Abstract
Hantaan virus A9 strain (HTNV A9) is an etiologic agent of hemorrhagic fever with renal syndrome in China. The virulence of the pathogenic hantaviruses is determined by their ability to alter key signaling pathways of early interferon (IFN) induction within cells. The potential role of HTNV A9 structural proteins, such as nucleocapsid (N) and envelope glycoproteins (Gn and Gc), in regulating human's innate antiviral immune response has not yet been clarified. In this study, we investigated the effect of HTNV A9 N protein on the regulation of the IFN pathway. We found that A9 N protein can influence the host innate immune response by regulating the activation of IFNβ. The A9 N protein stimulates IFN response in low doses, whereas significantly inhibits IFNβ production at high doses. Furthermore, A9 N protein constitutively inhibits nuclear factor kappa B activation. A high dose of A9 N protein could inhibit either Poly IC-induced IFNβ or vesicular stomatitis virus-induced IFNβ and interferon-stimulated gene production. Our results indicate that HTNV A9 N protein helps virus establish successful infection by downregulating the IFN response and shed new light to the understanding of the interaction between the host innate immunity and virus during Hantaan virus infection.
Introduction
Hantaviruses [belonging to the Bunyaviridae family (24)] can cause serious diseases such as hemorrhagic fever with renal syndrome (HFRS) and the hantavirus pulmonary syndrome (HPS) among humans (7,25,30,31). HFRS is primarily caused by several hantaviruses in Eurasia, such as the Hantaan virus (HTNV), Seoul virus (SEOV), Puumala virus (PUUV), and Dobrava virus (DOBV). Meanwhile, Sin Nombre virus (SNV), Andes virus (ANDV), and New York virus (NY-V) cause HPS throughout the Americas. The severe forms of HFRS cause 5–12% case fatality rate, whereas HPS can cause 50% case fatality rate (7,30,31).
Like other viruses in the family, hantaviruses are enveloped viruses that contain a trisegmented, single-stranded, and negative sense RNA genome (7). The small segment (S) encodes the nucleocapsid (N); the medium segment (M) encodes the viral glycoprotein precursor (Gn-Gc) that is cleaved into two mature glycoproteins Gn and Gc; and the large segment (L) encodes RNA-dependent RNA polymerase (RdRp or L protein) (5). N protein encapsidates viral genomic RNAs to form ribonucleoprotein and is the highly expressed in the cytoplasm of the infected cells. Gn and Gc are type I integral membrane proteins and form viral spikes on the virion surface, which are required for virus entry into cells and virus assembly in the Golgi (5). Although rodents are the major reservoir of hantaviruses, antibodies against hantaviruses are also present in domestic and wild animals like cats, dogs, pigs, cattle, and deer (31). Much effort has been exerted to develop safe and effective vaccines against hantaviruses, such as the attenuated virus, virus-like particles (16), viral proteins (9), and DNA vaccines (6,17).
Mammals execute an immediate innate immune response specialized to rapid virus detection (1,10,29). Viral dsRNA or RNA elements are both recognized through either toll-like receptors or intracellular RNA helicases. The retinoic acid-inducible gene I (RIG-I)-mediated type I interferon (IFN) pathways are triggered upon the infection of hantaviruses (12,14). The activation of RIG-I results in the binding of mitochondrially located adaptor protein mitochondrial antiviral signaling. After recruiting further cofactors, the complex activates the transcription factors IRF3/7 and nuclear factor kappa B (NFκB), which initiate the transcriptional activation of type I IFNs (alpha/beta) and other proinflammatory cytokines (8,29). IFNs can activate numerous interferon-stimulated genes (ISGs); many of them encode antiviral restriction factors that inhibit different procedures of the life cycle of the virus, such as entry, transcription, replication, translation, assembly, and release from infected cells (11,23,29). Thus, the spreading of virus is either slowed down or entirely blocked. Pathogenic viruses would then develop strategies to escape from the innate immune response of the body (18).
