Background: BCO2 converts xanthophylls in rodents, but it is controversial whether this role is conserved in primates.
Results: Recombinant primate BCO2 displays enzymatic activity and is expressed as an oxidative stress-induced mitochondrial protein.
Conclusion: Primate BCO2 displays a conserved structural fold and enzymatic function.
Significance: Our data suggest that inducible carotenoid breakdown systems are conserved in primates.
Keywords: carotenoid, dioxygenase, enzyme, mitochondria, vision
Abstract
A family of enzymes collectively referred to as carotenoid cleavage oxygenases is responsible for oxidative conversion of carotenoids into apocarotenoids, including retinoids (vitamin A and its derivatives). A member of this family, the β-carotene 9,10-dioxygenase (BCO2), converts xanthophylls to rosafluene and ionones. Animals deficient in BCO2 highlight the critical role of the enzyme in carotenoid clearance as accumulation of these compounds occur in tissues. Inactivation of the enzyme by a four-amino acid-long insertion has recently been proposed to underlie xanthophyll concentration in the macula of the primate retina. Here, we focused on comparing the properties of primate and murine BCO2s. We demonstrate that the enzymes display a conserved structural fold and subcellular localization. Low temperature expression and detergent choice significantly affected binding and turnover rates of the recombinant enzymes with various xanthophyll substrates, including the unique macula pigment meso-zeaxanthin. Mice with genetically disrupted carotenoid cleavage oxygenases displayed adipose tissue rather than eye-specific accumulation of supplemented carotenoids. Studies in a human hepatic cell line revealed that BCO2 is expressed as an oxidative stress-induced gene. Our studies provide evidence that the enzymatic function of BCO2 is conserved in primates and link regulation of BCO2 gene expression with oxidative stress that can be caused by excessive carotenoid supplementation.
Introduction
Carotenoids, a class of isoprenoid lipids, affect a rich variety of physiological functions in nature and are crucial for human health (1). For instance, the carotenoids zeaxanthin, lutein, and meso-zeaxanthin accumulate in high concentrations in the primate retina (2) where these macular pigments (MPs)2 lessen chromatic aberration and filter phototoxic blue light. Certain carotenoids also are the major dietary source for retinoids, which encompass all derivatives of vitamin A (all-trans-retinol) (3). These carotenoid derivatives exert vital physiological functions as visual chromophore (11-cis-retinal) (4) and vitamin A hormone (all-trans-retinoic acid) (5).
There is interest to enhance macular pigment levels because of emerging evidence that these compounds are beneficial to eye health (6). In general, a relatively linear increment of xanthophyll plasma levels is achieved during supplementation and that eventually reaches a plateau. Additionally, the density and concentration of macular carotenoids vary more than 10-fold among individuals (7). Moreover, plasma and tissue levels of xanthophylls may decline in certain disease states that are associated with oxidative stress such as cardiovascular disease and certain forms of cancer (6). However, the mechanism(s) that controls plasma and tissue levels of these compounds remains to be defined.
Genetic studies in animals suggest that the β-carotene 9,10-dioxygenase (BCO2) plays a critical role in controlling carotenoid tissue levels and preventing excess accumulation of these compounds. Eriksson et al. (8) associated the yellow skin (carotenoids) in domesticated chickens with regulatory mutations that inhibit expression of the BCO2 enzyme in skin. In sheep, the yellow fat phenotype is caused by mutations in the BCO2 gene, and a null mutation in the bovine BCO2 gene causes a change in the β-carotene content in the cow's milk (9, 10). Similarly, genetic disruption of BCO2 function in mice results in increased plasma and blood accumulation of these dietary pigments. Prolonged carotenoid supplementation of Bco2 knock-out mice caused oxidative stress in tissues and the induction of stress-associated signaling pathways. In wild-type animals, a 7-fold increase in Bco2 mRNA expression averted this scenario (11).
For humans, conflicting results are reported in the literature on whether the function of BCO2 was conserved during evolution (12, 13). A major drawback of a rigorous biochemical analysis of this enzyme is the lack of methodology allowing for expression of recombinant BCO2s in active forms. Additionally, little is known about the regulation of BCO2 gene expression, although the protein is differentially expressed in various human tissues (14). Hence, we have established novel methods and tools to enzymatically characterize BCO2s from different mammalian species. Additionally, we utilized human cell lines and mouse models, respectively, to study the transcriptional regulation and function of this protein in carotenoid homeostasis of blood and tissues. The picture that emerges verifies the critical role that BCO2s play in this process and indicates that oxidative stress in chronic disease induces BCO2 and carotenoid breakdown in tissues and blood.
Experimental Procedures
Three-dimensional Structure Models
All protein models were prepared through the fully automated protein structure homology-modeling server SWISS-MODEL using retinal pigment epithelium protein of 65 kDa (RPE65) (Protein Data Bank code 4F2Z) as their template (15). Amino acids 109–126, which were not present in the deposited coordinates, were modeled in an α-helical conformation in accordance with the experimental electron density map. Molecular graphics and analyses were performed with the UCSF Chimera package (16).
Plasmid Constructs for Bacterial and Eukaryote Expression
Total RNA prepared according to Mustafi et al. (17) was a gift from Dr. Brian Kevany. Total RNA was isolated from ∼50 mg of fixed tissue using the RNAeasy Mini kit (Qiagen) and stored at −80 °C. 1 μg of total RNA was reverse transcribed with the SuperScript One-Step RT-PCR for Long Templates system. This cDNA library was used to amplify truncated macaque BCO2 with the following primers: 5′-GCC ATC TTT GGG CAG TGT CGG-3′ (forward) and 5′-TTA GAT GGG TAT GAA GGT ACC ATG G-3′ (reverse). This set of primers included the addition of a stop codon. To amplify full-length macaque BCO2 (MaBCO2) cDNA encoding the NP_001271880 variant of MaBCO2 with an N-terminal extension, the following PCR was carried out using the forward primer 5′-AAG GAG GAA TAA ACC-3′ and the reverse primer 5′-GAT GGG TAT GAA GGT ACC ATG GAA TCC-3′. The amplified macaque BCO2 cDNAs were then cloned in-frame into pBAD TOPO TA (Invitrogen) for bacterial expression and pcDNA3.1/V5-His TOPO (Invitrogen) for eukaryote expression. All PCRs were carried out with the Expanded High Fidelity PCR system (Roche Applied Science). All plasmid constructs were verified by sequence analysis (Genomics Core Sequencing Facility, Case Western Reserve University, Cleveland, OH).
Transient Transfection and Immunofluorescence
HepG2 cells (American Type Culture Collection) were transiently transfected with corresponding vectors carrying the various amplified BCO2 cDNA isoforms of murine, human, and macaque. All transfections and immunofluorescence were performed as described previously (13). HepG2 cells (American Type Culture Collection) were maintained in DMEM with 10% FBS and 10 units/ml penicillin-streptomycin antibiotics at 37 °C with 5% CO2. For immunofluorescence, HepG2 cells were grown on polylysine-treated glass coverslips to 50–70% confluence and then transfected with purified plasmid DNA using X-tremeGENE HP (Roche Applied Science) transfection reagent. 24 h post-transfection, cells were fixed in a solution of 10% formalin phosphate in phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.4 mm KH2PO4, pH 7.4; PBS) overnight at 4 °C. Afterward, cells were washed and permeabilized with 0.1% Triton X-100 (Roche Applied Science) in PBS (PBS-T) and blocked with 10% bovine albumin and 5% goat serum (Sigma Life Science) in PBS-T (blocking buffer). Cells were then incubated overnight at 4 °C in blocking buffer containing a mouse anti-V5 serum (to detect BCO2; Invitrogen) and rabbit anti-cytochrome c oxidase subunit IV serum (Cell Signaling Technology, Boston, MA) diluted 1:200. Then cells were washed and incubated at room temperature with anti-mouse and anti-rabbit secondary antibodies conjugated to Alexa Fluor 594 and Alexa Flour 488 (Life Technologies), respectively, diluted 1:400 in blocking buffer. DAPI was used to stain nuclei. Confocal images were acquired with a Zeiss LSM 510 META laser scanning confocal microscope (Carl Zeiss MicroImaging, Jena, Germany) by using a multiline argon laser (excitation, 488 and 594 nm) or a 405 diode laser (excitation, 405 nm) with a 63× C-Apochromat (numerical aperture, 1.2; oil objective).
Expression of Murine and Macaque BCO2 in E. coli
Protein expression was performed as described (18) with some modifications. BL21(DE3) competent Escherichia coli (New England BioLabs, Ipswich, MA) were transformed with the expression vectors for full-length and truncated macaque BCO2 as described above. These bacterial cells were also transformed with the expression vectors for murine BCO2 as reported previously (1). Bacteria were grown at room temperature with constant shaking at 200 rpm until an A600 of 0.4 was reached. Protein expression was induced with l-arabinose at a final concentration of 0.02%. At the point of induction, FeSO4 and ascorbic acid were added to a final concentration of 5 μm and 10 mm, respectively, and the temperature was decreased to 10 °C. Protein expression was allowed to proceed for 2 days, cells were then collected by centrifugation at 4500 × g for 15 min, and cell pellets were stored at −80 °C until needed or immediately submitted for in vitro enzyme assays.
