Background: The substrates of Polo-like kinase in Trypanosoma brucei are mostly not identified.
Results: A basal body protein was identified as a substrate of Polo-like kinase and is required for basal body segregation and flagellum adhesion.
Conclusion: Polo-like kinase regulates a basal body protein.
Significance: Dissecting the Polo-like kinase pathway is crucial for understanding its cellular functions.
Keywords: gene silencing, molecular cell biology, parasite, parasitology, Trypanosoma brucei, basal body, Polo-like kinase, flagellum, flagellum attachment zone
Abstract
The Polo-like kinase (PLK) in Trypanosoma brucei plays multiple roles in basal body segregation, flagellum attachment, and cytokinesis. However, the mechanistic role of TbPLK remains elusive, mainly because most of its substrates are not known. Here, we report a new substrate of TbPLK, SPBB1, and its essential roles in T. brucei. SPBB1 was identified through yeast two-hybrid screening with the kinase-dead TbPLK as the bait. It interacts with TbPLK in vitro and in vivo, and is phosphorylated by TbPLK in vitro. SPBB1 localizes to both the mature basal body and the probasal body throughout the cell cycle, and co-localizes with TbPLK at the basal body during early cell cycle stages. RNAi against SPBB1 in procyclic trypanosomes inhibited basal body segregation, disrupted the new flagellum attachment zone filament, detached the new flagellum, and caused defective cytokinesis. Moreover, RNAi of SPBB1 confined TbPLK at the basal body and the bilobe structure, resulting in constitutive phosphorylation of TbCentrin2 at the bilobe. Altogether, these results identified a basal body protein as a TbPLK substrate and its essential role in promoting basal body segregation and flagellum attachment zone filament assembly for flagellum adhesion and cytokinesis initiation.
Introduction
Polo-like kinases (PLKs)2 are evolutionarily conserved serine/threonine protein kinases, which are characterized by the C-terminal Polo box domain (PBD) and play essential roles in mitosis and cytokinesis in eukaryotes (1, 2). The localization of PLK to various subcellular structures is known to be mediated by the PBD via association with different substrate proteins located at specific subcellular structures (3–6).
Trypanosoma brucei, an early branching unicellular eukaryote and the causative agent of human sleeping sickness, expresses a single PLK homolog, TbPLK, which displays a subcellular localization pattern different from its yeast and animal homologs (7–9). Early in the cell cycle, TbPLK is localized to the flagellar basal body and the bilobe structure adjacent to the proximal region of the flagellum attachment zone (FAZ) filament. During G2 and mitotic phases, TbPLK is enriched at the anterior tip of the new FAZ filament. The anterior tip of the new FAZ filament is believed to constitute the cytokinesis initiation site in trypanosomes (10–13). Throughout the cell cycle, TbPLK is excluded from the nucleus, despite the presence of a nuclear localization signal sequence between the kinase domain and the first PBD (14). Consistent with its localization at the basal body, bilobe, and the new FAZ tip, TbPLK is required for basal body segregation, bilobe duplication, FAZ filament assembly, flagellum adhesion, and cytokinesis, but plays no essential role in mitosis (7, 8, 15).
Despite the essential function of TbPLK in trypanosomes, the mechanistic roles of TbPLK remain poorly understood, mainly because most of its substrates have not been identified. TbCentrin2 is the first identified substrate of TbPLK, which localizes to the basal body, the bilobe, and the flagellum (8, 16). Phosphorylation of TbCentrin2 by TbPLK occurs only on the bilobe structure and appears to be required to maintain flagellum attachment (16). However, RNAi of TbCentrin2 does not cause flagellum detachment (17). To better understand the function of TbPLK in regulating basal body segregation, flagellum attachment, and cytokinesis, we sought to identify additional substrates of TbPLK and characterize their function. Through yeast two-hybrid screening using TbPLK-K70R, a kinase-dead mutant of TbPLK (14), and the PBD as baits, we identified 12 proteins as potential TbPLK-associated proteins and substrates. In this study, we report the functional characterization of one of these proteins, SPBB1, and its interplay with TbPLK. Through biochemical approaches, we demonstrate the in vitro and in vivo interactions between SPBB1 and TbPLK and verify SPBB1 as an in vitro TbPLK substrate. We also demonstrate that SPBB1 is a novel basal body protein and co-localizes with TbPLK during early cell cycle stages. Importantly, RNAi against SPBB1 recapitulates the defects caused by TbPLK depletion, i.e. inhibition of basal body segregation, detachment of the flagellum, and defective cytokinesis. Finally, we show that RNAi of SPBB1 restricts TbPLK at the basal body and the bilobe structure, where the latter constitutively phosphorylates TbCentrin2 at the bilobe.
Experimental Procedures
Yeast Two-hybrid Screening and Directional Yeast Two-hybrid Assay
To construct the Gal4 activation domain (AD) fusion library for two-hybrid screening, trypanosome total RNA was purified and used to generate a cDNA library cloned in the pGADT7 vector using the MatchmakerTM library construction and screening kit (Clontech). The full-length coding sequence of the kinase-dead mutant TbPLK-K70R and the sequence encoding the PBD of TbPLK (PBDTbPLK) were each cloned into pGBKT7 vector for expression of Gal4 binding domain fusion proteins (bait). The Gal4 AD fusion library was transformed into strain AH109 (mating type a), whereas the bait plasmids (pGBK-TbPLK-K70R and pGBK-PBDTbPLK) were transformed into strain Y187 (mating type α). After mating the haploids, the diploids were plated on SD-Leu-Trp-His plates to screen for positive clones.
For directional yeast two-hybrid assay, the full-length coding sequence of SPBB1 was cloned into the pGADT7 vector for expression of Gal4 AD-fused SPBB1 (prey). Full-length TbPLK, TbPLK-K70R, and the PBD alone were each cloned into the pGBKT7 vector to express Gal4 binding domain-fused proteins (bait). The prey plasmid was transformed into stain AH109, and the bait plasmids were transformed into strain Y187. The yeast strains carrying both the bait and the prey plasmids were obtained by mating the two haploids at 30 °C overnight, plating the diploid on SD-Leu-Trp plates, and incubating them at 30 °C for 2–3 days. Each combination strain was spotted in three 10-fold serial dilutions onto SD-Leu-Trp and SD-Leu-Trp-His plates, and the growth of yeast on SD-Leu-Trp-His plate indicates the interaction between the bait and the prey proteins.
Purification of GST Fusion Proteins, GST Pulldown, and in Vitro Kinase Assay
The full-length coding sequence of SPBB1 was cloned into the pGEX-4T-3 vector for expression of recombinant GST-SPBB1 in bacteria. However, the recombinant protein was insoluble. We therefore expressed GST-fused SPBB1 truncations, SPBB1-N (amino acids 1–500) and SPBB1-C (amino acids 501–980), in bacteria. Recombinant GST-SPBB1-N and GST-SPBB1-C were expressed in Escherichia coli BL21 cells and purified through a column of glutathione-Sepharose 4B beads (GE Healthcare). For GST pulldown, trypanosome cells overexpressing TbPLK-3HA or TbPLK-K70R-3HA were lysed in trypanosome lysis buffer (25 mm Tris-HCl, pH 7.6, 500 mm NaCl, 1 mm DTT, 1% Nonidet P-40, and protease inhibitor cocktail) on ice for 30 min and cleared by centrifugation at the highest speed in a microcentrifuge. The cleared lysate (500 μl) was then incubated with GST-fused SPBB1-N or SPBB1-C or GST bound to glutathione-Sepharose 4B beads at room temperature for 1 h. The beads were then washed six times with the lysis buffer, and bound proteins were eluted by boiling the beads in SDS-PAGE sampling buffer for 5 min and separated on SDS-PAGE. Western blotting was then carried out with anti-HA antibody to detect TbPLK-3HA and TbPLK-K70R-3HA.
