Background: Mitochondrial dynamics is important for regulating cellular physiological function.
Results: Depletion of DAP3 impairs mitochondrial-encoded protein synthesis and leads to mitochondrial fission.
Conclusion: DAP3 is essential for maintaining mitochondrial function.
Significance: DAP3 is the first mitochondrial ribosomal protein to be characterized that regulates mitochondrial dynamics.
Keywords: autophagy; cell death; cell signaling; metabolism; mitochondria; Drp1, DAP3
Abstract
Mitochondrial morphologies change over time and are tightly regulated by dynamic machinery proteins such as dynamin-related protein 1 (Drp1), mitofusion 1/2, and optic atrophy 1 (OPA1). However, the detailed mechanisms of how these molecules cooperate to mediate fission and fusion remain elusive. DAP3 is a mitochondrial ribosomal protein that involves in apoptosis, but its biological function has not been well characterized. Here, we demonstrate that DAP3 specifically localizes in the mitochondrial matrix. Knockdown of DAP3 in mitochondria leads to defects in mitochondrial-encoded protein synthesis and abnormal mitochondrial dynamics. Moreover, depletion of DAP3 dramatically decreases the phosphorylation of Drp1 at Ser-637 on mitochondria, enhancing the retention time of Drp1 puncta on mitochondria during the fission process. Furthermore, autophagy is inhibited in the DAP3-depleted cells, which sensitizes cells to different types of death stimuli. Together, our results suggest that DAP3 plays important roles in mitochondrial function and dynamics, providing new insights into the mechanism of a mitochondrial ribosomal protein function in cell death.
Introduction
Mitochondria are highly important organelles in the life and death of eukaryotic cells. They provide energy in the form of ATP via oxidative phosphorylation, which is critical for cellular metabolism and biosynthesis. Mitochondrial dynamics refers to the opposing fusion and fission processes that enable cells to respond and adapt to their ever changing physiological conditions (1, 2). Such processes are known to be controlled by several machinery proteins that are highly evolutionarily conserved (1, 3–5). However, the exact mechanisms responsible for their activity remain elusive. The fusion of mitochondria involves the merging of both outer and inner mitochondrial membranes of two or more mitochondria, which is controlled by three large GTPases. Specifically, they are outer mitochondrial membrane proteins Mfn12 and Mfn2 (6, 7) and the inner mitochondrial membrane anchored protein OPA1 (6, 8). In contrast, mitochondrial fission is highly controlled by another large dynamin-related GTPase Drp1, in which the mode of action has been intensively studied (9, 10). Drp1 exists as oligomers and mainly presents in the cytoplasm, but there is still a small portion of Drp1 located on the mitochondria in the form of foci (5). In the current model, cytosolic Drp1 is recruited onto mitochondria and forms spirals, resulting in the constriction and division of both outer and inner membranes (11). The regulation of Drp1 fission activity involves several post-translational modifications such as ubiquitination (12–14), sumoylation (15), and phosphorylation (16–19), among which the phosphorylation of Drp1 has been studied most intensively. For example, the phosphorylation status of Drp1 at Ser-637 is highly dynamic and reversibly regulated by cyclic AMP-dependent kinase (PKA) and calcineurin, a Ca2+ and calmodulin-dependent phosphatase (16–19). More recently, a novel mitochondrial phosphatase PAGM5 was reported to dephosphorylate Drp1 at Ser-637 and mediate mitochondrial fission during necrosis (20).
Autophagy is a highly conserved process for self-degradation of cellular components including long-lived proteins, damaged organelles and pathogens (21). When autophagy occurs, double membrane vesicles termed phagophore are initiated, and they expand to engulf cytosolic content to form the so-called autophagosomes (22). The autophagosomal outer membrane docks and fuses with the lysosomal membrane and the cargo is subsequently degraded by the lysosomal hydrolases (21). Not only can the organelles be removed by selective autophagy, but they can also contribute to the process of autophagy. For example, mitochondria can supply a membrane source (23, 24), generate ROS to induce autophagy (25), and tether endoplasmic reticulum to provide docking sites for autophagosome formation (23, 26, 27). However, at the molecular level, the underlying mechanism of the cross-talk between mitochondria and autophagy remains to be further elucidated.
DAP3 is a pro-apoptosis protein that involves in various types of stimuli-induced apoptosis, such as interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and TNF-related apoptosis-inducing ligand (28, 29). However, due to its subcellular localization, its precise role in cell death has been challenged (30, 31). Loss of DAP3 in mice leads to lethal embryonic mutations at E9.5, suggesting that it is essential to embryonic development (32). More importantly, the mitochondria in embryos lacking DAP3 contain swollen cristae and shrunken morphologies, indicating that DAP3 may play a role in regulating mitochondrial function (32). Furthermore, overexpression of DAP3 leads to an obvious increase in the rate of mitochondrial fission (33). However, the role of DAP3 in regulating mitochondrial fission or fusion has not yet been well studied. Here, we show that DAP3 is critical for maintaining mitochondrial fusion and fission homeostasis via regulating the phosphorylation of Drp1 at Ser-637 and is required for mitochondrial-encoded protein synthesis. In addition, autophagy is shown to be inhibited by DAP3 depletion, which subsequently sensitizes cells to intrinsic death stimuli. Our results reveal that DAP3 plays a key role in regulating mitochondrial function, providing new insights into a role of the mitochondrial ribosomal protein function in cell death.
Experimental Procedures
Reagents
DRP1 cDNA was kindly provided by Dr. Victor Chun-Kong Yu (National University of Singapore, Singapore). Mito-RFP (mitochrondrial targeted red fluorescent protein) plasmid was kindly provided by Dr. Quan Chen (Chinese Academy of Sciences, China). DAP3 gene was amplified from a cDNA library extracted from HEK-293T cells was inserted into an EGFP-N1 vector (Clontech) to fuse with a GFP tag at the C terminus of DAP3. The antibodies used in this study were as follows: anti-DAP3 (BD BioScience, 610662), anti-Tom20 (Santa Cruz, sc-17764), anti-cytochrome c (BD Bioscience, 556433), anti-Tim23 (BD BioScience, 611222), anti-VDAC1 (Santa Cruz, sc-8828), anti-Hsp60 (Santa Cruz, sc-1052), anti-Drp1 (BD BioScience, 611112), anti-Phospho-Drp1Ser−637 (Cell Signaling, 4867), anti-Phospho-Drp1Ser-616 (Cell Signaling, 3455), anti-Mff (Abcam, 139026), anti-Fis1 (Enzo, ALX-210-1037-0100), anti-OPA1 (BD BioScience, 612606), anti-Mfn1 (Santa Cruz, sc-50330), anti-Mfn2 (Abcam, ab50838), anti-LC3-II (Cell Signaling, 3868 and MBL, PM036), anti-MT-ND5 (Abcam, ab92624), anti-cytochrome c oxidize subunit II (Abcam, ab79393), anti-tubulin (Sigma, T5168), anti-FLAG (Sigma, F3165) and anti-GAPDH (Santa Cruz, sc-47724).
Cell Culturing and Treatment
HeLa, HEK-293T, and mouse embryo fibroblast cells were from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM) (Hyclone), supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco) and 10 units/ml of penicillin-streptomycin (Hyclone) unless otherwise noted. SH-SY5Y cells were maintained in a 1:1 mixture of Eagle's minimum essential medium (Hyclone) with non-essential amino acid and Ham's F-12 medium supplemented with 10% fetal bovine serum. All cultures were maintained at 37 °C, 5% CO2.
Plasmid and siRNA Transfection
For HeLa cells, EffecteneTM (Qiagen) was used to transfect plasmids. At 70–80% confluence, cells were washed by 1× PBS and transfected with the indicated plasmids according to the manufacturer's instructions. The transfection of siRNA into HeLa or SH-SY5Y cells using OligofectamineTM (Invitrogen) or RNAimax (Invitrogen) was performed according to the manufacturer's protocols. Oligonucleotides for siRNA were synthesized by Invitrogen and the sequences were as follows: DAP3 siRNA#1, 5′-AGGCUUCAACCUGGCUGAAGAAUUU-3′; DAP3 siRNA#2, 5′-CCUAGUGGCCGUGGAUGGAAUCAAU-3′; and DAP3 siRNA#3, 5′-GGCUUAUCUCUAGGAUCCAUAAGUU-3′; Mff siRNA, 5′-AACGCUGACCUGAACAAGGA-3′; and Drp1 siRNA, 5′-AGAAGCAGAAGAAUGGGGUAAAUUU-3′. The control siRNA sequence was: 5′-UUCUCCGAACGUGUCACGUTT-3′.