Purification and Quantification of BCO2
Cell pellets obtained from the procedure as described above were thawed on ice. Cells pellets were resuspended in buffer containing 20 mm Tricine, pH 7.5, 1 mm tris(2-carboxyethyl)phosphine hydrochloride (TCEP/HCl) (Hampton Research) and one cOmplete EDTA-free protease inhibitor mixture tablet (Roche Applied Science) at 4 ml/g. Lysis was performed by three passages on a cold French press while collecting lysates on iced tubes. The cell lysate was then subjected to centrifugation at 100,000 × g at 4 °C for 30 min. The supernatant was collected, transferred to 50-ml tubes, and kept on ice. Then it was loaded onto a 10-ml column containing 1.5 ml of Talon Co2+-resin suspension (Clontech) pre-equilibrated with 5 column volumes of ice cold buffer containing 250 mm NaCl, 20 mm Tricine, pH 7.5, and 1 mm TCEP. After flow-through collection, the Talon column was first washed with 5 column volumes of ice-cold buffer containing 250 mm NaCl, 20 mm Tricine, pH 7.5, and 1 mm TCEP and then with 5 column volumes of buffer containing 250 mm NaCl, 20 mm Tricine, pH 7.5, 1 mm TCEP, and 1 mm imidazole. Finally, BCO2 was eluted in ice-cold buffer containing 250 mm NaCl, 20 mm Tricine, pH 7.5, 1 mm TCEP, and either 5 or 50 mm imidazole. Eluted BCO2 fractions were pooled and concentrated in a 30,000-molecular weight cutoff Amicon® Ultra Centrifugal Filter (Millipore) before being loaded onto a SuperdexTM 200 10/300 GL size exclusion column (GE Healthcare). Enzyme fractions were eluted from the column in 0.5-ml fractions at a flow rate of 0.4 ml/min with buffer containing 100 mm NaCl, 20 mm Tricine, pH 7.5, and 1 mm TCEP. Fractions containing purified enzyme were then pooled, concentrated to desired concentrations in a 30,000-molecular weight cutoff Amicon Ultra Centrifugal Filter and stored on ice until further use. Western blotting of this purified enzyme at varying concentrations was run concurrently with Talon Co2+- purified murine and primate BCO2s for quantification. Quantification of expressed BCO2 protein was performed using ImageJ and the known concentrations of the purified standards as well as with Bradford (53) assays.
In Vitro Enzyme Activity Assay
Cell pellets were thawed on ice, and their net weight was determined. Lysing reagent was prepared by combining 50 ml of B-PER Bacterial Protein Extraction Reagent (Life Technologies) with one protease inhibitor cOmplete ULTRA Mini EDTA-free tablet and final concentrations of 2 mm ascorbic acid solution (Sigma Life Science), 1 mm TCEP (Thermo Scientific, Rockford, IL), and 20 units of recombinant, RNase-free DNase I. To lyse cells and solubilize recombinant proteins, 4 ml of lysing reagent/g of bacterial pellet was used. Cell pellets were gently vortexed with lysing reagent and allowed to incubate at room temperature for 10 min. The cell lysate was then cooled on ice and subjected to centrifugation at 100,000 × g at 4 °C for 30 min for the purpose of Western blotting analysis. For enzymatic activity assays, whole cell lysates were used, and assays were carried out as previously described (19, 20) but with the following modifications. 2, 2-Dioctylpropane-1,3-bis-β-d-maltopyranoside (decyl maltose neopentyl glycol (DMN)) micelles loaded with zeaxanthin were prepared as follows. 33 μl of 3% DMN detergent solution was mixed with 10 μm (final concentration) of zeaxanthin dissolved in acetone in a 2-ml Eppendorf tube. This mixture (substrate) was then dried in an Eppendorf Vacufuge plus. To the substrate, 100 μl of cell lysate was added, vortexed vigorously for 20 s, and placed on an Eppendorf thermal shaker for 15 min at 300 rpm. Control assays were performed with uninduced E. coli cell pellet lysates. The reaction was stopped by adding 100 μl of water and 400 μl of acetone. Lipids were extracted by adding 400 μl of diethyl ether and 100 μl of petroleum ether, vortexed (3 × 10-s period), and centrifuged at 15,000 × g for 1 min. Finally, the resulting organic phase was collected. The extraction was performed twice, and the collected organic phase was dried by Vacufuge. Dried supernatant was redissolved in mobile phase (90:10 hexane:ethyl acetate) and subjected to either HPLC or LC-MS analysis.
HPLC and LC-MS System
HPLC analysis was carried out with an Agilent 1260 Infinity Quaternary HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a pump (G1312C) with an integrated degasser (G1322A), a thermostat column compartment (G1316A), an autosampler (G1329B), a diode array detector (G1315D), and online analysis software (Chemstation). The analyses were carried out at 25 °C using a normal phase Zorbax SIL (5 μm; 4.6 × 150 mm) column (Agilent Technologies) protected with a guard column with the same stationary phase. Carotenoid and apocarotenoid separation was achieved using an isocratic composition of 70:30 (v/v) hexane:ethyl acetate. For β-carotene and xanthophyll separation from animal tissues, a step gradient of 1% ethyl acetate in hexane over 5 min followed by 10 min with 10% ethyl acetate in hexane and then 18 min with 30% ethyl acetate in hexane was used. The flow rate for all systems was 1.4 ml/min. Detection of carotenoids and apocarotenoids was performed at 420-nm wavelength. For LC-MS analyses, the eluate was directed into an LXQ linear ion trap mass spectrometer (Thermo Scientific, Waltham, MA) through an atmospheric pressure chemical ionization source working in the positive mode. To ensure optimal sensitivity, the instrument was tuned with zeaxanthin as well as apocarotenoids. Identification of BCO2 cleavage products was based on retention time, mass, and spectral characteristics compared with those of a known standard, 10′-β-apocarotenal (BASF, Ludwigshafen, Germany), and previously published reports (21, 22).
Animals, Husbandry, and Experimental Diets
Animal procedures and experiments were approved by the Case Western Reserve University Animal Care Committee and conformed to recommendations of both the American Veterinary Medical Association Panel on Euthanasia and the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. Animal experiments were carried out using β-carotene 15,15′-dioxygenase (Bco1) knock-out, Bco2−/−, and Bco1−/−/Bco2−/− double knock-out (DKO) mice with a C56/BL6; 129Sv mixed genetic background. Feeding experiments were performed as described previously (23). In all experiments, mice were maintained at 24 °C in a 12:12-h light-dark cycle and had free access to food and water. During the breeding and weaning periods (up to 5 weeks of age), mice were maintained on breeder chow containing ∼29,000 IU vitamin A/kg of diet (Prolab RMH 3000, LabDiet, St. Louis, MO). After 5 weeks of age, mice were fed with an experimental diet containing a mixture of β-carotene and zeaxanthin (25 and 75 mg/kg of diet for each carotenoid, respectively) for 10 weeks. The diet contained no other source of vitamin A except for β-carotene. After 10 weeks of dietary intervention, mice were fasted overnight and anesthetized by intraperitoneal injection of a mixture containing 15 mg of ketamine, 3 mg of xylazine, 0.5 mg of acepromazine, and sterile water or saline at a dose of 0.2 ml/25 g of mouse body weight. Blood was drawn directly from the heart by cardiac puncture under deep anesthesia. Mice were then perfused with 20 ml of PBS, pH 7.3 and killed by cervical dislocation for further tissue collection.
Extraction of Carotenoids from Animal Tissues
Carotenoids were extracted from tissues of mice (n = 5 per genotype and diet) under a dim red safety light (600 nm) and quantified by HPLC as described previously (23). 100 μl of serum in 200 μl of PBS or one whole eye homogenized in 200 μl of PBS and 100 μl of hydroxylamine was extracted using 300 μl of methanol, 600 μl of acetone, 300 μl of diethyl ether, and 400 μl of hexane. The organic phase was removed, and the extraction was repeated with an additional 500 μl of hexanes. After centrifugation, organic layers were collected, pooled, dried in a Vacufuge at 30 °C, and redissolved in HPLC mobile phase solvent. For saponification, gonadal white adipose tissues were homogenized in 200 μl of 30% KOH in water and then incubated with 100 μl of 12% pyrogallol (Sigma-Aldrich) in ethanol and 1 ml of ethanol for 1 h at 60 °C. After saponification, 2 ml of ethanol and 2 ml of H2O were added, and samples were extracted twice with 3 ml of diethyl ether:hexane (2:1; stabilized with 1% ethanol). The organic layers were collected, pooled, and evaporated in a Vacufuge until nearly dry. The second extraction solution was then added to the nearly dry solution. The extraction solution was composed of 200 μl of water, 200 μl of methanol, 400 μl of acetone, 250 μl of diethyl ether, and 400 μl of hexane. The organic phase was then removed, and the extraction was repeated with 500 μl of hexanes. After centrifugation, organic layers were collected, pooled, dried in a Vacufuge at 30 °C, and redissolved in corresponding HPLC mobile phase solvent.
Induction of BCO2 and Real Time PCR
HepG2 cells (American Type Culture Collection) were maintained in DMEM with 10% FBS and 10 units/ml penicillin-streptomycin antibiotics at 37 °C with 5% CO2. Cells were grown in polylysine-treated dishes until 50–70% confluence. To measure relative BCO2 levels, cells were washed with PBS and then incubated in 5 mm hydrogen peroxide in PBS for 5 min. Then cells were washed in DMEM to remove excess hydrogen peroxide. Cells were then maintained in DMEM and 1 μm zeaxanthin, 0.1% dimethyl sulfoxide in DMEM, or 0.5 mm hydrogen peroxide in DMEM for 4 h. Media were then aspirated, and RNA was collected using TRIzol reagent (Life Technologies). RNA was transcribed to cDNA using a High-Capacity RNA-to-cDNA kit (Life Technologies). This cDNA was then used for quantitative real time PCR using TaqMan Gene Expression Master Mix (Life Technologies) and GAPDH (control) and BCO2 probes (Life Technologies) as described previously (13). Measurements in relative levels of BCO2 mRNA levels were normalized to levels of GAPDH and were taken as biological duplicates and technical quadruplets.
