway-level analyses.
mRNA expression data
Paired tumour (ESCC and GC) and histologically confirmed normal squamous oesophageal and gastric mucosa tissues were collected from a subset of Shanxi cases enrolled between 1998 and 2001. Cases enrolled had no prior therapy for their cancer and all underwent surgical resection at the time of hospitalization. Selection of subjects for tissue study was based solely on the availability of appropriate tissues for RNA testing [i.e. consecutive testing of cases with available frozen tissue, tumour samples that were predominantly (>50%) tumour, normal paired tissues that were dissected at least 2 cm distant regions from the tumours and RNA quality/quantity adequate for testing]. All tissues were snap-frozen in liquid nitrogen and stored at −130°C until required for RNA extraction. RNA extraction and mRNA microarray analyses have been described elsewhere.42,43 Among the subjects with mRNA expression data available for paired tumour/normal tissues (133 ESCC cases, 62 GCA cases and 72 GNCA cases), 100 ESCC, 34 GCA and 56 GNCA cases also had GWAS germline scan data available to be used for eQTL analyses. Gene expression data are publically available from Gene Expression Omnibus [http://www.ncbi.nlm.nih.gov/geo/] with accession numbers of GSE2340021 for ESCC and GSE2927222 for GCA and GNCA.
Functional annotation of genes and SNPs
We used custom tracks on the UCSC Genome browser [http://genome.ucsc.edu] to screen NIH Roadmap and ENCODE data containing the SNP region for evidence of regulatory relevance44 in oesophageal and gastric tissues/cells. We also used the online tools HaploReg [http://www.broadinstitute.org/mammals/haploreg/haploreg.php] and RegulomeDB [http://regulome.stanford.edu] as complementary analyses to confirm the location of each SNP in relation to protein-coding and/or non-coding RNA genes.
Expression quantitative trait loci (eQTLs) analysis
To assess the potential of a risk variant to regulate mRNA expression, we conducted eQTL analysis in normal tissues: 100 normal squamous oesophageal, 34 normal cardia, and 56 normal non-cardia tissues. Normal cardia and non-cardia were also combined to evaluate 90 normal gastric tissues. Candidate SNPs were selected as the SNP with the lowest P-valueSNP among all SNPs tested within a gene; genes evaluated were limited to those with a P-valueGENE <0.05. Based on available probes from the U133A Affymetrix array, we had 16, 20, 23 and 23 SNP:probe pairs(or tests) available in normal oesophageal, gastric, cardia and noncardia tissues, respectively (Supplementary Table 6, available as Supplementary data at IJE online). Correlation coefficients (rho) between SNPs and mRNA expression level were estimated using the nonparametric Spearman rank correlation test. For selected SNPs (i.e. rs12724079 and rs1879271) where fewer than five cases were homozygous for the minor allele, we combined heterozygote cases with minor allele homozygote cases and applied the dominant model to increase power. We used data from GTEx [http://www.gtexportal.org/home/] as an alternative measure for unavailable gene probes from our array (e.g. MDB3L1) and as a source for independent validation of potential eQTLs. GTEx eQTL results were based on RNA-sequencing data, and the relevant tissues available included normal oesophageal mucosa (n = 95), normal oesophageal muscularis (n = 91) and normal gastric (n = 76) tissues. Based on the number of SNPs tested as eQTLs, our Bonferroni-corrected significance threshold for eQTL testing was P = 0.002 (0.05/23 number of pairs).
Results
Population characteristics
A total of 1942 ESCC cases, 1758 GC (1126 GCA and 632 GNCA cases) and 2111 controls from the combined studies were analysed in this study. Demographic and risk factor information for both individual studies and the combined population are shown in Supplementary Table 2 (available as Supplementary data at IJE online).
Pathway-based analyses
Associations were observed for the overall epigenetic pathway and risk of GCA (P-valuePATH = 0.034) and for the chromatin remodelling subpathway with risk of GCA (P-valuePATH = 0.039) (Table 1). However, these associations did not reach the Bonferroni P-value threshold for multiple comparison adjustment.
Table 1.
Associations between epigenetic machinery pathways and the risk of UGI cancer
| Pathway/Subpathway | No of genes | ESCC |
GC |
GCA |
GNCA |
||||
|---|---|---|---|---|---|---|---|---|---|
| No of SNPs (total/unique) | P-value PATH | No of SNPs (total/unique) | P-value PATH | No of SNPs (total/unique) | P-value PATH | No of SNPs (total/unique) | P-value PATH | ||
| Epigenetic pathway | 192 | 2181/2174 | 0.219 | 2181/2174 | 0.267 | 2173/2166 | 0.034 | 2225/2218 | 0.607 |
| Chromatin remodelling | 142 | 1731/1731 | 0.174 | 1728/1728 | 0.349 | 1724/1724 | 0.039 | 1727/1727 | 0.432 |
| DNA methylation | 15a | 126/126a | 0.441 | 124/124a | 0.245 | 123/123a | 0.081 | 172/172 | 0.899 |
| microRNA biosynthesis | 35 | 324/324 | 0.677 | 328/328 | 0.471 | 326/326 | 0.392 | 327/327 | 0.612 |
Abbreviations: ESCC, oesophageal squamous cell carcinoma; GC, gastric cancer; GCA, gastric cardia adenocarcinoma; GCNA, gastric non-cardia adenocarcinoma; UGI, upper gastrointestinal.
aMBD5 was excluded due to gene maximum missing rates >10%.
