Summary
Black raspberry treatment modulates multiple metabolic pathways, including amino acid, glutathione, lipid and nucleotide metabolisms. Putrescine and linolenate might be markers of BRB intervention.
Abstract
Freeze-dried black raspberries (BRBs) have demonstrated chemopreventive effects in a dietary intervention trial with human colorectal cancer patients. The aim of this study was to investigate BRB-caused metabolite changes using the ApcMin/+ mouse as a model of human colorectal cancer. Wild-type (WT) mice were fed control diet, and ApcMin/+ mice were fed either control diet or control diet supplemented with 5% BRBs for 8 weeks. Colonic and intestinal polyp size and number were measured. A non-targeted metabolomic analysis was conducted on colonic mucosa, liver and fecal specimens. Eight weeks of BRB treatment significantly decreased intestinal and colonic polyp number and size in ApcMin/+ mice. The apc gene mutation significantly changed 52 metabolites in colonic mucosa associated with increased amino acid and decreased lipid metabolites, as well as 39 liver and 8 fecal metabolites. BRBs significantly reversed 23 apc-regulated metabolites, including 13 colonic mucosa, 8 liver and 2 fecal metabolites that were involved in amino acid, glutathione, lipid and nucleotide metabolism. Of these, changes in eight metabolites were linearly correlated with decreased colonic polyp number and size in BRB-treated ApcMin/+ mice. Elevated levels of putrescine and linolenate in ApcMin/+ mice were significantly decreased by BRBs. Ornithine decarboxylase expression, the key enzyme in putrescine generation, was fully suppressed by BRBs. These results suggest that BRBs produced beneficial effects against colonic adenoma development in ApcMin/+ mice and modulated multiple metabolic pathways. The metabolite changes produced by BRBs might potentially reflect the BRB-mediated chemopreventive effects in colorectal cancer patients.
Introduction
Colorectal cancer is the third most common cancer in the world (1). Mutation in the Adenomatous polyposis coli (apc) gene, a tumor suppressor gene that functions to inhibit the Wnt signaling pathway by promoting β-catenin degradation, is the strongest contributing factor to colorectal cancer (2,3). Multiple intestinal neoplasia mice (ApcMin/+) carry a truncating mutation at codon 850 of the apc gene, which results in an enhanced activation of the Wnt signaling pathway. ApcMin/+ mice develop numerous colonic and intestinal polyps within a short period, making it a widely used model of human colorectal cancer phenotype (3).
Dietary factors can strongly influence the risk of colorectal cancer development. Methylation on the apc gene promoter region, which is frequently observed in colorectal cancer patients, has been linked to a high intake of red meat and processed meat and with low levels of folate and fiber (4). In contrast, many bioactive foods, such as berries (5), tea (6), broccoli (7) and beans (8), have demonstrated beneficial effects against the development of colorectal cancer. Black raspberries (BRBs), which contain high levels of multiple chemopreventive compounds, such as anthocyanins and ellagitannins (5), have been shown to inhibit cell transformation, proliferation and tumor-specific gene expression, as well as to promote apoptosis and differentiation (9). Cyanidin-3-O-glucoside, the most abundant anthocyanin in our diet, inhibited adenoma development in ApcMin/+ mice (10).
Treating rats with 5% BRBs in the diet has been shown to have a greater inhibition against azoxymethane-induced colon adenocarcinomas than 2.5% BRBs. Increasing to 10% BRBs, however, showed no benefit over the 5% dose (11). Similar effects have been observed between 5% BRBs and 10% BRBs against N-nitrosomethylbenzylamine-induced rat esophageal tumors (12). In addition, our previous human clinical trial with a group of 28 colorectal cancer patients demonstrated that 60g of BRBs daily, which is equivalent to a rodent diet of 7% BRBs, significantly suppressed colon cancer cell proliferation (13). BRBs exhibited protective effects against colorectal cancer cells through suppressing β-catenin and reducing chronic inflammation (14). Therefore, in the current study, we used the ApcMin/+ mouse model to recapitulate the phenotype of human colorectal cancer and aimed to address potential metabolite changes caused by 5% BRBs. A non-targeted metabolomic study was performed to investigate if BRB-mediated metabolite changes are associated with their chemopreventive effects against colorectal cancer development in ApcMin/+ mice. An 8-week dietary BRB intervention significantly inhibited colonic adenoma development and reversed 13 colonic mucosa, 8 liver and 2 fecal apc-induced metabolites. Among these, BRB-caused changes in 8 metabolites were linearly correlated with decreased colonic polyp size and number. Importantly, two of these metabolites, putrescine (10,15) and linolenate (16) have been linked to the development of human colorectal cancer. Changes in their levels might serve as potential biomarkers of BRB intervention. In summary, our data suggest that BRBs produce systemic effects and modulate multiple metabolic pathways that may reflect their ability to protect against the development of colorectal cancer in human patients.