Results
Structural Comparison between Rodent and Primate BCO2s
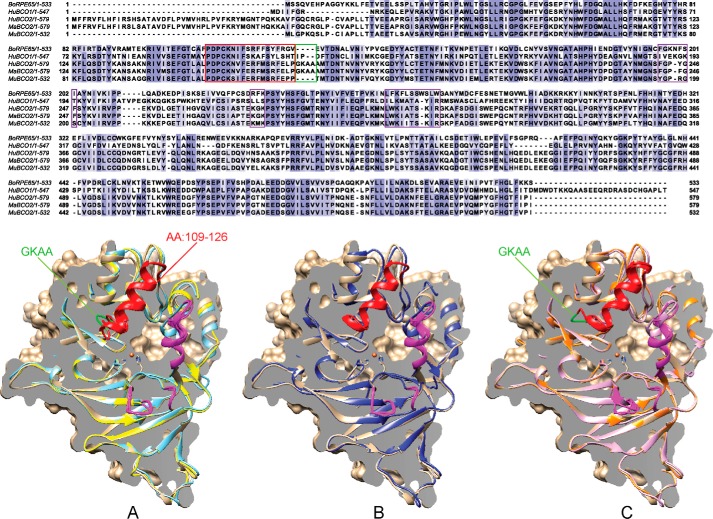
Besides BCO2, the human genome encodes two other carotenoid cleavage oxygenase (CCO) family members, RPE65 and BCO1, both of which are critical for human retinoid metabolism (24). Sequence alignments revealed that the most pronounced difference between human BCO2 and other family members is a long N-terminal leader sequence for mitochondrial import (13) (Fig. 1). This N-terminal extension is also present in other primate BCO2s, e.g. the predicted MaBCO2, but is absent in murine BCO2 (MuBCO2) (11, 13). Pairwise alignment scores performed using ClustalW (25) reveal that the primate BCO2s share a 96% amino acid sequence identity with each other and an 81% amino acid sequence identity with MuBCO2 when excluding the N-terminal leader sequence (Fig. 1). The primate BCO2s also share an average ∼42% sequence identity with human RPE65 and ∼41% sequence identity with human BCO1 (Fig. 1). Four key histidine residues (RPE65 His180, His241, His313, and His527) responsible for the ligation of an iron cofactor are conserved in all CCOs (Fig. 1). Excluding the N-terminal extension, the major difference between primate BCO2s and other family members, including rodent BCO2s, is the presence of a four-amino acid-long insertion, 169GKAA172. Recently, it was proposed that this primate-specific insertion is responsible for BCO2 inactivation by sterically interfering with substrate binding.
FIGURE 1.
Sequence and structure homology of CCOs. Top, alignment of bovine RPE65, human BCO1, human BCO2, macaque BCO2, and murine BCO2 is shown. The GKAA primate insertion is boxed in green, membrane binding domains are in magenta, and the previously unresolved amino acid (AA) 109–121 region is boxed in red. All structures are modeled using bovine RPE65 (tan) as template and are aligned facing into the active site with the Fe2+ iron cofactor denoted as an orange sphere. A comparison of three-dimensional structures of human BCO2 (light blue) and murine BCO2 (yellow) (A), chimeric human BCO2 (blue) (B), and wild-type (light purple) and chimeric (orange) macaque BCO2s (C) is shown. The four-amino acid insertion (GKAA) in primate BCO2s is colored green. The previously unresolved amino acid 109–126 region is colored red, and the presumed plasma membrane binding sites are highlighted in magenta.
To investigate the putative structural changes brought on by the absence or presence of these four amino acids, we modeled wild-type and chimeric (with deleted insertion) human and macaque BCO2s as well as MuBCO2 (SWISS-MODEL) using the recently solved crystal structure of enzymatically active bovine RPE65 as template (26). The electron density maps calculated from this new data set indicate that the region containing the insertion (amino acids 109–126 of bovine RPE65) adopts an α-helical conformation (Fig. 1A). Upon homology modeling of BCO2 based on this extended template, no marked structural differences between HuBCO2 and MuBCO2, including the region with the GKAA insertion, were observed for the proteins (Fig. 1A). The predicted basic structural fold was a rigid seven-bladed β-propeller covered by a half-dome. A highly conserved structural space in the active site domain near the four conserved histidines (only two are shown in the figure) ligated the iron cofactor. The GKAA extension (green) in HuBCO2 is shown here as a loop that did not significantly change the overall fold of the α-helix depicted in red or any of the other features noted here at the opening of the substrate tunnel. As can be seen in our model, this loop was instead localized away from the substrate tunnel and would not pose an obstruction to the substrate entrance of the enzyme as proposed previously (12).
Notably, the integrity of all these portions of the human enzyme was retained in the chimeric model with the deleted GKAA insertion (HuBCO2ΔGKAA) (Fig. 1B, blue). Overall, the same observations hold true in the predicted structures of MaBCO2 (Fig. 1C) and the chimeric protein with the deleted GKKA insertion (MaBCO2ΔGKAA) (Fig. 1C). Slight differences in the positioning of the membrane binding domains became detectable but did not play a role in altering the main structure of the substrate tunnel leading to the active site. The positioning of the GKAA portion for the macaque enzyme differed slightly from the human but did not alter the α-helical structure, which corresponded to macaque amino acids 151–168 and 109–126 for bovine RPE65. Together, modeling of different BCO2s using the novel RPE65 template did not reveal any structural differences that would suggest that the presence of the GKAA insertion in primate BCO2s causes an inactivation of the enzymes. Homology models for RPE65 and MaBCO2 have been deposited as supplemental Data 1 and 2, respectively.
Effect of Detergents on Enzyme Activity
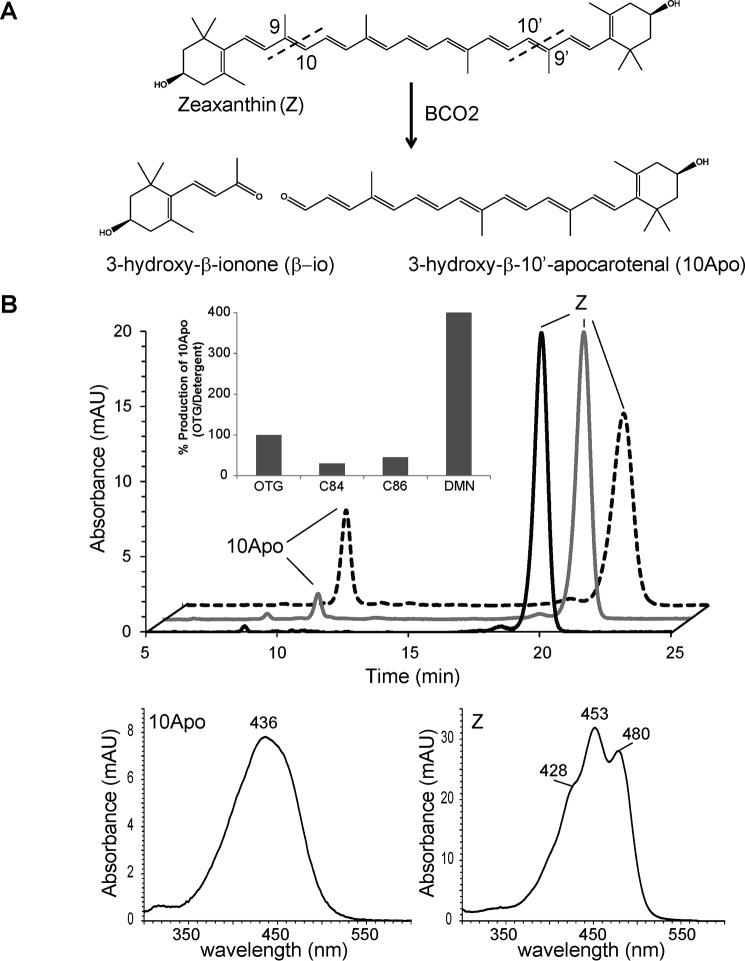
We and others showed that the assay conditions significantly influence the activity of RPE65 and BCO1 (19, 27, 28). Therefore we turned our attention to improve the fundamental enzyme activity assay for recombinant BCO2 expressed in an E. coli system. To do so, we focused on recombinant MuBCO2, which previously showed convincing turnover of carotenoid substrates in such tests (11). Here, we were able to recapitulate similar results under the previously established conditions as noted in Fig. 2. It has been our experience that the activity of CCO greatly relies on the manner in which the enzyme is solubilized and how substrate is delivered to the enzyme. For this purpose, CCOs are solubilized during cell lysis with detergent and carotenoids delivered to the enzyme via detergent micelles (18, 29, 30). Predominantly, we have used β-d-1-thioglucopyranoside for this multiple task as the use of other detergents has been shown to cause a decrease in enzymatic activity (27, 28). We wondered whether an alternate detergent would improve MuBCO2 activity. A screen of several detergents showed a decrease in enzyme activity (Fig. 2B, inset), but the use of one detergent, DMN, resulted in a 4-fold increased production of the cleavage product, 3-hydroxy-β-10′-apocarotenal (10Apo), from zeaxanthin cleavage (Fig. 2B, dashed trace). Conversely, the substrate level strongly decreased when the enzyme was assayed in this manner. Thus, the use of the detergent DMN to solubilize protein and create substrate delivery micelles significantly increased the enzymatic activity of recombinant MuBCO2.
FIGURE 2.