Hantaviruses primarily infect human endothelial cells (ECs) and, therefore, cause hemorrhagic diseases. The induction of type I IFN among ECs typically restricts viral replication (18). Previous reports suggest that the virulence of hantaviruses is determined by their ability to alter key signaling pathways of early IFN induction within cells (18–20,26,30). For example, pathogenic hantaviruses, such as ANDV, HTNV, and NY-1V, inhibit early ISG induction and successfully replicate among ECs, whereas a nonpathogenic virus, like Prospect Hill virus (PHV), cannot replicate among ECs (3). Gn and N proteins from various pathogenic viruses inhibit IFN production through different mechanisms. For example, the Gn protein from NY-1V disrupts the binding of TBK1 with TRAF3 and, thereby, inhibits the production of IFNβ (2). The N protein of HTNV binds with importin α and inhibits tumor necrosis factor-alpha (TNFα)-induced NFκB activation (28). Thus, hantaviruses must interfere with the production of IFN to replicate among ECs. Distinct hantaviruses, however, have different strategies to modulate their immune signaling pathways (18).
The HTNV strain A9 was first isolated in China from Apodemus agrarius in 1982. It is an etiologic agent of HFRS that causes severe and even fatal HFRS (21,27). The potential role of structure proteins of Hantaan virus A9 strain (HTNV A9) in regulating an innate antiviral immune response has not been clarified. In this study, we investigated the effect of the expression of HTNV A9 N protein on regulating type I IFN signaling in human embryonic kidney 293T cells.
Materials and Methods
Cells and virus
The human embryonic kidney cell line 293T and monkey kidney epithelial cells Vero E6 were maintained in Dulbecco's Minimum Essential Medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in humidified air containing 5% CO2 at 37°C according to ATCC's guidelines.
A recombinant vesicular stomatitis virus expressing green fluorescent protein (VSV-GFP) was kindly provided by Dr. Dong Chunsheng, Soochow University. VSV-GFP virus was propagated and titrated in Vero E6 cells and used to infect 293T cells at a multiple of infection (MOI) of 2.0.
Plasmid construction and transfection
A construct expressing HTNV A9 N protein (pCMV-A9S) was generated in accordance with the standard protocol. Briefly, the coding region of A9 virus S segment was polymerase chain reaction amplified with gene-specific primers that contained the BglII and SalI restriction sites (shown in Table 1) and cloned into the pCMV expression vector. Transfections were performed on subconfluent monolayers of 293T cells plated onto a 96-well plate, using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Table 1.
Sequences of Oligonucleotides for the Polymerase Chain Reaction Used in the Present Study
| No. | Sequence (5′-3′) | Note |
|---|---|---|
| 1 | GACAGATCTACGATGGCAACTATGGAGGAATTAC | Forward primer for A9 S coding region |
| 2 | TGCGTCGACTTATAGTTTTAAAGGCTCTTGGTTG | Reverse primer for A9 S coding region |
| 3 | GGGCATGGAGTCCTGTGGCA | Forward primer for qPCR of human β-actin gene |
| 4 | GGGTGCCAGGGCAGTGATCTC | Reverse primer for qPCR of human β-actin gene |
| 5 | CATTACCTGAAGGCCAAGGA | Forward primer for qPCR of human IFNβ gene |
| 6 | CAGCATCTGCTGGTTGAAGA | Reverse primer for qPCR of human IFNβ gene |
| 7 | ACGCCTTCCAGCAGCGTCTG | Forward primer for qPCR of human ISG15 gene |
| 8 | CGCATTTGTCCACCACCAGCA | Reverse primer for qPCR of human ISG15 gene |
| 9 | AAGAGCCGGCTGTGGATATG | Forward primer for qPCR of human MxA gene |
| 10 | TTTGGACTTGGCGGTTCTGT | Reverse primer for qPCR of human MxA gene |
| 11 | GATCTCAGTGCAGAGGCTCG | Forward primer for qPCR of human MCP-1 gene |
| 12 | TGCTTGTCCAGGTGGTCCAT | Reverse primer for qPCR of human MCP-1 gene |
| 13 | GGTTTCTGCAGCGCTTCTGT | Forward primer for qPCR of human MCP-2 gene |
| 14 | CTTCATGGAATCCCTGACCC | Reverse primer for qPCR of human MCP-2 gene |
| 15 | ACCACACCCTGCTGCTTTGCC | Forward primer for qPCR of human RANTES gene |
| 16 | CTCCCGAACCCATTTCTTCTC | Reverse primer for qPCR of human RANTES gene |
| 17 | CCACGTGTTGAGATCATTGC | Forward primer for qPCR of human CXCL10 gene |
| 18 | CCTCTGTGTGGTCCATCCTT | Reverse primer for qPCR of human CXCL10 gene |
qPCR, quantitative polymerase chain reaction.