Subcellular Fractionation
HEK-293T, HeLa, or mouse embryo fibroblast cells cultured in 10-cm dishes were washed with 1× PBS before being harvested with a pre-cold mitochondrial extraction buffer (220 mm mannitol, 70 mm sucrose, 20 mm Hepes-KOH, pH 7.5, 1 mm EDTA, 0.5 mm PMSF, and 2 mg/ml of BSA) and supplemented with protease inhibitors including 10 μg/ml of aprotinin, 1 mm PMSF, 1 μm pepstatin, and 10 μm leupeptin. The cells were scraped down and transferred to a new 1.5-ml tube, then passed through a 25-gauge syringe (BD) 10 times on ice. The homogenized cells were centrifuged with a speed of 1000 × g for 15 min at 4 °C. The supernatant was then transferred into a new tube followed by another 20-min centrifugation at 4 °C, 10,000 × g, to pellet the mitochondria. The supernatant fraction was the cytosolic fraction.
For the mitochondrial membrane analysis, the mitochondria of HeLa cells were pelleted using the method described previously. The mitochondrial samples were then resuspended in freshly prepared 0.1 m Na2CO3, pH 11.5, and incubated on ice for 30 min with vortexing every 10 min. After the incubation, the membranes were centrifuged down to a pellet at 100,000 × g for 30 min at 4 °C, and the supernatant was collected as inter-mitochondrial membrane space and matrix proteins. For the proteinase K digestion assay, the isolated mitochondria were resuspended in a mitochondria-isolation buffer and incubated with different proteinase K concentrations on ice for 30 min. PMSF was added to stop the digestion and the samples were precipitated by TCA. The pellets were resuspended in RIPA buffer (20 mm Tris-Cl, pH 8.0, 125 mm NaCl, 0.5% Nonidet P-40, 5% glycerol) with phosphatases inhibitors including 20 mm NaF, 0.2 mm Na3VO4, protease inhibitors and 2 mm EDTA for 30 min and subjected to SDS-PAGE and Western blotting.
Immunofluorescence
For immunofluorescence, cells were seeded on sterilized glass coverslips in 12-well plates for 24 h before transfection with the indicated plasmids or siRNAs. After transfection, the cells were cultured for the indicated time and fixed by freshly prepared 4% paraformaldehyde for 15 min at room temperature. Paraformaldehyde was removed after fixation, followed by a 2 times wash with PBS and the coverslips were then incubated with 3% BSA + 0.1–0.5% Triton X-100 in PBS for 30 min at room temperature for blocking and permeabilization. After the incubation, the coverslips underwent a 3 times wash by PBS, followed by incubation with primary antibodies diluted in 3% BSA for 1 h at room temperature. After the primary antibody incubation, the coverslips were washed again and incubated with fluorescence-conjugated secondary antibodies for 45 min at room temperature. After incubation, the coverslips were washed twice by PBS and the fluorescent dye Hoechst 33342 was applied to stain DNA (Invitrogen) for 15 min at room temperature. Cells stained by the various dyes were mounted onto glass slides using FluorSaveTM reagent (Calbiochem) and preserved at 4 °C for further analysis.
Live Cell Imaging and Fluorescence Recovery After Photobleaching (FRAP)
For live cell imaging, HeLa cells were seeded in a 35-mm glass-bottom dish with different treatments and incubated at 37 °C, 5% CO2 in a chamber equipped with a microscope. Time-lapse imaging was conducted using an Ultraview Vox spinning disc confocal system (PerkinElmer). VolocityTM (PerkinElmer Life Sciences) was used to control all of the parameters used for image acquisition.
For FRAP analysis, HeLa cells stably expressing mito-RFP were treated by different siRNAs until the indicated time. The cells were monitored on the live-cell imaging system as described above, and a laser line of 561 nm was used to bleach a 2 × 2-μm2 area placed on the mitochondrial network fiber. Twenty s of recovery time for each bleaching was used to make sure the recovery had reached the plateau. Thirty FRAP curves for each siRNA transfection experiment were analyzed by measuring the intensities of mito-RFP fluorescence in the photobleached area. Mobile fraction and turnover half-life (t½) for mito-RFP were calculated by fitting a normalized recovery curve into a constrained exponential formula (34).
Metabolic Labeling of Mitochondrial Translation Products Using AHA
The pulse labeling of mitochondrial translation products in the control and siRNA-treated HeLa cells was performed as a reported protocol with modifications (35). Specifically, cells at 80% confluence were incubated with 30 μm AHA in DMEM and lack methionine and cysteine containing 100 μg/ml of cytosolic translation inhibitor emetine for 3 h. After AHA labeling, the cells were lysed and a “click” reaction was performed by incubating the cell lysates with fluorescent dye-conjugated TAMRA for 2 h at room temperature. The proteins were precipitated by ice-cold acetone and air-dried before being subjected to NuPAGE® Novex® 4–12% Bis-Tris Protein Gel (Invitrogen) separation. The gel was visualized using a Typhoon 9410 laser scanner (GE HealthCare) and the images were analyzed by ImageQuantTM software (GE HealthCare).
Immunoblotting and Immunoprecipitation
Immunoblotting was performed as previously described (36). For immunoprecipitation, a total of 5 μl of FLAG M2 beads (Sigma) were equilibrated in 1 ml of mammalian cell lysis buffer (50 mm Tris-HCl, pH 7.4, 10% glycerol, 1% Triton X-100, 100 mm NaCl, and 0.5 mm MgCl2 supplemented with protease inhibitors, including 10 μg/ml of aprotinin, 1 mm PMSF, 1 μm pepstatin, and 10 μm leupeptin). The equilibrated FLAG M2 beads were added into the cleared cytosol fraction and resolved in a mitochondrial fraction harvested from two 10-cm cell culture dishes and incubated for 3–5 h at 4 °C. After incubation, the immunocomplexes were washed 3–4 times in mammalian cell lysis buffer and the bead-conjugated proteins were denatured by incubation with 2× SDS loading buffer for 15 min at 95 °C. Protein samples were then separated by polyacrylamide gel electrophoresis and detected by Western blotting.
ATP Production Assay
ATP production was measured using an ATP determination kit (Molecular Probes) according to the manufacturer's instructions. In brief, HeLa cells transfected with control- or DAP3-siRNAs were washed with PBS and harvested using ice-cold ATP buffer (20 mm Tris-HCl, PH7.5, 0.5% Nonidet P-40, 25 mm NaCl, and 2.5 mm EDTA) for 5 min. The lysates were then centrifuged at 13,000 × g for 30 min at 4 °C, and the supernatants were collected and protein concentrations were measured using the Bradford Protein Assay reagent (Bio-Rad). The ATP levels were determined using 0.5 μg of proteins for each reaction and every sample was measured in triplicate.
Biochemical Assays
To inhibit ATP synthase activity, 10 μg/ml of oligomycin was used to treat HeLa cells for 3 h. To inhibit calcineurin activity or stimulate PKA activity, SH-SY5Y cells were treated with 200 nm FK506 (InvivoGen; dissolved in DMSO) or 20 μm forskolin (Enzo; dissolved in DMSO) for 2 h, respectively. To investigate the autophagy process, EBSS (minimum essential medium) was used to treat HeLa cells for 2 h. To induce intrinsic and extrinsic cell death, 20 ng/ml of TNF-α (Sigma; dissolved in DMSO) plus 3 μg/ml of cycloheximide (Sigma; dissolved in DMSO), 20 μm CCCP (Sigma; dissolved in DMSO), and 100 nm staurosporine (Sigma; dissolved in DMSO) were applied for 24 h, respectively.
Flow Cytometry Analysis
HeLa cells transfected with DAP3 siRNAs for 48 h were trypsinized and washed twice by PBS. JC-1 dye (Invitrogen) for measuring mitochondrial membrane potential (ΔΨm) was diluted in medium at 1 μg/ml to resuspend and stain the cells. The resuspended cells were put in a cell culture incubator for 15 min for the staining, after which the JC-1 medium was replaced by fresh medium. The labeled samples were then analyzed using Dako flow cytometry (Dako) and the results were analyzed using SummitTM 4.3. Ten thousand events were recorded for each experiment.