Detergent affects CCO enzyme activity. A, schematic of BCO2 oxidative cleavage of zeaxanthin. B, HPLC chromatogram comparing the in vitro activity of murine BCO2 when performed with n-octyl β-d-thioglucopyranoside (OTG) (gray trace) or DMN (dashed trace) detergent. The black trace shows the control. Relative activities (measured as the production of 10Apo) comparing different detergents is shown in the inset. C, UV/visible absorbance spectra of the product 10Apo and substrate zeaxanthin. C84, tetraethylene glycol monooctyl ether; C86, hexaethylene glycol monooctyl ether; mAU, milli-absorbance units.
Biochemical Characterization of Macaque BCO2
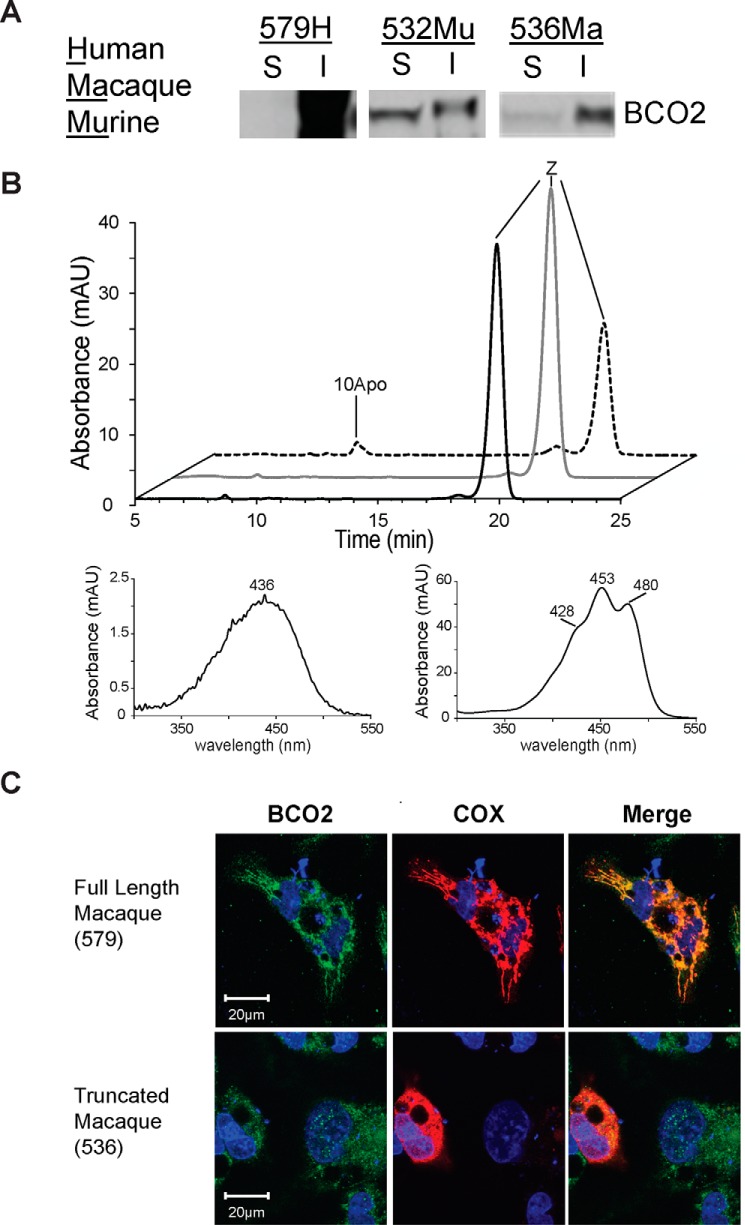
Most recently (13), our laboratory provided evidence of an active, recombinantly expressed HuBCO2 in β-carotene-producing E. coli (18). Albeit the experimental conditions resulted in very modest turnover of this enzyme requiring mass spectrometry for detection of the β-10′-apocarotenal product, it did provide proof of the activity of the human enzyme. However, our attempts to assay recombinant HuBCO2 or other primate BCO2 proteins in cell-free systems have failed thus far. Successful expression of active recombinant enzymes in E. coli systems must overcome a multitude of hurdles (31). Perhaps the greatest obstacle has been the production of soluble, hence functional, proteins and the avoidance of the formation of insoluble aggregates known as inclusion bodies (32). We speculated that this problem could explain the lack of activity when trying to characterize enzymatic properties of the HuBCO2 recombinant protein expressed in E. coli. To investigate this possibility, we expressed the human 579-amino acid-long protein variant (579H) in E. coli and found upon centrifugal fractionation and Western blotting analysis that the enzyme was present in the insoluble fraction (Fig. 3A). The same protocol applied to MuBCO2 revealed that expression of this enzyme produced equal quantities of soluble and insoluble proteins (Fig. 3A). Enzyme activity assays of the collected insoluble MuBCO2 fraction incubated with substrate failed to produce cleavage products that were detected from the soluble fraction (Fig. 3B, black trace). Resuspended insoluble HuBCO2 also failed to cleave zeaxanthin in vitro (Fig. 3B, gray trace). The production of a significant amount of soluble, functional murine protein and the exclusive production of insoluble HuBCO2 in this E. coli system may explain in part the discrepancy between their reported enzymatic activities (12, 13, 18). Although refolding procedures of inclusion bodies have been successfully used to study a bacterial CCO (28, 33), so far our best attempts at refolding HuBCO2 did not produce functional protein. Therefore, we focused our efforts on characterization of MaBCO2 activity.
FIGURE 3.
Recombinant expression, enzyme activity, and cellular localization of BCO2s. A, Western blot of soluble (S) and insoluble (I) centrifugal fractions from lysed bacteria expressing recombinant (from left to right) human, murine, and macaque BCO2. B, insoluble fraction of HuBCO2 expressed in E. coli is inactive. HPLC of lipid extracts of in vitro enzyme activity assays of soluble MuBCO2 (dashed trace), insoluble MuBCO2 (black trace), and insoluble HuBCO2 (gray trace) on zeaxanthin was carried out. UV spectra of 10Apo and zeaxanthin are shown. C, confocal images of HepG2 cells transfected with full-length (top panel) and truncated (bottom panel) macaque BCO2-encoding plasmids. Immunostaining was performed with anti-V5 antibody for BCO2 (red) and anti-cytochrome c oxidase subunit IV (COX) antibody (green). Nuclei were stained with DAPI (blue). Only the full-length MaBCO2 shows co-localization with cytochrome c oxidase subunit IV in merged images. mAU, milli-absorbance units.
Previously, we showed that the N-terminal leader sequence of HuBCO2 is removed during mitochondrial import and not required for enzyme activity (13). To confirm this observation for the macaque enzyme, we expressed the full-length MaBCO2 and an N-terminally truncated MaBCO2 (trMaBCO2) in the human hepatoma cell line HepG2 as C-terminally V5-tagged proteins. Immunostaining and confocal imaging revealed that the MaBCO2 variant was distributed in similar patterns as the mitochondrial marker protein cytochrome c oxidase subunit IV (Fig. 3C). In contrast, staining for trMaBCO2 did not merge with the mitochondrial marker protein, indicating that the N-terminal leader is mandatory for the import process. Thus, to better mimic the native protein and the murine enzyme, which lacks the N terminus, we expressed MaBCO2 in E. coli without the N-terminal domain. Notably, this truncated enzyme also contained vector-derived fusion tags similar to the previously tested recombinant murine BCO2. To increase its solubility, we additionally expressed trMaBCO2 at low temperature (10 °C) for an extended period of 2 days. Western blotting analysis showed that using this technique produced soluble trMaBCO2 protein (Fig. 2A). Although levels of soluble murine protein were not achieved, our efforts relayed a more promising result than that obtained above with the largely insoluble recombinant HuBCO2.
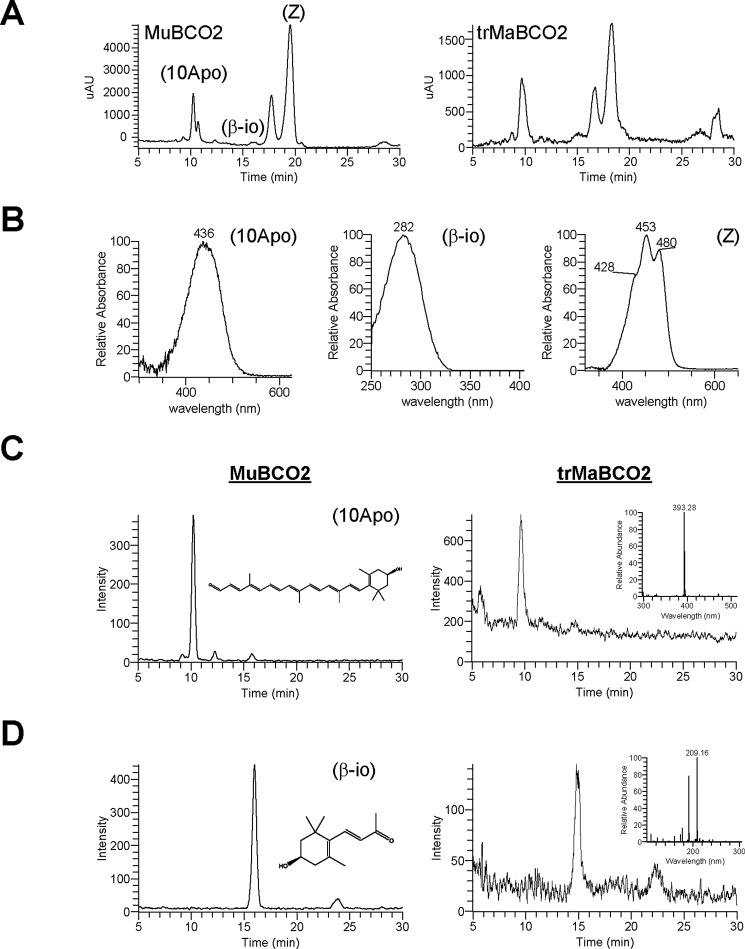
Having established that the trMaBCO2 enzyme can be expressed in E. coli in a soluble form, we set out to test its enzymatic activity. As a control, we tested MuBCO2 in parallel. To identify trMaBCO2 and MuBCO2 activity, we used an LC-MS-based method that directly detects products of BCO2-catalyzed oxidative cleavage of zeaxanthin. UV-visible HPLC analyses revealed the presence of two distinct products generated by both enzymes, one at retention time ∼10 min and the second at ∼15 min (Fig. 4A, left and right, MuBCO2 and trMaBCO2 respectively). Spectral characteristics (Fig. 4B) and retention time analysis indicated that the first product was 10Apo as expected from our initial trials with MuBCO2 (Fig. 2B). The spectral characteristic of the second product (Fig. 4B) is consistent with that of 3-hydroxy-β-ionone and was present in both MuBCO2 and trMaBCO2 trials. Mass spectra from extracted ion chromatograms (Fig. 4C) provided further evidence that the peaks at 10 min in the chromatogram can be attributed to 10Apo as the m/z of 393.28 was consistent with the calculated [MH]+ ion (Fig. 4C, left, inset). Extracted ion chromatograms for m/z 209.18 [MH]+ (Fig. 4, D and inset) provided confirmation that the peak at 16 min corresponded to 3-hydroxy-β-ionone.
