Abstract
Introduction. Given that influenza A(H9N2) is recognized as a pandemic threat, we evaluated the overall burden of influenza A(H9N2) infections among avian-exposed human populations.
Methods. We performed a systematic search of PubMed, AGRICOLA, and CAB Abstracts databases for literature published during 1997–2013. Studies reporting serological evidence of human influenza A(H9N2) infection among avian-exposed populations were included. We used a World Health Organization (WHO)–recommended case definition for serological evidence of infection based on results of hemagglutination inhibition (HI) and microneutralization (MN) assays. We calculated overall seroprevalence through a random effects meta-analysis model.
Results. Seroprevalence data reported by the studies ranged from 1% to 43% (median, 9%) by HI, which was not significantly different from the seroprevalence estimated through the WHO-recommended case definition (median, 1.3%; range, 0.5%–42.6%). Reported seroprevalence by MN ranged from 0.6% to 9% (median, 2.7%), which was greater than the seroprevalence estimated through the WHO-recommended case definition (median, 0.3%; range, 0.1%–1.4%).
Conclusions. A small proportion of avian-exposed humans had evidence of influenza A(H9N2) infection. As the virus has a near global distribution in poultry, it seems likely that present surveillance efforts are missing mild or asymptomatic infections among avian-exposed persons. It seems prudent to closely monitor avian-exposed populations for influenza A(H9N2) infection to provide prepandemic warnings.
Keywords: influenza A virus, H9N2 subtype, seroprevalence, systematic review, meta-analysis, hemagglutination inhibition test, microneutralization test
The first known infection by influenza A(H9N2) occurred in 1966 among turkeys in Wisconsin. Since the 1990s, the virus has readily circulated among domestic poultry populations in several Asian countries and is now considered to have a near global, albeit sporadic distribution among poultry. Influenza A(H9N2) has also been identified in wild birds, domestic mammals, and, occasionally, in humans [1, 2]. Zoonotic transmission of influenza A(H9N2) was not considered a concern until 1998–1999, when the virus was first isolated from pig samples [2]; seropositive human cases were detected in Guangdong Province, China [3]; and the virus was isolated from hospitalized patients in Hong Kong [1]. Although human influenza A(H9N2) cases have only been reported in Hong Kong and mainland China, and more recently in Bangladesh [1, 4, 5], serological evidence of human infection has been reported in Asia, the Middle East, Africa, and parts of North America (Figure 1).
Figure 1.
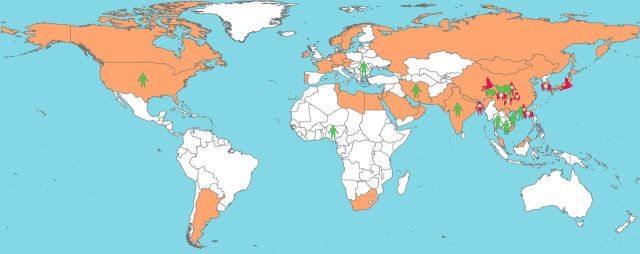
Reported global distribution for influenza A(H9N2) in humans and animals from 1997 to 2013. Countries that detected influenza A(H9N2) in poultry have an orange background. Species-specific symbols were assigned to each country. When a human or animal was found to be seropositive for influenza A(H9N2), the species was colored green; if a virological sample from humans or animals tested positive, the species was colored red.
Human infection with influenza A(H9N2) generally results in mild or asymptomatic illness and often goes unnoticed [6]. However, influenza A(H9N2) is a potential pandemic threat because of its rapid evolution, ability to acquire and transfer genetic materials from other pathogenic subtypes, and efficiency in poultry-to-human transmission [7, 8]. Currently, influenza A(H9N2) infections in poultry have led to multiple reassortments, resulting in many novel genotypes of influenza A(H9N2) with gene segments from various lineages and serving as a constant reminder of its ability to acquire and transfer pandemic potential among the circulating influenza strains [9, 10]. Molecular analyses of the recently emerged influenza A(H7N9) and influenza A(H10N8) suggest that they have acquired gene segments from influenza A(H9N2) [11, 12], which probably enabled them to survive and be repeatedly transmitted among poultry before adapting to humans. Unlike other avian influenza viruses (eg, influenza A[H5N1]) that chiefly bind to human receptors in the lower respiratory tract, influenza A(H9N2) binds to α-2,6 sialic acid receptors that are abundant in the human upper respiratory tract [13], allowing for much greater infection efficiency.