The full-length coding sequence of TbPLK was cloned into pET41 (18), and recombinant GST-TbPLK was purified from the soluble fraction. Purified recombinant proteins (GST-TbPLK and GST-SPBB1-C) were dialyzed against 50 mm Tris-Cl, pH 7.6, and 50 mm NaCl. Purified GST fusion proteins were incubated in kinase buffer (10 mm HEPES, pH 7.5, 50 mm NaCl, 10 mm MgCl2, and 1 mm DTT) containing 1 μCi of [γ-32P]ATP at room temperature for 60 min. Reactions were stopped by adding 1× SDS-PAGE sampling buffer and boiling for 5 min. Proteins were separated on SDS-PAGE, and the gel was exposed to x-ray film. GST-SPBB1-C was detected by Coomassie Blue staining of the SDS-PAGE gel after exposure. GST-TbPLK was detected by Western blotting with anti-GST antibody due to its low abundance.
Trypanosome Cell Culture and RNAi
The procyclic trypanosome strain 29-13 (19) was cultured at 27 °C in SDM-79 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Inc.), 15 μg/ml G418, and 50 μg/ml hygromycin B. The procyclic trypanosome strain 427 was maintained in SDM-79 medium containing 10% fetal bovine serum. Cells were routinely diluted when the density reached 5 × 106/ml.
To silence SPBB1 by RNAi, a 530-bp DNA fragment corresponding to the N-terminal coding region of SPBB1 was PCR-amplified and cloned into the pZJM vector (20). The resulting plasmid was linearized with NotI digestion and transfected into the 29-13 cell line. Transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution on a 96-well plate. To induce RNAi, the transfectants were incubated with 1.0 μg/ml tetracycline, and cell growth was monitored daily by counting the cell number with a hemocytometer.
Epitope Tagging of Endogenous Proteins
A 500-bp DNA fragment corresponding to the N-terminal coding region of SPBB1 was cloned into pN-3HA-PAC and pN-PTP-PAC for N-terminal tagging of SPBB1 at the endogenous locus. The resulting constructs were linearized with appropriate restriction enzymes and transfected into the 427 cell line. Transfectants were selected under 1.0 μg/ml puromycin and cloned by limiting dilution in a 96-well plate. Subsequently, the pC-TbPLK-3HA-Neo vector (18) was linearized and transfected into the cells harboring the pN-PTP-SPBB1-PAC construct, and the transfectants were selected under 40 μg/ml G418 in addition to 1 μg/ml puromycin and cloned by limiting dilution.
Generation of Anti-TbPLK Antibody
To generate anti-TbPLK polyclonal antibody, DNA sequence encoding the Polo box domain of TbPLK was cloned into pET26 for expression of recombinant His-tagged PBDTbPLK in bacteria. The recombinant protein was purified under denaturing conditions and used to immunize rabbits to generate polyclonal antibody at Cocalico Biologicals, Inc. (Reamstown, PA). The anti-TbPLK serum was used directly for Western blotting and immunofluorescence.
Co-Immunoprecipitation and Western Blotting
Cells (107) expressing N-terminal PTP-tagged SPBB1 were harvested by centrifugation, lysed in 1 ml of lysis buffer (25 mm Tris-HCl, pH 7.6, 500 mm NaCl, 1 mm DTT, 1% Nonidet P-40, and protease inhibitor cocktail), and incubated with 10 μl pf settled IgG-Sepharose 6 Fast Flow beads (GE HealthCare) at 4 °C for 1 h. The beads were then washed six times with the immunoprecipitation buffer, and bound proteins were eluted with 10% SDS, separated on SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with anti-TbPLK pAb and anti-protein A to detect TbPLK and PTP-SPBB1, respectively. Wild-type 427 cells were included as the negative control.
Inhibition of TbPLK Activity by a Small Molecule Inhibitor
To inhibit TbPLK activity in trypanosomes, 5 μm GW843286X (Tocris Bioscience), an inhibitor of human Plk1 (21) and trypanosome TbPLK (18), was added to trypanosome cell culture and incubated for 24 h. The phenotype caused by GW843286X treatment is similar to that caused by TbPLK RNAi (18).
Immunofluorescence Microscopy
Cells were washed with PBS, adhered onto coverslips, and fixed in cold (−20 °C) methanol for 30 min. Fixed cells were then rehydrated with PBS and incubated in blocking buffer (1% BSA and 0.1% Triton X-100 in PBS) for 1 h at room temperature. Cells were then incubated with the primary antibody diluted in PBS containing 1% BSA at room temperature for 1 h. The following primary antibodies were used: FITC-conjugated anti-HA mAb (1:400 dilution, Sigma-Aldrich), anti-protein A pAb (1: 400 dilution, Sigma-Aldrich), anti-TbPLK pAb (1:1,000 dilution), L3B2 (1:20 dilution) (22), YL 1/2 mAb (1:1,000 dilution) (23), 20H5 (anti-Centrin mAb, 1:400 dilution) (24), PS54 (anti-phospho-Ser-54 of TbCentrin2, 1: 30,000 dilution) (16), and anti-TbSAS-6 pAb (1:1,000 dilution) (25). After three washes with wash buffer (0.1% Triton X-100 in PBS), cells were incubated with FITC-conjugated or Cy3-conjugated secondary antibody at room temperature for another hour. The following secondary antibodies were used: FITC-conjugated anti-mouse IgG (Sigma-Aldrich), FITC-conjugated anti-rat IgG (Sigma-Aldrich), and Cy3-conjugated anti-rabbit IgG (Sigma-Aldrich). After three more washes with the wash buffer, the slides were mounted in VECTASHIELD mounting medium (Vector Laboratories) containing DAPI and examined with an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a Plan Apo N 60 × 1.42-NA differential interference contrast objective. Images were acquired using the Slidebook5 software (Intelligent Imaging Innovations).
Statistical Analysis
Statistical analysis was performed using the t test provided in the Microsoft Excel software. For immunofluorescence microscopy experiments, slides were blinded to the investigator who took images under the fluorescent microscope. Images were randomly taken. Counting of cells with different fluorescence patterns was then carried out by another investigator, and all of the cells in each image were counted.
Results
Identification of TbPLK-associated Proteins and Substrates by Yeast Two-hybrid Screening
To identify potential substrates of TbPLK, we employed yeast two-hybrid screening using a kinase-dead mutant TbPLK (TbPLK-K70R) and the PBD as baits. There are advantages of using kinase-dead mutant and the PBD as baits in yeast two-hybrid screening for identification of TbPLK substrates. First, because kinase-substrate interaction is transient and the substrates often dissociate from the kinase immediately after being phosphorylated, using kinase-dead mutant kinase as the bait in yeast two-hybrid screening may trap the substrate on the kinase, thus allowing detection of kinase-substrate interaction. Second, the PBD is known to mediate interaction between PLK and its substrates (3–6). Therefore, using PBD as the bait may identify potential TbPLK substrates. However, it should be noted that some proteins identified in this screen might be TbPLK-interacting proteins instead of substrates.
A total of 43 positive clones were identified, of which 35 clones were identified by interaction with the PBD and 8 clones were identified by interaction with TbPLK-K70R. PCR amplification of the DNA insert in these clones identified 12 distinct genes, eight of which encode proteins of unknown function (Table 1). Because TbPLK plays unusual roles in trypanosomes, we focused on the eight novel proteins. Our preliminary RNAi experiments showed that Tb927.6.900 is essential in procyclic trypanosomes and, therefore, it was characterized in detail. Tb927.6.900 encodes an ∼112-kDa protein of 980 residues, and was named SPBB1 for substrate of Polo-like kinase in the basal body 1. It contains mostly the coiled-coil motifs (from amino acids 140 to 740), and is only conserved in the kinetoplastida parasites (T. brucei, Trypanosoma cruzi, and Leishmania).
TABLE 1.