Materials and methods
Animals and BRBs
All protocols were carried out in accordance with institutional guidelines for animal care dictated by the Medical College of Wisconsin Animal Care and Use Committee. Wild-type (WT) and ApcMin/+ mice were purchased from the Jackson Laboratory (Bar Harbor, ME) when they were 3–4 weeks old. The animals were placed in the protocol 1 week after they arrived from the vendor, and the study was of 8 weeks duration. WT mice (n = 20) were fed control diet, and ApcMin/+ mice were fed either control diet (n = 19) or control diet supplemented with 5% BRBs (n = 24). The control diet was the American Institute of Nutrition synthetic diet 76A (AIN-76A; Dyets, Bethlehem, PA). Procedures for the preparation of BRB powder have been described by Montrose et al. (17); the same batch of BRB powder used by Montrose and colleagues was used in this study. Mice were euthanized by CO2 asphyxiation. Colonic and intestinal polyp number and size were measured. Liver, colonic mucosa and fecal specimens were collected from a subgroup of control diet-treated WT mice (n = 7), control diet-treated ApcMin/+ mice (n = 6) and 5% BRB-treated ApcMin/+ mice (n = 9) for metabolomic profiling.
Metabolomic profiling
Mass spectrometer platforms, sample extraction and preparation, instrument settings and conditions and data handling were performed at Metabolon, Inc. (Research Triangle Park, NC) and have been described previously in detail (18–20). The major components of the process are summarized briefly as follows: The sample preparation process was carried out using an automated MicroLab STAR® system (Hamilton, Reno, NV). Recovery standards were added prior to the first step in the extraction process for QC purposes. Homogenized liver and colonic mucosa samples were extracted using 5 µl methanol per mg tissue. Homogenized fecal samples were extracted using 25 µl methanol per mg feces. The resulting extract was divided into three fractions for non-targeted metabolic profiling and randomized for analysis. Each sample was dried under vacuum to remove the organic solvent. Samples were characterized using three independent platforms: ultra-high performance liquid chromatography/tandem mass spectrometry (UHPLC-MS/MS) in the negative ion mode, UHPLC-MS/MS in the positive ion mode and gas chromatography-mass spectrometry (GC-MS) after sialylation. The reproducibility of the extraction protocol was assessed by the recovery of the xenobiotic compounds spiked in every sample prior to extraction. Identification of known chemical entities was based on comparison to metabolomic library entries of purified standards based on chromatographic properties and mass spectra. The GC-MS-MS data and the LC-MS-MS data for each tissue matrix were listed in Supplementary Tables 1–6, available at Carcinogenesis Online.
Immunohistochemistry and computer-assisted image analysis
Pre- and post-treatment colonic specimens were cut into 4 μm sections and placed on slides. The slides were placed in a 60°C oven for 1h, cooled, deparaffinized and rehydrated through xylenes and graded ethanol solutions to water. All slides were treated for 5min with a 3% H2O2 solution in water to block endogenous peroxidase. The slides were stained using a Dako Autostainer with a primary antibody to ornithine decarboxylase (ODC) (1:300, ab193338, Abcam, Cambridge, MA) for 1h at room temperature. Stained slides were viewed and photographed at ×200 magnification with a bright-field microscope mounted with a high-resolution spot camera. The camera was interfaced with a computer containing a Matrox frame grabber board (Matrox Electronic Systems Dorval, Canada) and image analysis software (Simple PCI Imaging Systems, Compix). All staining was quantified by the image analysis software program.
Measurements of fecal microbial populations by quantitative real-time PCR
Fecal specimens were collected from control diet-treated WT mice, control diet-treated ApcMin/+ mice and BRB-treated ApcMin/+ mice at the end of the study. Fecal DNA samples were isolated using PowerSoil® DNA Isolation Kit (MO Bio laboratories, Carlsbad, CA). Quantitative real-time PCR was performed to measure the populations of Lactobacillus (Forward: AGCAGTAGGGAATCTTCCA; Reverse: CACCGCTACACATGGAG), Bacteroidiaceae (Forward: GGTTCTGAGAGGAGGTCCC; Reverse: GCTGCCTCC CGTAGGAGT), Bifidobacteriales (Forward: CTCCTGGAAACGGGTGGT; Reverse: GCTGCCTCCCGTAGGAGT) and Ruminococcus (Forward: ACTCCTAC GGGAGGCAGC; Reverse: GCTTCTTAGTCAGGTACCGTCAT).
Statistical analysis
Welsh’s two sample t-test was employed in R version 2.14.2 (21) to identify statistically significant metabolite differences in the different matrices, between micefed BRB diet and control diet. For all analyses, missing values (if any) were imputed with the observed minimum for that particular compound (imputed values were added after block-normalization). The statistical analyses were performed on natural log-transformed data to reduce the effect of any potential outliers in the data. One-way analysis of variance and linear regression tests were performed using SigmaStat3.5 to determine the changes of ODC, the association between metabolites and colonic polyp number/size, and the changes of fecal microbial populations. A P < 0.05 was considered statistically significant.