Note: Pathway-based p-values (P-valuePATH) are shown for the overall epigenetic pathway and three subpathways in each cancer.
Gene-based analyses
Overall, 37 genes were associated with at least one type of cancer risk (P-valueGENE < 0.05, Table 2): 11 with ESCC, 11 with GC, 14 with GCA and 12 with GNCA risk. Among these 37 genes, 14 (ASH1L, EED, EZH1, KAT2A, KAT6A, KAT8, KDM5A, PRMT1, PRMT7, RING1, SETD1A, SETD1B, SMARCC, and SMARCD1) were chromatin remodelling genes (Supplementary Table 7, available as Supplementary data at IJE online). The genes with the lowest P-values for each site were SETD1B in ESCC (P-valueGENE = 0.006), SUV420H1 in GC (P-valueGENE = 0.006), GSG2 in GCA (P-valueGENE = 0.003) and SMARCC1 in GNCA (P-valueGENE = 0.012). ASH1L was the only gene associated with risk for three cancer sites: overall GC (P-valueGENE = 0.010), GCA (P-valueGENE = 0.018) and GNCA risk (P-valueGENE = 0.019). None of the gene associations had P-values below the Bonferroni threshold following correction for multiple comparisons. Results for all 192 genes evaluated are shown in Supplementary Table 1 (available as Supplementary data at IJE online).
Table 2.
Top-ranked SNPs in genes (P-valueGENE < 0.05) associated with risk of UGI cancers
| Oesophageal squamous cell carcinoma (ESCC) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene (n = 11) | Gene function | Genomic location | P-valueGENE | Top-ranked SNP | Major/minor | MAF Cases | MAF Controls | OR (95% CI) | P-valueSNP |
| SETD1B | Chromatin remodelling | 12q24.31 | 0.006 | rs2242259 | C/T | 0.39 | 0.35 | 1.14 (1.04–1.24) | 0.006 |
| CBX4 | Chromatin remodelling | 17q25.3 | 0.020 | rs4889898 | C/A | 0.33 | 0.30 | 1.15 (1.05–1.27) | 0.003 |
| KAT5 | Chromatin remodelling | 11q13 | 0.021 | rs1151500 | C/T | 0.13 | 0.11 | 1.21 (1.06–1.39) | 0.006 |
| ZGPAT | Chromatin remodelling | 20q13.3 | 0.022 | rs8957 | C/A | 0.34 | 0.36 | 0.93 (0.84–1.02) | 0.104 |
| BTD | Chromatin remodelling | 3p25 | 0.025 | rs2455823 | G/A | 0.50 | 0.47 | 1.14 (1.04–1.24) | 0.005 |
| EED | Chromatin remodelling | 11q14.2-q22.3 | 0.026 | rs10898459 | C/T | 0.47 | 0.50 | 0.88 (0.81–0.96) | 0.005 |
| SMARCD1 | Chromatin remodelling | 12q13-q14 | 0.027 | rs836178 | G/T | 0.14 | 0.16 | 0.86 (0.76–0.97) | 0.013 |
| POLE3 | Chromatin remodelling | 9q33 | 0.030 | rs8177812 | C/T | 0.22 | 0.20 | 1.17 (1.05–1.30) | 0.005 |
| AGO2 | microRNA biosynthesis | 8q24.3 | 0.031 | rs6983924 | A/G | 0.50 | 0.47 | 1.16 (1.06–1.27) | 0.001 |
| RING1 | Chromatin remodelling | 6p21.3 | 0.046 | rs213194 | G/A | 0.08 | 0.07 | 1.18 (1.00–1.40) | 0.051 |
| MBD3L1 | DNA methylation | 19p13.2 | 0.047 | rs10412487 | T/C | 0.32 | 0.34 | 0.88 (0.80–0.97) | 0.008 |
| Gastric cancer (GC) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene (n = 11) | Gene function | Genomic location | P-valueGENE | Top-ranked SNP | Major/minor | MAF Cases | MAF Controls | OR (95% CI) | P-valueSNP |
| SUV420H1 | Chromatin remodelling | 11q13.2 | 0.006 | rs10896300 | G/A | 0.11 | 0.13 | 0.86 (0.75–0.99) | 0.036 |
| ASH1L | Chromatin remodelling | 1q22 | 0.010 | rs12724079 | T/C | 0.17 | 0.20 | 0.84 (0.75–0.94) | 0.003 |
| KAT5 | Chromatin remodelling | 11q13 | 0.011 | rs1151500 | C/T | 0.13 | 0.11 | 1.23 (1.07–1.42) | 0.004 |
| JARID2 | Chromatin remodelling | 6p24-p23 | 0.023 | rs7769291 | C/T | 0.26 | 0.24 | 1.14 (1.02–1.26) | 0.016 |
| PRMT1 | Chromatin remodelling | 19q13 | 0.029 | rs3745469 | C/T | 0.06 | 0.08 | 0.79 (0.66–0.95) | 0.011 |
| MBD3L1 | DNA methylation | 19p13.2 | 0.029 | rs10412487 | T/C | 0.31 | 0.34 | 0.87 (0.79–0.96) | 0.005 |
| SRRT | microRNA biosynthesis | 7q21 | 0.030 | rs13245899 | A/G | 0.42 | 0.39 | 1.14 (1.04–1.25) | 0.006 |
| SIRT3 | Chromatin remodelling | 11p15.5 | 0.033 | rs1045454 | C/T | 0.18 | 0.16 | 1.17 (1.03–1.32) | 0.013 |