For a dose-dependent expression of HTNV A9 N proteins, the concentrations of expression plasmids pCMV-A9S were set to 0, 2, 10, 50 ng per 96-well plate well, and the total amount of DNA was kept constant by supplementing with empty pCMV vector. The dose-dependent expression of A9 N proteins was confirmed by Western blotting using the anti-HTNV N protein antibody (Cat. 10R-H104a; Fitzgerald Industries International).
Reporter assays
Luciferase reporter plasmids pIFNβ-Luc (p125-Luc) and pIRF3-Luc [pPRD(III–I)-Luc, based on an IRF3-specific reporter (4)] were kindly provided by Dr. Rongtuan Lin, McGill University, Canada (32). pNFκB-Luc was purchased from Clontech.
The transcriptional activation of IFNβ gene was measured using IFNβ promoter-driven luciferase reporter assays. Briefly, subconfluent 293T cells (in 96-well plates) were transfected with 10 ng of pRL-TK reporter (herpes simplex virus thymidine kinase promoter-driven Renilla luciferase; internal control), 100 ng of IFNβ luciferase reporter (firefly luciferase; experimental reporter) plasmid, various doses (0, 2, 10, 50, and 100 ng) of recombinant expressing plasmids (pCMV-A9S or pCMV control), along with 50 ng of expression plasmids of RIG-I, MDA5, TBK, or IRF3. For Poly IC stimulation, cells were not cotransfected with plasmids expressing RIG-I-related proteins, instead, treated with Poly IC at 1 μg/mL (Poly ICs were transfected into cells with Lipofectamine 2000).
For measuring the activation of transcription factor NFκB and IRF3, NFκB and IRF3 responsive element-specific reporter plasmids were used in the luciferase reporter assays. Subconfluent 293T cells were transfected with 10 ng of pRL-TK reporter, 100 ng of NFκB (pNFκB-Luc) or IRF3 [pPRD(III–I)-Luc] luciferase reporter plasmid, various doses of recombinant expression plasmids (pCMV-A9S or pCMV control), along with 50 ng of expression plasmids of RIG-I. At 24 h post-transfection, the cells were lysed and the luciferase activity was measured using a Dual Glow kit according to the manufacturer's instructions (Promega).
mRNA expression of IFNβ and ISG
The effects of viral proteins on the mRNA expressions of both IFNβ and ISGs were measured through quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The subconfluent 293T cells on the 48-well plates were transfected with a high dose of pCMV-A9S or control plasmids (1 μg/well). At 24 h post-transfection, the cells were either stimulated for 16 h with 1 μg/mL poly I:C (by transfection) or infected for 8 h with VSV-GFP (13) virus at MOI=2. The cells were lysed and their RNAs were extracted, in accordance to the manufacturer's instructions (OMEGA). Reverse transcription was performed through the PrimeScript RT reagent Kit (TAKARA) using random hexamers and 0.5 μg of total RNA in a 10 μL reaction. The expression levels of mRNAs of human IFNβ, ISG15, MxA, and chemokine genes (MCP-1, MCP-2, RATENS, and CXCL10) were measured through SYBRGreen-based qPCR (Roche) using gene-specific primers (Table 1) and were normalized using the human β-actin gene.