Results
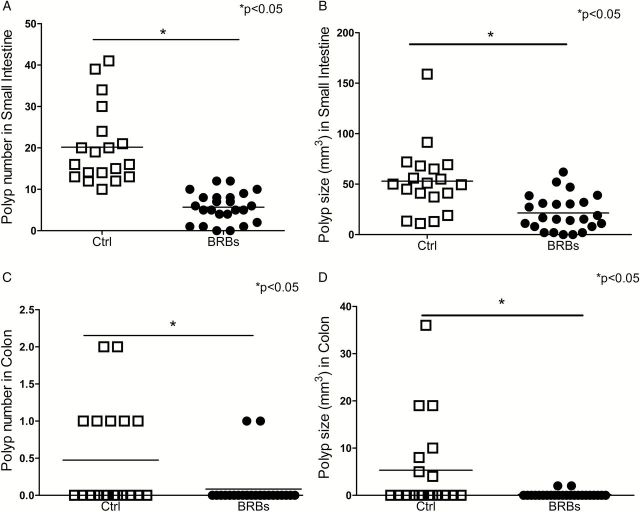
The ApcMin/+ mice were given either AIN-76A control diet or control diet containing 5% BRBs for 8 weeks. Control diet-treated age-matched WT mice were included as non-cancer controls. The ApcMin/+ mice primarily develop small intestine polyps and less frequently develop colonic polyps (22,23). By the end of this study, 100% of the ApcMin/+ mice on the control diet had developed multiple polyps in the small intestine, while only 88% of the mice fed the BRB diet developed intestinal polyps. More importantly, there was a significant 78% decrease in colonic polyps in BRB-treated ApcMin/+ mice. 37% (7 of 19) of control diet-treated ApcMin/+ mice developed colonic polyps, whereas only 8% (2 of 24) of BRB-treated ApcMin/+ mice developed colonic polyps. The intestinal (Figure 1A and B) and colonic (Figure 1C and D) polyps that developed in BRB-treated mice were significantly smaller than those in control diet-treated mice.
Figure 1.
Dietary supplementation with 5% BRBs significantly decreased intestinal and colonic polyp number and size in ApcMin/+ mice. (A) Polyp number in small intestine; (B) polyp size in small intestine; (C) polyp number in colon; (D) polyp size in colon. Open squares represent control diet-treated ApcMin/+ mice, dots represent 5% BRB diet-treated ApcMin/+ mice. *P < 0.05.
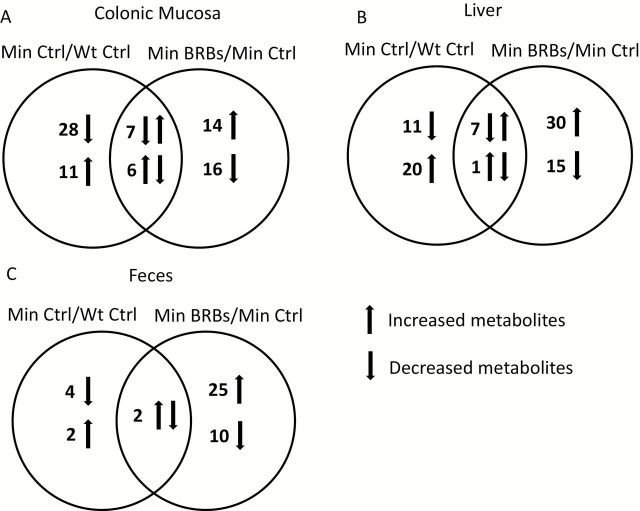
To determine metabolite changes caused by a mutated apc gene, we collected colonic mucosa, liver and fecal specimens from a subgroup of ApcMin/+ and WT mice. A MS-based non-targeted metabolomic analysis was conducted and a total of more than 400 metabolites were annotated in each specimen. Of these, the levels of 52 colonic mucosa, 39 liver and 8 fecal metabolites were significantly changed in control diet-treated ApcMin/+ mice compared to control diet-treated WT mice (Min Ctrl versus WT Ctrl) (Figure 2 and Supplementary Table 7, available at Carcinogenesis Online). Treatment of ApcMin/+ mice with 5% BRBs resulted in a significant modulation of 43 colonic mucosa, 53 liver and 37 fecal metabolites when compared with control diet-treated ApcMin/+ mice (Min BRBs versus Min Ctrl) (Figure 2 and Supplementary Table 8, available at Carcinogenesis Online). Of these, 13 colonic mucosa, 8 liver and 2 fecal apc-induced metabolites were reversed by BRBs. Significantly changed metabolites were annotated using the Koto Encyclopedia of Genes and Genomes (KEGG) database to identify specific metabolic pathways associated with the apc gene mutation and modulated by BRBs. Potential metabolic pathways were associated with amino acid, glutathione, lipid and nucleotide metabolism (Table 1).
Figure 2.
Number of metabolites that are significantly changed by the apc gene mutation or BRBs in colonic mucosa (A), liver (B) and feces (C). Left circle represents metabolites that are significantly changed by the apc gene mutation (Min Ctrl/WT Ctrl); right circle represents metabolites that are significantly regulated by BRBs (Min BRBs/Min Ctrl); overlapped area represents metabolites that are significantly changed by the apc gene mutation and reversed by BRBs.
Table 1.