| KDM5A | Chromatin remodelling | 12p13.33 | 0.033 | rs527118 | T/C | 0.31 | 0.34 | 0.87 (0.79–0.96) | 0.004 |
| DROSHA | microRNA biosynthesis | 5q11.2 | 0.035 | rs7735863 | G/A | 0.41 | 0.43 | 0.89 (0.81–0.97) | 0.011 |
| KAT6A | Chromatin remodelling | 8p11 | 0.040 | rs7008906 | G/A | 0.42 | 0.39 | 1.14 (1.04–1.24) | 0.006 |
| Gastric cardia adenocarcinoma (GCA) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene (n = 14) | Gene function | Genomic location | P-valueGENE | Top-ranked SNP | Major/minor | MAF Cases | MAF Controls | OR (95% CI) | P-valueSNP |
| GSG2 | Chromatin remodelling | 17p13 | 0.003 | rs7220026 | G/A | 0.28 | 0.25 | 1.19 (1.05–1.33) | 0.005 |
| MBD3L1 | DNA methylation | 19p13.2 | 0.004 | rs10412487 | T/C | 0.30 | 0.34 | 0.82 (0.74–0.92) | 0.001 |
| SIRT6 | Chromatin remodelling | 19p13.3 | 0.009 | rs352493 | T/C | 0.27 | 0.24 | 1.21 (1.08–1.37) | 0.002 |
| KDM4A | Chromatin remodelling | 1p34.1 | 0.013 | rs304303 | C/A | 0.18 | 0.22 | 0.81 (0.71–0.93) | 0.002 |
| PRMT7 | Chromatin remodelling | 16q22.1 | 0.016 | rs2863973 | T/G | 0.07 | 0.09 | 0.73 (0.60–0.89) | 0.002 |
| SUV420H1 | Chromatin remodelling | 11q13.2 | 0.016 | rs10896300 | G/A | 0.11 | 0.13 | 0.87 (0.74–1.03) | 0.105 |
| ASH1L | Chromatin remodelling | 1q22 | 0.018 | rs8179271 | A/G | 0.23 | 0.26 | 0.85 (0.75–0.96) | 0.007 |
| SRRT | microRNA biosynthesis | 7q21 | 0.021 | rs13245899 | A/G | 0.42 | 0.39 | 1.16 (1.05–1.29) | 0.005 |
| SMARCD1 | Chromatin remodelling | 12q13-q14 | 0.026 | rs836178 | G/T | 0.14 | 0.16 | 0.83 (0.71–0.96) | 0.011 |
| DROSHA | microRNA biosynthesis | 5q11.2 | 0.034 | rs7735863 | G/A | 0.40 | 0.43 | 0.86 (0.77–0.95) | 0.004 |
| EZH1 | Chromatin remodelling | 17q21.1-q21.3 | 0.034 | rs4792953 | T/C | 0.41 | 0.44 | 0.87 (0.78–0.97) | 0.011 |
| KAT2A | Chromatin remodelling | 17q12-q21 | 0.034 | rs1122326 | A/C | 0.08 | 0.10 | 0.80 (0.66–0.96) | 0.015 |
| DICER1 | microRNA biosynthesis | 14q32.13 | 0.036 | rs7157322 | A/C | 0.42 | 0.38 | 1.17 (1.05–1.30) | 0.004 |
| CARM1 | Chromatin remodelling | 19p13.2 | 0.048 | rs1541596 | G/A | 0.20 | 0.23 | 0.86 (0.75–0.97) | 0.019 |
|
Gastric non-cardia adenocarcinoma (GNCA) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene (n = 12) | Gene function | Genomic location | P-valueGENE | Top-ranked SNP | Major/minor | MAF Cases | MAF Controls | OR (95% CI) | P-valueSNP |
| SMARCC1 | Chromatin remodelling | 3p21.31 | 0.012 | rs13094264 | G/A | 0.32 | 0.28 | 1.18 (1.02–1.35) | 0.022 |
| PPP1CA | Chromatin remodelling | 11q13 | 0.016 | rs1790733 | T/C | 0.32 | 0.29 | 1.16 (1.01–1.33) | 0.038 |
| RPS6KA5 | Chromatin remodelling | 14q31-q32.1 | 0.017 | rs3783834 | C/T | 0.42 | 0.37 | 1.25 (1.10–1.43) | 0.001 |
| GEMIN4 | microRNA biosynthesis | 17p13.3 | 0.018 | rs910924 | C/T | 0.10 | 0.13 | 0.73 (0.59–0.89) | 0.002 |
| KAT8 | Chromatin remodelling | 16p11.1 | 0.018 | rs2855475 | C/T | 0.10 | 0.07 | 1.34 (1.07–1.68) | 0.010 |
| ASH1L | Chromatin remodelling | 1q22 | 0.019 | rs12724079 | T/C | 0.16 | 0.20 | 0.79 (0.67–0.94) | 0.007 |
| KAT5 | Chromatin remodelling | 11q13 | 0.029 | rs1151500 | C/T | 0.14 | 0.11 | 1.29 (1.07–1.56) | 0.009 |
| PRMT1 | Chromatin remodelling | 19q13 | 0.033 | rs3745469 | C/T | 0.06 | 0.08 | 0.74 (0.57–0.96) | 0.023 |
| KDM8 | Chromatin remodelling | 16p12.1 | 0.037 | rs12051243 | T/G | 0.17 | 0.20 | 0.80 (0.68–0.95) | 0.011 |
| SETD7 | Chromatin remodelling | 4q31.1 | 0.042 | rs720257 | G/A | 0.36 | 0.32 | 1.22 (1.07–1.40) | 0.003 |
| SETD1A | Chromatin remodelling | 16p11.2 | 0.046 | rs897986 | G/A | 0.08 | 0.06 | 1.33 (1.04–1.70) | 0.021 |
| SMAD3 | microRNA biosynthesis | 15q21-q22 | 0.047 | rs1465842 | G/A | 0.21 | 0.17 | 1.27 (1.08–1.50) | 0.003 |
Abbreviations: CI, confidence intervals; MAF, minor allele frequency; OR, odds ratio; UGI, upper gastrointestinal.