VSV infection and microscopy analysis
The monolayers of 293T cells were prepared and transfected with a high dose of plasmid of pCMV-A9S or vector control. At 24 h post-transfection, the cells were infected with VSV-GFP virus at MOI=2. The viral replication level was monitored by imaging the recombinant GFP signal using a Nikon inverted fluorescence microscope. The number of VSV-GFP-positive cells was counted using ImageJ software (http://imagej.nih.gov/ij/index.html).
Results
Effects of various doses of HTNV A9 N protein on IFNβ transcription
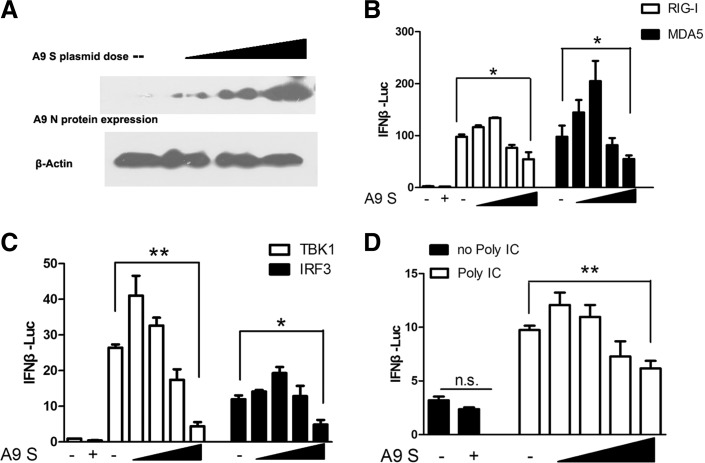
RIG-I, MDA5, TBK1, and IRF3 signaling axes are needed in inducting IFNβ onECs. To determine whether the A9 N protein influences RIG-I/MDA5/TBK1/IRF3-directed IFN responses, we transfected 293T cells with a plasmid expressing A9 N proteins (A9 S plasmid) in various doses, as shown in Figure 1A, together with plasmids encoding RIG-I/MDA5/TBK1 or IRF3, and assessed the IFNβ transcriptional responses using a luciferase reporter assay. The overexpression of RIG-I/MDA5/TBK1/IRF3 significantly activated IFNβ reporter, as shown in Figure 1B and C.
FIG. 1.
Various doses of HTNV A9 N protein regulated RIG-I- or Poly IC-induced IFNβ promoter activation. (A) Dose-dependent expression of A9 N protein (encoded by A9 S plasmid) in the transfection experiments, as shown by Western blot. (B, C) Various doses of A9 N protein regulated IFNβ promoter activation directed by RIG-I (B), MDA5 (B), TBK1 (C), and IRF3(C). (D) Various doses of HTNV A9 N protein regulated IFNβ promoter activation induced by Poly IC at 1 μg/mL (Poly ICs were transfected into cells by Lipofectamine 2000). The first two bars in each panel of (B–D) reflected the basal level of IFNβ promoter activity (without overexpression of RIG-I-related plasmids or without Poly IC stimulation). Results are expressed as mean±SEM. *p<0.05 and **p<0.01 (t-test). Representative results from at least three independent experiments. HTNV A9, Hantaan virus A9 strain; IFN, interferon; RIG-I, retinoic acid-inducible gene I.
The final effect of A9 N protein on the activation of IFNβ induced by RIG-I/MDA5/TBK1 or IRF3 is related with its doses. On one hand, the coexpression of A9 N protein at a low dose promoted RIG-I/MDA5/TBK1/IRF3-directed IFNβ transcription. On the other hand, the N expression at a high dose started to inhibit the IFNβ transcription (Fig. 1B, C). Similar results were observed for the effect of A9 N proteins on the Poly IC-induced IFNβ production (Fig. 1D). These results suggested that the regulation of A9 N protein on IFN signaling was related to its protein levels in the cells.