List of metabolites that are significantly modulated by the apc gene mutation and significantly reversed by BRBs
| Colon mucosa | Metabolic pathways | Metabolite | Min Ctrl/WT Ctrl | Min BRBs/Min Ctrl |
|---|---|---|---|---|
| Amino acid | Phenylalanine and tyrosine metabolism | N-acetylphenylalanine | 1.54 | 0.76 |
| Phenylacetylglycine | 12.50 | 0.11 | ||
| p-cresol sulfate | 4.35 | 0.28 | ||
| Lipid | Polyunsaturated fatty acid (n3 and n6) | Stearidonate (18:4n3) | 0.29 | 2.86 |
| Linolenate [alpha or gamma; (18:3n3 or 6)] | 0.49 | 1.99 | ||
| Lysolipid | 2-docosahexaenoylglycerophosphocholine | 0.37 | 3.84 | |
| 2-docosahexaenoylglycerophosphoethanolamine | 0.33 | 3.34 | ||
| Monoacylglycerol | 1-Docosahexaenoylglycerol (1-monodocosahexaenoin) | 0.54 | 3.5 | |
| Sphingolipid metabolism | N-palmitoyl-D-erythro-sphingosine | 0.57 | 1.91 | |
| Primary bile acid metabolism | Tauro(alpha + beta)muricholate | 1.79 | 0.5 | |
| Nucleotide | Purine metabolism | Inosine | 0.25 | 8.56 |
| Cofactor and Vitamin | Nicotinate and nicotinamide metabolism | Nicotinamide | 1.27 | 0.71 |
| Xenobiotics | Benzoate metabolism | 4-methylcatechol sulfate | 1.96 | 0.47 |
| Liver | Metabolic pathways | Metabolite | Min Ctrl/WT Ctrl | Min BRBs/Min Ctrl |
| Amino acid | Glutamate metabolism | Gamma-aminobutyrate (GABA) | 0.67 | 1.73 |
| Glutathione metabolism | S-methylglutathione | 1.30 | 0.78 | |
| S-lactoylglutathione | 0.42 | 1.65 | ||
| 5-Oxoproline | 0.75 | 1.31 | ||
| Ophthalmate | 0.37 | 2.16 | ||
| Lipid | Fatty acid metabolism | Hydroxybutyrylcarnitine | 0.54 | 2.47 |
| Ketone bodies | 3-hydroxybutyrate (BHBA) | 0.56 | 2 | |
| Feces | Metabolic pathways | Metabolite | Min Ctrl/WT Ctrl | Min BRBs/Min Ctrl |
| Amino acid | Polyamine metabolism | Putrescine | 7.69 | 0.22 |
| Lipid | Polyunsaturated fatty acid (n3 and n6) | Docosapentaenoate (n6 DPA; 22:5n6) | 2.44 | 0.37 |
Fold change is calculated as the ratio of Min Ctrl versus WT Ctrl and Min BRBs versus Min Ctrl.
Amino acid metabolism
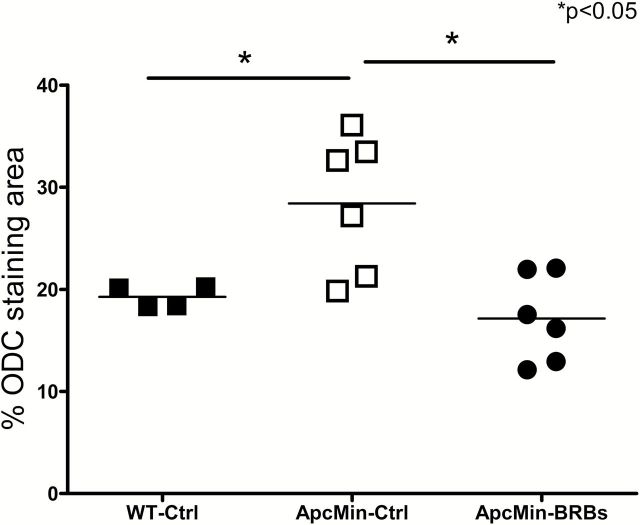
The apc gene mutation induced significant changes in amino acid metabolism in control diet-treated ApcMin/+ mice compared to WT mice (Supplementary Table 7, available at Carcinogenesis Online). Most of these changes in amino acid metabolites were associated with lysine, phenylalanine, tyrosine, tryptophan and polyamine metabolism. Interestingly, 8-week BRB intervention significantly reversed 6 amino acid metabolites in BRB-treated ApcMin/+ mice (Table 1), i.e. N-acetylphenylalanine, phenylacetylglycine and p-cresol sulfate in colonic mucosa, gamma-aminobutyrate and 2-aminobutyrate in liver, and putrescine in feces. Expression of ODC, which catalyzes the decarboxylation of ornithine to form putrescine, was significantly increased in colonic crypts in control diet-treated ApcMin/+ mice compared to WT mice (Figure 3). More importantly, BRBs significantly reversed the enhanced expression of colonic ODC levels in ApcMin/+ mice (Figure 3 and Supplementary Figure 1, available at Carcinogenesis Online).
Figure 3.
BRBs significantly reversed the increased levels of ODC in ApcMin/+ mice. Solid squares represent control diet-treated WT mice, open squares represent control diet-treated ApcMin/+ mice, dots represent 5% BRB diet-treated ApcMin/+ mice. *P < 0.05.
Glutathione metabolism
The apc gene mutation caused unique changes in glutathione metabolism in liver with significantly increased levels of S-methylglutathione and decreased levels of S-lactoylglutathione, 5-oxoproline and ophthalmate (Table 1). BRBs fully reversed all four metabolite changes in livers of BRB-treated ApcMin/+ mice (Table 1).