Putative TbPLK-associated proteins and substrates identified by yeast two-hybrid screening
MM, molecular mass; FKBP, FK506-binding protein.
| Accession No. | MM | Protein description | Positive clones |
||
|---|---|---|---|---|---|
| K70R | PBD | Total | |||
| kDa | |||||
| Tb927.10.4620 | 11.6 | FKBP-type peptidyl-prolyl cis-trans isomerase | 2 | 12 | 14 |
| Tb927.8.6510 | 22.9 | Ubiquitin-conjugating enzyme | 2 | 2 | |
| Tb927.6.2790 | 36 | l-Threonine 3-dehydrogenase | 1 | 1 | |
| Tb927.8.7730 | 45.1 | Hypothetical protein | 2 | 2 | |
| Tb927.10.14030 | 50.4 | Hypothetical protein | 8 | 8 | |
| Tb927.11.6590 | 55.3 | Aminopeptidase, putative | 1 | 1 | |
| Tb927.5.2150 | 60 | Hypothetical protein | 1 | 1 | |
| Tb927.7.3100 | 69.7 | Hypothetical protein | 4 | 4 | |
| Tb927.11.15800 | 89.6 | Hypothetical protein | 1 | 1 | |
| Tb927.6.900 | 112 | SPBB1 | 1 | 1 | |
| Tb927.10.15080 | 180.2 | Hypothetical protein | 1 | 1 | |
| Tb927.6.620 | 301.8 | Hypothetical protein | 4 | 3 | 7 |
SPBB1 Is an in Vitro Substrate of TbPLK
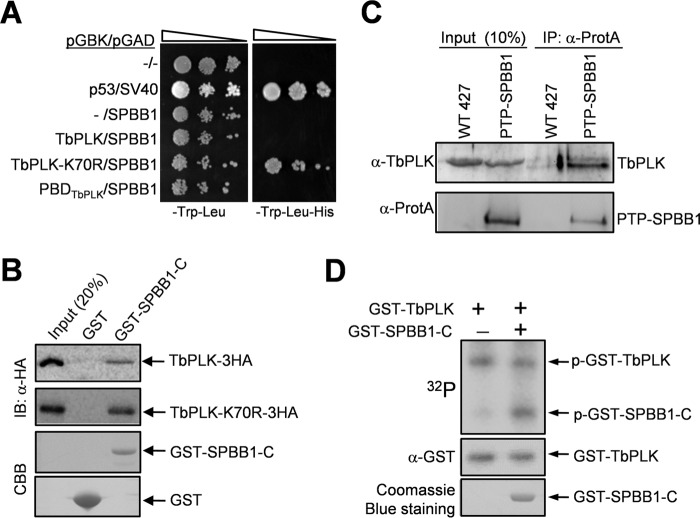
In the yeast two-hybrid screen, SPBB1 was identified through interaction with TbPLK-K70R, but not the PBD (Table 1). To confirm the interaction between SPBB1 and TbPLK-K70R and to test whether SPBB1 interacts with wild-type TbPLK, we carried out directional yeast two-hybrid experiments. The results showed that SPBB1 only interacts with TbPLK-K70R, but not wild-type TbPLK and the PBD of TbPLK in yeast (Fig. 1A), which further validated our yeast two-hybrid screen result. We next carried out GST pulldown experiments to examine the in vitro interaction between SPBB1 and both TbPLK and TbPLK-K70R. Because the recombinant full-length SPBB1 was insoluble in E. coli, we purified truncations of SPBB1 and found that the C-terminal fragment (amino acids 501–980) of SPBB1 (designated SPBB1-C) was able to pull down both TbPLK and TbPLK-K70R, with more TbPLK-K70R precipitated by GST-fused SPBB1-C (Fig. 1B). This result suggests that the C-terminal fragment of SPBB1 mediates the interaction with TbPLK and confirms that SPBB1 interacts with wild-type TbPLK in vitro. To investigate whether SPBB1 and TbPLK interact in vivo in trypanosomes, we carried out co-immunoprecipitation. To this end, SPBB1 was endogenously tagged with a PTP (protein A-TEV-protein C) epitope (26) at its N terminus. Immunoprecipitation of PTP-SPBB1 was capable of pulling down TbPLK from the trypanosome cell lysate as detected by anti-TbPLK antibody (Fig. 1C), indicating that SPBB1 interacts with TbPLK in vivo. We then investigated whether SPBB1 is phosphorylated by TbPLK in vitro by kinase assay with GST fusion proteins purified from bacteria. The results showed that TbPLK was capable of phosphorylating SPBB1-C (Fig. 1D), suggesting that SPBB1 is an in vitro substrate of TbPLK.
FIGURE 1.
SPBB1 interacts with TbPLK in vivo and is phosphorylated by TbPLK in vitro. A, yeast two-hybrid assays to detect the interaction between SPBB1 and TbPLK. p53/SV40 served as the positive control. −, empty vector. B, GST pulldown to detect the in vitro interaction between the C-terminal fragment of SPBB1 (SPBB1-C) and TbPLK. GST was included as the negative control. IB, immunoblot; CBB, Coomassie Brilliant Blue. C, co-immunoprecipitation (IP) of SPBB1 and TbPLK from trypanosome cell lysate. D, in vitro kinase assay to investigate the phosphorylation of SPBB1 by TbPLK. A C-terminal truncation of SPBB1 was purified as GST fusion protein for kinase assay with purified GST-fused TbPLK. GST-TbPLK was detected by Western blotting with anti-GST antibody, whereas GST-SPBB1-C was detected by Coomassie Blue staining. p-GST-TbPLK, phospho-GST-TbPLK; p-GST-SPBB1-C, phospho-GST-SPBB1-C.
SPBB1 Co-localizes with TbPLK in the Basal Body during Early Cell Cycle Stages
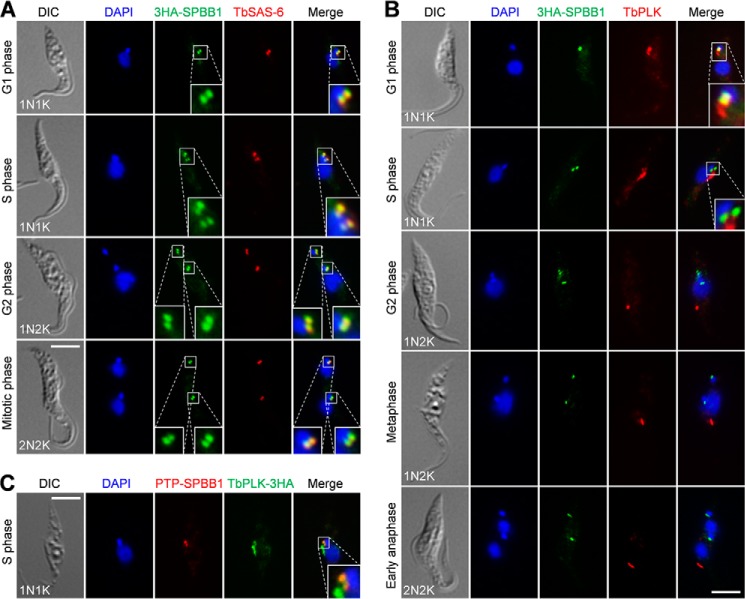
To determine the subcellular localization of SPBB1, we tagged the endogenous SPBB1 with a triple HA epitope at the N terminus for immunofluorescence microscopy. The results showed that 3HA-SPBB1 was localized to the basal body as verified by co-localization with TbSAS-6, the basal body cartwheel protein in both the mature basal body and the probasal body (25) (Fig. 2A). In G1 cells, SPBB1 was detected as two closely associated bright fluorescence spots, which represent a mature basal body and its associated probasal body as marked by TbSAS-6 (Fig. 2A). Starting from S-phase to late mitotic phases, SPBB1 remained at the mature basal body and the probasal body (Fig. 2A). These results suggest that SPBB1 is a novel basal body protein and is localized to the basal body throughout the cell cycle.