Given the pandemic potential of influenza A(H9N2), it is important to know the burden of human infections. However, the variation attributed to serological assays makes it difficult to accurately estimate the overall burden of influenza A(H9N2) infections. Hence, for this systematic review we sought to evaluate the overall burden of influenza A(H9N2) infection among avian-exposed human populations by summarizing serological data identified in the published literature. In addition, we compared the disease burden revealed by the 2 assays with that yielded by a standardized case definition for serological evidence of influenza A(H9N2) infection.
METHODS
Search Strategy
We performed a systematic, Internet-based search using PubMed, AGRICOLA, and CAB Abstracts databases. The following keywords were used: “influenza H9N2” AND “serological surveys” OR “seroprevalence” OR “sero-prevalence” OR “seroepidemiology” OR “sero-epidemiology”. To minimize publication bias, we retrieved the reference lists and manually searched for relevant studies that met our inclusion criteria. Since the initial serological and virological evidences of first human influenza A(H9N2) infections were reported between 1997 and 1999 [1, 3, 14], our temporal limit for the search was set from January 1997 to December 2013. If data were missing, we contacted the corresponding authors of the studies. We followed PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines for this systematic review and used meta-analysis to aggregate the data [15]. Our protocol for the systematic review will be made available upon request.
Inclusion and Exclusion Criteria
After receiving training, 2 individuals (each with a Masters in Public Health degree) independently reviewed the abstracts and identified articles for detailed assessment. We considered studies regardless of their language, and abstracts that were not written in English were translated. Articles were selected only if an abstract contained data on serological assessment of human samples for evidence of influenza A(H9N2) infection and if the study involved a poultry and wild bird–exposed population. We included surveillance reports, cross-sectional studies, and prospective studies in our data extractions. Studies that were not done in avian-exposed populations, diagnostic evaluations, pathogenesis models, animal models, reviews, comments to editors, perspectives, and personal opinions were excluded because they did not provide primary data for a population-based estimate of influenza A(H9N2) seroprevalence in populations occupationally exposed to poultry and wild birds.
Data Abstraction and Study Outcome
We used a standardized method for extraction of data from the articles. For each of the studies, we first determined the type of serological test they used and the total population tested. Later, we extracted the number of end point–titer positives for each of the dilution cutoffs (1:10 to 1:640). For our analysis, we aggregated hemagglutination inhibition (HI) assay and microneutralization (MN) test positivity data at different dilution cutoffs, as well as data regarding seroconversion (Supplementary Table 1 and 2). For cohort studies, we counted the number of individuals followed up for a specific year and the number of individuals who tested positive by means of the assays or seroconverted during the follow-up period. Results from the cohort studies were broken down by the number of years studied.
Study Quality and Risk-of-Bias Assessment
We assigned 2 reviewers to critically review and assess the quality of the studies and reduced the risk of bias through a structured approach described elsewhere [15, 16]. We assessed each study for 11 assessment criterion: whether it was a population-based study; whether the sample size was ≥100; whether HI was performed; whether MN was performed; whether it used horse red blood cells (RBCs) for the HI assay; whether the World Health Organization (WHO) standard cutoffs for HI assays (an HI titer of ≥1:160 titer or a 4-fold rise in titer over time) were used to define seropositivity [17]; whether the WHO standard cutoffs for MN tests (an MN of ≥1:80 or a 4-fold rise in titer over time) were used to define seropositivity [17]; whether the study population had avian exposures; whether study participants' age distribution was mentioned; whether end point titers of the HI assay were provided in the results; and whether end point titers of the MN assay were provided in the results.