FIGURE 4.
Enzymatic activity assays of murine and macaque BCO2. A, HPLC analysis of lipid extracts isolated from in vitro enzyme assays of recombinant murine (left) and macaque (right) BCO2s and zeaxanthin (Z). B, the resulting oxidative cleavage products 10Apo and 3-hydroxy-β-ionone (β-io) as well as the substrate were identified in part by retention time and known absorption spectra. Mass spectra (left and right, murine and macaque, respectively) from extracted ion chromatograms identifying the peaks at ∼10 min with an m/z of 393.28 as 10Apo (right inset) (C) and the peaks at ∼16 min with an m/z os 209.18 as 3-hydroxy-β-ionone (right inset) (D) are shown. mAU, milli-absorbance units.
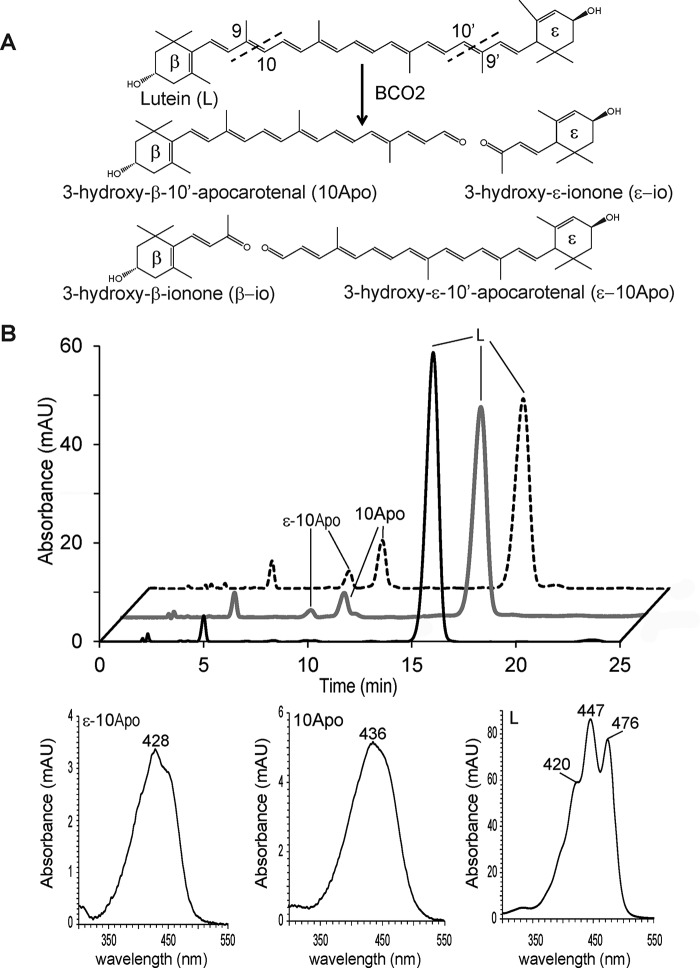
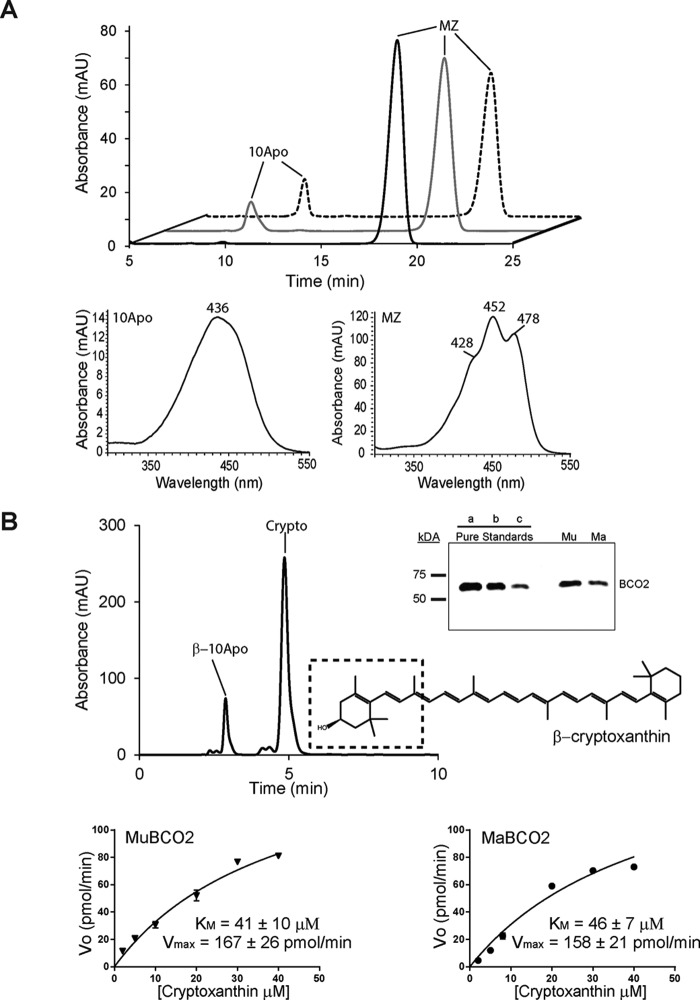
We proceeded to investigate the enzyme activity of both MuBCO2 and trMaBCO2 using lutein and meso-zeaxanthin as substrates. We anticipated that cleavage of lutein by BCO2 could produce five distinct products depending on the preference of the enzyme for the β-ionone or ϵ-ionone ring (Fig. 5A). These products could comprise 10Apo, 3-hydroxy-β-ionone, 3-hydroxy-ϵ-10′-apocarotenal (ϵ-10Apo), 3-hydroxy-ϵ-ionone, and rosafluene (not depicted) (Fig. 5A). The enzymatic assays for both the murine and primate BCO2s produced two detectable products, 10Apo and ϵ-10Apo (Fig. 5B). The identity of 10Apo was deciphered from retention time and UV/visual spectral characteristics. Although a standard of ϵ-10Apo was not available for comparison, we previously detected this compound via MS analysis (11). Here, the retention time and the blue shift in the maximum absorbance of the spectrum (Fig. 5C, left), indicating a break in the double bond conjugation across the apocarotenoid backbone, are consistent with our previous observations. The production of 10Apo and ϵ-10Apo by MuBCO2 was greater than that of trMaBCO2 by roughly 2.5 times. Both MuBCO2 and trMaBCO2 seem to preferentially cleave lutein on the ϵ-ring side to produce 10Apo as the production of 10Apo was 3 times the production of ϵ-10Apo (Fig. 5B). Next, incubation of E. coli cell lysate expressing MuBCO2 with meso-zeaxanthin produced one detectable product with our HPLC system. This product as identified by retention time and spectral characteristics was found to be 10Apo (Fig. 6A, dashed trace). The same product was observed when trMaBCO2-expressing E. coli lysates were incubated with meso-zeaxanthin (Fig. 6A, gray trace). With this detection method, it remains unclear as to whether the cleavage of meso-zeaxanthin is preferred on the 3R,3′R or 3R,3′S side. Under these experimental conditions, the production of 10Apo was 1.5 times greater by MuBCO2 cleavage than that of trMaBCO2. Finally, we attempted to use β-cryptoxanthin to determine Michaelis constants and turnover rates for the enzymes (Fig. 6B). This asymmetric carotenoid is preferentially converted to β-10′-apocarotenal through removal of 3-hydroxy-β-ionone. The initial rate of the enzymatic reaction measured at various substrate concentrations was used to assess the apparent Km and Vmax values. Both the macaque and murine enzymes revealed similar Km values of approximately 42 μm (Fig. 6B). For determining Kcat, we purified murine BCO2. We then used known amounts of this protein as a standard to quantify BCO2 amounts in cell lysates by quantitative Western blotting. This analysis revealed that the macaque BCO2 enzyme has a slightly higher turnover rate at 20 min−1 compared with the murine BCO2 enzyme at 14 min−1.