Note: Per-allele ORs and P-valueSNP were estimated for minor allele among controls in combined population adjusted for age, sex, study, and eigenvectors. The table was sorted by P-valueGENE.
SNP-based analyses
Overall, SNP associations with risk (P-valueSNP less than 0.05) were seen for 132 SNPs in 49 genes with ESCC and for 123 SNPs in 57 genes with GC (Supplementary Tables 3 and 4, available as Supplementary data at IJE online). However, none of the individual SNP P-values for associations with any of the four cancers examined here reached the Bonferroni correction threshold for multiple comparisons.
Table 2 shows the top-ranked SNP in each of genes with P-valueGENE < 0.05 for associations with risk of ESCC, GC, GCA and GNCA. Our top-ranked SNPs were: rs6983924 in AGO2 (OR = 1.16; 95% CI = 1.06–1.27; P-valueSNP = 9.7E-04) for ESCC; rs12724079 in ASH1L (OR = 0.84; 95% CI = 0.75–0.94; P-valueSNP = 0.003) for GC; rs10412487 in MBD3L1 (OR = 0.82; 95% CI = 0.74–0.92; P-valueSNP = 7.2E-04) in GCA; and rs3783834 in RPS6KA5 (OR = 1.25; 95% CI = 1.10–1.43; P-valueSNP = 6.3E-04) for GNCA. The top-ranked SNPs in ASH1L differed by GC subtype: ASH1L rs12724079 was the top-ranked SNP with GC (OR = 0.84; 95% CI = 0.75–0.94; P-valueSNP = 0.003) and GNCA (OR = 0.79; 95% CI = 0.67–0.94; P-valueSNP = 0.007) risk associations, whereas rs8179271 was the top-ranked SNP with GCA risk (OR = 0.85; 95% CI = 0.75–0.96; P-valueSNP = 0.007).
In silico functional annotation and eQTL analyses of SNPs in genes (P-valueGENE < 0.05)
Top-ranked SNPs (38 SNPs) in 37 genes (P-valueGENE < 0.05) were mapped for their putative functional/regulatory sites (Supplementary Table 5, available as Supplementary data at IJE online). There were 28 SNPs in intronic regions, 5 SNPs in 3’ UTRs and 4 SNPs in 5’ UTRs in these genes of interest. Rs352493 was the only non-synonymous SNP (pro456Arg) in SIRT6. Several SNPs, including rs4889898 (CBX4), rs1045454 (SIRT3), rs304303 (KDM4A), rs8179271 (ASH1L), rs910924 (GEMIN4) and rs2855475 (KAT8) were located proximal to non-coding RNA genes. A number of SNPs mapped to strong enhancer and transcription-like enhancer regions and DNaseI/open chromatin sites, including rs6983924 (AGO2) in normal oesophageal tissue and rs1541596 (CARM1), rs1465842 (SMAD3) and rs1151500 (KAT5) in stomach tissues. Many SNPs had the potential to alter the DNA-binding motifs of a number of proteins and transcription factors.
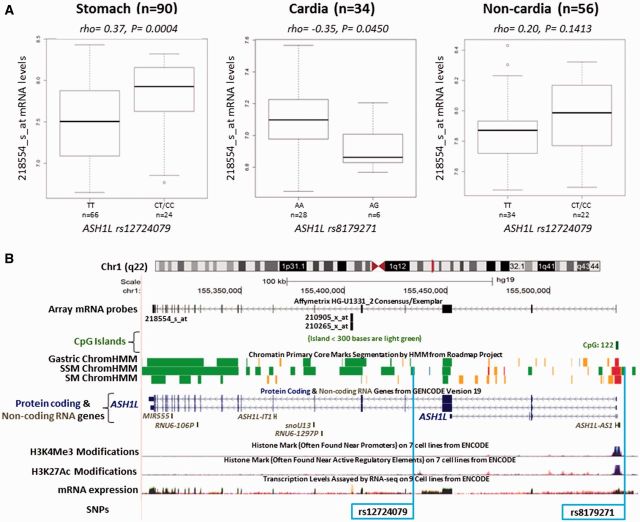
Table 3 shows results for eQTLs with P-values less than 0.05. Rs10898459 (variant T) in EED was associated with lower mRNA levels (rho = –0.22; P = 0.029) in normal oesophagus, and the same trend for T variant was seen in GTEx data (P = 0.060). In normal gastric tissue, rs12724079 (variant C) in ASH1L was associated with higher mRNA levels (rho = 0.35; P = 6.3E–04), a result which remained even after adjustment for multiple comparisons. In contrast, rs8179271 (variant G, pairwise LD r2 = 0.70 with rs12724079) was associated with lower mRNA levels of ASH1L (rho = –0.35; P = 0.045) in normal gastric cardia tissues. Figure 1A shows eQTL results from the dominant model for ASH1L SNPs, and indicated that correlation coefficients for normal gastric (rho = 0.37, P = 0.0004) and normal non-cardia gastric (rho = 0.20, P = 0.1413) were both positive, whereas the coefficient for normal cardia was negative (rho = –0.35, P = 0.045). We also found that rs7157322 (variant C) in DICER1 and rs1790733 (variant C) in PPP1CA were correlated with reduced mRNA levels of DICER1 in normal cardia (rho = –0.42; P = 0.014) and of PPP1CA in normal non-cardia gastric tissues (rho = –0.41; P = 0.002), respectively (Table 3). However, these associations were not replicated in GTEx data.