HeLa cells transfected with DAP3 siRNAs for 48 h were treated with 20 ng/ml of TNF-α + 3 μg/ml of cycloheximide, 20 μm CCCP, 100 nm staurosporine or EBSS as indicated for 24 h. Immediately after the treatment, the cells were harvested to a 15-ml Falcon tube (including floating cells). The cells were pelleted by centrifugation at (200 × g, 5 min) and the supernatant was discarded. The propidium iodide (10 μg/ml, Sigma) for staining dead cells was diluted in medium at 10 ng/ml to resuspend and stain the cells. Five min later, the labeled samples were analyzed using Dako flow cytometry (Dako) and the results were analyzed using summit 4.3. Ten thousand events were recorded for each experiment.
Results
DAP3 Localizes Inside Mitochondria
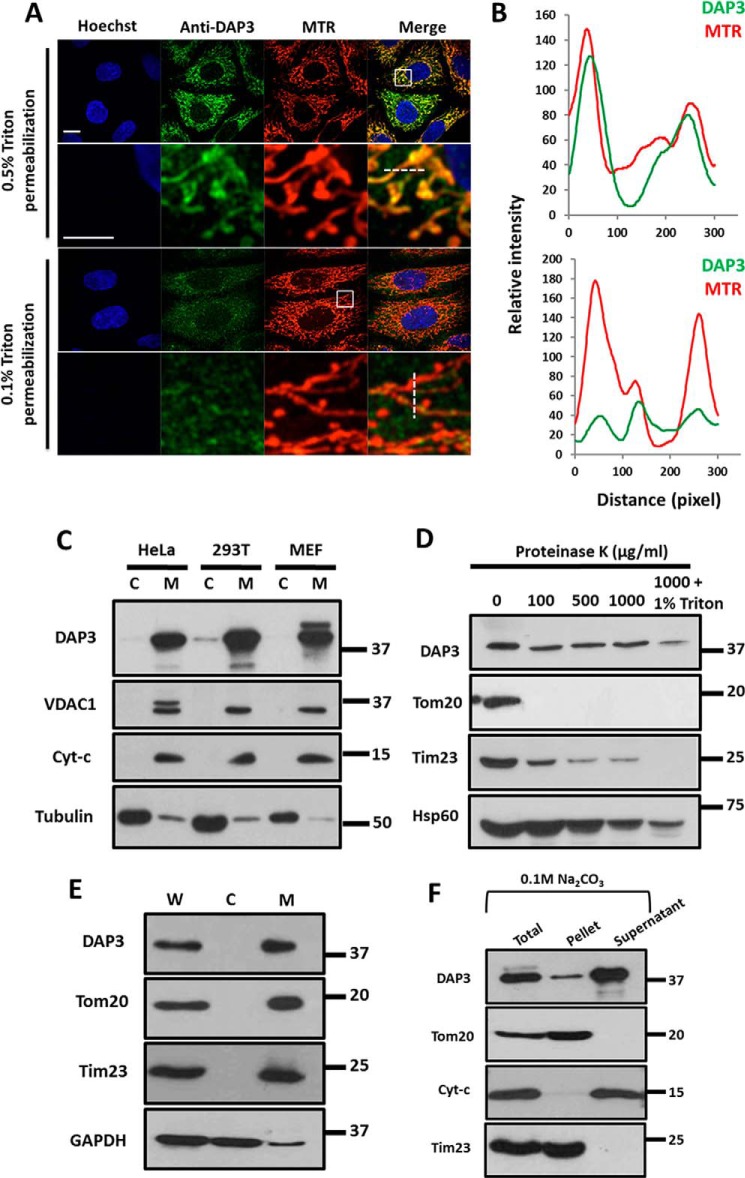
Previous studies suggest that DAP3 has a function in promoting cell death (28, 37), but the subcellular location of DAP3 remains controversial. A report indicated that DAP3 is located in mitochondria (29), whereas the other argued that there is also a cytosolic pool of DAP3 (30). To address this, we conducted immunostaining for endogenous DAP3 and Western blots to examine its subcellular location. Our result showed that endogenous DAP3 co-localized well with mitochondria when treated with 0.5% Triton X-100 (Fig. 1, A and B, upper panel). In contrast, the DAP3 signal was significantly decreased on mitochondria if treated with 0.1% Triton X-100 (Fig. 1, A and B, lower panel), suggesting that DAP3 was probably located inside of the mitochondria. Consistently, endogenous levels of DAP3 in different cell lines (HeLa, HEK-293T, and mouse embryo fibroblasts) were also detected in the mitochondrial fraction, but not in the cytosolic fraction (Fig. 1C). To further determine the submitochondrial location of DAP3, proteinase K protection assay was performed. As shown in Fig. 1D, proteinase K treatment resulted in the disappearance of the outer membrane protein Tom20 and the inner membrane protein Tim23 gradually with the proteinase K concentration going up. In contrast, DAP3 and Hsp60, a mitochondrial matrix protein, were protected from the proteolysis by proteinase K, unless Triton X-100 was added (Fig. 1D). In addition, the mitochondrial fraction applied to alkaline extraction suggested that DAP3, like cytochrome c, was predominantly retained in the supernatant fraction, whereas the membrane-integrated proteins Tom20 and Tim23 were mainly found in the mitochondrial membrane pellet (Fig. 1, E and F). Taken together, our data indicate that DAP3 mainly localizes in the matrix of mitochondria.
FIGURE 1.
DAP3 localizes to the mitochondrial matrix. A, HeLa cells were fixed with 4% paraformaldehyde and permeablized by 0.5 or 0.1% Triton X-100 for 30 min, respectively. The cells were stained with anti-DAP3 antibody and mitochondrial specific dye MitoTracker. The fluorescent Hoechst 33342 dye was used to stain DNA. The insets show enlarged views of the boxed regions. Scale bar, 10 μm; inset scale bar, 5 μm. B, graphs represent the relative fluorescence intensity along the line scans of the DAP3 signal and the mitochondrial signal, as indicated by a white dashed line in A. C, HeLa, HEK-293T, and mouse embryo fibroblast cells were fractionated and analyzed by immunoblotting using antibodies against the indicated proteins. C, cytosol; M, mitochondria. D, mitochondria of HeLa cells from panel C were digested by proteinase K with the indicated concentrations and analyzed by immunoblotting using antibodies against DAP3, Tom20, cytochrome c, Tim23, and HSP60. E, mitochondria of HeLa cells from panel C were analyzed by immunoblotting using antibodies against DAP3, Tom20, Tim23, and GAPDH. F, the mitochondrial fraction of HeLa cells from panel C was subjected to alkaline extraction using sodium carbonate and immunoblotted for DAP3, Tom20, cytochrome c, and Tim23.