Statistical Analysis
We calculated nonadjusted seroprevalence rates for the range of end point titers used for each diagnostic test. The rates were modeled by applying a random effects meta-analysis model that assumed heterogeneity across the aggregated study population. For each of the titer cutoffs for a test (either HI or MN), we generated a forest plot that showed individual prevalence estimates for each study, by study size.
We assessed heterogeneity by the Pearson χ2 test, which used the I2 index statistic to estimate the proportion of total variation due to heterogeneity [18]. An I2 score of <25% indicated low heterogeneity, and a score of ≥75% suggested a very high degree of heterogeneity.
We considered an HI titer of ≥1:160, an MN titer of ≥1:80, or seroconversion (ie, a 4-fold rise in HI or MN titer over time) as a standardized case definition for influenza A(H9N2) seropositivity, because it is generally recognized as such by the WHO and others [17]. We performed the meta-analysis of the seropositive results through this standard case definition and compared results with the reported influenza A(H9N2)–seropositive results from the selected studies. Because the mean values were markedly affected by outliers, we used median estimates and performed the Wilcoxon rank sum test to assess whether there was a significant difference in the influenza A(H9N2) seroprevalences calculated through the WHO standard case definition and the studies' individual case definitions.
Mapping Influenza A(H9N2) in Humans and Animals
We performed a database search on PubMed and CAB Abstracts to identify reports of human and animal cases of influenza A(H9N2) infection. We also reviewed reports from the WHO (available at: http://www.who.int), the World Organization for Animal Health (available at: http://www.oie.int), and the Food and Agriculture Organization of the United Nations (EMPRES-i data sets; available at: http://empres-i.fao.org/eipws3g/#h=1) to identify reported animal and human influenza A(H9N2) cases and mapped them to show their global distribution based on the serological and virological assessments.
RESULTS
Search Results and Study Selection
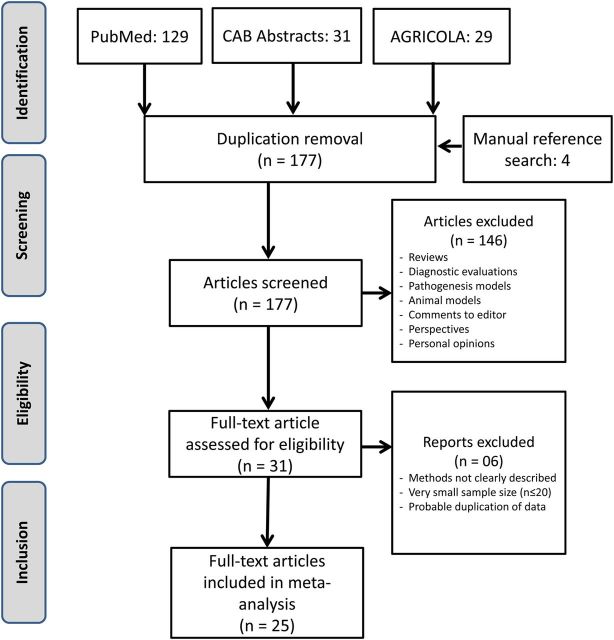
Our literature search identified 193 studies. Sixteen were duplicates and were removed. With the previously stated inclusion and exclusion criteria, we screened the remaining study abstracts and identified 31 articles for full review. We received replies from 5 of 6 authors of the selected articles. Details of the article-screening procedure and reasons for exclusion are summarized in Figure 2.
Figure 2.
Flow chart of the literature search, screening, assessing influenza A(H9N2) serological data for eligibility, and selecting articles for the meta-analysis.
Study Characteristics
Through screening, we identified data from 25 articles that were suitable for meta-analysis [3, 19–43]. The articles were published between 1999 and 2013 from 10 countries of Asia, Africa, the Middle East, and North America. In the majority (78%) of the studies reporting participants’ age, all participants were aged ≥18 years, and all of the reports included participants who had varying degrees of avian exposure (Supplementary Table 3). Avian exposure ranged from being in daily contact with poultry in the live bird markets to having occasional contact with poultry in farms and rural villages and with wild birds. The characteristics of the individual studies are detailed in Supplementary Table 4. Of the 25 articles, 14 (56%) reported results of HI assays, with influenza A(H9N2) seroprevalences ranging from 1% to 43%. MN assay data were available from 14 studies (56%), reported influenza A(H9N2) seroprevalences ranged from 0.0% to 9.1% (Supplementary Table 1 and 2).