Table 3.
Selected eQTL analysis results (P < 0.05) of genes (P-valueGENE < 0.05) in normal oesophageal and stomach tissues
| Shanxi normal esophagus (n = 100) |
GTEx normal esophagus mucosa (n = 95)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Top-ranked SNP | OR (95% CI) | U133A mRNA probe | Major/minor | rhoa | P-value | Ref/Alt | Direction | P-value |
| EED | rs10898459 | 0.88 (0.81–0.96) | 209572_s_at | C/T | −0.22 | 0.029 | T/C | + | 0.060 |
|
Shanxi normal stomach (n = 90) |
GTEx normal stomach (n = 76)b |
||||||||
| U133A mRNA probe | Major/minor | rhoa | P-value | Ref/Alt | Direction | P-value | |||
| ASH1L | rs12724079 | 0.84 (0.75–0.94) | 218554_s_at | T/C | 0.35 | 0.0006 | T/C | + | 0.800 |
|
Shanxi normal cardia stomach (n = 34) |
GTEx normal stomach (n = 76)b |
||||||||
| U133A mRNA probe | Major/minor | rhoa | P-value | Ref/Alt | Direction | P-value | |||
| DICER1 | rs7157322 | 1.17 (1.05–1.30) | 206061_s_at | A/C | −0.42 | 0.014 | A/C | ± | 0.200 |
| ASH1L | rs8179271 | 0.85 (0.75–0.96) | 218554_s_at | A/G | −0.35 | 0.045 | No SNP | No SNP | No SNP |
|
Shanxi normal non-cardia stomach (n = 56) |
GTEx normal stomach (n = 76)b |
||||||||
| U133A mRNA probe | Major/minor | rhoa | P-value | Ref/Alt | Direction | P-value | |||
| PPP1CA | rs1790733 | 1.16 (1.01–1.33) | 200846_s_at | T/C | −0.41 | 0.002 | A/G | ± | 0.400 |
Abbreviations: eQTL, expression quantitative trait loci; Ref/Alt, reference/alternative allele.
a Statistical significance for correlation (rho) was determined using nonparametric Spearman's rank test; Direction is based on minor allele.
b GTEx Portal [http://www.gtexportal.org/home/]; Direction is based on alternative allele.; Symbols: +, positive influence; ±, no change.
Figure 1.
Expression quantitative trait loci (eQTL) analysis of ASH1L mRNA expression in normal gastric tissues, and genomic locations for ASH1L and SNPs. A: eQTL analyses were conducted using dominant model for rs12724079 in 90 gastric tissues, rs8179271 in 34 cardia tissues and rs12724079 in 56 non-cardia tissues. ASH1L mRNA levels were assessed using the Affymetrix_U133A probe 218 554_s_at. The major/minor allele for each SNP in our population is shown. B: Genome Browser [http://genome.ucsc.edu/] image of ASH1L gene region on human assembly hg19 based on NIH Epigenomics Roadmap data [http://www.genboree.org/epigenomeatlas/] and ENCODE data [http://genome.ucsc.edu/ENCODE/]. The position of the Affymetrix_U133A probe 218 554_s_at, which detects variant mRNAs 1, 2 and 3 of ASH1L and CpG islands in this region are shown. Regulatory domains [chromatin state segmentation using a hidden Markov Model (ChromHMM)] and core histone marks for normal gastric tissue (Gastric), stomach mucosa (SM) and stomach smooth muscle (SSM) tissue are shown: Red, active transcriptional start site (TSS); Dark Green, transcription elongation/transition; Orange, active-to-weak enhancer. H3K4Me3 and H3K27Ac activation marks in this region and mRNA levels from a large number of ENCODE cells lines is also shown. SNPs rs12724079 and rs8179271 are highlighted by a light blue-coloured box. For clarity, not all SNPs contained in this region (dbSNP) are shown. ASH1L-AS1 and other non-coding RNA genes within ASH1L are indicated and text is coloured grey.
Discussion
In this study we systematically examined the cumulative effects of variation in multiple epigenetic machinery genes acting in functional mechanistic pathways as well as individual gene effects, to understand genetic susceptibility to ESCC and GC in a high-risk Chinese population.
We found pathway-based associations for the overall epigenetic pathway and the chromatin remodelling subpathway with GCA risk. Given the relatively large proportion of chromatin remodelling genes contributing to the overall pathway, the chromatin remodelling subpathway (including genes encoding enzymes involved in histone modifications) is likely driving the association of GCA risk with the overall epigenetic pathway. The absence of pathway-based associations for GC and GNCA may be due to reduced power from the smaller number of GNCA cases genotyped here. Alternatively, this result may reflect aetiological differences in the gastric carcinogenesis pathway11,45–47 and in epigenetic mechanisms important for the development of GC subtypes in this high-risk population. We found no evidence for epigenetic pathway-based associations with ESCC.