FIGURE 5.
BCO2 cleaves lutein from both the β and ϵ ring side at both the 9,10 and 9′,10′ double bonds. A, schematic of BCO2 oxidative cleavage of lutein showing four of the possible five products. Rosafluene is not depicted. B, HPLC chromatograms of lipid extracts of in vitro enzyme activity assays of MuBCO2 (dashed trace) and trMaBCO2 (gray trace) on lutein (L). The black trace shows the control. C, UV spectra of ϵ-10Apo, 10Apo, and lutein. mAU, milli-absorbance units.
FIGURE 6.
Comparison of MaBCO2 and MuBCO2 enzymatic activity on meso-zeaxanthin (MZ) and β-cryptoxanthin (Crypto). A, HPLC chromatograms of lipid extracts of in vitro enzyme activity assays of MuBCO2 (dashed trace) and trMaBCO2 (gray trace) on meso-zeaxanthin. The black trace shows the control. UV spectra of 10Apo and meso-zeaxanthin are shown. B, sample HPLC chromatogram displaying BCO2 cleavage activity on β-cryptoxanthin. The dashed box shows the BCO2 preference for cleaving on the 3-carbon hydroxylated ring side to produce β-10′-apocarotenal (β-10Apo) and 3-hydroxy-β-ionone. Inset, quantitative Western blotting analyses of BCO2 protein amounts in lysates. Three different amounts of purified MuBCO2 (a, b, and c correspond to 0.12, 0.06, and 0.03 μg of protein, respectively) were used to quantify MuBCO2 (Mu) and MaBCO2 (Ma) protein amounts in cell lysates. Lower panel, enzyme kinetic analyses of MuBCO2 and MaBCO2 comparing Km and Vmax values for β-cryptoxanthin. Values were calculated using Origin 9 software. Values represent means ± S.E. (error bars) of at least two independent assays. mAU, milli-absorbance units.
Knock-out of BCO2 Leads to Systemic Tissue Accumulation
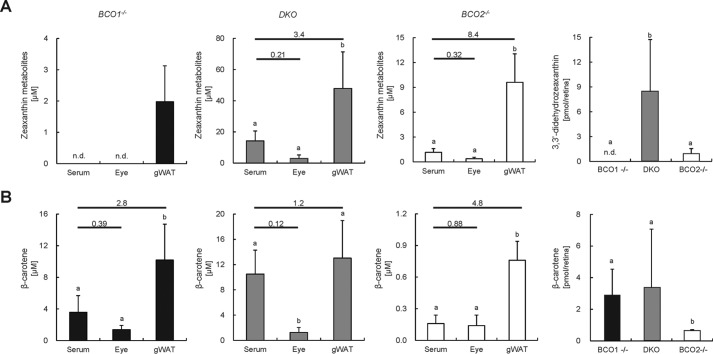
Recently, it has been proposed that inactivity of BCO2 provides a mechanism for the concentration of xanthophylls (1 mm versus 1–6 μm in serum and tissues) in the primate retina. This hypothesis was supported by the observation that BCO2-deficient mice display zeaxanthin in the retina, whereas wild-type mice do not despite abundant expression of putative zeaxanthin- and meso-zeaxanthin-binding proteins (12). To scrutinize this theory, we compared body distribution of zeaxanthin and β-carotene in Bco1−/−, Bco2−/−, and DKO mice. Mice (n = 5 per genotype) were subjected to feeding with a diet that contained β-carotene and zeaxanthin together (25 and 75 mg/kg, respectively). After 10 weeks of intervention, we determined carotenoid levels of serum and peripheral tissues (eyes and gonadal fat) in different mouse genotypes (Fig. 7). If as proposed by others (12) the BCO2 genotype determines ocular xanthophyll levels, then zeaxanthin but not β-carotene should specifically accumulate in the retina. Additionally, zeaxanthin concentration should be much higher in the eyes when compared with serum and other tissues. HPLC analyses revealed that both β-carotene and zeaxanthin accumulated in the murine retina, respectively, in a BCO1- and BCO2-dependent manner (Fig. 7, A and B, far right). As reported previously (23), zeaxanthin existed in the form of its oxidized didehydro metabolites and was highest in DKO mice, which display increased intestinal carotenoid absorption (Fig. 7A, far right). We obtained the same genotype-dependent carotenoid accumulation pattern when we analyzed gonadal white adipose pads. However, there was a marked difference in carotenoid levels. Although ocular levels were well below serum levels, carotenoids were highly concentrated in white adipocytes over serum (more than 10-fold in DKO mice). Thus, we conclude that adipose tissues concentrate carotenoids under conditions of CCO deficiency.
FIGURE 7.
Carotenoid accumulation in tissues of CCO knock-out mice. Total xanthophyll (A) and β-carotene (B) concentrations in serum, whole eyes, and gonadal white adipose tissue (gWAT) of CCO knock-out mice, Bco1−/−, Bco2−/−, and respective DKO kept on a diet containing β-carotene and zeaxanthin. Connecting cross-bars indicate ratios of eye and gonadal white adipose tissue carotenoid levels compared with serum. The far right charts in A and B show retinal concentrations of 3,3′-didehydrozeaxanthin and β-carotene, respectively. Values indicate means ± S.D. (error bars) from at least five female mice. Means with different letters (a and b) differ significantly. Statistical significance was assessed by one-way analysis of variance followed by Scheffe tests using Origin 9 software with the threshold of significance set at p <0.05. n.d., not detectable.
Human BCO2 Expression Is Oxidative Stress-responsive
Previously, we reported that carotenoid accumulation in cells and tissues can cause oxidative stress (11, 34). Endogenous BCO2 gene expression or forced expression of BCO2 transgene protects human cells against such insult (11, 34). Although BCO2 is tissue-specifically expressed in humans (14), little is known about how the gene is regulated. To investigate this, we used human HepG2 cells. This cell line has been successfully used to study carotenoid and retinoid metabolism in our and other laboratories (11, 13, 35). In a first set of experiments, we incubated cells in the presence of zeaxanthin dissolved in THF at a final concentration of 1 μm. After different time intervals, we harvested cells and isolated RNA for quantitative RT-PCR analyses. These analyses revealed that HuBCO2 expression was time-dependently increased. Thus, as reported previously for mouse liver (11), HuBCO2 mRNA expression was induced by carotenoids in this hepatic cell model (Fig. 8). We next wondered whether HuBCO2 mRNA expression might be responsive to reactive oxygen species. These damaging compounds are produced when the mitochondrial electron chain is perturbed by carotenoids (11). We treated HepG2 cells with hydrogen peroxide to mimic this stress and measured HuBCO2 mRNA expression. Quantitative RT-PCR analyses revealed that HuBCO2 mRNA levels increased up to 200-fold in HepG2 cells subjected to hydrogen peroxide treatment (Fig. 8). Thus, we have provided evidence that HuBCO2 mRNA expression is a regulated process and is responsive to oxidative stress.
FIGURE 8.
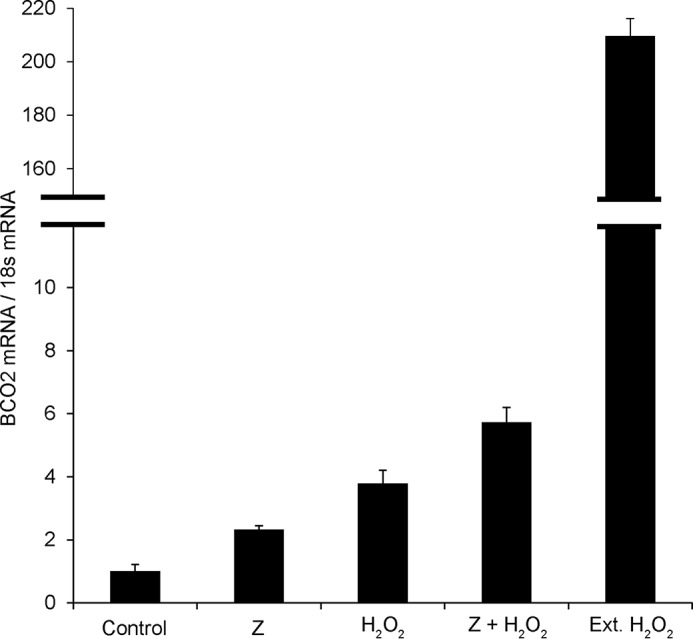
Oxidative stress induces BCO2 expression in human hepatic cells. Relative HuBCO2 mRNA levels measured when treated with vehicle control, zeaxanthin (Z), hydrogen peroxide, zeaxanthin in combination with hydrogen peroxide, and hydrogen peroxide for an extended (Ext.) period are presented. BCO2 transcript levels were weighted against 18S mRNA levels. Values indicate means ± S.D. (error bars) from at least four different measurements.