FIGURE 2.
SPBB1 co-localizes with TbPLK in the basal body during early cell cycle stages. A, subcellular localization of 3HA-SPBB1 during the cell cycle. SPBB1 was endogenously tagged with an N-terminal triple HA epitope. Cells were co-immunostained with the FITC-conjugated anti-HA mAb and anti-TbSAS-6 pAb, and then counterstained with DAPI for DNA. DIC, differential interference contrast. Scale bar: 5 μm. B, localization of 3HA-SPBB1 and TbPLK during the cell cycle. Cells were co-immunostained with the FITC-conjugated anti-HA mAb and anti-TbPLK pAb, and then counterstained with DAPI for DNA. Scale bar: 5 μm. C, co-localization of PTP-SPBB1 and TbPLK-3HA in the basal body. SPBB1 was endogenously tagged with the PTP epitope at the N terminus in the same cell line expressing endogenously 3HA-tagged TbPLK. Cells were co-immunostained with the FITC-conjugated anti-HA mAb and anti-protein A pAb to detect TbPLK-3HA and PTP-SPBB1, respectively. Scale bar: 5 μm.
Because SPBB1 interacts with TbPLK in vivo, we asked when the two proteins co-localize during the cell cycle. Co-immunostaining with anti-HA and anti-TbPLK antibodies showed that in G1 cells, SPBB1 and TbPLK co-localized in the basal body, although TbPLK was additionally localized to the bilobe structure (Fig. 2B). During S-phase, SPBB1 still co-localized with TbPLK in the basal body, although TbPLK level in the basal body appeared to be reduced and was highly enriched in the bilobe structure (Fig. 2B). From G2 phase and thereafter, TbPLK was only detected at the anterior tip of the new FAZ filament and did not co-localize with SPBB1, which remained in the basal body until the end of mitosis (Fig. 2B). The localization of SPBB1 to the basal body and the co-localization between SPBB1 and TbPLK in the basal body were further confirmed with PTP-tagged SPBB1 (Fig. 2C).
RNAi of SPBB1 Causes Defective Cytokinesis and Flagellum Detachment
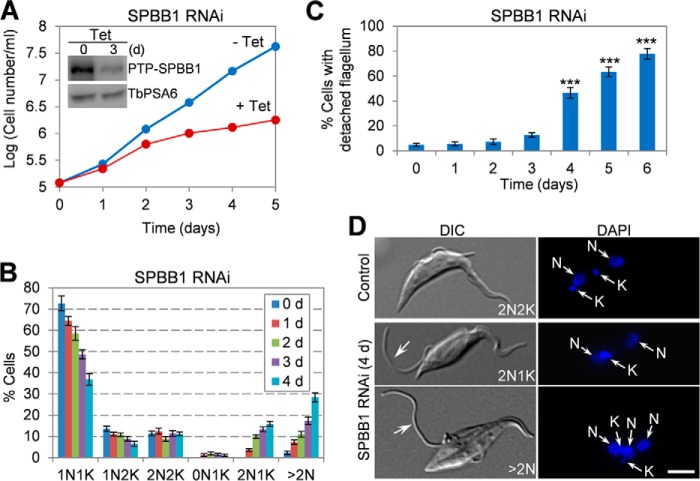
To investigate the function of SPBB1, RNAi was carried out in the procyclic form of T. brucei. To monitor the efficiency of RNAi, SPBB1 was endogenously tagged with an N-terminal PTP epitope in SPBB1 RNAi cells, and Western blotting showed that after tetracycline induction for 3 days, the level of PTP-SPBB1 was decreased to about 30% of that in the control cells (Fig. 3A, inset). This depletion of SPBB1 caused severe growth defect (Fig. 3A), suggesting that SPBB1 is essential for cell proliferation in the procyclic form. Non-induced control and RNAi-induced cells were tabulated for the numbers of cells with different numbers of nucleus (N) and kinetoplast (K), and the results showed that upon SPBB1 RNAi induction for up to 4 days, cells with two nuclei and one kinetoplast (2N1K) and cells with multiple nuclei (>2N) gradually emerged to ∼14 and ∼25%, respectively (Fig. 3B), suggesting an inhibition of kinetoplast duplication or segregation and defective cytokinesis. Additionally, flagellum detachment was clearly detectable in SPBB1 RNAi cells, and at day 6 of RNAi, ∼78% of the cells possessed at least one detached flagellum (Fig. 3C). It appeared that the new flagellum was often detached in 2N1K and multi-nucleated (>2N) cells (Fig. 3D).
FIGURE 3.
SPBB1 RNAi causes cytokinesis defects and flagellum detachment. A, RNAi of SPBB1 caused growth defects. Inset shows the Western blot to monitor the level of PTP-SPBB1, which was endogenously tagged in the SPBB1 RNAi cell line, before and after tetracycline (Tet) induction of RNAi. The level of the α-6 subunit of the 26S proteasome (TbPSA6) was included as the loading control. B, tabulation of cells with different numbers of kinetoplast (K) and nucleus (N) before and after SPBB1 RNAi for up to 4 days. About 200 cells were counted from each time point. Error bars represent S.D. calculated from three independent experiments. C, the percentage of cells with detached flagellum upon SPBB1 RNAi. About 200 cells were counted from each time point, and error bars represent S.D. calculated from three independent experiments. ***, p < 0.001. D, morphology of SPBB1 RNAi cells, showing the 2N1K and XN1K cells with a detached new flagellum (arrows). DIC, differential interference contrast. Scale bar: 5 μm.
SPBB1 Is Required for New FAZ Filament Assembly
The flagellum in a trypanosome cell is attached to the cell body via the FAZ filament (27), and defects in the FAZ filament cause flagellum detachment (28–31). To examine whether flagellum detachment in SPBB1 RNAi cells was caused by defective assembly of the FAZ filament, cells were immunostained with the L3B2 antibody, which labels the FAZ1 protein in the FAZ filament (22, 29), to monitor the integrity of the FAZ filament. In the non-induced control cells, all of the 2N2K cells contained two full-length FAZ filaments and two attached flagella. However, upon SPBB1 RNAi for 4 days, ∼42% of the 2N2K cells possessed a detached new flagellum and an associated short, new FAZ filament (Fig. 4, A and B). For those SPBB1-deficient 2N1K cells with a detached new flagellum, ∼25% of them contained only a full-length old FAZ filament, and the rest of the 2N1K cells contained a full-length old FAZ filament and a short, new FAZ (Fig. 4, A and B). For SPBB1-deficient multi-nucleated (>2N) cells, ∼12% of them contained only a full-length old FAZ filament, whereas ∼88% of them contained a full-length old FAZ and one or multiple (≥2) short, new FAZ (Fig. 4, A and B). Together, these results suggest that SPBB1 is required for assembly of the new FAZ filament to maintain flagellum adhesion.
FIGURE 4.