The functions of the 37 genes we observed as associated with UGI cancer risk are summarized in Supplementary Table 7, available as Supplementary data at IJE online. In relation to chromatin remodelling genes (histone and nucleosome modifiers), we identified six ESCC-associated genes encoding products involved in the methylation (SETD1B and EED), sumoylation (CBX4), ubiquitination (RING1), biotinylation (BTD) and acetylation (KAT5) of histone tails, and a further two associated genes that encode proteins involved in nucleosome (chromatin) remodelling/dynamics (POLE3 and SMARCD1).48,49 KAT5 was also associated with GC and GNCA risk, and SMARCD1 was associated with GNCA risk. Twelve different chromatin remodelling genes were associated with GC, CGA and GNCA risk. These genes encode unique histone modifying demethylases (KDM5A, KDM4A and KDM8), acetyltransferases (KAT2A, KAT6A and KAT8), and methyltransferases or protein units (EZH1 and SUV420H1) and a deacetylase enzyme (SIRT6), as well as arginine methyltransferases (PRMT1, PRMT7 and CARM1).48–50 Post-translational modification of histones and nucleosome remodelling are important epigenetic mechanisms regulating gene expression,49 and alterations of these genes have been associated with the development of many cancers.50
Interestingly, 14 of the ESCC- and GC-associated chromatin remodelling genes encode proteins whose function has been linked to the regulation of HOX genes (Supplementary Table 7, available as Supplementary data at IJE online), suggesting that genetic variation in these genes may result in altered HOX expression in oesophageal and gastric tissues. HOX genes encode important transcription factors involved in developmental processes and cell differentiation, and dysregulation of specific HOX genes has previously been reported in ESCC and GC.51,52
EED encodes a member of the polycomb repressive complex 2 (PRC2) that interacts with EZH2, a methyltransferase which negatively regulates HOX gene expression.49,50 In our study, EED was associated with risk of ESCC. The T variant of its top-ranked SNP (rs10898459) was associated with a protective effect for ESCC and correlated with reduced EED mRNA levels in normal oesophageal tissues. This finding is consistent with previous data that EED mRNA and protein are expressed at low to medium levels in normal oesophageal tissues, but are overexpressed in a number of cancers [http://www.proteinatlas.org/ENSG00000074266-EED/gene], including ESCC.53 In 133 tumour and normal paired oesophageal tissues from our study population, we also observed upregulation of EED mRNA in ESCC tumour compared with normal oesophageal tissues (fold-change = 1.34, P = 3.3E-17). Collectively, these data are consistent with the possibility that rs10898459 and low basal expression of EED may be protective against ESCC development.
ASH1L encodes a TrxG methyltransferase that is required for maximal expression and H3K4 methylation of HOX genes.54,55 In this study, ASH1L was the only gene associated with risk of all three gastric cancer sites examined. We also identified two potential eQTLs (rs12724079 and rs8179281) influencing ASH1L mRNA levels in normal gastric tissues. However, the putative protective genotypes of both SNPs had opposite effects on ASH1L mRNA in cardia and non-cardia normal gastric tissues (Figure 1A), suggesting the possibility that the effect of this SNP on ASH1L expression may differ by anatomical subtype. Interestingly, rs8179271 maps 3’ to ASH1L antisense RNA 1 (ASH1L-AS1), which encodes a long non-coding RNA (lncRNA) of unknown function (Figure 1B). Thus, confirming the influence of rs12724099 and rs8179271 on ASH1L (and ASH1L-AS1) expression in gastric tissue remains an interesting question for future research.
PPP1CA encodes a catalytic subunit of protein phosphatase 1 (PP1), whose activity is important for chromatin structure and inflammation. The GNCA risk-associated rs1790733 variant in PPP1CA was significantly associated with reduced levels of PPP1CA mRNA in normal noncardia gastric tissue. Downregulation of PPP1CA has been shown to promote proliferation, migration and invasion of gastric cancer cells in vitro,56 suggesting that dysregulation of PPP1CA may contribute to GNCA susceptibility.
Just one DNA methylation machinery gene (MBD3L1) was related to UGI cancer in our study, and it was associated with risks for ESCC, GC and GCA but not GNCA. MBD3L1 encodes a transcriptional repressor whose expression appears to be restricted to germ cells [http://www.proteinatlas.org/ENSG00000170948-MBD3L1/tissue], thus the significance of MBD3L1 in relation to ESCC and GC is unclear.
Lastly, six genes (AGO2, DICER1, DROSHA, GEMIN4, SMAD3 and SRRT) involved in microRNA biosynthesis were found to be associated with one or more UGI cancers. It has been suggested that the global downregulation of microRNAs in tumours compared with normal tissues57 may be due in part to reduced expression of DROSHA and DICER.58 In our study, DICER1 was associated with GCA risk and the risk allele of its top-ranked SNP (rs7157322) was correlated with decreased mRNA levels in normal cardia tissues, an observation consistent with a potential cis-regulatory effect of rs7157322 on DICER1 expression.