Quality and Heterogeneity of Selected Studies
We detailed the quality assessments of the individual studies in Supplementary Table 3. Of the 25 studies included for final analysis, 20 were cross-sectional surveys, 4 were prospective cohort studies, and 1 involved surveillance. Of the 14 articles that reported performing HI assays, 8 (57%) considered a titer cutoff of ≥1:20 for influenza A(H9N2) seropositivity, 4 (29%) had a titer cutoff of ≥1:40, and 2 (14%) had a titer cutoff of ≥1:160 (Supplementary Table 1). Of the 14 studies that reported performing MN assays, 9 (64%) considered a ≥1:10 titer cutoff for influenza A(H9N2) seropositivity, 2 (14%) had a titer cutoff of ≥1:20, and 2 (14%) had a titer cutoff of ≥1:40 (Supplementary Table 2).
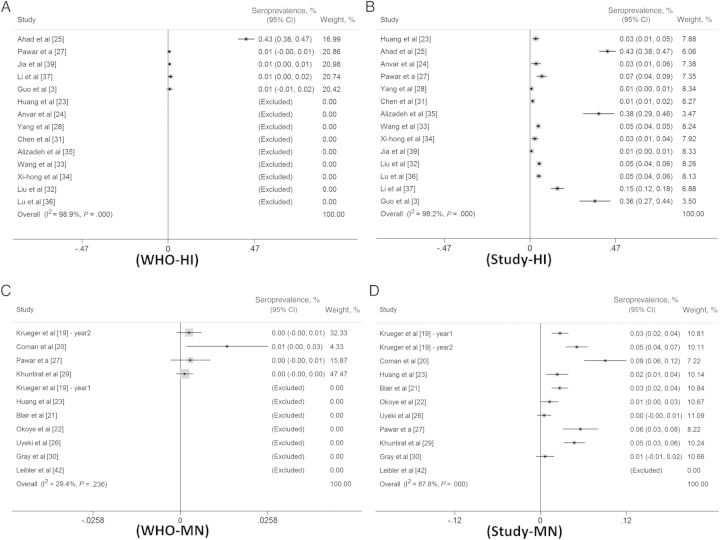
We identified a high degree of heterogeneity of the selected studies that performed HI assays (I2 = 0.98; P < .001) and MN assays (I2 = 0.88; P < .001) with their individually defined cutoffs for influenza A(H9N2) seropositivity (Figure 3).
Figure 3.
Forest plots of the overall seroprevalence calculated from hemagglutination inhibition (HI) and microneutralization (MN) assay results, using reported cutoff corresponded with the standard cutoffs recommended by the World Health Organization (WHO) (ie, HI assay titers of ≥1:160, MN assay titers of ≥1:80, or seroconversion, defined as a 4-fold rise in HI or MN titers over time). A, Forest plot of seroprevalence calculated from HI assay results, using the WHO-recommended cutoff (≥1:160). B, Forest plot of seroprevalence calculated from HI assay results, using study cutoffs. C, Forest plot of seroprevalence calculated from MN assay results, using the WHO recommended cutoff (≥1:80). D, Forest plot of seroprevalence calculated from MN assay results, using the study cutoff.
Influenza A(H9N2) Seroprevalence
The meta-analysis demonstrated that the overall median HI seroprevalence calculated using the antibody cutoffs reported by the studies was 4.9% (range, 0.6%–42.6%; I2 = 0.99), which was not significantly different from the standardized case definition (≥1:160) for seropositivity (median seroprevalence, 1.3%; range, 0.5%–42.6%; I2 = 0.99; Figure 3A and 3B). For the MN meta-analysis, the overall reported median seroprevalence was 2.7% (range, 0.5%–9%; I2 = 0.88), which was significantly greater (P < .05) than that yielded by the standardized case definition (≥1:80) for seropositivity (median seroprevalence, 0.3%; range, 0.1%–1.4%; I2 = 0.29; Figure 3C and 3D).