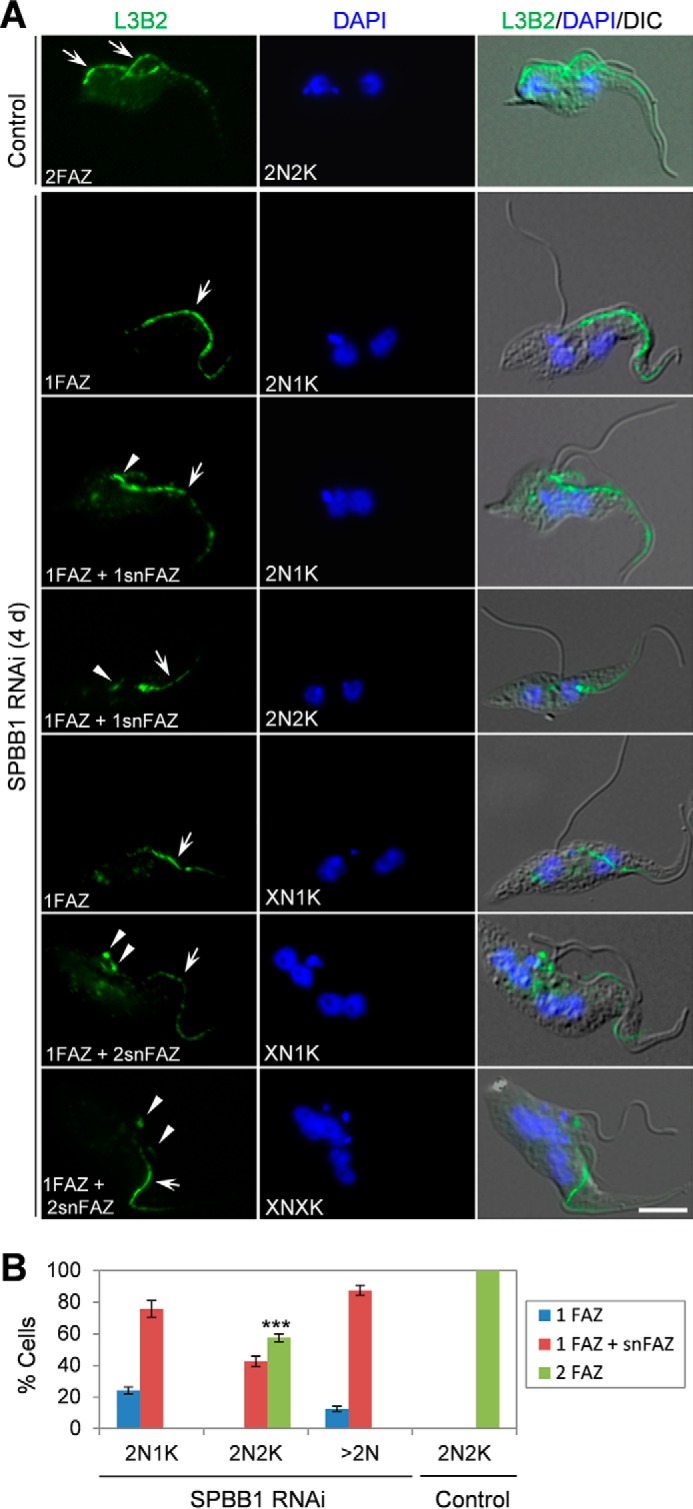
RNAi of SPBB1 disrupts the new FAZ filament. A, non-induced control and SPBB1 RNAi-induced cells (4 days) were immunostained with L3B2, which labels the FAZ1 protein in the FAZ filament. Arrows in the control 2N2K cells show the full-length new and old FAZ filaments. Arrows in SPBB1 RNAi cells indicate the full-length, old FAZ filament, whereas arrowheads show the short, new FAZ filament. Scale bar: 5 μm. B, quantification of cells with different numbers of full-length FAZ filament and short, new FAZ filament. About 300 cells were counted for each cell type (2N1K, 2N2K, >2N), and error bars represent S.D. calculated from three independent experiments. snFAZ: short, new FAZ. ***, p < 0.001.
RNAi of SPBB1 Inhibits Basal Body Separation
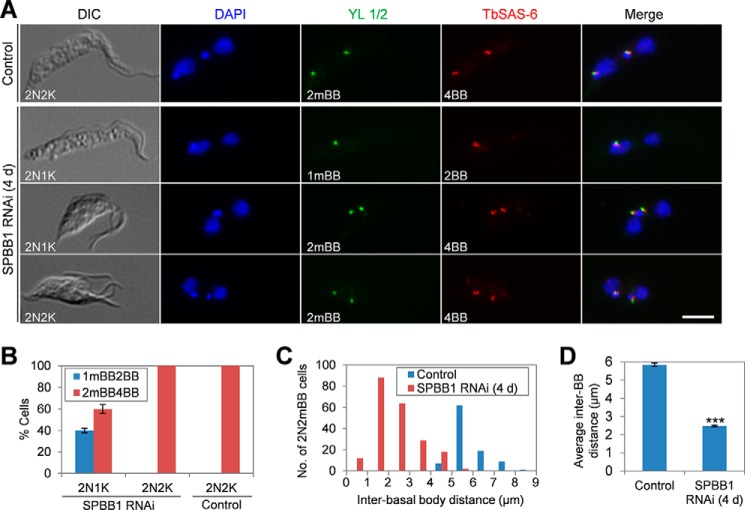
Because the kinetoplast is physically attached to the basal body (32) and segregation of the duplicated kinetoplasts is mediated by basal body separation (33), we reasoned that the emergence of 2N1K cells upon SPBB1 RNAi was due to defective duplication or segregation of basal bodies. To test this hypothesis, we co-immunostained the cells with the YL 1/2 antibody, which labels the transition fibers near the mature basal body and serves as a marker for the mature basal body (34, 35), and anti-TbSAS-6, which detects the basal body cartwheel protein in both the mature basal body and the probasal body (25). We then quantified the numbers of mature basal bodies (mBB) and total basal bodies (BB) in 2N1K, 2N2K, and multi-nucleated (>2N) cells from SPBB1 RNAi and in 2N2K cells from the non-induced control. As expected, the 2N2K cells from the non-induced control all contained two mature basal bodies labeled by YL 1/2 and four basal bodies (both mature and probasal bodies) labeled by TbSAS-6 (Fig. 5, A and B). The two pairs of basal bodies in control 2N2K cells were normally separated, with an average distance of ∼5.8 μm (Fig. 5, A, C, and D). In SPBB1-deficient 2N1K cells, ∼40% of them contained one mature basal body and an associated probasal body, whereas the rest of them contained two mature basal bodies and two associated probasal bodies, but the two pairs of basal bodies were not well separated in these cells (Fig. 5, A and B). In SPBB1-deficient 2N2K cells, all of them contained two mature basal bodies and two associated probasal bodies, which were also not well separated (Fig. 5, A and B). For the 2N1K and 2N2K cells with two mature basal bodies (2mBB), the average distance between the mature basal bodies was calculated to be ∼2.4 μm, which is less than half of the inter-basal body distance in the control 2N2K cells (Fig. 5, C and D). These results suggest that SPBB1 RNAi inhibited the separation of the duplicated basal bodies, thereby inhibiting kinetoplast segregation.
FIGURE 5.
SPBB1 is required for basal body segregation. A, non-induced control and SPBB1 RNAi-induced cells were immunostained with YL 1/2 for mature basal body and anti-TbSAS-6 antibodies for both the mature basal body and the pro-basal body. Cells were counterstained with DAPI for DNA. DIC, differential interference contrast; 2mBB, two mature basal bodies; 1mBB, one mature basal body; 4BB, four basal bodies; 2BB, two basal bodies. Scale bar: 5 μm. B, quantification of cells with different numbers of the mature basal body (mBB) and basal body (both the mature basal body and the probasal body, BB). About 200 cells were counted for each cell type (2N1K and 2N2K), and error bars represent S.D. calculated from three independent experiments. C, inter-basal body distances of 2N2mBB cells from the non-induced control cell line and the SPBB1 RNAi-induced (4-day) cell line. About 200 2N1K and 2N2K cells from the SPBB1 RNAi cell line and 100 2N2K cells from the non-induced control cell line were randomly selected to measure the distance between the two mature basal bodies. D, average inter-basal body distances of the control 2N2K cells and the 2N2K and 2N1K cells from SPBB1 RNAi. Error bars represent S.D. calculated from three independent experiments. ***, p < 0.001.