This study had several strengths and limitations. The literature on genetic variants in genes encoding epigenetic regulators and their association with UGI cancer is very limited25,59,60 (Supplementary Table 7, available as Supplementary data at IJE online). Our study is the first to perform a comprehensive examination of genetic variants in epigenetic machinery genes at multiple levels of association (i.e. pathways, genes and SNPs) using new state-of-the-art biostatistical methodologies. Additional in silico and eQTL analyses provided complementary functional characterization to support observed risk associations. The relatively large number of cases studied allowed us to assess associations with good power. However, examination of large numbers of genes and SNPs in multiple types of cancers raises concerns about multiple comparisons. Further, our sample size was only modest for eQTL analyses, which limited our power to see true associations. Lack of replication in GTEx data could be also partly due to limited statistical power as well as the lack of information on anatomical location (cardia versus non-cardia). Many associations were found with P-values less than the nominal 0.05 level, but none of our pathway, gene or SNP association P-values passed Bonferroni adjustment; thus, we are unable to exclude the potential role of chance as an explanation for our results. Furthermore, the lack of H. pylori data is a potential concern. However, the genetic loci that have previously been related to H. pylori status were not among the epigenetic pathway genes assessed here,61 thus confounding by H. pylori status seems unlikely. Finally, the generalizability of our findings to other ethnic populations remains to be determined.
In conclusion, our study provides suggestive but limited evidence for the involvement of epigenetic pathways and genes in UGI cancer susceptibility, particularly for the chromatin remodelling pathway and risk of gastric cardia cancer. Future studies are warranted to replicate our findings and to further address functional roles for potentially important epigenetic machinery genes in UGI cancers.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This research was funded by Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics.
Conflict of interest: None declared.
Supplementary Material
References
- 1.Ferlay J, Dikshit R, Eser S, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer, 2013. [Google Scholar]
- 2.Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970-90. Int J Cancer 2002;102:271–74. [DOI] [PubMed] [Google Scholar]
- 3.Lee CH, Wu DC, Lee JM, et al. Carcinogenetic impact of alcohol intake on squamous cell carcinoma risk of the oesophagus in relation to tobacco smoking. Eur J Cancer 2007;43:1188–99. [DOI] [PubMed] [Google Scholar]
- 4.Pandeya N, Williams G, Green AC, et al. Alcohol consumption and the risks of adenocarcinoma and squamous cell carcinoma of the esophagus. Gastroenterology 2009;136:1215–24, e1–2. [DOI] [PubMed] [Google Scholar]
- 5.Tran GD, Sun XD, Abnet CC, et al. Prospective study of risk factors for oesophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer 2005;113:456–63. [DOI] [PubMed] [Google Scholar]
- 6.Plummer M, Franceschi S, Vignat J, et al. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer 2014;136:487–90. [DOI] [PubMed] [Google Scholar]
- 7.Chang-Claude J, Becher H, Blettner M, et al. Familial aggregation of ooesophageal cancer in a high incidence area in China. Int J Epidemiol 1997;26:1159–65. [DOI] [PubMed] [Google Scholar]
- 8.Robertson EV, Jankowski JA. Genetics of gastrooesophageal cancer: paradigms, paradoxes, and prognostic utility. Am J Gastroenterol 2008;103:443–49. [DOI] [PubMed] [Google Scholar]
- 9.Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance oesophageal cancer risk. Gastroenterology 2009;137:1768–75. [DOI] [PubMed] [Google Scholar]
- 10.Wang LD, Zhou FY, Li XM, et al. Genome-wide association study of oesophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet 2010;42:759–63. [DOI] [PubMed] [Google Scholar]
- 11.Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and oesophageal squamous cell carcinoma. Nat Genet 2010;42:764–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C, Hu Z, He Z, et al. Genome-wide association study identifies three new susceptibility loci for oesophageal squamous-cell carcinoma in Chinese populations. Nat Genet 2011;43:679–84. [DOI] [PubMed] [Google Scholar]
- 13.Wu C, Kraft P, Zhai K, et al. Genome-wide association analyses of oesophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet 2012;44:1090–97. [DOI] [PubMed] [Google Scholar]
- 14.Wu C, Wang Z, Song X, et al. Joint analysis of three genome-wide association studies of oesophageal squamous cell carcinoma in Chinese populations. Nat Genet 2014;46:1001–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abnet CC, Wang ZM, Song X, et al. Genotypic variants at 2q33 and risk of oesophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Hum Mol Genet 2012;21:2132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin GF, Ma HX, Wu C, et al. Genetic variants at 6p21.1 and 7p15.3 are associated with risk of multiple cancers in Han Chinese. Am J Hum Genet 2012;91:928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi YY, Hu ZB, Wu C, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet 2011;43:1215–U66. [DOI] [PubMed] [Google Scholar]
- 18.Kunej T, Godnic I, Ferdin J, et al. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res 2011;717:77–84. [DOI] [PubMed] [Google Scholar]
- 19.Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: An intricate network. Biochim Biophys Acta 2010;1799:694–701. [DOI] [PubMed] [Google Scholar]
- 20.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature 2012;489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiklund ED, Kjems J, Clark SJ. Epigenetic architecture and miRNA: reciprocal regulators. Epigenomics 2010;2:823–40. [DOI] [PubMed] [Google Scholar]