The HI and MN assay titers at individual cutoffs were analyzed for meta-seroprevalence and plotted with their 95% confidence intervals in Supplementary Figure 1. Data presented in Supplementary Figure 1A demonstrated that the seroprevalence at an HI dilution of ≥1:10 was about 9%, which gradually decreased to 4% at a dilution of ≥1:160 and peaked to 8% at a dilution of ≥1:320 before diminishing at a ≥1:640 dilution. Supplementary Figure 1B demonstrated a gradual decrease in MN titer positivity, from 1.7% to 0.05%, from dilution 1:10 to dilution 1:320.
Global Distribution of Influenza A(H9N2) in Humans and Animals
Figure 1 shows the global distribution of reported influenza A(H9N2) infections in humans, poultry, pet birds, and other domestic animals. Influenza A(H9N2) was detected in humans chiefly in China, Hong Kong, and Bangladesh. However, serological evidence of human influenza A(H9N2) infection has been found in Asia, the Middle East, Africa, and North America. Thus far, influenza A(H9N2) has been detected in wild waterfowl, pigs, dogs, horses, and a companion bird in China, Hong Kong, and Japan. However, the virus had a near global distribution in domestic poultry populations.
DISCUSSION
We performed a systematic review and meta-analysis with the goal of estimating the global prevalence of influenza A(H9N2) infection among avian-exposed populations. Studies reporting seroconversion on the basis of HI and MN assay results identified that a relatively small proportion of the at-risk population had evidence of previous infection with influenza A(H9N2). Compared with the seroprevalence of influenza A(H9N2) reported by the studies, the WHO-recommended standard cutoffs resulted in an even lower seroprevalence through MN assays.
Although the prevalence of human infection is low, it may still pose a global pandemic threat because of the following factors. First, the virus has a near global distribution. Should these viruses become highly transmissible and virulent, they may cause marked worldwide morbidity. Although reports on influenza A(H9N2) antibody kinetics are scarce, kinetic data from influenza A(H5N1) infections suggest that individuals who had mild influenza A(H5N1) infection experienced reduced antibody titers, dropping from 4-fold to 32-fold 11 months after infection [44]. Second, there is considerable probability of the immune system failing to produce an anti-H9 neutralizing antibody response [44, 45]. Humans in general have poor immune responses to infections with avian viruses. For instance, studies have shown that individuals with mild or asymptomatic influenza A(H5N1) infection had a lower initial antibody titer than individuals who developed severe illness [44]. Third, because influenza A(H9N2) commonly causes mild or asymptomatic human infections, it seems reasonable to assume that the infection will trigger a weaker immune response and lower detectable antibody titers than other highly pathogenic influenza viruses. However, there is currently no universally agreed upon antibody titer cutoff for these assays, which explains why different antibody titer cutoffs were used in the included studies. Selecting a low titer cutoff can often lead to overestimation of the seroprevalence of influenza A virus infections, particularly due to cross-reactivity with commonly circulating seasonal influenza viruses, resulting in a low sensitivity for assays such as the HI assay [45–47]. To overcome such limitations of traditional MN and HI assays, newer diagnostic tests, such as the virus-specific T-cell response assay and the subtype-specific pseudotype particle-based MN assay, have been developed [44, 45]. However, one of the major limitations of these new assays is their time dependence. The assay results vary depending on when a case was sampled after the hosts' infection. This is particularly striking for the virus-specific T-cell response assay because T-cell responses are only detectable within a limited interval after an infection [48]. Overcoming these limitations may require the development of standardized sampling protocols specific to the assays or the development of a modified assay that minimizes these constraints.