Discussion
Absorption of dietary carotenoids and distribution to tissues exemplify the discriminatory nature of carotenoid metabolism. From the large number of dietary carotenoids, just about 10 are present in human plasma (36), and only two are selectively accumulated in the human retina (2). The published reports that MPs are inversely associated with the prevalence of age-related macular degeneration (37, 38) instigated trials seeking to increase their concentration in the human retina (39, 40). In general, it was reported that a relatively linear increment of xanthophyll serum levels was achieved during supplementation and that eventually these levels reached a plateau. When supplementation was discontinued, a steep decline in serum concentrations was reported with baseline levels ultimately being restored. In response to this supplementation, the macular pigment optical density also increased to varying degrees (41). If the trend observed in serum were to hold true, then clearance of the MPs until baseline levels were achieved would occur. This indicates a relatively fast clearance of excessive supplemented xanthophyll. Evidence from animal studies demonstrates that BCO1 and BCO2 play a critical role for this process. In humans, mutations in the BCO1 gene cause hyper-β-carotenemia (42), but patients with BCO2 mutations have not yet been reported. Recently, Li et al. (12) postulated that accumulation of the human macular pigment is caused by inactivation of BCO2 resulting from the loss of an alternate splice site in the human gene. This hypothesis was based on the observation that xanthophylls are present in the retina of BCO2-deficient but not wild-type mice and on the absence of HuBCO2 enzymatic activity in a zeaxanthin-accumulating E. coli test system. However, this proposal is surprising because the overall sequence and structure of human BCO2 is evolutionarily well conserved. In fact, our previous analysis showed that recombinant HuBCO2 in β-carotene-producing E. coli cells was able to produce low amounts of apocarotenoid metabolites (13). The discrepancy between the outcomes of these studies can possibly be explained by the inherent difficulty in studying this family of enzymes in vitro. We therefore set out to examine the probability that primate enzyme characterization is limited by the current expression methods and enzyme assays. Additionally, in mouse mutants, we analyzed how BCO2 deficiency affected carotenoid homeostasis of the eyes.
An in silico inquiry into the structural models of the murine and primate CCOs did not produce significant differences that indicate that the primate BCO2 enzymes would be rendered inactive. These findings gave credence to our hypothesis that the setback in primate BCO2 characterization lay in the biochemical methodology. The choice of detergents to properly solubilize CCOs in water-based buffers has been shown to be a critical factor as several detergents greatly inhibit CCO activity (27, 28). A screen of several detergents for MuBCO2 activity identified DMN as an optimal choice in maintaining a functionally, soluble protein. It was not surprising to find that our initial trials of primate protein expression in E. coli produced only insoluble protein as recombinant protein misfolding in bacterial cells seems to be the norm (31). The expression of many recombinant proteins at lower temperatures has been a successful technique at limiting their in vivo aggregation (43). Using this technique, we were able to produce soluble trMaBCO2 but not HuBCO2. However, given the high degree of homology between the primate enzymes, MaBCO2 characterization should provide an identical biochemical insight into HuBCO2. Using these newly developed techniques, we were able to assay cleavage of the MPs by trMaBCO2. Determination of apparent Km values with the model substrate β-cryptoxanthin revealed comparable values for primate and murine BCO2s. Similarly, the specific turnover rates were roughly 20 and 14 s−1 for primate and murine BCO2s, respectively. This finding suggests that the enzymes display comparable affinity to this substrate, assuming that Michaelis-Menten kinetics can be applied and the oxidative cleavage is the rate-limiting step of the reaction.
Despite positive associations to the health benefits of antioxidants and blue light filters, carotenoids have been reported to act as pro-oxidants under high oxygen tension and high concentrations (44). In acting as reactive oxygen species scavengers, carotenoids undergo oxidation and generate various oxidation products (45, 46). Carotenoid supplementation studies in humans and monkeys have demonstrated a significant increase of these metabolites, which include didehydro derivatives, in serum and ocular tissues (47, 48). Because carotenoids are ubiquitous and their amounts can be abundant in food, mechanisms must have evolved that counteract such a scenario. We showed previously in mice and human cell lines that carotenoid homeostasis is tightly regulated. The primary regulation takes place at the level of intestinal absorption via retinoid and intestine specific homeobox signaling (23, 49). The secondary regulation is performed at the tissue level and involves CCOs. Genetic ablation of these regulatory processes in Bco2−/− and DKO mice fed a combination of zeaxanthin and β-carotene resulted in an amassing (including the retina) of zeaxanthin mostly in its didehydro derivative form. If indeed BCO2 inactivation drives specific accumulation of xanthophylls to ocular tissues, then it would be expected that Bco2−/− and DKO mice would have selectively greater -fold increases of zeaxanthin levels in the retina over other tissues as observed in primates. In a study in which macaques were first deprived of xanthophylls and then resupplemented, zeaxanthin accumulated to a 4.6-fold greater level in the retina over serum, whereas in fat, a 2.2-fold increase over that in serum was reported (50). In primates, the distribution of the MPs within the retina is assumed to undergo an additional selective mechanism due to its ordered dispersion (2, 51). Also, it would be expected that this accumulation occurs singularly with xanthophylls, but Bco1−/− and DKO mice fed the same combination of zeaxanthin and β-carotene diet also amassed β-carotene in their retinas. Consequently, we showed that the unregulated intake of carotenoids, carotenes, and xanthophylls alike causes a systemically indiscriminate accumulation of these compounds. We have shown previously in mice that this accumulation can cause oxidative stress in tissue. Similarly, carotenoids and their metabolites, including those derived from zeaxanthin and lutein, can induce oxidative stress (34, 52). It is not known whether xanthophylls play a role in the mitochondria, but it is clear that mitochondrial BCO2 expression plays a role in their controlled accumulation in this organelle. When we treated hepatic human cells with zeaxanthin, HuBCO2 mRNA expression was significantly induced. Additionally, when these cells were exposed to hydrogen peroxide, a highly reactive oxygen species, HuBCO2 mRNA levels increased up to 200-fold, providing additional evidence that HuBCO2 expression is responsive to oxidative stress. This sophisticated regulation manages the chemistry and biology of compounds that act as anti- and pro-oxidants depending on the subcellular localization and concentration. This finding may provide an explanation for the low carotenoid status of patients affected by chronic disease because they generally are associated with inflammation and oxidative stress.
In summary, we provide evidence that primate BCO2s are active enzymes and that they are able to cleave all of the three major macular pigment xanthophylls. Also, further evidence that BCO2 expression is a regulated process controlled in part by induction of oxidative stress within mitochondria is provided. Together, these findings effectively postulate that, although carotenoids play physiologically beneficial roles in human health, their possible excessive accumulation, which has been shown to cause harmful cellular pathologies, is restrained by the actions of BCO2.
Author Contributions
J. V. L. and D. B. designed the study and wrote the paper. D. B. performed all enzymatic assays. G. P. and D. B. designed and constructed the plasmids used in this study. G. P. performed immunofluorescence and cell work. M. A. K. W.-A. performed all animal studies. M. G. performed mass spectrometry analysis. P. D. K. provided the RPE65 template for all three-dimensional structure models. All authors analyzed the data and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Brian Kevany for providing macaque BCO2 total RNA. We thank Dr. Adrian Wyss for the generous gift of meso-zeaxanthin and lutein. We also thank Maryanne Pendergast and the Neurosciences Imaging Center for assistance with confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants EY020551, EY023948, EY11373, and EY007157 from the NEI. This work was also supported by Department of Veterans Affairs Career Development Award IK2BX002683 (to P. D. K.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Data 1 and 2.
- MP
- macular pigment
- 10Apo
- 3-hydroxy-β-10′-apocarotenal
- BCO1
- β-carotene 15,15′-dioxygenase
- BCO2
- β-carotene 9,10-dioxygenase
- CCO
- carotenoid cleavage oxygenase
- DKO
- Bco1−/−/Bco2−/− double knock-out mice
- DMN
- decyl maltose neopentyl glycol
- ϵ-10Apo
- 3-hydroxy-ϵ-10′-apocarotenal
- RPE65
- retinal pigment epithelium protein of 65 kDa
- MaBCO2
- macaque BCO2
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- TCEP
- tris(2-carboxyethyl)phosphine
- MuBCO2
- murine BCO2
- HuBCO2
- human BCO2
- trMaBCO2
- N-terminally truncated MaBCO2.