DAP3 Depletion Results in Mitochondrial Fragmentation
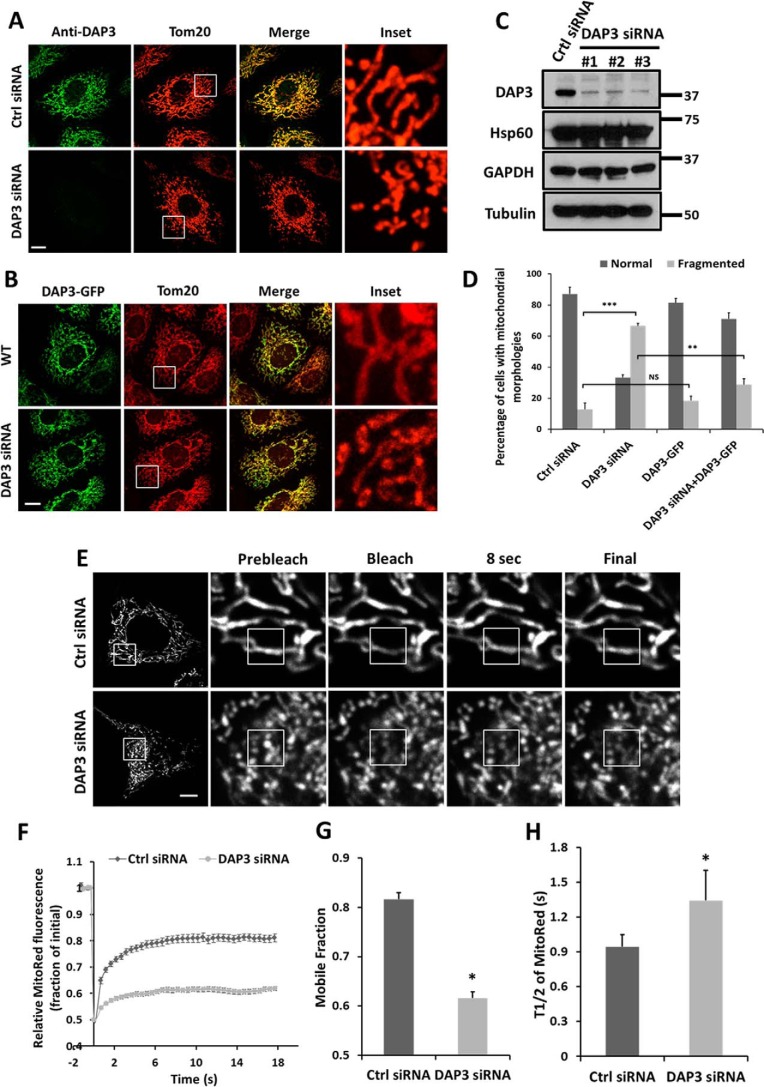
It has been reported that DAP3 is essential for embryonic development and mitochondrial function in the mouse (32). Mitochondria were shrunken and cristae were swollen in DAP3 knock-out mouse embryos (32). To investigate whether the developmental deficiency and abnormal mitochondrial morphology were related to mitochondrial dynamics, we designed three siRNAs to knockdown the endogenous DAP3 in HeLa cells and assessed mitochondrial morphology by immunostaining using Tom20. Knockdown levels of DAP3 were confirmed by immunostaining and Western blot analysis (Fig. 2, A and C). Depletion of DAP3 led to a significant increase of cells with fragmented mitochondria to around 70%, compared with about 10% in control cells (Fig. 2, A and D). On the contrary, mitochondrial morphology was not significantly changed upon overexpression of DAP3-GFP (Fig. 2, B and D). In addition, when DAP3-GFP was re-introduced back to DAP3-depleted cells, the fragmented mitochondria were rescued back to a tubular structure (Fig. 2, B and D), indicating the specific effect of DAP3 knockdown. To further examine the extent of fission induced by DAP3 depletion, a quantitative FRAP assay was conducted to measure the interconnectivity of mitochondria. The mitochondrial fluorescence recovery levels were lower and the recovery rate was slower in the DAP3-depleted cells than that in control cells (Fig. 2, E and F). Specifically, the average mobile fraction was ∼0.8 in control cells, whereas the average mobile fraction was decreased to around 0.6 in DAP3-depleted cells (Fig. 2G). Furthermore, the average recovery half-time (t½) of the DAP3-depleted cells was 1.34 s, which was much slower than that of 0.94 s in control cells (Fig. 2H). Taken together, these results indicate that DAP3 is essential for maintaining the network of mitochondria.
FIGURE 2.
Depletion of DAP3 results in mitochondrial fragmentation. A, HeLa cells transfected with control or DAP3-siRNA were stained with Tom20 (red) and DAP3 (green) antibodies. Scale bar, 10 μm. B, wild-type (WT) and DAP3-siRNA treated HeLa cells were transfected with DAP3-GFP and immunostained with Tom20. Scale bar, 10 μm. C, whole cell lysates of HeLa cells treated with control- or DAP3-siRNA for 48 h were collected and subjected to SDS-PAGE. The knockdown levels of DAP3 were detected using an anti-DAP3 antibody. D, the bar chart represents the percentage of different types of mitochondrial morphology of cells as treated in A and B (n = 100 cells quantified from three independent experiments). Statistical significance was calculated using Student's t test. **, p < 0.01; ***, p < 0.001; NS indicates not significant (p > 0.05). Error bars represent mean ± S.D. E, HeLa cells stably expressing mito-RFP treated as indicated were applied to FRAP analysis. The images were acquired in 0.5-s intervals. A 2 × 2-μm2 square region of interest (ROI) was placed on the mitochondrial fiber and photobleached with a 561-nm laser. Scale bar, 10 μm. F, normalized recovery curves of FRAP assay in E. The data were collected from 30 ROIs in 20 cells in 3 independent experiments. Error bars represent mean ± S.E. G and H, bar charts represent the mobile fraction and the recovery half-life (t½) of mito-RFP fluorescent signal in the photobleached area. Statistical significance was calculated using Student's t test. *, p < 0.05. Error bars represent mean ± S.E.
DAP3 Depletion Impairs Mitochondrial Protein Synthesis and Function
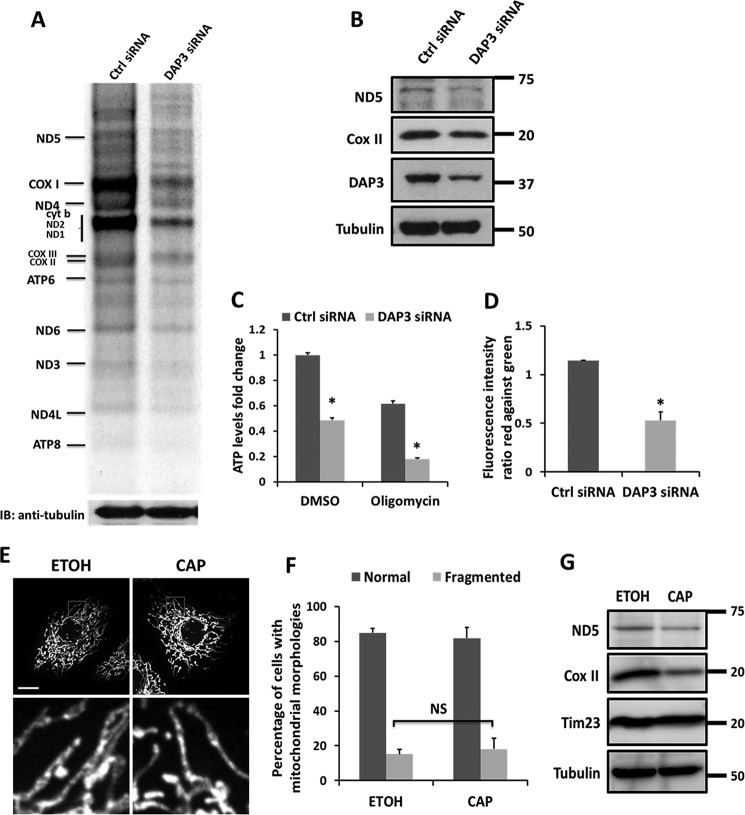
Because DAP3 is known as a mitochondrial ribosomal protein (31), to assess whether it plays a role in synthesizing mitochondrial proteins, we performed a metabolic pulse labeling assay using the methionine surrogate AHA in the presence of emetine, which can inhibit cytosolic protein synthesis. Our result showed that upon knockdown of DAP3, the translation of mitochondrial-encoded proteins was globally decreased compared with that in control cells (Fig. 3A). Furthermore, the steady-state levels of mitochondrial-encoded proteins, the complex I subunit ND5, and the complex IV subunit COX II were also attenuated in DAP3-depleted cells (Fig. 3B). These data suggest that as a mitochondrial ribosomal protein, DAP3 is essential for the de novo mitochondrial-encoded protein synthesis. Because all 13 of the polypeptides are components of the mitochondrial respiratory chain, we further examined whether DAP3 affected ATP production. Knockdown of DAP3 decreased cellular ATP production to about 50%, compared with that in control cells (Fig. 3C). Moreover, blocking the ATP synthase activity by oligomycin further disrupted the ATP level in DAP3-depleted cells to less than 20% compared with 60% in control cells (Fig. 3C), suggesting that the knockdown of DAP3 may also affect other respiratory chain complexes in addition to ATP synthase. It is known that mitochondrial ribosome intactness is important for ΔΨm (38), we then investigated the ΔΨm using a cationic dye, JC-1. Our quantitative analysis by FACS showed that upon DAP3 depletion Δψm decreased significantly (Fig. 3D). Taken together, our results indicate that DAP3 is required for maintaining the physiological function of mitochondria.
FIGURE 3.