Lipid metabolism
The apc gene mutation substantially decreased 32 lipid metabolite levels in colonic mucosa, including medium-chain and long-chain fatty acids, polyunsaturated fatty acids (PUFAs), monohydroxy fatty acids, inositols, phospholipids, lysolipids, monoacylglycerols and sphingolipids compared to WT mice (Supplementary Table 7, available at Carcinogenesis Online). Of these, BRBs significantly increased 6 metabolites, stearidonate (18:4n3), linolenate (18:3n3/6), 2-docosahexaenoylglycerophosphocholine, 2-docosahexaenoylglycerophosphoethanolamine, 1-docosahexaenoylglycerol and N-palmitoyl-D-erythro-sphingosine in colonic mucosa (Table 1). In addition, two bile acid metabolites, deoxycholate and tauro (α + β) muricholate, were significantly increased in colonic mucosa of control diet-treated ApcMin/+ mice (Supplementary Table 7, available at Carcinogenesis Online), and BRBs significantly reduced the levels of tauro (α + β) muricholate in ApcMin/+ mice (Table 1). In liver, the apc gene mutation significantly increased 11 and decreased 4 lipid metabolites (Supplementary Table 7, available at Carcinogenesis Online). Among these metabolites, decreased levels of hydroxybutyrylcarnitine and 3-hydroxybutyrate were reversed by BRBs (Table 1). In feces, BRB intervention downregulated docosapentaenoate (n6 DPA; 22:5n6) levels (Table 1), which were increased in control diet-treated ApcMin/+ mice relative to WT controls (Table 1).
Nucleotide metabolism
Inosine can be converted to hypoxanthine, which can be further converted to xanthine and urate through xanthine oxidase. ApcMin/+ mice had lower levels of inosine and xanthosine in colonic mucosa (Supplementary Table 7, available at Carcinogenesis Online), while BRBs elevated inosine levels in ApcMin/+ mice (Table 1).
Correlation between metabolites and colon polyps
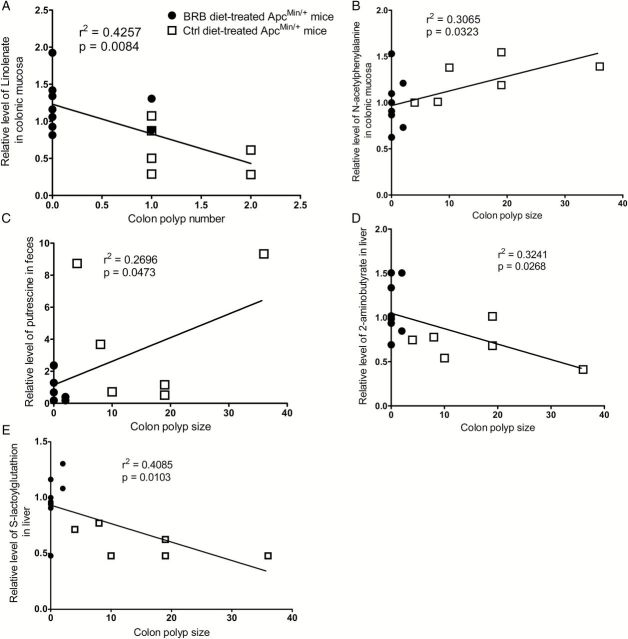
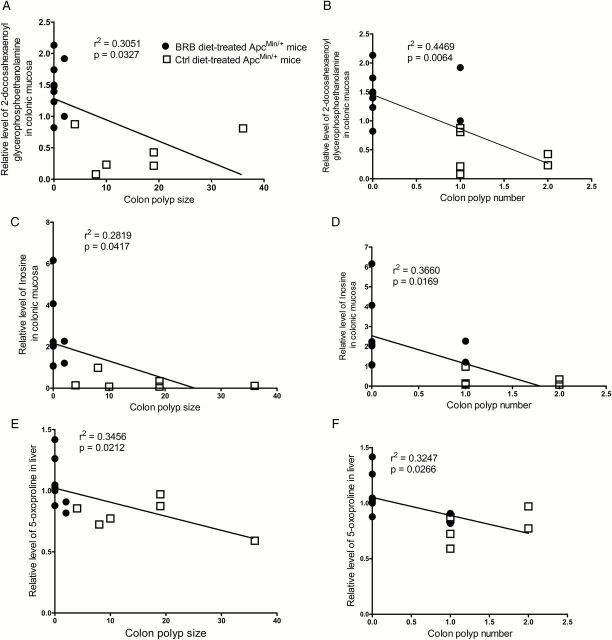
BRB caused changes in several metabolites were linearly correlated with decreased colonic polyp number and size in BRB-treated ApcMin/+ mice. For example, increased levels of linolenate in colonic mucosa (Figure 4A) correlated with decreased colonic polyp number in BRB-treated ApcMin/+ mice. In addition, decreased colonic polyp size in BRB-treated ApcMin/+ mice correlated with decreased levels of N-acetylphenylalanine in colonic mucosa (Figure 4B) and putrescine in feces (Figure 4C), and increased levels of 2-aminobutyrate (Figure 4D) and S-lactoylglutathione (Figure 4E) in liver. Furthermore, increased levels of 2-docosahexaenoyl glycerophosphoethanolamine (Figure 5A and B) and inosine (Figure 5C and D) in colonic mucosa, and 5-oxoproline (Figure 5E and F) in liver correlated with both decreased colonic polyp number and size in BRB-treated ApcMin/+ mice.
Figure 4.