Low rates of influenza A(H9N2) infection in poultry-exposed individuals may cause an epidemic with a longer duration or a greater magnitude than if the virus was introduced to a completely unexposed population [49]. The virus also demonstrates its potential to efficiently transmit itself from avian hosts and share genetic materials with other highly pathogenic influenza A virus subtypes of public health significance (eg, H5N1, H7N9, and H10N8) [11, 12, 50]. These phenomena raise the possibility that influenza A(H9N2) may acquire genes from highly pathogenic influenza virus subtypes and cause large epidemics. It also seems possible that influenza A(H9N2) may transfer genes to other low-pathogenic influenza viruses to cause sporadic human infections, as has occurred with influenza A(H7N9) and influenza A(H10N8).
Our study has multiple limitations. First, we had to calculate the nonadjusted seroprevalence of influenza A(H9N2) infection in avian-exposed populations, because not all studies reported demographic characteristics (eg, age distribution) for their sampled populations (Supplementary Table 3). Second, only a handful of the studies included in our analysis used horse RBCs for their HI assay; not doing so is thought to reduce the sensitivity of the outcome. Third, we generally expected to identify fewer individuals to have higher antibody titers, because titers wane over time. Our analysis identified a nonlinear decrease in seroprevalence over increasing titer cutoffs through HI assays (Supplementary Figure 1A). This is not surprising, because many of the studies included for meta-analysis were cross-sectional in nature, and the individuals were expected to have elevated titers of antibodies against influenza A(H9N2). This may also be the result of lower sensitivity and specificity of the HI assay, which is supported by Supplementary Figure 1B, which shows a linear decreasing trend of influenza A(H9N2) antibody titers measured by the MN assay. Finally, a few of the studies used noncirculating influenza A(H9N2) strains in performing the MN assay, which might have reduced the sensitivity for detecting cases. However, because the virus has a near global distribution in poultry and efficient surveillance tools are not always in place, the studies might have identified evidence of infections that were missed by routine surveillance efforts.
Findings from the meta-analysis suggest that few avian-exposed humans are infected with influenza A(H9N2). Because many of the influenza virus subtypes, including influenza A(H9N2), produces mild or asymptomatic infections in humans, the current serological assays have a limited sensitivity and specificity to identify evidence of infections. Developing standardized sampling protocols specific to the assays or development of a modified assay that could detect serological evidence of mild or asymptomatic infections could minimize the current constraints. However, should influenza A(H9N2) further adapt to human hosts and develop more-virulent characteristics, they may spread quite rapidly among humans, causing significant morbidity and mortality. Efforts should be increased to conduct more-aggressive surveillance for influenza A(H9N2) strains, such that genetic changes might be identified in time to provide prepandemic warnings.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgment. We thank Robert Albury for reviewing the manuscript and adding his grammatical edits.
Financial support. This work was supported by the Department of Defense Armed Forces Health Surveillance Center's Global Emerging Infections Surveillance and Response Program (multiple grants), the National Institute of Allergy and Infectious Diseases (grant R01 AI108993), the Department of Agriculture's National Institute of Food and Agriculture (grant 2011-51110-31199), and the University of Florida Emerging Pathogens Institute.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Peiris M, Yuen KY, Leung CW, et al. Human infection with influenza H9N2. Lancet 1999; 354:916–7. [DOI] [PubMed] [Google Scholar]
- 2.Guo YJ, Krauss S, Senne DA, et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 2000; 267:279–88. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, Li J, Cheng X. Discovery of men infected by avian influenza A (H9N2) virus [in Chinese]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 1999; 13:105–8. [PubMed] [Google Scholar]