Knockdown of DAP3 impairs mitochondrial function. A, pulse labeling mitochondrial translation assay for control- or DAP3-siRNA-transfected HeLa cells by AHA. B, steady-state levels of mitochondrial respiratory components from the mitochondrial translation products were subjected to SDS-PAGE and immunoblotted (IB) with antibodies against ND5, COX II, DAP3, and Tubulin, respectively. C, HeLa cells were transfected with control- or DAP3-siRNA for 48 h. Immediately, the cells were treated with DMSO or 10 μg/ml of oligomycin for 3 h, respectively. ATP levels were measured by luciferin-luciferase bioluminescence assay. The bar chart represents the fold-changes in normalized ATP values to the control cells. Each treatment was measured in triplicates. Statistical significance was calculated using Student's t test. *, p < 0.05. Error bars represent mean ± S.D. D, quantitative analysis of the ΔΨm using JC-1 dye in control- or DAP3-siRNA transfected HeLa cells was performed using FACS by three independent experiments. Statistical significance was calculated using Student's t test. *, p < 0.05. Error bars represent mean ± S.E. E, HeLa cells were treated with vehicle (EtOh, ethanol) or chloramphenicol for 48 h. Immediately, cells were fixed and immunostained by Tom20 antibody. The insets show enlarged views of the boxed regions. Scale bar, 10 μm. F, the bar chart represents the percentage of different types of mitochondrial morphology of cells as treated in E (n = 100 cells quantified from three independent experiments). Statistical significance was calculated using Student's t test. NS indicated not significant (p > 0.05). Error bars represent mean ± S.D. Scale bar, 10 μm. G, whole cell lysates of HeLa cells treated as in E were collected and subjected to immunoblotting using indicated antibodies.
To test whether the inhibition of mitochondrial protein synthesis is responsible for mitochondrial fragmentation, chloramphenicol was used to block the translation of the mitochondrial-encoded proteins and examine mitochondrial dynamics. There was no marked difference on mitochondrial fusion or fission observed between the vehicle and chloramphenicol (Fig. 3, E and F). The inhibition effect of chloramphenicol was evaluated by immunoblotting with mitochondrial-encoded ND5 and COX II (Fig. 3G).
DAP3-induced Mitochondrial Fragmentation Is Dependent on Mff-Drp1 Activity
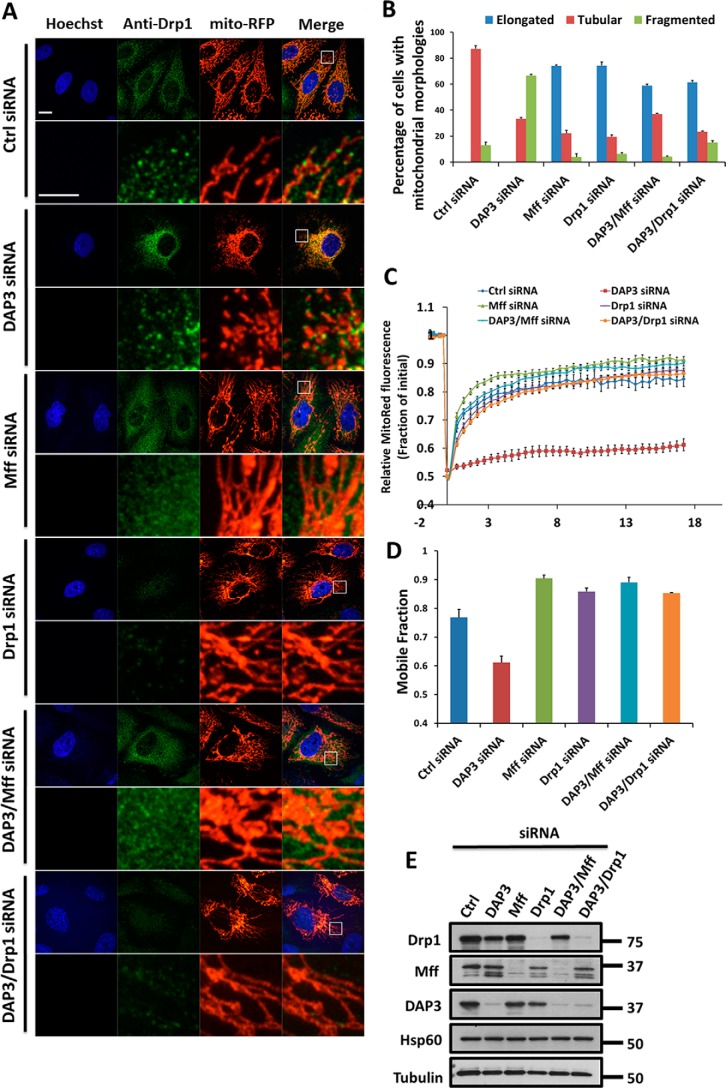
To understand the mechanism of DAP3 in regulating mitochondrial fragmentation, we next examined the effect of DAP3 knockdown on mitochondrial fission machinery. Mitochondrial fission is known to be controlled by Drp1 and its receptor Mff (39); knockdown of either Drp1 or Mff yielded more than 70% of cells with elongated mitochondria (Fig. 4, A and B). To further test whether the effect of DAP3 on mitochondrial morphology was associated with the function of Drp1 and Mff, we co-depleted DAP3 with Drp1 or Mff and found that the number of fragmented mitochondria decreased drastically from about 60% in the DAP3-depleted cells to ∼5–10% in the Mff + DAP3 or Drp1 + DAP3 co-depleted cells (Fig. 4, A and B). Moreover, to quantitate fusion and fission status, we utilized a FRAP assay to monitor the effect of different siRNA-depleted cells and found that the fluorescence recovery levels as indicated by the mobile fraction were much more elevated in the DAP3 + Mff or DAP3 + Drp1 co-depleted cells (Fig. 4, C and D). The depletion effectiveness of various siRNA was confirmed by Western blot analysis (Fig. 4E). Collectively, these data suggest that depletion of DAP3-mediated mitochondrial fragmentation is dependent on Mff-Drp1 fission activity.
FIGURE 4.
DAP3-depletion-induced mitochondrial fission is dependent on Drp1. A, representative images of mitochondrial morphology in HeLa cells treated as indicated are shown. Scale bar, 10 μm. Endogenous Drp1 was stained with anti-Drp1 antibody. The DNA was stained with Hoechst 33342. Mitochondria are indicated by mito-RFP. The lower panel is an enlargement of the boxed inset of the corresponding panel above. Scale bar, 10 μm; inset scale bar, 5 μm. B, the bar chart represents the percentage of different types of mitochondrial morphology for HeLa cells treated as in A (n = 100 cells quantified from three independent experiments). C, normalized recovery curves of FRAP assay as described in the legend to Fig. 2F. Cells were treated as indicated. D, a bar chart represents the mobile fraction of mito-RFP fluorescent signal from the curves in C. E, whole cell lysates of HeLa cells treated as in A were collected and subjected to immunoblotting. The knockdown levels of DAP3, Mff, and Drp1 were detected using the indicated antibodies. Hsp60 and Tubulin are loading controls.
It has been reported that the cytosolic Drp1 is recruited onto mitochondria, forming puncta to execute mitochondrial division (11). To further explore the regulatory mechanism of DAP3 on Drp1 function, the localization of endogenous Drp1 was examined by immunostaining (Fig. 4A). We observed that Drp1 was presented in puncta structures on each fragmented mitochondria in the DAP3-depleted cells, whereas Drp1 puncta in the control cells were located on mitochondrial fibers (Fig. 4A). Interestingly, the co-depletion of DAP3 with Mff or Drp1 could either remove Drp1 from the mitochondria or eliminate Drp1, resulting in mitochondrial elongation (Fig. 4A), further confirming that Drp1 activity, especially the puncta form, is required for DAP3 depletion-induced mitochondrial fragmentation.