Linear regression between metabolites and colonic polyp number and size. (A) linolenate in colonic mucosa; (B) N-acetylphenylalanine in colonic mucosa; (C) putrescine in feces; (D) 2-aminobutyrate in liver; (E) S-lactoylglutathione in liver. Open squares represent control diet-treated ApcMin/+ mice, dots represent 5% BRB diet-treated ApcMin/+ mice.
Figure 5.
Linear regression between metabolites and colonic polyp number and size. (A, B) 2-docosahexaenoyl glycerophosphoethanolamine in colonic mucosa; (C, D) inosine in colonic mucosa; (E, F) 5-oxoproline in liver. Open squares represent control diet-treated ApcMin/+ mice, dots represent 5% BRB diet-treated ApcMin/+ mice.
Discussion
We previously reported that dietary intervention of 28 colorectal cancer patients with 60g/day BRB powder for an average of 4 weeks resulted in a significant decrease in cancer cell proliferation (13). Our current study used ApcMin/+ mice as a model for human colorectal cancer and demonstrated that 8 weeks of 5% BRB treatment resulted in a significant decrease in adenoma development in small intestine and colon of ApcMin/+ mice. In addition, BRB-treated ApcMin/+ mice had a significantly reduced incidence of colonic polyp development. Using non-targeted metabolomic study, we investigated BRB-mediated metabolite changes in colonic mucosa, liver and fecal specimens in ApcMin/+ mice. The apc gene mutation significantly changed 52 colonic mucosa, 39 liver and 8 fecal metabolites relative to WT mice, and BRBs significantly modulated 43 colonic mucosa, 53 liver and 37 fecal metabolites relative to control diet-fed ApcMin/+ mice. More importantly, BRBs reversed 13 colonic mucosa, 8 liver and 2 fecal metabolites in BRB-treated ApcMin/+ mice. These metabolites were associated with amino acid, glutathione, lipid and nucleotide metabolism.
Elevated amino acid metabolism in colon tumor tissues has been reported in both human colorectal cancer patients (15,24–26) and in animal models of colorectal cancer, including azoxymethane-induced model in rats (3,15) and the ApcMin/+ mouse model (14,15,27). Similarly, we observed eight significantly increased amino acid metabolites in colonic mucosa of ApcMin/+ mice, which were associated with lysine, phenylalanine, tyrosine and tryptophan metabolism. For example, phenylacetylglycine was increased 12.5-fold in ApcMin/+ mice compared to WT mice and this increase was completely reversed by BRB treatment. Phenylacetylglycine has been positively associated with fatty acid β-oxidation (28). Our results showed accumulation of phenylacetylglycine and an overall decrease in lipid metabolites in colonic mucosa of ApcMin/+ mice, suggesting an enhanced fatty acid β-oxidation. BRBs decreased phenylacetylglycine and increased several lipid metabolites. Whether BRBs decreased phenylacetylglycine levels through regulating the overall fatty acid β-oxidation warrants further investigation. In addition, the apc gene mutation induced a significant increase in p-cresol sulfate (4.35-fold) in colonic mucosa. P-cresol sulfate has been reported to increase the expression of DNA methyltransferases in a kidney disease model (7). Our previous study has shown that BRB-derived anthocyanins suppressed activity and protein expression of DNA methyltransferase enzymes (DNMT1 and DNMT3B) in colon cancer cell lines (29). BRBs inhibited expression of DNMT3B in a mouse model of ulcerative colitis, which is an intermediate stage of colorectal cancer (30). In the current study, we observed that BRBs significantly downregulated p-cresol sulfate level in colonic mucosa, which might directly contribute to BRB-induced suppression of DNMT3B expression in colorectal cancer. Furthermore, we observed a significant increase in colonic ODC expression among colonic crypts of ApcMin/+ mice and a significant elevation in excretion of putrescine in the feces of ApcMin/+ mice. Increased expression of ODC, which catalyzes the decarboxylation of ornithine to form putrescine and other polyamines, has been detected in colonic mucosa of human colorectal cancer patients (10,15) and can promote colon carcinogenesis in ApcMin/+ mice (9,17). Interestingly, BRBs fully reversed the colonic ODC protein expression and putrescine levels in feces, which was positively correlated with a decrease in the size of colonic polyps. Given that ornithine levels were increased in colonic mucosa and liver of BRB-treated ApcMin/+ mice, these results suggest that BRBs suppressed polyamine generation and excretion in ApcMin/+ mice through inhibiting ODC activities. Fecal putrescine level may serve as a noninvasive biomarker for BRB intervention.
Glutathione plays a critical role in maintaining the redox status in cells (31). We observed a significant alteration in glutathione metabolism in livers of ApcMin/+ mice relative to WT controls, which were fully reversed by BRBs. More importantly, increased hepatic levels of 2-aminobutyrate, S-lactoylglutathione and 5-oxoproline by BRBs were correlated with decreased colon polyp size and number in BRB-treated ApcMin/+ mice. Therefore, our results indicate that dietary BRBs might cause protective effects through restoring the balance of glutathione metabolism.