- 4.International Centre for Diarrhoeal Disease Research, Bangladesh/Government of The People's Republic of Bangladesh. Outbreak of mild respiratory disease caused by H5N1 and H9N2 infections among young children in Dhaka, Bangladesh. Health Sci Bull 2011; 9:5–12. [Google Scholar]
- 5.Cheng VC, Chan JF, Wen X, et al. Infection of immunocompromised patients by avian H9N2 influenza A virus. J Infect 2011; 62:394–9. [DOI] [PubMed] [Google Scholar]
- 6.Malik Peiris JS. Avian influenza viruses in humans. Rev Sci Tech 2009; 28:161–73. [DOI] [PubMed] [Google Scholar]
- 7.Lam TT-Y, Wang J, Shen Y, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 2013; 502:241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashashati M, Vasfi Marandi M, Sabouri F. Genetic diversity of early (1998) and recent (2010) avian influenza H9N2 virus strains isolated from poultry in Iran. Arch Virol 2013; 158:2089–100. [DOI] [PubMed] [Google Scholar]
- 9.Iqbal M, Yaqub T, Reddy K, McCauley JW. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS One 2009; 4:e5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li KS, Xu KM, Peiris JS, et al. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J Virol 2003; 77:6988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Yuan H, Gao R, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 2014; 383:714–21. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Shi W, Shi Y, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 2013; 381:1926–32. [DOI] [PubMed] [Google Scholar]
- 13.Shelton H, Ayora-Talavera G, Ren J, et al. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J Virol 2011; 85:1875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uyeki TM, Chong YH, Katz JM, et al. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg Infect Dis 2002; 8:154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009; 6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayden JA, Cote P, Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med 2006; 144:427–37. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization (WHO). Recommendations and laboratory procedures for detection of avian influenza A(H5N1) virus in specimens from suspected human cases. Geneva: WHO, 2007. http://www.who.int/influenza/resources/documents/RecAIlabtestsAug07.pdf Accessed 17 June 2014. [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003; 327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueger WS, Khuntirat B, Yoon IK, et al. Prospective study of avian influenza virus infections among rural thai villagers. PLoS One 2013; 8:e72196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coman A, Maftei DN, Krueger WS, et al. Serological evidence for avian H9N2 influenza virus infections among Romanian agriculture workers. J Infect Public Health 2013; 6:438–47. [DOI] [PubMed] [Google Scholar]
- 21.Blair PJ, Putnam SD, Krueger WS, et al. Evidence for avian H9N2 influenza virus infections among rural villagers in Cambodia. J Infect Public Health 2013; 6:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okoye J, Eze D, Krueger WS, Heil GL, Friary JA, Gray GC. Serologic evidence of avian influenza virus infections among Nigerian agricultural workers. J Med Virol 2013; 85:670–6. [DOI] [PubMed] [Google Scholar]
- 23.Huang R, Wang AR, Liu ZH, et al. Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol 2013; 32:1347–51. [DOI] [PubMed] [Google Scholar]
- 24.Anvar E, Hosseini SM, Kheiri MT, et al. Serological survey of avian influenza (H9N2) among different occupational groups in Tehran and Qazvin provinces in IR Iran. Jundishapur J Microbiol 2013; 6:e5441. [Google Scholar]
- 25.Ahad A, Rabbani M, Yaqub T, et al. Serosurveillance to H9 and H7 avian influenza virus among poultry workers in Punjab Province, Pakistan. Pak Vet J 2013; 33:107–12. [Google Scholar]