- 22.Liu XL, Chen XY, Yu XF, et al. Regulation of microRNAs by epigenetics and their interplay involved in cancer. J Exp Clin Cancer Res 2013;32:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang D, Meyer L, Chang DW, et al. Genetic variants in microRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res 2010;70:9765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan H, Liu DS, Qiu XM, et al. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in oesophageal carcinoma. BMC Med 2010;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu QY, Lu S, Wang L, et al. DNMT3A rs36012910 A > G polymorphism and gastric cancer susceptibility in a Chinese population. Mol Biol Rep 2012;39:10949–55. [DOI] [PubMed] [Google Scholar]
- 26.Shen HB, Wang L, Spitz MR, et al. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res 2002;62:4992–95. [PubMed] [Google Scholar]
- 27.Lao Y, Wu H, Zhao C, et al. Promoter polymorphisms of DNA methyltransferase 3B and risk of hepatocellular carcinoma. Biomed Rep 2013;1:771–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Rodriguez M, Kim ES, et al. A novel C/T polymorphism in the core promoter of human de novo cytosine DNA methyltransferase 3B6 is associated with prognosis in head and neck cancer. Int J Oncol 2004;25:993–99. [PubMed] [Google Scholar]
- 29.Ye YQ, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for oesophageal cancer risk. Cancer Prev Res. 2008;1:460–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang HS, Dinney CP, Ye YQ, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res 2008;68:2530–37. [DOI] [PubMed] [Google Scholar]
- 31.Horikawa Y, Wood CG, Yang HH, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res 2008;14:7956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clague J, Lippman SM, Yang HS, et al. Genetic variation in microRNA genes and risk of oral premalignant lesions. Mol Carcinog 2010;49:183–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung H, Lee KM, Choi JY, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: a case-control study in Korea. Breast Cancer Res Treat 2011;130:939–51. [DOI] [PubMed] [Google Scholar]
- 34.Gao Y, Hu N, Han XY, et al. Risk factors for oesophageal and gastric cancers in Shanxi Province, China: a case-control study. Cancer Epidemiol 2011;35:e91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Taylor PR, Li JY, et al. Linxian nutrition intervention trials. Design, methods, participant characteristics, and compliance. Ann Epidemiol 1993;3:577–85. [DOI] [PubMed] [Google Scholar]
- 36.Murgatroyd C, Spengler D. Genetic variation in the epigenetic machinery and mental health. Curr Psychiatry Rep 2012;14:138–49. [DOI] [PubMed] [Google Scholar]
- 37.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 2012;22:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol 2010;28:1057–68. [DOI] [PubMed] [Google Scholar]
- 40.Yu K, Li QZ, Bergen AW, et al. Pathway analysis by adaptive combination of p-values. Genetic Epidemiology 2009;33:700–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Shi J, Liang F, et al. A fast multilocus test with adaptive SNP selection for large-scale genetic-association studies. Eur J Hum Genet 2014;22:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su H, Hu N, Yang HH, et al. Global gene expression profiling and validation in oesophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res 2011;17:2955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G, Hu N, Yang HH, et al. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS One 2013;8:e63826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res 2013;41:D56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao D, Wang Y, Xue L, et al. Different expression pattern and significance of p14ARF-Mdm2-p53 pathway and Bmi-1 exist between gastric cardia and distal gastric adenocarcinoma. Hum Pathol 2012;44:844–51. [DOI] [PubMed] [Google Scholar]
- 46.Shah MA, Khanin R, Tang L, et al. Molecular classification of gastric cancer: a new paradigm. Clin Cancer Res 2011;17:2693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stocks SC, Pratt N, Sales M, et al. Chromosomal imbalances in gastric and oesophageal adenocarcinoma: specific comparative genomic hybridization-detected abnormalities segregate with junctional adenocarcinomas. Genes Chromosomes Cancer 2001;32:50–58. [DOI] [PubMed] [Google Scholar]
- 48.Oh J, Sohn DH, Ko M, et al. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem 2008;283:11924–34. [DOI] [PubMed] [Google Scholar]
- 49.Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene 2011;30:3391–403. [DOI] [PubMed] [Google Scholar]
- 50.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 2010;10:669–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossi Degl'Innocenti D, Castiglione F, Buccoliero AM, et al. Quantitative expression of the homeobox and integrin genes in human gastric carcinoma. Int J Mol Med 2007;20:621–29. [PubMed] [Google Scholar]
- 52.Chen KN, Gu ZD, Ke Y, et al. Expression of 11 HOX genes is deregulated in oesophageal squamous cell carcinoma. Clinical Cancer Research 2005;11:1044–49. [PubMed] [Google Scholar]
- 53.Tzao C, Tung HJ, Jin JS, et al. Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod Pathol 2009;22:252–60. [DOI] [PubMed] [Google Scholar]
- 54.Gregory GD, Vakoc CR, Rozovskaia T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol 2007;27:8466–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyazaki H, Higashimoto K, Yada Y, et al. Ash1l methylates Lys36 of histone H3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet 2013;9:e1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu JG, Wang JJ, Jiang X, et al. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer 2014, 21 Sept. PMID: 25240408. [Epub ahead of print.} [DOI] [PubMed] [Google Scholar]
- 57.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–38. [DOI] [PubMed] [Google Scholar]
- 58.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol 2014;24:R762–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan S, Greco D, Michailidou K, et al. MicroRNA related polymorphisms and breast cancer risk. PLoS One 2014;9:e109973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang XX, He XQ, Li FX, et al. Risk-association of DNA methyltransferases polymorphisms with gastric cancer in the Southern Chinese population. Int J Mol Sci 2012;13:8364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mayerle J, den Hoed CM, Schurmann C, et al. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA 2013;309:1912–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.