References
- 1. Moise A. R., Al-Babili S., Wurtzel E. T. (2014) Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bone R. A., Landrum J. T., Fernandez L., Tarsis S. L. (1988) Analysis of the macular pigment by HPLC: retinal distribution and age study. Invest. Ophthalmol. Vis. Sci. 29, 843–849 [PubMed] [Google Scholar]
- 3. Sommer A., Vyas K. S. (2012) A global clinical view on vitamin A and carotenoids. Am. J. Clin. Nutr. 96, 1204S–1206S [DOI] [PubMed] [Google Scholar]
- 4. Palczewski K. (2006) G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chambon P. (1996) A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954 [PubMed] [Google Scholar]
- 6. Krinsky N. I., Johnson E. J. (2005) Carotenoid actions and their relation to health and disease. Mol. Aspects Med. 26, 459–516 [DOI] [PubMed] [Google Scholar]
- 7. Bhosale P., Zhao da Y., Bernstein P. S. (2007) HPLC measurement of ocular carotenoid levels in human donor eyes in the lutein supplementation era. Invest. Ophthalmol. Vis. Sci. 48, 543–549 [DOI] [PubMed] [Google Scholar]
- 8. Eriksson J., Larson G., Gunnarsson U., Bed'hom B., Tixier-Boichard M., Strömstedt L., Wright D., Jungerius A., Vereijken A., Randi E., Jensen P., Andersson L. (2008) Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4, e1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Våge D. I., Boman I. A. (2010) A nonsense mutation in the β-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berry S. D., Davis S. R., Beattie E. M., Thomas N. L., Burrett A. K., Ward H. E., Stanfield A. M., Biswas M., Ankersmit-Udy A. E., Oxley P. E., Barnett J. L., Pearson J. F., van der Does Y., Macgibbon A. H., Spelman R. J., Lehnert K., Snell R. G. (2009) Mutation in bovine β-carotene oxygenase 2 affects milk color. Genetics 182, 923–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amengual J., Lobo G. P., Golczak M., Li H. N., Klimova T., Hoppel C. L., Wyss A., Palczewski K., von Lintig J. (2011) A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 25, 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li B., Vachali P. P., Gorusupudi A., Shen Z., Sharifzadeh H., Besch B. M., Nelson K., Horvath M. M., Frederick J. M., Baehr W., Bernstein P. S. (2014) Inactivity of human β,β-carotene-9′,10′-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl. Acad. Sci. U.S.A. 111, 10173–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palczewski G., Amengual J., Hoppel C. L., von Lintig J. (2014) Evidence for compartmentalization of mammalian carotenoid metabolism. FASEB J. 28, 4457–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindqvist A., He Y. G., Andersson S. (2005) Cell type-specific expression of β-carotene 9′,10′-monooxygenase in human tissues. J. Histochem. Cytochem. 53, 1403–1412 [DOI] [PubMed] [Google Scholar]
- 15. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 17. Mustafi D., Kevany B. M., Bai X., Maeda T., Sears J. E., Khalil A. M., Palczewski K. (2013) Evolutionarily conserved long intergenic non-coding RNAs in the eye. Hum. Mol. Genet. 22, 2992–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kiefer C., Hessel S., Lampert J. M., Vogt K., Lederer M. O., Breithaupt D. E., von Lintig J. (2001) Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 276, 14110–14116 [DOI] [PubMed] [Google Scholar]
- 19. Lindqvist A., Andersson S. (2002) Biochemical properties of purified recombinant human β-carotene 15,15′-monooxygenase. J. Biol. Chem. 277, 23942–23948 [DOI] [PubMed] [Google Scholar]
- 20. Voolstra O., Kiefer C., Hoehne M., Welsch R., Vogt K., von Lintig J. (2006) The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry 45, 13429–13437 [DOI] [PubMed] [Google Scholar]
- 21. Amengual J., Widjaja-Adhi M. A., Rodriguez-Santiago S., Hessel S., Golczak M., Palczewski K., von Lintig J. (2013) Two carotenoid oxygenases contribute to mammalian provitamin A metabolism. J. Biol. Chem. 288, 34081–34096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maeda T., Perusek L., Amengual J., Babino D., Palczewski K., von Lintig J. (2011) Dietary 9-cis-β,β-carotene fails to rescue vision in mouse models of Leber congenital amaurosis. Mol. Pharmacol. 80, 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Widjaja-Adhi M. A., Lobo G. P., Golczak M., Von Lintig J. (2015) A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 24, 3206–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lobo G. P., Amengual J., Palczewski G., Babino D., von Lintig J. (2012) Mammalian carotenoid-oxygenases: key players for carotenoid function and homeostasis. Biochim. Biophys. Acta 1821, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 26. Kiser P. D., Farquhar E. R., Shi W., Sui X., Chance M. R., Palczewski K. (2012) Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc. Natl. Acad. Sci. U.S.A. 109, E2747–E2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kowatz T., Babino D., Kiser P., Palczewski K., von Lintig J. (2013) Characterization of human β,β-carotene-15,15′-monooxygenase (BCMO1) as a soluble monomeric enzyme. Arch. Biochem. Biophys. 539, 214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sui X., Kiser P. D., Che T., Carey P. R., Golczak M., Shi W., von Lintig J., Palczewski K. (2014) Analysis of carotenoid isomerase activity in a prototypical carotenoid cleavage enzyme, apocarotenoid oxygenase (ACO). J. Biol. Chem. 289, 12286–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. von Lintig J., Vogt K. (2000) Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving β-carotene to retinal. J. Biol. Chem. 275, 11915–11920 [DOI] [PubMed] [Google Scholar]
- 30. Hu K. Q., Liu C., Ernst H., Krinsky N. I., Russell R. M., Wang X. D. (2006) The biochemical characterization of ferret carotene-9′,10′-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo. J. Biol. Chem. 281, 19327–19338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baneyx F. (1999) Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10, 411–421 [DOI] [PubMed] [Google Scholar]
- 32. Baneyx F., Mujacic M. (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22, 1399–1408 [DOI] [PubMed] [Google Scholar]
- 33. Kloer D. P., Ruch S., Al-Babili S., Beyer P., Schulz G. E. (2005) The structure of a retinal-forming carotenoid oxygenase. Science 308, 267–269 [DOI] [PubMed] [Google Scholar]
- 34. Lobo G. P., Isken A., Hoff S., Babino D., von Lintig J. (2012) BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway. Development 139, 2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Amengual J., Golczak M., Palczewski K., von Lintig J. (2012) Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J. Biol. Chem. 287, 24216–24227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khachik F., Spangler C. J., Smith J. C., Jr., Canfield L. M., Steck A., Pfander H. (1997) Identification, quantification, and relative concentrations of carotenoids and their metabolites in human milk and serum. Anal. Chem. 69, 1873–1881 [DOI] [PubMed] [Google Scholar]
- 37. Seddon J. M., Ajani U. A., Sperduto R. D., Hiller R., Blair N., Burton T. C., Farber M. D., Gragoudas E. S., Haller J., Miller D. T., Yannuzzi L. A., Willett W. (1994) Dietary carotenoids, vitamins A, C, and E, and advanced age-related macular degeneration. Eye Disease Case-Control Study Group. JAMA 272, 1413–1420 [PubMed] [Google Scholar]
- 38. Bone R. A., Landrum J. T., Mayne S. T., Gomez C. M., Tibor S. E., Twaroska E. E. (2001) Macular pigment in donor eyes with and without AMD: a case-control study. Invest. Ophthalmol. Vis. Sci. 42, 235–240 [PubMed] [Google Scholar]
- 39. Landrum J. T., Bone R. A., Joa H., Kilburn M. D., Moore L. L., Sprague K. E. (1997) A one year study of the macular pigment: the effect of 140 days of a lutein supplement. Exp. Eye Res. 65, 57–62 [DOI] [PubMed] [Google Scholar]
- 40. Schalch W. (1992) Carotenoids in the retina—a review of their possible role in preventing or limiting damage caused by light and oxygen. EXS 62, 280–298 [DOI] [PubMed] [Google Scholar]
- 41. Schalch W., Cohn W., Barker F. M., Köpcke W., Mellerio J., Bird A. C., Robson A. G., Fitzke F. F., van Kuijk F. J. (2007) Xanthophyll accumulation in the human retina during supplementation with lutein or zeaxanthin—the LUXEA (LUtein Xanthophyll Eye Accumulation) study. Arch. Biochem. Biophys. 458, 128–135 [DOI] [PubMed] [Google Scholar]
- 42. Lindqvist A., Sharvill J., Sharvill D. E., Andersson S. (2007) Loss-of-function mutation in carotenoid 15,15′-monooxygenase identified in a patient with hypercarotenemia and hypovitaminosis A. J. Nutr. 137, 2346–2350 [DOI] [PubMed] [Google Scholar]
- 43. Vasina J. A., Baneyx F. (1997) Expression of aggregation-prone recombinant proteins at low temperatures: a comparative study of the Escherichia coli cspA and tac promoter systems. Protein Expr. Purif. 9, 211–218 [DOI] [PubMed] [Google Scholar]
- 44. Palozza P., Serini S., Di Nicuolo F., Piccioni E., Calviello G. (2003) Prooxidant effects of β-carotene in cultured cells. Mol. Aspects Med. 24, 353–362 [DOI] [PubMed] [Google Scholar]
- 45. Hurst J. S., Saini M. K., Jin G. F., Awasthi Y. C., van Kuijk F. J. (2005) Toxicity of oxidized β-carotene to cultured human cells. Exp. Eye Res. 81, 239–243 [DOI] [PubMed] [Google Scholar]
- 46. Prasain J. K., Moore R., Hurst J. S., Barnes S., van Kuijk F. J. (2005) Electrospray tandem mass spectrometric analysis of zeaxanthin and its oxidation products. J. Mass Spectrom. 40, 916–923 [DOI] [PubMed] [Google Scholar]
- 47. Khachik F., Bernstein P. S., Garland D. L. (1997) Identification of lutein and zeaxanthin oxidation products in human and monkey retinas. Invest. Ophthalmol. Vis. Sci. 38, 1802–1811 [PubMed] [Google Scholar]
- 48. Khachik F., de Moura F. F., Chew E. Y., Douglass L. W., Ferris F. L., 3rd, Kim J., Thompson D. J. (2006) The effect of lutein and zeaxanthin supplementation on metabolites of these carotenoids in the serum of persons aged 60 or older. Invest. Ophthalmol. Vis. Sci. 47, 5234–5242 [DOI] [PubMed] [Google Scholar]
- 49. Lobo G. P., Amengual J., Baus D., Shivdasani R. A., Taylor D., von Lintig J. (2013) Genetics and diet regulate vitamin A production via the homeobox transcription factor ISX. J. Biol. Chem. 288, 9017–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson E. J., Neuringer M., Russell R. M., Schalch W., Snodderly D. M. (2005) Nutritional manipulation of primate retinas, III: effects of lutein or zeaxanthin supplementation on adipose tissue and retina of xanthophyll-free monkeys. Invest. Ophthalmol. Vis. Sci. 46, 692–702 [DOI] [PubMed] [Google Scholar]
- 51. Bone R. A., Landrum J. T., Friedes L. M., Gomez C. M., Kilburn M. D., Menendez E., Vidal I., Wang W. (1997) Distribution of lutein and zeaxanthin stereoisomers in the human retina. Exp. Eye Res. 64, 211–218 [DOI] [PubMed] [Google Scholar]
- 52. Kalariya N. M., Ramana K. V., Srivastava S. K., van Kuijk F. J. (2008) Carotenoid derived aldehydes-induced oxidative stress causes apoptotic cell death in human retinal pigment epithelial cells. Exp. Eye Res. 86, 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.