Knockdown of DAP3 Affects Drp1 Phosphorylation
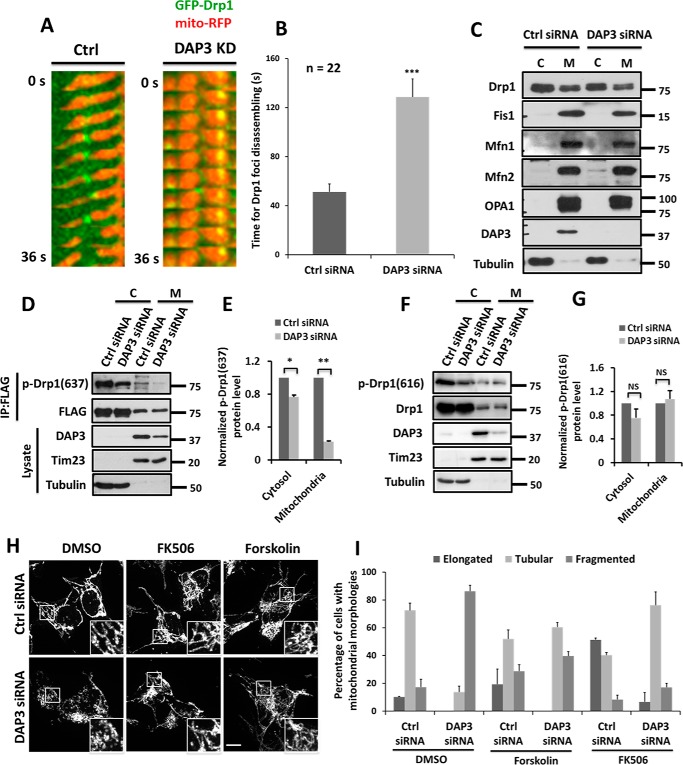
To further monitor Drp1 puncta movement on mitochondria, a live-cell imaging approach was utilized. Our results showed that in the control cells, GFP-Drp1 puncta were formed on mitochondrial fibers and mediated fission with an average lifetime of 51 s (Fig. 5, A and B, and supplemental Movie S1). Conversely, in the DAP3-depleted cells, GFP-Drp1 puncta remained on mitochondria for ∼129 s and could not be disassembled in a short time (Fig. 5, A and B, and supplemental Movie S2), suggesting that DAP3 might regulate Drp1 puncta dynamics. However, there is no significant change of the protein levels of Drp1 in either mitochondrial or cytosol fractions in DAP3-depleted cells compared with that in control cells (Fig. 5C). Because the localization of Drp1 on mitochondria is tightly regulated by its phosphorylation (16, 18, 40, 41), to further investigate whether the longer lifetime of Drp1 puncta on mitochondria in the DAP3-depleted cells is due to the regulation of Drp1 phosphorylation, immunoblotting assays were performed by using antibodies that specifically recognize phosphorylated Drp1 at Ser-637 or at Ser-616. Surprisingly, we found that phosphorylation of Drp1 at Ser-637 was significantly decreased in DAP3-depleted cells in both mitochondrial and cytosol fractions (Fig. 5, D and E), whereas the phosphorylation status of Drp1 at Ser-616 was not significantly changed (Fig. 5, F and G). In addition, the protein levels of other mitochondrial machinery proteins such as Fis1, Mfn1, Mfn2, and OPA1 had no significant change (Fig. 5C), suggesting that DAP3 may specifically regulate Drp1 function by mediating its phosphorylation at Ser-637.
FIGURE 5.
Knockdown of DAP3 affects Drp1 phosphorylation. A, representative kymographs of GFP-Drp1 and mito-RFP dynamics. Kymographs of the selected 30 × 18 pixel square area from HeLa cells treated as indicated. B, a bar chart represents the time for Drp1 foci disassembling. Statistical significance was calculated using Student's t test. ***, p < 0.001. Error bars represent mean ± S.E. C, HeLa cells treated as indicated were fractionated and analyzed by immunoblotting using antibodies against the indicated proteins. C, cytosol; M, mitochondria. D, HeLa cells transfected with control or DAP3-siRNA for 48 h were transfected with FLAG-Drp1 for another 24 h. After the treatment, cells were fractionated to cytosol and mitochondria. Both fractions from the same batch of cells were immunoprecipitated (IP) by FLAG-M2 beads. Lysate and beads were subjected to immunoblotting using antibodies against the indicated proteins. E, the bar chart represents the normalized phosphor-Drp1(Ser-637) protein levels in cells treated as in D. F, HeLa cells transfected with control or DAP3-siRNA for 48 h were fractionated to cytosol and mitochondria. Both fractions from the same batch of cells were subjected to immunoblotting using antibodies against the indicated proteins. G, the bar chart represents the normalized phosphor-Drp1(Ser-616) protein levels in cells treated as in F. H, SH-SY5Y cells transfected with control or DAP3-siRNA were treated with DMSO, 20 μm forskolin, and 200 nm FK506 for 2 h, respectively. Mitochondrial morphology was highlighted by Tom20. Scale bar, 10 μm. I, the bar chart represents the percentage of different types of mitochondrial morphology for SH-SY5Y cells treated as in H (n = 100 cells quantified from 3 independent experiments). Error bars represent mean ± S.D.
The phosphorylation of Drp1 at Ser-637 was reversibly regulated by PKA kinase and calcineurin phosphatase (16, 41), we hypothesized that the markedly reduced phosphorylation of Drp1 at Ser-637 in DAP3-depleted cells is related to the PKA-calcineurin pathway. To verify this, PKA was activated by treating with forskolin, or the activity of calcineurin was inhibited by adding FK506 in control and DAP3-depleted cells. Both agents could effectively rescue fragmented mitochondria to tubular fibers (Fig. 5, H and I). Taken together, these results suggest that DAP3 depletion attenuates the phosphorylation of Drp1 at Ser-637 via altering the antagonistic effect of PKA-calcineurin pathway on mitochondrial biogenesis, resulting in mitochondrial fragmentation.
Loss of DAP3 Function Inhibits Autophagy and Sensitizes Cell to Death
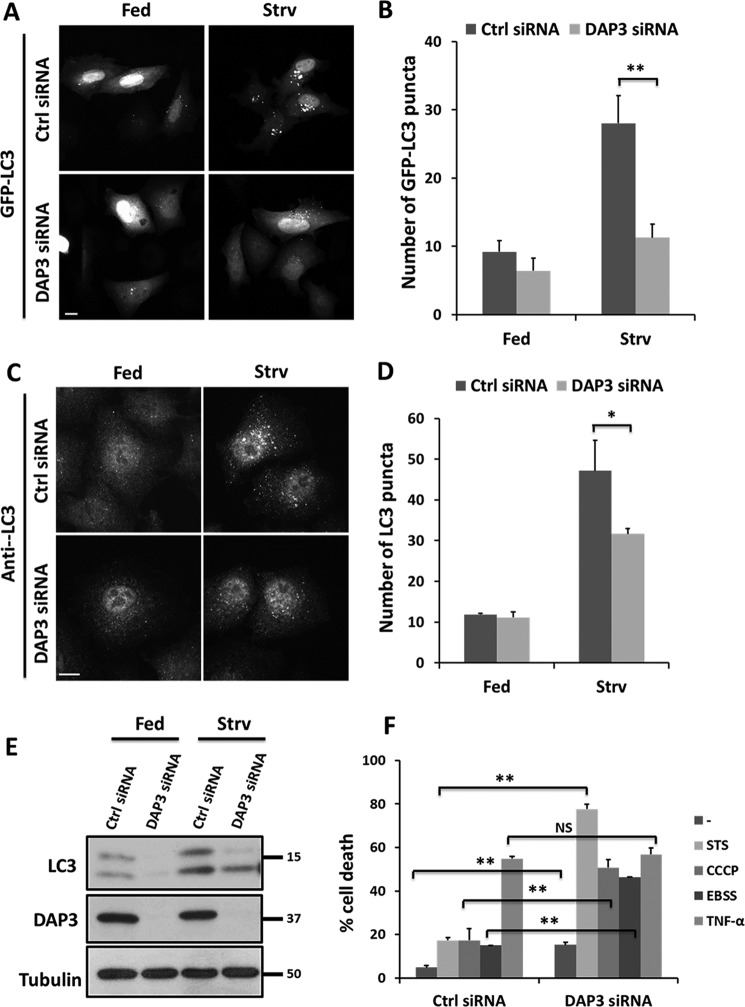
Accumulated evidence has suggested that mitochondria play a critical role in the autophagy process (19, 23). To investigate insights into the biological function of DAP3, the effect of DAP3 knockdown was examined on autophagic activity. Our results showed that starvation by EBSS treatment caused a marked increase of GFP-LC3 puncta in control cells, whereas depletion of DAP3 significantly reduced the number of GFP-LC3 puncta (Fig. 6, A and B). Consistently, we observed similar results that DAP3 depletion could significantly reduce the LC3 puncta under starvation conditions by using an anti-LC3 antibody (Fig. 6, C and D). Moreover, the basal and starvation-induced expression levels of LC3-II were lower in DAP3-depleted cells (Fig. 6E). All these results reveal that loss of DAP3 might inhibit starvation-induced autophagy.
FIGURE 6.