- 26.Uyeki TM, Nguyen DC, Rowe T, et al. Seroprevalence of antibodies to avian influenza A (H5) and A (H9) viruses among market poultry workers, Hanoi, Vietnam, 2001. PLoS ONE 2012; 7:e43948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawar SD, Tandale BV, Raut CG, et al. Avian influenza H9N2 seroprevalence among poultry workers in Pune, India, 2010. PLoS One 2012; 7:e36374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang P, Ma C, Shi WX, et al. A Serological Survey of antibodies to H5, H7 and H9 avian influenza viruses amongst the duck-related workers in Beijing, China. PLoS ONE 2012; 7:e50770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khuntirat BP, Yoon IK, Blair PJ, et al. Evidence for subclinical avian influenza virus infections among rural Thai villagers. Clin Infect Dis 2011; 53:e107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray GC, Ferguson DD, Lowther PE, Heil GL, Friary JA. A national study of US bird banders for evidence of avian influenza virus infections. J Clin Virol 2011; 51:132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Zheng Q, Yang K, et al. Serological survey of antibodies to influenza A viruses in a group of people without a history of influenza vaccination. Clin Microbiol Infect 2011; 17:1347–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Lu EJ, Wang YL, et al. Avian influenza virus infection in people occupied in poultry fields in Guangzhou city [in Chinese]. Zhonghua Liu Xing Bing Xue Za Zhi 2009; 30:1111–3. [PubMed] [Google Scholar]
- 33.Wang M, Fu CX, Zheng BJ. Antibodies against H5 and H9 avian influenza among poultry workers in China. New Engl J Med 2009; 360:2583–4. [DOI] [PubMed] [Google Scholar]
- 34.Lv XH, Jiang CY, Zhou YB. Serological survey on antibodies to influenza A viruses subtype HI, H3, H5 and H9 of population in Shanghai. Chin J Epidemiol 2009; 30:302. [PubMed] [Google Scholar]
- 35.Alizadeh E, Hosseini SM, Kheiri MT, Mazaheri V, Bashar R, Tabatabaeian M. Avian Influenza (H9N2) among poultry workers in Iran. Iran J Microbiol 2009; 1:3–6. [Google Scholar]
- 36.Lu CY, Lu JH, Chen WQ, et al. Potential infections of H5N1 and H9N2 avian influenza do exist in Guangdong populations of China. Chinese Med J-Peking 2008; 121:2050–3. [PubMed] [Google Scholar]
- 37.Li CH, Zhou XZ, Li MX. Discoveries of avian influenza A(H9N2) virus in chickens and men infected by H9N2 virus in Guangzhou area [in Chinese]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2004; 18:213–4. [PubMed] [Google Scholar]
- 38.Cheng X, Liu J, He J, Shan F. Virological and serological surveys for H9N2 subtype of influenza A virus in chickens and men in Shenzhen city [in Chinese]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2002; 16:319–21. [PubMed] [Google Scholar]
- 39.Jia N, de Vlas SJ, Liu YX, et al. Serological reports of human infections of H7 and H9 avian influenza viruses in northern China. J Clin Virol 2009; 44:225–9. [DOI] [PubMed] [Google Scholar]
- 40.Gray GC, McCarthy T, Capuano AW, Setterquist SF, Alavanja MC, Lynch CF. Evidence for avian influenza A infections among Iowa's agricultural workers. Influenza Other Respir Viruses 2008; 2:61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kayali G, Ortiz EJ, Chorazy ML, Gray GC. Evidence of previous avian influenza infection among US turkey workers. Zoonoses Public Hlth 2010; 57:265–72. [DOI] [PubMed] [Google Scholar]
- 42.Leibler JH, Silbergeld EK, Pekosz A, Gray GC. No Evidence of infection with avian influenza viruses among US poultry workers in the Delmarva Peninsula, Maryland and Virginia, USA. J Agromedicine 2011; 16:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers KP, Setterquist SF, Capuano AW, Gray GC. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin Infect Dis 2007; 45:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchy P, Vong S, Chu S, et al. Kinetics of neutralizing antibodies in patients naturally infected by H5N1 virus. PLoS ONE 2010; 5:e10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powell TJ, Fox A, Peng Y, et al. Identification of H5N1-specific T-cell responses in a high-risk cohort in vietnam indicates the existence of potential asymptomatic infections. J Infect Dis 2012; 205:20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowe T, Abernathy RA, Hu-Primmer J, et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol 1999; 37:937–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia JM, Pepin S, Lagarde N, et al. Heterosubtype neutralizing responses to influenza A (H5N1) viruses are mediated by antibodies to virus haemagglutinin. PLoS ONE 2009; 4:e7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMichael AJ, Gotch FM, Dongworth DW, Clark A, Potter CW. Declining T-cell immunity to influenza, 1977–82. Lancet 1983; 2:762–4. [DOI] [PubMed] [Google Scholar]
- 49.Pulliam JR, Dushoff JG, Levin SA, Dobson AP. Epidemic enhancement in partially immune populations. PLoS ONE 2007; 2:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA 1999; 96:9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.