Deregulated lipid metabolism has been reported in many types of cancer, including bladder (32), prostate (33), ovarian (34), breast (35) and colon (25,26). Increased fatty acid β-oxidation could provide energy to support more rapid cell proliferation (36). We observed a significant decrease in lipid metabolite levels in apc-induced colon adenomas with 32 lipid metabolites reduced in colonic mucosa specimens. This decrease in fatty acid levels suggests a higher rate of consumption of fatty acid for β-oxidation as an energy source. Alternatively, the free fatty acid pool may be depleted to support phospholipid synthesis for membrane expansion during cell duplication. BRBs significantly reversed seven metabolites in colonic mucosa, two metabolites in liver and one metabolite in feces of ApcMin/+ mice. Of these, increased levels of linolenate and 2-docosahexaenoyl glycerophosphoethanolamine were correlated with colonic polyp size and number. Lower levels of linolenate have been reported in subcutaneous adipose tissues of colorectal cancer patients (16). Given that linolenate could inhibit colon cancer cell proliferation (37), BRB-induced increases in linolenate level in colonic mucosa may suppress colonic polyp development, making linolenate a direct indicator of BRB intervention and reduced colon cancer cell proliferation.
Microbial dysbiosis has been associated with altered biochemical metabolites in colorectal tumor tissue (38,39). The elevated levels of p-cresol sulfate and phenylacetylglycine, which are derived from bacterial metabolism, suggest that BRB alters the gut microbiota. BRBs significantly increase Lactobacillus (Supplementary Figure 2A, available at Carcinogenesis Online) and Bacteroidiaceae (Supplementary Figure 2B, available at Carcinogenesis Online) populations in feces of ApcMin/+ mice, though they were not significantly altered in feces of ApcMin/+ mice compared to WT mice. BRBs did not significantly change Bifidobacteriales (Supplementary Figure 2C, available at Carcinogenesis Online) and Ruminococcus (Supplementary Figure 2D, available at Carcinogenesis Online) populations. Specific pathogen-free mice would be needed to investigate if dietary BRBs protect against colorectal cancer development through changing Lactobacillus and Bacteroidiaceae in the gut.
We previously observed significant changes in 34 urinary and 6 plasma metabolites in human colorectal cancer patients after BRB consumption (40). These BRB-caused significant metabolite changes, including increased amino acid and benzoate metabolism, might be directly attributed to BRB intake or the metabolism of BRBs. More importantly, the metabolite changes significantly correlated with the chemopreventive effects in these patients, such as antiproliferation, antiangiogenesis and proapoptosis (13). Consistently, the current study observed similar results as those from human colorectal cancer patients. For example, 3-[3-(sulfooxy)phenyl]propanoic acid was significantly increased in human post-BRB urine samples (40) and colon mucosa/liver/feces of BRB-treated ApcMin/+ mice. In addition, methyl-beta-glucopyranoside, a carbohydrate metabolite, was significantly increased in human post-BRB urine/plasma samples (40) and colon mucosa/liver of BRB-treated ApcMin/+ mice.
In summary, we demonstrated that 8-week BRB treatment significantly suppressed intestinal and colonic adenoma development in ApcMin/+ mice. More importantly, using non-targeted metabolomic analysis, we observed that BRBs significantly reversed 13 colonic mucosa, 8 liver and 2 fecal metabolites that have been altered by the apc gene mutation. Of these, changes in 8 metabolites were linearly correlated with the decrease in colonic polyp size and number, indicating that changes in these metabolites reflect BRB-mediated suppression of colon cancer cell proliferation. Our data suggest that fecal putrescine level could serve as a non-invasive biomarker, and colonic linolenate level could be a direct indicator of BRB-induced chemoprevention. Collectively, these results suggest that BRB treatment produces systemic effects, which impact multiple metabolic pathways. Furthermore, this beneficial modulation of metabolite profiles produced by BRBs in an ApcMin/+ mouse model might be reflective of changes in BRB-treated human colorectal cancers.
Supplementary material
Supplementary Table 1–8 and Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
NIH (5 R01 CA148818); American Cancer Society (RSG-13-138-01—CNE) to L.-S.W.
Supplementary Material
Acknowledgement
We thank Chieh-Ti Kuo for her assistance in this study.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BRB
black raspberry
- ODC
ornithine decarboxylase
- WT
wild type
References
- 1.World Health Organization. (2014) World cancer factsheet. http://publications.cancerresearchuk.org/downloads/product/CS_REPORT_WORLD.pdf (27 April 2015, date last accessed).