Knockdown of DAP3 affects mitochondrial function and cell death. A, HeLa cells were transfected with control- or DAP3-siRNA. After 48 h, the cells were transfected with GFP-LC3 for another 24 h and immediately starved with EBSS for 2 h. Scale bar, 10 μm. B, the bar chart represents the number of GFP-LC3 puncta in HeLa cells treated as in A (n = 30 cells quantified from 3 independent experiments). Statistical significance was calculated using Student's t test. **, p < 0.01. Error bars represent mean ± S.D. C, HeLa cells transfected with control- or DAP3-siRNA for 48 h were starved with EBSS for 2 h. LC3 puncta were indicated by anti-LC3 antibody. Scale bar, 10 μm. D, the bar chart represents the number of autophagosomes in HeLa cells treated as in C (n = 30 cells quantified from 3 independent experiments). Statistical significance was calculated using Student's t test. *, p < 0.05. Error bars represent mean ± S.D. E, HeLa cells treated as indicated were harvested and subjected to SDS-PAGE. Proteins were immunoblotted with antibodies as indicated. F, HeLa cells were transfected with control- or DAP3-siRNA for 48 h. Immediately, the cells were treated for 24 h with the following agents: 20 ng/ml of TNF-α + 3 μg/ml of cycloheximide, 20 μm CCCP, 100 nm staurosporine and EBSS. Percent cell death was assayed by propidium iodide uptake and quantitated using flow cytometry. Error bars represent mean ± S.E.
As mitochondrial dynamics is important in regulating cell death (42), DAP3 is reported to mediate apoptosis via an extrinsic pathway (28, 29), we examined whether DAP3 knockdown influenced cell death. As shown in Fig. 6F, the reduction of endogenous DAP3 yielded about 15% dead cells, significantly more than the 4% in control cells, suggesting that DAP3 depletion is able to induce cell death. Moreover, more cell death in the DAP3-depleted cells was markedly observed in response to intrinsic stress agents such as staurosporine (a pan-kinase inhibitor), mitochondrial uncoupler CCCP, and EBSS starvation. Specifically, DAP3-depleted cells exhibited cell death at about 77, 50, and 46%, in response to staurosporine, CCCP, or EBSS treatments, respectively, compared with ∼20% in control cells (Fig. 6F). In contrast, both DAP3-depleted and control cells displayed a similar rate of cell death to TNF-α treatment (Fig. 6F), indicating that DAP3 may not contribute to the extrinsic stimuli induced cell death. Taken together, these results demonstrate that the loss of DAP3 function sensitizes cells to the intrinsic mitochondrial mediated death pathway but not extrinsic death receptor-mediated cell death.
Discussion
Previous studies identified DAP3 as an apoptosis-promoting protein but raised a controversial issue on its localization of the cytosol or mitochondria (28, 29, 33, 37)? In this study, we used both imaging and biochemical fractionation approaches to show that DAP3 localizes not only to mitochondria, but also inside of mitochondria (Fig. 1), consistent with the report that DAP3 is a component of the mitochondrial ribosomal small subunit (31). In addition, we show that knockdown of DAP3 leads to mitochondrial protein synthesis defects and mitochondrial dissipation, which may alter the antagonistic activities of PKA and calcineurin on mitochondria (41, 43), resulting in the dephosphorylation of Drp1 at Ser-637 and mitochondrial fragmentation. It has been reported that ectopic overexpression of DAP3 caused mitochondrial fragmentation (33). Conversely, to our surprise, we observed that DAP3-GFP-expressing cells behaved with little or no difference from control cells on mitochondrial dynamics (Fig. 2, B and D). We could not explain the discrepancy between our findings from the previous report. The possible speculation is that it could be due to the different overexpression level of DAP3-GFP. As a physiological consequence of mitochondrial dysfunction, autophagy is inhibited, leading to sensitization to cell death induced by the intrinsic apoptotic pathway.
Mitochondrial ribosomes have been reported to associate with the inner mitochondrial membrane, linked by prohibitins (44). The depletion of prohibitins leads to mitochondrial fragmentation (45), indicating that an interaction between mitochondrial ribosomes and the inner membrane may play a role in regulating mitochondrial dynamics. As a component of mitochondrial ribosomes, DAP3 could interact with the inner membrane protein hNOA1, a binding partner for complex I of the respiratory chain (46), suggesting that DAP3 may exert a similar function as that of prohibitins to bridge mitochondrial ribosomes and inner membranes. Therefore, it is possible that DAP3 depletion-induced mitochondrial fission is due to disrupting the association between mitochondrial ribosomes and mitochondrial inner membranes. Our data also suggest that, in contrast to DAP3 knockdown, pharmacologically blocking the mitochondrial protein synthesis using chloramphenicol may not affect the mitochondrial dynamics, which is consistent with the result reported previously (38). On the other hand, disrupting the mitochondrial ribosome intact by actinonin can significantly induce mitochondrial fragmentation (38). This notion is in good agreement with our experimental results; because DAP3 itself is a mitochondrial ribosomal component; depletion of DAP3 might directly disrupt the mitochondrial ribosomal structure or function as the effect of actinonin, resulting in mitochondrial fragmentation.
As mitochondrial homeostasis is carefully balanced and integrated by the fusion and fission processes (2), it is reasonable for us to hypothesize that the loss of DAP3-induced mitochondrial fission could be due to enhanced fission, reduced fusion, or both. Multiple lines of evidence have indicated that during mitochondrial fission, Drp1 translocates from the cytosol to mitochondria, assembling to oligomers and mediating mitochondrial division (2, 10, 47). We show that under normal conditions, Drp1 puncta moves rapidly on mitochondria, mediating division at marked sites and disappearing (supplemental Movie S1). This notion provides a dynamic lifetime for Drp1 and supports the current model, in which only puncta forms of Drp1 exist on mitochondria (15, 47). DAP3 depletion largely disrupted the dynamic function cycle of Drp1 puncta, resulting in relatively stable Drp1 foci associated with fragmented mitochondria after division (supplemental Movie S2), suggesting that the stabilization of Drp1 foci was stuck on mitochondria block mitochondrial fusion.
Recent studies suggest that the phosphorylation of Drp1 at Ser-637 is critical in regulating mitochondrial dynamics (16, 41). It has also been speculated that phosphorylated Drp1 at Ser-637 cannot stay on mitochondria and consequently leads mitochondria toward elongation (19, 48). However, other groups have reported that there is an inactive form of Drp1 on mitochondria that, in contrast to mediating fission, induces mitochondrial hyperfusion (49–51). Accordingly, our data in this study suggest that the inactive form of Drp1 on mitochondria might be the phosphorylated Drp1 at Ser-637, as the loss of DAP3 dramatically decreases phosphorylated Drp1 at Ser-637 not only in the cytosol, but also on mitochondria, accompanied by mitochondrial fragmentation (Fig. 5). Because Drp1 phosphorylation at Ser-637 is reversibly regulated by the cyclic AMP-dependent protein kinase PKA and phosphatase calcineurin (16, 41), we examined the effects of specific pharmacologic agents (the PKA activator forskolin and calcineurin inhibitor FK506) on rescuing mitochondrial phenotypes induced by DAP3 depletion. Indeed, the fragmented mitochondria could be reversed to a tubular structure. Our findings provide, in part, the molecular mechanism for the effect of DAP3 on the regulation of mitochondrial dynamics.
Unbalanced mitochondrial fission and fusion confers cells to an enhanced or reduced sensitivity to cell death (11, 42). In this study, we demonstrated for the first time that the loss of the mitochondrial matrix protein DAP3 promotes mitochondrial fission and facilitates cell death; specifically, increases the sensitivity to the intrinsic death agents (Fig. 6F). Further studies are required to clarify the molecular mechanisms of effects of DAP3 in different cell death pathways. In addition to cell death, a growing number of reports have demonstrated that mitochondria are recognized as a core regulator of autophagy (23, 52–54). We found that DAP3, as a mitochondrial protein, is essential to autophagy induction. As a cell protective process, autophagy is important in maintaining cell function and viability under stress (55, 56), thus, reduced autophagy activity makes cells more susceptible to various types of stimuli. This scenario may contribute to the mechanisms of the enhanced cell death in DAP3 knockdown cells when treated with intrinsic agents. Accordingly, forced starvation led to more cell death in DAP3-depleted cells, further demonstrating the inhibitory effect on autophagy in DAP3-depleted cells.
Taken together, our results show that DAP3, as a mitochondrial ribosomal component, is critical for regulating mitochondrial dynamics and function by affecting the mitochondrial protein synthesis and phosphorylation status of Drp1. In addition, we demonstrate that loss of DAP3 function leads to the inhibition of starvation-induced autophagy and increases susceptibility to the intrinsic death agents. Our findings delineate broader questions about the mutual regulation among mitochondrial dynamics, autophagy, and cell death, which allow us to understand the role of DAP3 in broader aspects of pathologies, including neurodegenerative diseases, tumorigenesis, and diabetes.