TbPLK Is Not Required for SPBB1 Localization to the Basal Body
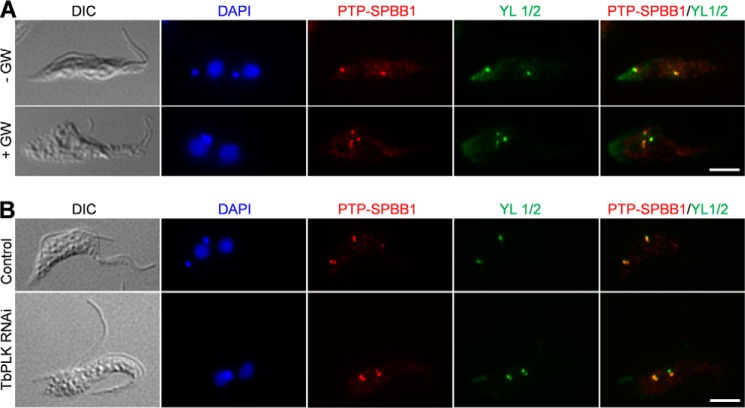
Given that SPBB1 is phosphorylated by TbPLK, we asked whether phosphorylation of SPBB1 by TbPLK is required for SPBB1 localization. To test this possibility, we treated the cells stably expressing PTP-tagged SPBB1 with GW843286X, a small molecule inhibitor developed against human Polo-like kinase 1 (21). This molecule also showed potent inhibition of trypanosome TbPLK, and treatment of T. brucei cells by GW843286X recapitulated the defects caused by TbPLK RNAi (18). We confirmed that GW843286X treatment caused the inhibition of basal body segregation, defective assembly of the new FAZ filament, and cytokinesis arrest, reminiscent of TbPLK RNAi (Fig. 6A). However, localization of SPBB1 to the basal body appeared to be unaffected by GW843286X treatment (Fig. 6A), suggesting that TbPLK activity is not required for targeting SPBB1 to the basal body. Furthermore, to test whether the interaction with TbPLK recruits SPBB1 to the basal body, PTP-SPBB1 was expressed endogenously in TbPLK RNAi cells, and immunostaining showed that PTP-SPBB1 was still localized to the basal body (Fig. 6B), suggesting that depletion of TbPLK did not abolish SPBB1 localization. Together, these results suggest that localization of SPBB1 to the basal body is independent of TbPLK activity and interaction with TbPLK.
FIGURE 6.
TbPLK deficiency does not abolish SPBB1 localization to the basal body. A, inhibition of TbPLK activity by a small molecule inhibitor of TbPLK, GW843682X (abbreviated as GW), and its effect on SPBB1 localization. Cells expressing endogenously PTP-tagged SPBB1 were treated with 5 μm GW843682X for 24 h and then co-immunostained with anti-protein A pAb and YL 1/2 mAb. Scale bar: 5 μm. B, effect of TbPLK depletion on SPBB1 localization. SPBB1 was endogenously tagged with the PTP epitope in cells harboring the TbPLK RNAi construct. TbPLK RNAi was induced for 24 h, and cells were co-immunostained with anti-protein A pAb and YL 1/2 mAb. Scale bar: 5 μm.
Depletion of SPBB1 Restricts TbPLK in the Basal Body and the Bilobe Structure
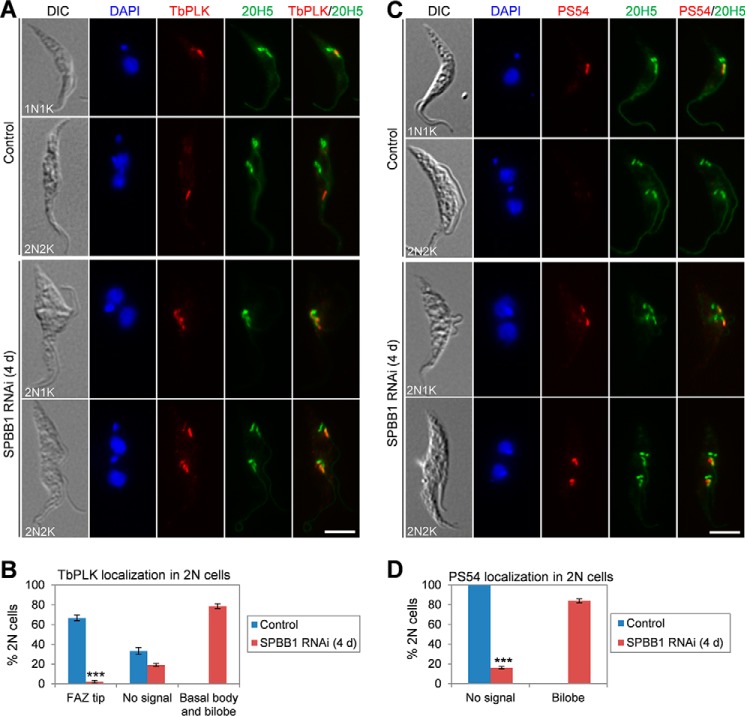
To test whether SPBB1 RNAi might affect the localization of TbPLK, non-induced control and SPBB1 RNAi cells induced for 4 days were co-immunostained with anti-TbPLK antibody and the 20H5 antibody, which stains the basal body and the bilobe structure in T. brucei (17). In non-induced control cells, TbPLK was localized to the basal body and the bilobe structure in G1 cells, and was enriched at the anterior tip of the new FAZ filament during G2 to early mitotic phases (metaphase and early anaphase) (Fig. 7A), similar to a previous study (36). In the control 2N2K cells, TbPLK was detected at the new FAZ tip in ∼65% of the 2N2K cells, which were at the early stages of mitosis (prophase to early anaphase), but was not detectable in the rest of the 2N2K cells, which were at the late stages of mitosis (late anaphase and telophase) (Fig. 7B). In the 2N cells (2N1K and 2N2K) from SPBB1 RNAi, however, TbPLK was detected in the basal body and the bilobe structure in the majority (∼79%) of these 2N cells, with only a very small number (<2%) of the 2N2K cells containing new FAZ tip-localizing TbPLK (Fig. 7, A and B). These results suggest that TbPLK was restricted at the basal body and the bilobe structure when SPBB1 was depleted.
FIGURE 7.
SPBB1 RNAi restricts TbPLK in the basal body and the bilobe, leading to constitutive TbCentrin2 phosphorylation by TbPLK. A, effect of SPBB1 RNAi on TbPLK localization. Non-induced control and SPBB1 RNAi-induced (4-day) cells were immunostained with anti-TbPLK pAb and 20H5, which labels the basal body, the bilobe structure, and the flagellum in trypanosomes. Scale bar: 5 μm. B, quantification of 2N cells (2N2K cells from the control and 2N1K and 2N2K cells from SPBB1 RNAi) with different TbPLK localization patterns. About 200 cells were counted, and error bars represent S.D. calculated from three independent experiments. ***, p < 0.001. C, effect of SPBB1 RNAi on TbPLK-mediated TbCentrin2 phosphorylation. Non-induced control and SPBB1 RNAi-induced (4-day) cells were immunostained with PS54 for Ser-54-phosphorylated TbCentrin2 and with 20H5 for the bilobe structure. Scale bar: 5 μm. D, quantification of 2N cells (2N2K cells from the control and 2N1K and 2N2K cells from SPBB1 RNAi) with different PS54 (Ser-54-phosphorylated TbCentrin2) localization patterns. About 200 cells were counted, and error bars represent S.D. calculated from three independent experiments. ***, p < 0.001.