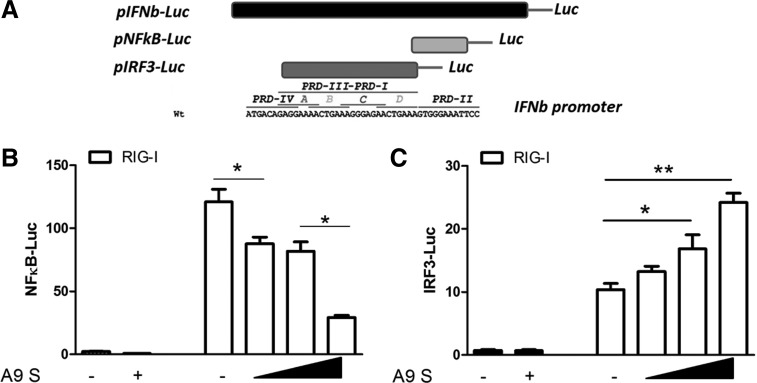
HTNV A9 N protein inhibited the RIG-I-directed NFκB activation, but upregulated the RIG-I-directed IRF3 activation
The induction of IFNβ requires both IRF3 phosphorylation and NFκB activation (4). We tested if the expression of the A9 N protein specifically affects the directed transcriptional responses of either NFκB or IRF3. We found that A9 N protein inhibited the RIG-I-directed NFκB activation in a dose-dependent manner (Fig. 2A). However, in contrast, A9 N upregulates the RIG-I-directed IRF3 activation in a dose-dependent manner [as measured using an IRF3-specific luciferase reporter, pRD(III–I)-Luc] (Fig. 2B). Thus, the A9 N protein may play different roles on the two branches of IFNβ signaling (IRF3 and NFκB). On one hand, the N protein constitutively inhibited the activation of NFκB, which is consistent with previous reports (28). On the other hand, the N protein also potentially activated the transcription activity of IRF3.
FIG. 2.
HTNV A9 N protein differentially regulated the transcriptional activity of NFκB and IRF3. (A) Schematic picture of reporter plasmids used in our study. (B) HTNV A9 N protein (encoded by A9 S plasmid) inhibited RIG-I-induced NFκB activation in a dose-dependent manner. (C) HTNV A9 N protein enhanced RIG-I-induced IRF3 activation. The first two bars in (B, C) reflected the basal level of promoter activity (without overexpression of RIG-I). Results are expressed as mean±SEM. *p<0.05 and **p<0.01 (t-test). Representative results from at least three independent experiments. NFκB, nuclear factor kappa B.
Overexpression of A9 virus N protein in a high dose downregulated IFNβ and ISG production in cells treated with Poly IC or VSV and promoted VSV replication
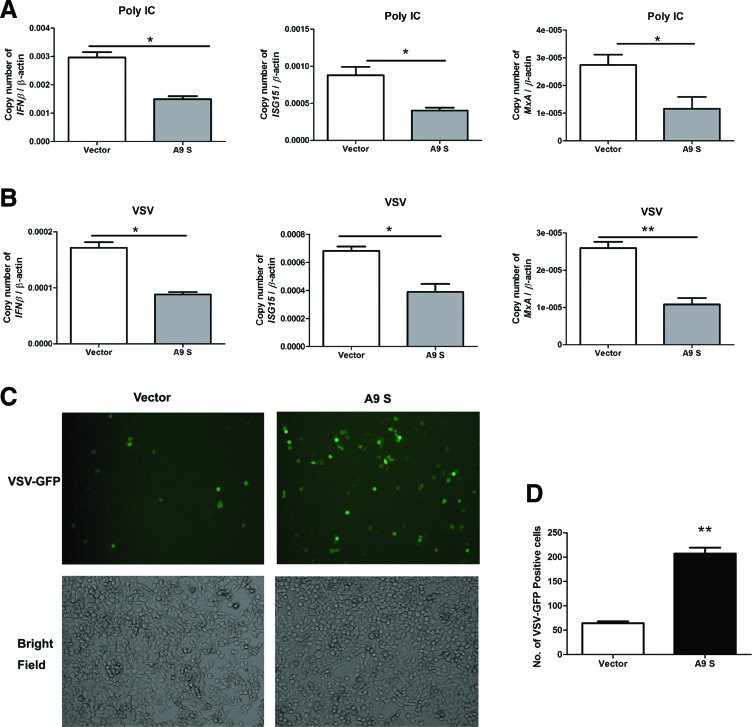
To test the effect of HTNV A9 N protein on the production of IFNβ and ISGs mRNAs, we transfected 293T cells with a high dose of A9 N protein expression plasmid or control plasmid and stimulated cells with Poly IC or VSV. We found that a high dose of A9 N protein downregulated the IFNβ mRNA expression in the overexpressed cells in comparison with controls (Fig. 3A, B). The A9 N protein also inhibited the expression of typical ISGs: ISG15 and MxA (Fig. 3A, B) and chemokines: MCP-2 (CCL8), RATENS (CCL5), and CXCL10 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/vim) in cells treated with Poly IC or VSV.