- 2. Weitz J., et al. (2005) Colorectal cancer. Lancet, 365, 153–165. [DOI] [PubMed] [Google Scholar]
- 3. McCart A.E., et al. (2008) Apc mice: models, modifiers and mutants. Pathol. Res. Pract., 204, 479–490. [DOI] [PubMed] [Google Scholar]
- 4. Gay L.J., et al. (2012) Dietary, lifestyle and clinicopathological factors associated with APC mutations and promoter methylation in colorectal cancers from the EPIC-Norfolk study. J. Pathol., 228, 405–415. [DOI] [PubMed] [Google Scholar]
- 5. Stoner G.D. (2009) Foodstuffs for preventing cancer: the preclinical and clinical development of berries. Cancer Prev. Res. (Phila)., 2, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oh S., et al. (2014) Green tea polyphenol EGCG suppresses Wnt/beta-catenin signaling by promoting GSK-3beta- and PP2A-independent beta-catenin phosphorylation/degradation. BioFactors, 40, 586–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tse G., et al. (2014) Cruciferous vegetables and risk of colorectal neoplasms: a systematic review and meta-analysis. Nutr. Cancer, 66, 128–139. [DOI] [PubMed] [Google Scholar]
- 8. Bobe G., et al. (2008) Dietary flavonoids and colorectal adenoma recurrence in the Polyp Prevention Trial. Cancer Epidemiol. Biomarkers Prev., 17, 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoner G.D., et al. (2008) Laboratory and clinical studies of cancer chemoprevention by antioxidants in berries. Carcinogenesis, 29, 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cooke D., et al. (2006) Effect of cyanidin-3-glucoside and an anthocyanin mixture from bilberry on adenoma development in the ApcMin mouse model of intestinal carcinogenesis–relationship with tissue anthocyanin levels. Int. J. Cancer, 119, 2213–2220. [DOI] [PubMed] [Google Scholar]
- 11. Harris G.K., et al. (2001) Effects of lyophilized black raspberries on azoxymethane-induced colon cancer and 8-hydroxy-2’-deoxyguanosine levels in the Fischer 344 rat. Nutr. Cancer, 40, 125–133. [DOI] [PubMed] [Google Scholar]
- 12. Reen R.K., et al. (2006) Modulation of N-nitrosomethylbenzylamine metabolism by black raspberries in the esophagus and liver of Fischer 344 rats. Nutr. Cancer, 54, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang L.S., et al. (2011) Modulation of genetic and epigenetic biomarkers of colorectal cancer in humans by black raspberries: a phase I pilot study. Clin. Cancer Res., 17, 598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bi X., et al. (2010) Black raspberries inhibit intestinal tumorigenesis in apc1638+/- and Muc2-/- mouse models of colorectal cancer. Cancer Prev. Res. (Phila)., 3, 1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manna S.K., et al. (2014) Biomarkers of coordinate metabolic reprogramming in colorectal tumors in mice and humans. Gastroenterology, 146, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cottet V., et al. (2015) Fatty acid composition of adipose tissue and colorectal cancer: a case-control study. Am. J. Clin. Nutr., 101, 192–201. [DOI] [PubMed] [Google Scholar]
- 17. Montrose D.C., et al. (2011) Anti-inflammatory effects of freeze-dried black raspberry powder in ulcerative colitis. Carcinogenesis, 32, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans A.M., et al. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem., 81, 6656–6667. [DOI] [PubMed] [Google Scholar]
- 19. Dehaven C.D., et al. (2012) Software techniques for enabling high-throughput analysis of metabolomic datasets. http://www.intechopen.com/books/metabolomics/software-techniques-for-enabling-high-throughput-analysis-on-metabolomic-datasets (27 April 2015, date last accessed). [Google Scholar]
- 20. Dehaven C.D., et al. (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform., 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. RC Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 22. Tammariello A.E., et al. (2010) Mouse models for unraveling the importance of diet in colon cancer prevention. J. Nutr. Biochem., 21, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCart A.E., et al. (2008) Apc mice: models, modifiers and mutants. Pathol. Res. Pract., 204, 479–490. [DOI] [PubMed] [Google Scholar]
- 24. Denkert C., et al. (2008) Metabolite profiling of human colon carcinoma–deregulation of TCA cycle and amino acid turnover. Mol. Cancer, 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chan E.C., et al. (2009) Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res., 8, 352–361. [DOI] [PubMed] [Google Scholar]
- 26. Ong E.S., et al. (2010) Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol. Cell Proteomics. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 27. Su L.K., et al. (1992) Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science, 256, 668–670. [DOI] [PubMed] [Google Scholar]
- 28. Connor S.C., et al. (2004) Effects of feeding and body weight loss on the 1H-NMR-based urine metabolic profiles of male Wistar Han rats: implications for biomarker discovery. Biomarkers, 9, 156–179. [DOI] [PubMed] [Google Scholar]
- 29. Wang L.S., et al. (2013) Black raspberry-derived anthocyanins demethylate tumor suppressor genes through the inhibition of DNMT1 and DNMT3B in colon cancer cells. Nutr. Cancer, 65, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L.S., et al. (2013) Black raspberries protectively regulate methylation of Wnt pathway genes in precancerous colon tissue. Cancer Prev. Res. (Phila)., 6, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balendiran G.K., et al. (2004) The role of glutathione in cancer. Cell Biochem. Funct., 22, 343–352. [DOI] [PubMed] [Google Scholar]
- 32. Wittmann B.M., et al. (2014) Bladder cancer biomarker discovery using global metabolomic profiling of urine. PLoS One, 9, e115870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y. (2006) Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis., 9, 230–234. [DOI] [PubMed] [Google Scholar]
- 34. Fong M.Y., et al. (2011) Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One, 6, e19963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qiu Y., et al. (2013) Mass spectrometry-based quantitative metabolomics revealed a distinct lipid profile in breast cancer patients. Int. J. Mol. Sci., 14, 8047–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang F., et al. (2012) Dysregulated lipid metabolism in cancer. World J. Biol. Chem., 3, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chamberland J.P., et al. (2015) Down-regulation of malignant potential by alpha linolenic acid in human and mouse colon cancer cells. Fam. Cancer, 14, 25–30. [DOI] [PubMed] [Google Scholar]
- 38. Nugent J.L., et al. (2014) Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J. Proteome Res., 13, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang T., et al. (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J., 6, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan P., et al. (2015) Beneficial Regulation of Metabolic Profiles by Black Raspberries in Human Colorectal Cancer Patients. Cancer Prev Res (Phila), 8, 743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.