In the bilobe, TbPLK is known to phosphorylate the bilobe-resident TbCentrin2 at serine 54 during late G1 and early S-phase of the cell cycle, but not at late S-phase and beyond (16). Importantly, expression of phospho-deficient and phospho-mimic mutants of TbCentrin2, TbCentrin2-S54A and TbCentrin2-S54E, caused defective FAZ filament assembly and flagellum detachment (16). The enrichment of TbPLK in the bilobe in 2N1K and 2N2K cells upon SPBB1 RNAi (Fig. 7A) prompted us to investigate whether TbCentrin2 was still phosphorylated in these cells. To this end, non-induced control and SPBB1 RNAi cells were immunostained with the PS54 antibody, which detects the TbPLK-phosphorylated Ser-54 residue in TbCentrin2 (16). We confirmed that in the non-induced control, TbCentrin2 was phosphorylated in the bilobe in G1/S (1N1K) cells but not in mitotic (2N2K) cells (Fig. 7, C and D). However, in the majority (∼81%) of the SPBB1-deficient 2N1K and 2N2K cells, TbCentrin2 was phosphorylated at Ser-54 in the two segregated bilobe structures (Fig. 7, C and D), presumably because TbPLK remained at the bilobe structures (Fig. 7A) and phosphorylated TbCentrin2 at Ser-54 in these cells. Together, these results suggest that RNAi of SPBB1 confined TbPLK to the basal body and the bilobe structure, where TbPLK constitutively phosphorylated TbCentrin2 in mitotic and post-mitotic cells.
Discussion
In this study, we reported the identification and functional characterization of a new TbPLK substrate, SPBB1, in T. brucei. SPBB1 is the first basal body protein identified as a TbPLK substrate. TbCentrin2, a bilobe protein involved in bilobe biogenesis, Golgi duplication, and flagellum adhesion, is also a TbPLK substrate (8, 16, 17). TbPLK is known to differ significantly from its yeast and animal homologs in terms of subcellular localization and cellular functions (7–9, 15), and previous studies have demonstrated that TbPLK is required for basal body segregation, bilobe duplication, flagellum attachment, and cytokinesis initiation (7, 8, 14, 15, 36). Given its localization to multiple subcellular structures and its multiple roles in organelle biogenesis, TbPLK may phosphorylate proteins located in the basal body, the bilobe structure, and the anterior tip of the new FAZ filament. Although our yeast two-hybrid screening with TbPLK-K70R and PBD as baits has identified several TbPLK-interacting partners (Table 1), their candidacy as TbPLK substrates remains to be experimentally verified. Using proximity-dependent biotin identification (BioID) and stable isotope labeling in the cell culture (SILAC) coupled with quantitative mass spectrometry, a recent study identified a few basal body proteins as TbPLK close neighbors and identified several other proteins as TbPLK substrates (37). This study confirms that TbPLK regulates many downstream factors in the basal body, the bilobe, and the FAZ filament.
RNAi against SPBB1 recapitulated the defects caused by TbPLK RNAi in procyclic trypanosomes, although the defects appeared to be less severe than that caused by TbPLK RNAi, presumably because TbPLK has additional downstream targets, including TbCentrin2, which may play redundant roles in executing TbPLK functions. As a support of this assumption, mutation of the TbPLK-phosphorylated Ser-54 in TbCentrin2 disrupted the assembly of the new FAZ filament, leading to detachment of the new flagellum, and caused defects in bilobe duplication and cytokinesis in T. brucei (16). It is notable that expression of TbCentrin2-S54A or TbCentrin2-S54E also caused less severe defects than TbPLK RNAi (16). It is also interesting to note that despite the distinct subcellular localizations of SPBB1 and TbCentrin2, deficiency in SPBB1 and deficiency in TbPLK-mediated TbCentrin2 phosphorylation caused similar phenotypes, i.e. malformation of the new FAZ filament, flagellum detachment, inhibition of basal body segregation, and defective cytokinesis (Figs. 3–5). However, SPBB1 RNAi appeared to exert little effect on bilobe duplication, although bilobe segregation appeared to be inhibited in the 2N1K cells (Fig. 7, A and C). In contrast, expression of TbCentrin2-S54A and TbCentrin2-S54E inhibited bilobe duplication (16). This difference in bilobe duplication/segregation defects between SPBB1 RNAi and TbCentrin2-S54A/S54E mutants is likely to be attributed to the fact that TbCentrin2 is localized in the bilobe, in addition to the basal body and the flagellum (16, 36), whereas SPBB1 is localized only in the basal body (Fig. 2). Therefore, defective segregation of the duplicated bilobe structures in SPBB1 RNAi cells is likely an indirect effect, presumably due to the inhibited segregation of basal bodies.
Duplication of the flagellar basal body represent one of the earliest cytoskeletal events in the trypanosome cell cycle. A number of proteins have been localized to the basal body in T. brucei, but only a few of them are localized to both the probasal body and the mature basal body, which includes TbCentrin2 (17, 36), TbCentrin4 (38), TbSAS-6 (25), and an unknown protein that is recognized by a monoclonal antibody BBA4 (39). Centrins and SAS-6 are evolutionarily conserved components of the centriole/basal body, and are present in all of the eukaryotic organisms investigated so far (40, 41). However, SPBB1 appears to be unique to the kinetoplastida parasites; therefore, its function might also be specific to these early diverged organisms. Unlike TbCentrin2, which is required for basal body biogenesis (17), SPBB1 is required for segregation of the duplicated basal bodies (Fig. 5), but its precise role in basal body segregation remains unclear. In T. brucei, segregation of basal bodies appears to be coupled with the elongation of the new FAZ filament during the cell cycle (42). SPBB1 RNAi also caused a defective assembly/elongation of the new FAZ filament (Fig. 4) in addition to the inhibition of basal body segregation (Fig. 5). These results further confirmed that basal body segregation and new FAZ elongation are well coordinated in T. brucei.
An unexpected but intriguing observation made in this study is the restriction of TbPLK in the basal body and the bilobe in SPBB1 RNAi cells, and as a consequence, constitutive phosphorylation of the bilobe-resident TbCentrin2 after the S-phase of the cell cycle (Fig. 7). Constitutive phosphorylation of TbCentrin2 by TbPLK also appears to cause defective FAZ filament assembly, leading to flagellum detachment (16). Therefore, constitutive phosphorylation of TbCentrin2 in SPBB1 RNAi cells may exert an additive effect on FAZ filament assembly, leading to greater FAZ filament assembly defects than SPBB1 RNAi alone. However, given that SPBB1 is not localized to the anterior tip of the new FAZ filament to meet with TbPLK after S-phase, it is unclear how depletion of SPBB1 prevented TbPLK from being targeted from the basal body and the bilobe to the anterior tip of the new FAZ filament. It is likely that the failure for TbPLK to localize to the anterior tip of the new FAZ was due to the absence of new FAZ filament assembly, thus restricting TbPLK at the basal body and the bilobe structure. In this regard, this defect in TbPLK localization could be an indirect effect.
Although we have proven the candidacy of SPBB1 as an in vitro TbPLK substrate and have demonstrated the essential role of SPBB1 in basal body segregation and FAZ filament assembly, the mechanistic role(s) of SPBB1 in regulating these cellular processes and how TbPLK regulates SPBB1 to execute its function remain unclear. Through immunoprecipitation and mass spectrometry, we have identified nine in vivo phosphorylation sites in SPBB1 (Ser-265, Ser-276, Thr-647, Thr-659, Ser-742, Ser-818, Ser-892, Ser-900, and Ser-901). Notably, seven out of the nine phosphosites are located in the C terminus of SPBB1, which was used for the in vitro kinase assay (Fig. 1D). Future work will be directed to investigate whether phosphorylation of SPBB1 by TbPLK is required for SPBB1 function. Given that SPBB1 only contains the coiled-coil motifs, which are known to be involved in protein-protein interactions (43), SPBB1 is unlikely to possess any catalytic activity. It could serve as a scaffold in a multi-protein complex, although this still needs further investigation. Moreover, because inhibition of TbPLK activity by a small molecule inhibitor or depletion of TbPLK by RNAi did not abolish SPBB1 targeting to the basal body (Fig. 6), this suggests that the physical interaction with TbPLK and the activity of TbPLK are not required for SPBB1 localization. SPBB1 might just serve as one of the downstream effectors of TbPLK to execute the function of TbPLK in controlling basal body segregation and assembling the new FAZ filament to maintain flagellum adhesion.