FIG. 3.
A high dose of HTNV A9 N protein inhibited Poly IC- or VSV-induced IFNβ and ISGs mRNA production and promoted VSV replication. (A) HTNV A9 N proteins (encoded by A9 S plasmid) at a high dose inhibited VSV-induced mRNA production of IFNβ, ISG15, and MxA. (B) HTNV A9 N proteins at high doses inhibited Poly IC-induced IFNβ, ISG15, and MxA mRNA production. Results are expressed as mean±SEM. *p<0.05 and **p<0.01 (t-test). Representative results from at least three independent experiments. (C) High doses of HTNV A9 N protein enhanced VSV replication in 293T cells. The viral replication level of VSV-GFP was monitored by imaging the recombinant GFP signal using a Nikon inverted fluorescence microscope. (D) Number of VSV-GFP-positive cells per field as counted by ImageJ software (http://imagej.nih.gov/ij/index.html). **p<0.01 (t-test). GFP, green fluorescent protein; ISGs, interferon-stimulated genes; VSV, vesicular stomatitis virus. Color images available online at www.liebertpub.com/vim
Since the HTNV A9 N protein inhibited a VSV-induced IFNβ response, we further explored the effect of N protein on VSV replication in 293T cells. A recombinant VSV virus expressing GFP reporter (VSV-GFP) was used in this assay, and the expression of GFP was correlated with the level of viral replication. As shown in Figure 3C and D, the expression of A9 N proteins significantly promoted VSV replication in 293T cells, suggesting that the A9 N protein may promote the VSV replication by downregulating the type I IFN response.
Discussion
Different hantaviruses have different mechanisms to encounter the signaling of IFN (18,19). Taylor et al. reported that the N protein of HTNV inhibited the TNFα-induced activation of NFκB, whereas the G protein did not (28). PHV was found to highly induce IFN and many ISGs among human ECs at early times after infection, which is in contrast to pathogenic HTNV, NY-1V, and ANDV hantaviruses (19). Levine et al. suggested that the coexpression of G and N proteins of ANDV and SNV modulates both the early induction of IFN and the downstream of JAK/STAT signaling pathway (15). The Gn proteins of ANDV and NY-1V inhibit TBK1-mediated IFN signaling (2,3). The N protein from HTNV 76-118 strain could inhibit nuclear transfer of NFκB and, thereby, modulate the apoptosis and immune signaling in infected cells (22). The ANDV N protein and NY-1Y GnT protein both inhibit RIG-I-directed IFN signaling by blocking IRF3 phosphorylation (20,30). In this report, we reported that HTNV A9 strain has a distinct manner on regulating IRF3- and NFκB-directed IFNβ production.