Author Contributions
L. X. and Y.-C. L. conceived and designed the research. L. X., H. X., K. Y. L., and B. X. performed the experiments. L. X., H. W., F. Y., H.-M. S., and Y.-C. L. analyzed data and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank all members in the Y.-C. L laboratory for valuable discussions. We thank the Center of BioImaging Sciences (CBIS) at the National University of Singapore, especially to Yan Tong for technical support. We particularly thank Victor Chun-Kong Yu and Lih Wen Deng for kindly providing several plasmids and helpful discussions and suggestions. In addition, we thank Jigang Wang for help in the AHA-“click” assay.
This work was supported in part by Grants MOE-T2-1-153 and Tier 1 from the Ministry of Education, and NMRC-CBRG13nov097, Singapore (to Y.-C. L.) The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movies S1 and S2.
- Mfn1
- mitofusin 1
- Mfn2
- mitofusin 2
- Drp1
- dynamin-related protein 1
- OPA1
- optic atrophy 1
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- FRAP
- fluorescence recovery after photobleaching
- PMSF
- phenylmethylsulfonyl fluoride
- AHA
- l-azidohomoalanine
- Δψm
- mitochondrial membrane potential
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DMSO
- dimethyl sulfoxide
- EBSS
- Earle's balanced salt solution.
References
- 1. Westermann B. (2010) Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 [DOI] [PubMed] [Google Scholar]
- 2. Chan D. C. (2012) Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 [DOI] [PubMed] [Google Scholar]
- 3. Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mozdy A. D., McCaffery J. M., Shaw J. M. (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santel A., Fuller M. T. (2001) Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867–874 [DOI] [PubMed] [Google Scholar]
- 7. Yue W., Chen Z., Liu H., Yan C., Chen M., Feng D., Yan C., Wu H., Du L., Wang Y., Liu J., Huang X., Xia L., Liu L., Wang X., Jin H., Wang J., Song Z., Hao X., Chen Q. (2014) A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 24, 482–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexander C., Votruba M., Pesch U. E., Thiselton D. L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., Bhattacharya S. S., Wissinger B. (2000) OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215 [DOI] [PubMed] [Google Scholar]
- 9. Hoppins S., Lackner L., Nunnari J. (2007) The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 [DOI] [PubMed] [Google Scholar]
- 10. Smirnova E., Shurland D. L., Ryazantsev S. N., van der Bliek A. M. (1998) A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Youle R. J., van der Bliek A. M. (2012) Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. (2006) MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 7, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karbowski M., Neutzner A., Youle R. J. (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 178, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang H., Song P., Du L., Tian W., Yue W., Liu M., Li D., Wang B., Zhu Y., Cao C., Zhou J., Chen Q. (2011) Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J. Biol. Chem. 286, 11649–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wasiak S., Zunino R., McBride H. M. (2007) Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 177, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang C. R., Blackstone C. (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 282, 21583–21587 [DOI] [PubMed] [Google Scholar]
- 17. Chang C. R., Blackstone C. (2007) Drp1 phosphorylation and mitochondrial regulation. EMBO Rep. 8, 1088–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cereghetti G. M., Stangherlin A., Martins de Brito O., Chang C. R., Blackstone C., Bernardi P., Scorrano L. (2008) Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 105, 15803–15808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gomes L. C., Di Benedetto G., Scorrano L. (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell. Biol. 13, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z., Jiang H., Chen S., Du F., Wang X. (2012) The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 [DOI] [PubMed] [Google Scholar]
- 21. He C., Klionsky D. J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng Y., He D., Yao Z., Klionsky D. J. (2014) The machinery of macroautophagy. Cell Res. 24, 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hailey D. W., Rambold A. S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P. K., Lippincott-Schwartz J. (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tooze S. A., Yoshimori T. (2010) The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835 [DOI] [PubMed] [Google Scholar]
- 25. Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Brito O. M., Scorrano L. (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 27. Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 [DOI] [PubMed] [Google Scholar]
- 28. Kissil J. L., Deiss L. P., Bayewitch M., Raveh T., Khaspekov G., Kimchi A. (1995) Isolation of DAP3, a novel mediator of interferon-γ-induced cell death. J. Biol. Chem. 270, 27932–27936 [DOI] [PubMed] [Google Scholar]
- 29. Miyazaki T., Reed J. C. (2001) A GTP-binding adapter protein couples TRAIL receptors to apoptosis-inducing proteins. Nat. Immunol. 2, 493–500 [DOI] [PubMed] [Google Scholar]
- 30. Berger T., Kretzler M. (2002) Interaction of DAP3 and FADD only after cellular disruption. Nat. Immunol. 3, 3–5 [DOI] [PubMed] [Google Scholar]
- 31. Cavdar Koc E., Ranasinghe A., Burkhart W., Blackburn K., Koc H., Moseley A., Spremulli L. L. (2001) A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 492, 166–170 [DOI] [PubMed] [Google Scholar]
- 32. Kim H. R., Chae H. J., Thomas M., Miyazaki T., Monosov A., Monosov E., Krajewska M., Krajewski S., Reed J. C. (2007) Mammalian dap3 is an essential gene required for mitochondrial homeostasis in vivo and contributing to the extrinsic pathway for apoptosis. FASEB J. 21, 188–196 [DOI] [PubMed] [Google Scholar]
- 33. Mukamel Z., Kimchi A. (2004) Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. J. Biol. Chem. 279, 36732–36738 [DOI] [PubMed] [Google Scholar]
- 34. Ye F., Tan L., Yang Q., Xia Y., Deng L. W., Murata-Hori M., Liou Y. C. (2011) HURP regulates chromosome congression by modulating kinesin Kif18A function. Curr. Biol. 21, 1584–1591 [DOI] [PubMed] [Google Scholar]
- 35. Dieterich D. C., Lee J. J., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. (2007) Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protocols 2, 532–540 [DOI] [PubMed] [Google Scholar]
- 36. Xia Y., Ongusaha P., Lee S. W., Liou Y. C. (2009) Loss of Wip1 sensitizes cells to stress- and DNA damage-induced apoptosis. J. Biol. Chem. 284, 17428–17437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kissil J. L., Cohen O., Raveh T., Kimchi A. (1999) Structure-function analysis of an evolutionary conserved protein, DAP3, which mediates TNF-α- and Fas-induced cell death. EMBO J. 18, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richter U., Lahtinen T., Marttinen P., Myöhänen M., Greco D., Cannino G., Jacobs H. T., Lietzén N., Nyman T. A., Battersby B. J. (2013) A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr. Biol. 23, 535–541 [DOI] [PubMed] [Google Scholar]
- 39. Otera H., Wang C., Cleland M. M., Setoguchi K., Yokota S., Youle R. J., Mihara K. (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kashatus D. F., Lim K. H., Brady D. C., Pershing N. L., Cox A. D., Counter C. M. (2011) RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cribbs J. T., Strack S. (2007) Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suen D. F., Norris K. L., Youle R. J. (2008) Mitochondrial dynamics and apoptosis. Genes Dev. 22, 1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dickey A. S., Strack S. (2011) PKA/AKAP1 and PP2A/Bβ2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J. Neurosci. 31, 15716–15726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He J., Cooper H. M., Reyes A., Di Re M., Sembongi H., Litwin T. R., Gao J., Neuman K. C., Fearnley I. M., Spinazzola A., Walker J. E., Holt I. J. (2012) Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 40, 6109–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., Langer T. (2008) Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang T., Zheng B., Chen S. H., Murphy A. N., Kudlicka K., Zhou H., Farquhar M. G. (2009) hNOA1 interacts with complex I and DAP3 and regulates mitochondrial respiration and apoptosis. J. Biol. Chem. 284, 5414–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cecconi F., Levine B. (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rambold A. S., Kostelecky B., Elia N., Lippincott-Schwartz J. (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 108, 10190–10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., Nistér M. (2011) Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30, 2762–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palmer C. S., Osellame L. D., Laine D., Koutsopoulos O. S., Frazier A. E., Ryan M. T. (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Losón O. C., Song Z., Chen H., Chan D. C. (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singh S. B., Ornatowski W., Vergne I., Naylor J., Delgado M., Roberts E., Ponpuak M., Master S., Pilli M., White E., Komatsu M., Deretic V. (2010) Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat. Cell Biol. 12, 1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Graef M., Nunnari J. (2011) Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 30, 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Codogno P., Meijer A. J. (2005) Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12, 1509–1518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.