Author Contributions
Z. L. conceived and designed the experiments; H. H. and Q. Z. performed the experiments; H. H., Q. Z., and Z. L. analyzed the data; and Z. L. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to Dr. George Cross of Rockefeller University for providing the 29-13 cell line, Dr. Paul Englund of Johns Hopkins Medical School for providing the pZJM vector, Dr. Arthur Günzl of University of Connecticut Health Center for providing the pN-PTP-PAC vector, Dr. Keith Gull of the University of Oxford for the L3B2 antibody, and Dr. Christopher de Graffenried of Brown University for the PS54 antibody.
This work was supported by the National Institutes of Health Grants AI101437 and AI108657 (to Z. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- PLK
- Polo-like kinase
- TbPLK
- Trypanosoma brucei PLK homolog
- PBD
- Polo box domain
- FAZ
- flagellum attachment zone
- AD
- activation domain
- SD
- synthetic defined
- TEV
- tobacco etch virus
- N
- nucleus
- K
- kinetoplast
- mBB
- mature basal bodies.
References
- 1. Barr F. A., Silljé H. H., Nigg E. A. (2004) Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 2. Archambault V., Glover D. M. (2009) Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10, 265–275 [DOI] [PubMed] [Google Scholar]
- 3. Jang Y. J., Lin C. Y., Ma S., Erikson R. L. (2002) Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc. Natl. Acad. Sci. U.S.A. 99, 1984–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee K. S., Grenfell T. Z., Yarm F. R., Erikson R. L. (1998) Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc. Natl. Acad. Sci. U.S.A. 95, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song S., Grenfell T. Z., Garfield S., Erikson R. L., Lee K. S. (2000) Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell. Biol. 20, 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reynolds N., Ohkura H. (2003) Polo boxes form a single functional domain that mediates interactions with multiple proteins in fission yeast polo kinase. J. Cell Sci. 116, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 7. Kumar P., Wang C. C. (2006) Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell 5, 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Graffenried C. L., Ho H. H., Warren G. (2008) Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J. Cell Biol. 181, 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Umeyama T., Wang C. C. (2008) Polo-like kinase is expressed in S/G2/M phase and associated with the flagellum attachment zone in both procyclic and bloodstream forms of Trypanosoma brucei. Eukaryot. Cell 7, 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vaughan S., Gull K. (2003) The trypanosome flagellum. J. Cell Sci. 116, 757–759 [DOI] [PubMed] [Google Scholar]
- 11. Briggs L. J., McKean P. G., Baines A., Moreira-Leite F., Davidge J., Vaughan S., Gull K. (2004) The flagella connector of Trypanosoma brucei: an unusual mobile transmembrane junction. J. Cell Sci. 117, 1641–1651 [DOI] [PubMed] [Google Scholar]
- 12. Li Z., Lee J. H., Chu F., Burlingame A. L., Günzl A., Wang C. C. (2008) Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One 3, e2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z., Umeyama T., Wang C. C. (2009) The Aurora kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu Z., Liu Y., Li Z. (2012) Structure-function relationship of the Polo-like kinase in Trypanosoma brucei. J. Cell Sci. 125, 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammarton T. C., Kramer S., Tetley L., Boshart M., Mottram J. C. (2007) Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol. Microbiol. 65, 1229–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Graffenried C. L., Anrather D., Von Raußendorf F., Warren G. (2013) Polo-like kinase phosphorylation of bilobe-resident TbCentrin2 facilitates flagellar inheritance in Trypanosoma brucei. Mol. Biol. Cell 24, 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He C. Y., Pypaert M., Warren G. (2005) Golgi duplication in Trypanosoma brucei requires Centrin2. Science 310, 1196–1198 [DOI] [PubMed] [Google Scholar]
- 18. Li Z., Umeyama T., Li Z., Wang C. C. (2010) Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryot. Cell 9, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 20. Wang Z., Morris J. C., Drew M. E., Englund P. T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174–40179 [DOI] [PubMed] [Google Scholar]
- 21. Lansing T. J., McConnell R. T., Duckett D. R., Spehar G. M., Knick V. B., Hassler D. F., Noro N., Furuta M., Emmitte K. A., Gilmer T. M., Mook R. A., Jr., Cheung M. (2007) In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol. Cancer Ther. 6, 450–459 [DOI] [PubMed] [Google Scholar]
- 22. Kohl L., Sherwin T., Gull K. (1999) Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46, 105–109 [DOI] [PubMed] [Google Scholar]
- 23. Kilmartin J. V., Wright B., Milstein C. (1982) Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93, 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paoletti A., Moudjou M., Paintrand M., Salisbury J. L., Bornens M. (1996) Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109, 3089–3102 [DOI] [PubMed] [Google Scholar]
- 25. Hu H., Liu Y., Zhou Q., Siegel S., Li Z. (2015) The centriole cartwheel protein SAS-6 in Trypanosoma brucei is required for probasal body biogenesis and flagellum assembly. Eukaryot. Cell 14, 898–907, 10.1128/EC.00083-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schimanski B., Nguyen T. N., Günzl A. (2005) Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 4, 1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gull K. (1999) The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 53, 629–655 [DOI] [PubMed] [Google Scholar]
- 28. Zhou Q., Liu B., Sun Y., He C. Y. (2011) A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J. Cell Sci. 124, 3848–3858 [DOI] [PubMed] [Google Scholar]
- 29. Vaughan S., Kohl L., Ngai I., Wheeler R. J., Gull K. (2008) A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159, 127–136 [DOI] [PubMed] [Google Scholar]
- 30. LaCount D. J., Barrett B., Donelson J. E. (2002) Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277, 17580–17588 [DOI] [PubMed] [Google Scholar]
- 31. Zhou Q., Hu H., He C. Y., Li Z. (2015) Assembly and maintenance of the flagellum attachment zone filament in Trypanosoma brucei. J. Cell Sci. 128, 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogbadoyi E. O., Robinson D. R., Gull K. (2003) A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 14, 1769–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson D. R., Gull K. (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352, 731–733 [DOI] [PubMed] [Google Scholar]
- 34. Sherwin T., Schneider A., Sasse R., Seebeck T., Gull K. (1987) Distinct localization and cell cycle dependence of COOH terminally tyrosinolated α-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 104, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stephan A., Vaughan S., Shaw M. K., Gull K., McKean P. G. (2007) An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic 8, 1323–1330 [DOI] [PubMed] [Google Scholar]
- 36. Ikeda K. N., de Graffenried C. L. (2012) Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J. Cell Sci. 125, 3173–3184 [DOI] [PubMed] [Google Scholar]
- 37. McAllaster M. R., Ikeda K. N., Lozano-Núñez A., Anrather D., Unterwurzacher V., Gossenreiter T., Perry J. A., Crickley R., Mercadante C. J., Vaughan S., de Graffenried C. L. (2015) Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in T. brucei. Mol. Biol. Cell 26, 3013–3029, 10.1091/mbc.E15-04-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi J., Franklin J. B., Yelinek J. T., Ebersberger I., Warren G., He C. Y. (2008) Centrin4 coordinates cell and nuclear division in T. brucei. J. Cell Sci. 121, 3062–3070 [DOI] [PubMed] [Google Scholar]
- 39. Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., Gull K. (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491–500 [DOI] [PubMed] [Google Scholar]
- 40. Carvalho-Santos Z., Machado P., Branco P., Tavares-Cadete F., Rodrigues-Martins A., Pereira-Leal J. B., Bettencourt-Dias M. (2010) Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123, 1414–1426 [DOI] [PubMed] [Google Scholar]
- 41. Hodges M. E., Scheumann N., Wickstead B., Langdale J. A., Gull K. (2010) Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Absalon S., Kohl L., Branche C., Blisnick T., Toutirais G., Rusconi F., Cosson J., Bonhivers M., Robinson D., Bastin P. (2007) Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS One 2, e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burkhard P., Stetefeld J., Strelkov S. V. (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 [DOI] [PubMed] [Google Scholar]