HTNV A9 virus, being a pathogenic virus, can resist the host innate immune response by regulating the type I IFN activation, at least through its N proteins. Our results suggested that during the early stage of infection, a low dose of N proteins could inhibit NFκB, but stimulate IRF3 activation. While there were abundant A9 N proteins (due to the initial viral replication) in host cells, these viral proteins continued to inhibit NFκB, with a higher potential, and turned to inhibit IFNβ production (because the induction of IFNβ needs activation of both NFκB and IRF3). At this stage, we could not completely rule out the possibility that mRNA of virus N gene could activate RIG-I signaling, because in the process of natural viral infection, the N protein and mRNA will both exist in host cells. We hypothesized that the inhibitory effect seen with higher doses of transfected plasmid might be due to the N protein interfering with components of NFκB branch downstream of RIG-I. The final inhibition of IFNβ by N proteins may help the A9 virus to successfully replicate among host cells.
Our results can also explain the previous report from certain aspects that hantaviruses were sensitive to the addition of IFN at early time points postinfection and resistant to IFN addition at later time points (19). It seems that the IFN signaling pathway would be blocked by the abundant viral proteins, such as N proteins, at the later stage of virus replication within infected cells. The dose-related manner on the regulation of IFN pathway by HTNV A9 N protein revealed a dynamic interaction between the host cell and virus.
Authors' Contributions
W.P. and J.D. designed the experiments and prepared the article. G.B., W.P., K.W., and T.F. performed all the experiments. All authors read and approved the final article.
Supplementary Material
Acknowledgments
This work was supported by a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT); the National Natural Science Foundation of China (NSFC) (No. 81172812, 81271792, 31200648, 31300714, 81471571, and 31400737), and the Jiangsu Natural Science Foundation (BK2012180). J.D. is a member of the Jiangsu Provincial Innovative Research Team.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Akira S, Uematsu S, and Takeuchi O. Pathogen recognition and innate immunity. Cell 2006;124:783–801 [DOI] [PubMed] [Google Scholar]
- 2.Alff PJ, Gavrilovskaya IN, Gorbunova E, et al. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J Virol 2006;80:9676–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alff PJ, Sen N, Gorbunova E, Gavrilovskaya IN, and Mackow ER. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J Virol 2008;82:9115–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante CR, Nistal-Villan E, Shen L, Garcia-Sastre A, and Aggarwal AK. Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer. Mol Cell 2007;26:703–716 [DOI] [PubMed] [Google Scholar]
- 5.Hepojoki J, Strandin T, Lankinen H, and Vaheri A. Hantavirus structure—molecular interactions behind the scene. J Gen Virol 2012;93:1631–1644 [DOI] [PubMed] [Google Scholar]
- 6.Hooper JW, Moon JE, Paolino KM, et al. A Phase 1 clinical trial of Hantaan virus and Puumala virus M-segment DNA vaccines for haemorrhagic fever with renal syndrome delivered by intramuscular electroporation. Clin Microbiol Infect 2014;20 Suppl 5:110–117 [DOI] [PubMed] [Google Scholar]
- 7.Jonsson CB, Figueiredo LT, and Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev 2010;23:412–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato H, Sato S, Yoneyama M, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005;23:19–28 [DOI] [PubMed] [Google Scholar]
- 9.Klingstrom J, Maljkovic I, Zuber B, Rollman E, Kjerrstrom A, and Lundkvist A. Vaccination of C57/BL6 mice with Dobrava hantavirus nucleocapsid protein in Freund's adjuvant induced partial protection against challenge. Vaccine 2004;22:4029–4034 [DOI] [PubMed] [Google Scholar]
- 10.Koyama S, Ishii KJ, Coban C, and Akira S. Innate immune response to viral infection. Cytokine 2008;43:336–341 [DOI] [PubMed] [Google Scholar]
- 11.Kunzi MS, and Pitha PM. Interferon targeted genes in host defense. Autoimmunity 2003;36:457–461 [DOI] [PubMed] [Google Scholar]
- 12.Lalwani P, Raftery MJ, Kobak L, et al. Hantaviral mechanisms driving HLA class I antigen presentation require both RIG-I and TRIF. Eur J Immunol 2013;43:2566–2576 [DOI] [PubMed] [Google Scholar]
- 13.Lawson ND, Stillman EA, Whitt MA, and Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A 1995;92:4477–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MH, Lalwani P, Raftery MJ, et al. RNA helicase retinoic acid-inducible gene I as a sensor of Hantaan virus replication. J Gen Virol 2011;92:2191–2200 [DOI] [PubMed] [Google Scholar]
- 15.Levine JR, Prescott J, Brown KS, Best SM, Ebihara H, and Feldmann H. Antagonism of type I interferon responses by new world hantaviruses. J Virol 2010;84:11790–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Liu F, Liang M, et al. Hantavirus-like particles generated in CHO cells induce specific immune responses in C57BL/6 mice. Vaccine 2010;28:4294–4300 [DOI] [PubMed] [Google Scholar]
- 17.Lindkvist M, Lahti K, Lilliehook B, Holmstrom A, Ahlm C, and Bucht G. Cross-reactive immune responses in mice after genetic vaccination with cDNA encoding hantavirus nucleocapsid proteins. Vaccine 2007;25:1690–1699 [DOI] [PubMed] [Google Scholar]
- 18.Mackow ER, Dalrymple NA, Cimica V, Matthys V, Gorbunova E, and Gavrilovskaya I. Hantavirus interferon regulation and virulence determinants. Virus Res 2014;187:65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthys V, and Mackow ER. Hantavirus regulation of type I interferon responses. Adv Virol 2012;2012:524024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthys VS, Cimica V, Dalrymple NA, Glennon NB, Bianco C, and Mackow ER. Hantavirus GnT elements mediate TRAF3 binding and inhibit RIG-I/TBK1-directed beta interferon transcription by blocking IRF3 phosphorylation. J Virol 2014;88:2246–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mou DL, Wang YP, Huang CX, et al. Cellular entry of Hantaan virus A9 strain: specific interactions with beta3 integrins and a novel 70 kDa protein. Biochem Biophys Res Commun 2006;339:611–617 [DOI] [PubMed] [Google Scholar]
- 22.Ontiveros SJ, Li Q, and Jonsson CB. Modulation of apoptosis and immune signaling pathways by the Hantaan virus nucleocapsid protein. Virology 2010;401:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindler C, and Plumlee C. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol 2008;19:311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmaljohn C, and Hooper JW. Bunyaviridae: the viruses and their replication. In: Knipe DM. and Howley PM, eds. Fields Virology, 4th ed. Philadephia: Lippincott Willians & Wilkins, 2001:1581–1602 [Google Scholar]
- 25.Schmaljohn CS, Hasty SE, Dalrymple JM, et al. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science 1985;227:1041–1044 [DOI] [PubMed] [Google Scholar]
- 26.Shim SH, Park MS, Moon S, et al. Comparison of innate immune responses to pathogenic and putative non-pathogenic hantaviruses in vitro. Virus Res 2011;160:367–373 [DOI] [PubMed] [Google Scholar]
- 27.Song G, Hang CS, Qiu XZ, et al. The etiological studies of epidemic hemorrhagic fever. I. Isolation and characterization of a virus strain using the Apodemus agrarius from the unendemic area. Acta Acad Med Sinica 1982;4:66a–73a [PubMed] [Google Scholar]
- 28.Taylor SL, Frias-Staheli N, Garcia-Sastre A, and Schmaljohn CS. Hantaan virus nucleocapsid protein binds to importin alpha proteins and inhibits tumor necrosis factor alpha-induced activation of nuclear factor kappa B. J Virol 2009;83:1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkins C, and Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 2010;22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanagihara R, Gu SH, Arai S, Kang HJ, and Song JW. Hantaviruses: rediscovery and new beginnings. Virus Res 2014;187:6–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeier M, Handermann M, Bahr U, et al. New ecological aspects of hantavirus infection: a change of a paradigm and a challenge of prevention—a review. Virus Genes 2005;30:157–180 [DOI] [PubMed] [Google Scholar]
- 32.Zhao T, Yang L, Sun Q, et al. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol 2007;8:592–600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.